Abstract
Clear organizational patterns on the genome have emerged from the statistics of population studies of fixed cells. However, how these results translate into the dynamics of individual living cells remains unexplored. We use statistical mechanics models derived from polymer physics to inquire into the effects that chromosome properties and dynamics have in the temporal and spatial behavior of the genome. Overall, changes in the properties of individual chains affect the behavior of all other chains in the domain. We explore two modifications of chain behavior: single chain motion and chain-chain interactions. We show that there is not a direct relation between these effects, as increase in motion, doesn’t necessarily translate into an increase on chain interaction.
Keywords: polymer models, chromosome territories, interphase dynamics, chromosome tethers, DNA damage
Introduction
The organization of chromosomes in the nucleus is intimately coupled with the regulatory machinery responsible for DNA transactions, including transcription, replication, and segregation. Recent developments in techniques to capture the conformation of chromosomes (3C and higher iterations) reveal with no ambiguity that the spatial distribution is non-random.1-6 The most parsimonious interpretation is one of neighborhoods, in which individual chromosomes reside. There are interactions with the nuclear periphery as well as preferred interaction between specific loci, such as highly transcribed regions,7 or regions predisposed to translocation.8 The challenge is deconvolving the organizational principles that emerge from the statistics of population studies of fixed cells into an individual living cell, where the chromosome is constantly wiggling and writhing within a very active nucleoplasm.
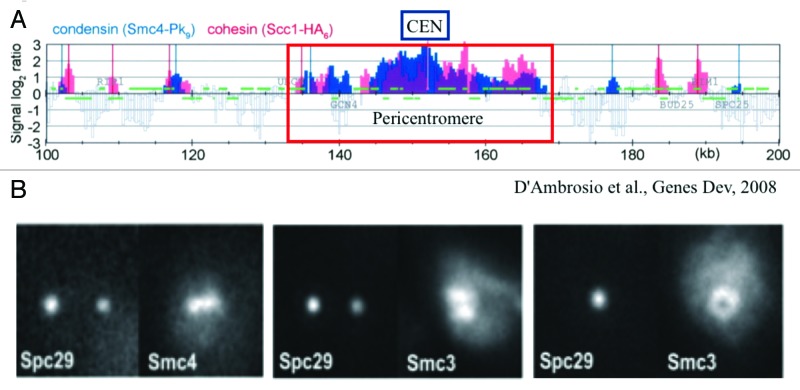
The complexity in deducing mechanism from population statistics is exemplified in the analysis of the Structural Maintenance of Chromosome (SMC) proteins from population studies vs. live cell analysis.9 As shown in Figure 1, the population analysis reveals highly overlapping patterns of condensin (Smc4) and cohesin (Scc1) throughout the centromere and surrounding region (Fig. 1A). In contrast, single cell analysis (Fig. 1B) reveals a non-overlapping distribution with condensin (Smc4) along the spindle axis and cohesin (Smc3) radially displaced from the spindle axis (the spindle axis indicated by the spindle pole marker [Spc29]).10,11 This apparent discrepancy reflects the dynamics of DNA. In all cells, DNA is in constant thermal fluctuation. Even though cohesin and condensin in the pericentromere are spatially segregated, they bind on average to all DNA segments in the region. The behavior of single chromatin chains in live cells is then necessary to interpret statistical data from population studies.
Figure 1. Population distribution vs. single cell distribution of cohesin (Scc1 and Smc3 subunits) and condensin (Smc4). (A) Smc4 and Scc1 are enriched in the pericentromere (~35 kb) surrounding the centromere (CEN). The distribution patterns appear similar in range and intensity. Adapted from reference 9. (B) Fluorescence images of Smc4 and Smc3 in single cells. Three pairs of cells are shown. (left panel) The mitotic spindle is parallel to the plane of focus and the two spindle pole bodies appear as two diffraction spots due to labeling the spindle pole protein Spc29 with RFP. Condensin (Smc4-GFP) appears as a line along the spindle axis between the two spindle poles. (middle panel) The mitotic spindle viewed from the perspective as left. Cohesin (Smc3-GFP) appears as a bi-lobed structure radially displaced from the spindle axis. (right panel) The mitotic spindle is perpendicular to the plane of focus. Only one spindle pole body is in focus (Spc29-RFP). Smc3-GFP is concentrated around the spindle, and appears as a hollow cylinder. Pericentric cohesin (Smc3-GFP) is best modeled as a hollow cylinder surrounding the mitotic spindle (Stephens et al., 2013). The diameter of the cohesin cylinder is ~500 nm, the cylindrical spindle ~250 nm. Adapted from reference 11.
Single chain dynamics are impacted by thermal as well as active non-directed processes. Both cohesin and condensin hydrolyze ATP and slide along the helical backbone.12-14 Cohesin holds sister chromatids together either as a ring entrapping two strands or as a bracelet in which two rings each entrap one DNA strand, while condensin can supercoil and compact DNA. Chromatin remodelers and chaperones are also sliding, removing, re-organizing, and re-assembling nucleosomes. The transcription and replication machinery is a large source of motion; DNA and RNA polymerases generate considerable force (up to 40 pN) during processes of replication and transcription. Thus mechanical force plays a critical role in DNA metabolic processes.15-17 However excessive force (>10 pN) can inhibit chromatin assembly in S-phase18 or lead to chromosome breakage in mitosis (in chromosomes with two centromeres, known as dicentric chromosomes).19,20 It is therefore likely that forces on chromosomes are spatially as well as temporally regulated throughout the cell cycle.
On a macromolecular scale (visible in the light microscope), there are topologically associated domains (TADs)21,22 and a variety of “nuclear bodies” in which transcription and RNA modification factors are concentrated for specific loci such as the rDNA (nucleolus) and histone genes (Cajal bodies).23,24 Many of the nuclear domains are fluid in the sense that they are transiently assembled and disassembled through the course of the cell cycle. Through examination and modeling the dynamics of specific chromatin domains in cells we provide a statistical mechanical view of a living chromosome that will enable our understanding of how energy and structural information is stored and accessed in our genome.
Mathematical Model
Long chain polymers such as chromosomes are challenging to model, due to the wide range of length and time scales involved in their dynamics.25 One of the problems for modeling is the vast number and breadth of polymer configurations, even though the large number of configurations contributes significantly to the entropy and therefore the elastic restoring force characteristic of polymeric systems. Even with robust algorithms, the complexity in the models must necessarily be reduced relative to what is present in vivo. Representation of the restoring force with springs, and modeling the polymer as a bead-spring chain has proven valuable to understanding chromosome behavior. These coarse grained polymer models do remarkably well in capturing the large scale folding and organizational principles derived from population studies.
We develop a statistical mechanics model derived from polymer physics that contains the essential physics necessary to reproduce experimental observations in single cell experiments. In previous work,26 we found that coarse graining of the dynamics of each chromosome arm into a double tethered bead spring chain, with the addition of excluded volume interactions and confinement by the cell wall, accurately recapitulates the thermal, and random ATP-dependent motion of chromosome spots observed experimentally.
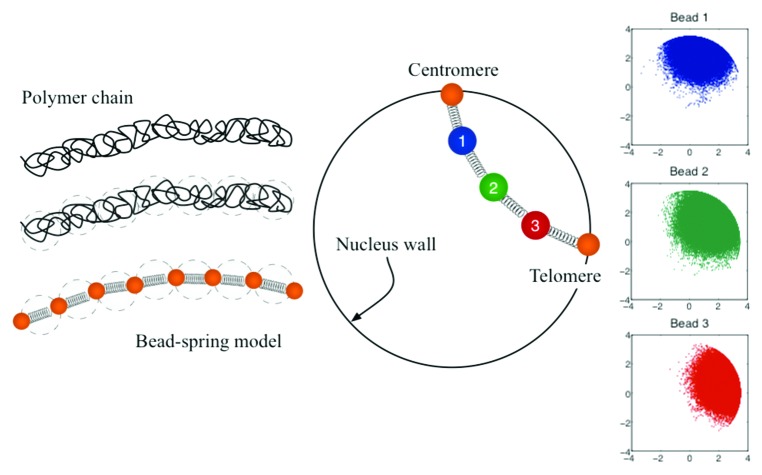
We model each chromosome arm as a bead-spring chain (as depicted in the Fig. 2). In the zero-mass limit, a balance of forces in bead i gives,
Figure 2. Coarse-grained representation of chromosome arms using bead-spring models. Since entropy is proportional to the log of the number of the microscopic states;32 in this model the entropy comes from the number of possible configurations. Configuration space is then a function of the number of beads, the spring constant of the individual springs, the confinement of the nucleus wall, and the excluded volume interactions.
Using linear spring and drag laws, together with a excluded volume potential described by a Gaussian distribution, gives the following evolution equation for the position of bead i,
The term in the left hand side comes from the drag force. The first term in the right hand side represents the intra-molecular forces, in this case captured by a Hookean spring. The second term captures random motion due to thermal fluctuations. It is understood that because of ATP related mechanism inside the cell, the effective temperature differs from the room temperature. The last term in the right hand side describes the excluded volume interaction among the beads. The parameters z and d are nondimensional quantities that characterize the strength (z) and range (d) of excluded volume interaction, which takes the form of a Gaussian potential.27 This term in equation (2) not only accounts for intra-chain repulsion but also for inter-chain coupling due to the repulsion of beads belonging to different chains.
Variations in the chromosome properties can be described by different models parameters (Fig. 5 in ref. 26). For instance, nucleosome depletion is captured by the persistence length of the chains, Lp, which in turns modifies the spring constant, defined as
In addition, loss of cohesin was captured by an increase on the number of beads in each chain.26
In this study we further explore changes in the properties of individual chromosomes and their effects in the overall organization and interaction of four chromosome arms. We consider the following model parameters: Lp = 50 nm, z = 0.5, d = 1, 52 beads per chain, and the radius of the confining circle equal to 1 micron. While Eqn. (2) governs the motion of beads 2 to 51, beads 1 and 52 are fixed at the boundary and represent, respectively, the centromere and telomere sites. Since a chromosome arm is on average 250 kb, using 52 beads is equivalent to have beads separated by roughly 5 kb.
Chromosome Motion
We consider four different ways to analyze the dynamics of individual chromosomes and their interactions with the other chromosomes. Mean-square displacement (MSD) and effective spring constants give insight into the dynamics of individual foci and variations within a chain. Mean square distance between beads and contact maps show spatial organization and probe chain-chain interactions.
Mean-squared displacement
The short persistence length of a chromosome (on the order of 50–200nm) relative to cytoskeletal polymers such as actin (Lp = 15 µm) and microtubules (Lp = 6 mm), results in a trajectory that is best described as a random walk. The chromosome is floppy and constantly stepping forward, backward, up and down due to thermal and non-thermal, ATP dependent fluctuations. Mean-square displacement measurements are a quantitative tool to deduce the nature of the driving force (random or directed) and the area of exploration (Rc).
The mean squared displacement (MSD) for a given lag time, τ, is defined as
It can be shown that if the movement is a random walk, a log-log plot of MSD vs. lag time is a straight line with slope 1, MSD ~ τ.28 However, live cell experiments have reported mean square displacements that behave as MSD ~ τα, with α ≈ 0.4.26,29,30 Processes that exhibit this type of behavior, α < 1, are called sub-diffusive. In the cell, this sub-diffusive behavior is due in part to molecular crowding.31 In addition, there is also confinement of motion due to cell walls and tethering, resulting in a MSD plot that “plateaus” at large lag times. This plateau gives an estimate of the total area that can be explored, or radius of confinement, Rc.26
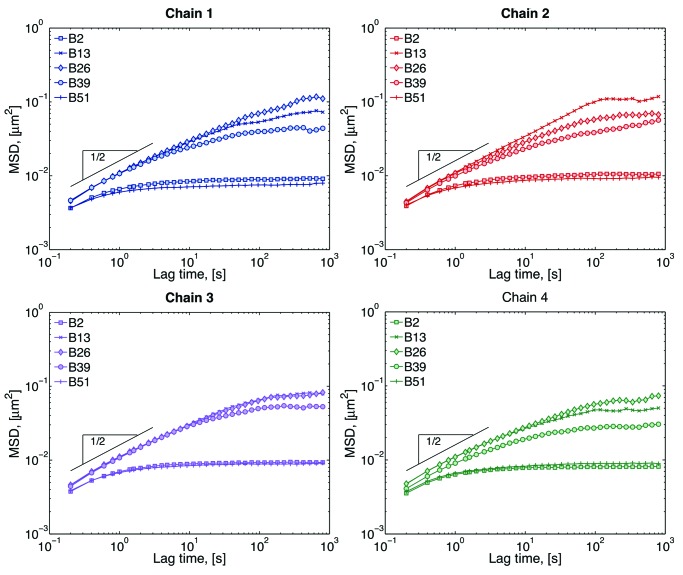
MSD data from our modeling is shown in Figure 3. The MSD for the beads close to the tethers (beads 2 and 51) show a smaller radius of confinement, while beads in the middle (bead 26) explore a much larger space. The exact MSD behavior depends not only on the chromosome (chain 1, 2, 3, or 4), but also on the position within a given chromosome (bead number). This difference is only evident at large lag times, which means that, at small lag times, all beads are behaving sub-diffusively and they are moving in the same medium. Single Rouse chains (our model without excluded volume, wall interactions and tethers) have an MSD scaling of MSD ~ τ1/2.32 The fact that all chains follow this scaling, suggest that chromosome motion at these short time scales is independent of chain-chain and chain-wall interactions. At large lag times, different beads have different radius of confinement depending on their position within a chain. One way of differentiating these results is by calculating the plateau value; however the lack of a well-defined plateau in the MSD curves sometimes hinders this approach. Instead we use the effective spring constant as explained in reference 26, which roughly relates inversely to the plateau value.
Figure 3. Mean-squared displacement (MSD) of selected beads within each chain. Short-lag time behavior is sub-diffusive, while the plateau a large lag times comes from the confinement of the nucleus wall and tethering of the chains. The inverse of the plateau value gives an approximation of the range of motion of the beads.26 Beads close to the tethers explore smaller spaces than beads in the middle of the chain.
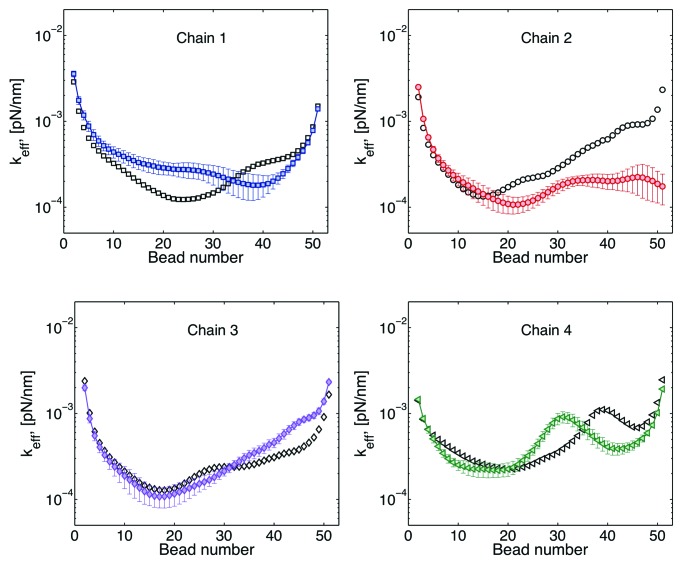
Effective spring constant
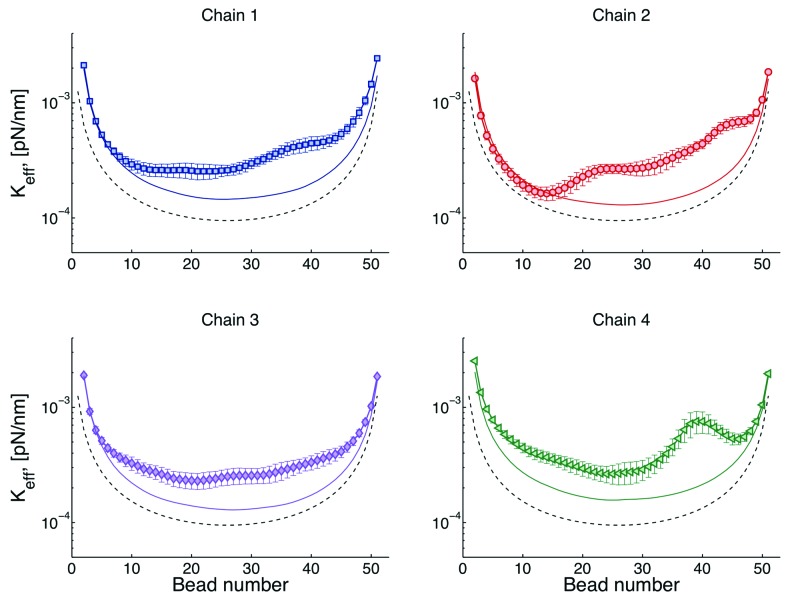
The effective spring constant of a given bead can be obtained from the standard deviation of the distribution of the bead positions.26 Typical results for effective spring constants are shown Figure 4. There is roughly an inverse relation between this curves and the plateau of the MSD curves discussed above. Large spring constants imply a smaller radius of confinement, Rc, while a small spring constant is equivalent to loci that explores a larger space.
Figure 4. Effective spring constants for four chains. The simplest model is that of a chain tethered at both ends, this model is known a doubly tethered Rouse chain (dashed lines). In the Rouse model32 the only forces considered in Eqn. (1) are the drag, spring, and thermal forces. By adding interactions with the wall (Fwall), chains behave as if they were “stiffer” (solid lines). Since there is not interaction among chains, both the dashed and solid lines are symmetric with respect to the distance from the tether sites. Finally, the symbols correspond chains interacting through excluded volume. In this case, heterogeneity of motion is observed for beads as a function of their distance with respect to the centromere (bead 1).
A critical insight to biological function is the contribution of “tethers” to chromosome motion. If we have a single chain, tethered at both ends and without nuclear confinement (doubly tethered Rouse chain), its behavior is described by the dash lines in Figure 4. Note that chromosome loci close to the tethers explore less space than loci in the middle. If in addition one includes a confining domain, like the nuclear membrane (solid lines), there is an increase of the “stiffness” of the chain, or a decrease on the radius of confinement. This indicates that the chains want to explore a space larger than that allowed by the dimensions of the cell. Finally, if we allow a chain to interact with other chains through excluded volume (symbols), there is a breaking of the symmetry, since different regions of the chains interact differently with other chains. For instance, the region around the centromere (beads 1) is more crowded than any other region. Due to this crowding the behavior of beads is mainly governed by cell confinement (symbols overlap with solid lines).
Mean-squared distance
There are several ways to quantify the intermediate-scale properties within interphase chromosomes. One such way is to measure physical distances between pairs of fluorescently marked specific DNA sequences (see ref. 33). These data are then used to construct a graph of mean-square 2D distance, <r2>, between two DNA probes as a function of their genomic separation along a chromosome.
Figure 5A shows the resulting <r2> for adjacent beads, which corresponds to an average separation between DNA spots of ~5 kb in our model. Figure 5C gives <r2> for a 200 kb separation and similar graphs can be constructed for other separations. To obtain a value of <r2> for a given separation, say, 5 kb (first data point in the Fig. 5B), one will take an average of the plots in Figure 5A (solid line). Note that this averaging will obscure the intra-chain dynamics observed in the figure, where is evident that <r2> depends on the distance of the probe from the centromere. Recall that the motion of beads close to the tethers is relatively similar, as shown in Figures 3 and 4. However, Figure 5 suggests that the interaction between beads is different if they are close to the centromere, than if they are close to the telomere. This difference arises from the “crowding” of chains near the centromere (as shown in Fig. 7A). Data from Figures 5A and C suggest that there exist other factors, besides motion, that dictate chain-chain interaction dynamics.
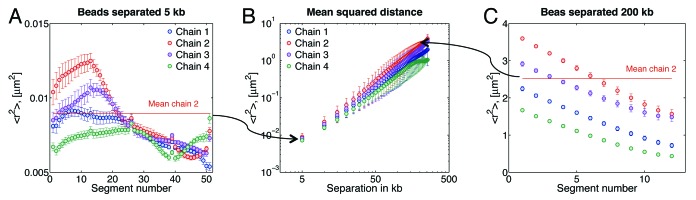
Figure 5. Mean-squared distance, <r2>, between beads. (A) Adjacent beads, which corresponds to an average separation between DNA spots of ~5 kb in our model. Segment numbers are given with respect to the centromere; segment 1 is between beads 1 and 2, segment 2 between beads 2 and 3, etc. (B) Average <r2>. Note that this averaging will obscure intra-chain dynamics observed in the figures (Fig. 5A and C), where it is evident that bead separation depends on the particular location of the bead. For instance, in chain 2, Figure 5A indicates that the separation between beads 47–48 (segment 47) is in average 77 nm, and separation of beads 13–14 is 112 nm. While the average separation for adjacent beads reported in Figure 5B (first data point) is 94 nm. (C) <r2> for 200 kb separation, i.e., every 40 beads, in this case segment 1 is for beads 1 and 41, segment 2 for beads 2 and 42, etc. Every data point in (B) corresponds to the average of the curves of figures like (A) and (B) for the given separation in kb. Note the difference in <r2> values between (A) and (B).
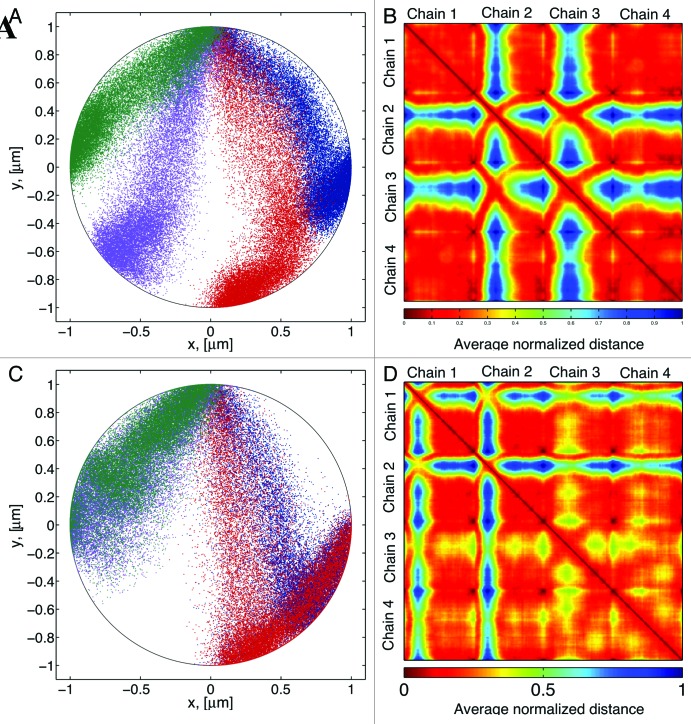
Figure 7. (A) Distribution of bead positions for chains tethered at different points in the cell boundary. Tether sites are located at π/2, 3π/4, 5π/4, and 3π/2 in the circumference. (B) Interaction heat maps corresponding to bead positions in A, dark red corresponds small separations between beads (contact), blue corresponds to the maximum separation observed between two beads (no contact). (C) Distribution of bead positions when two chains are tethered at the same point. (D) Interaction heat map corresponding to (C).
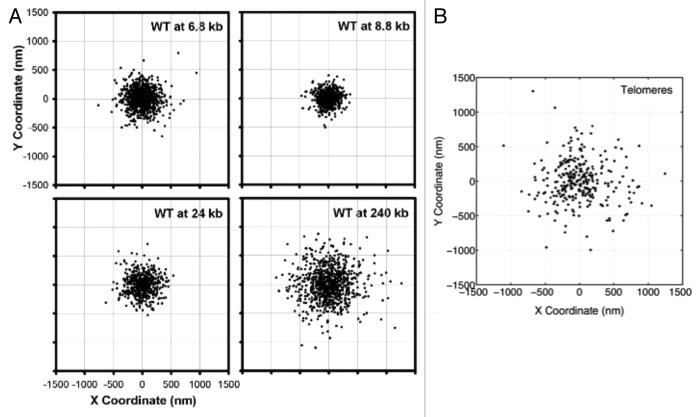
In addition, experimental data indicates that the mechanism of tethering is different in the centromere and telomeres. Figure 6A reproduced from reference 26 shows loci positions for wild type cells at different locations in the arm. Figure 6B shows positions for telomere sites. The range of motion for the telomere is larger than predicted for centromere-like tethers. Centromeres are tethered via microtubules to the spindle pole body, which is a large macromolecular structure (100 × 150 nm) embedded in the nuclear envelope. Telomere association with the nuclear envelope is dependent on protein-protein interactions, whose dissociation rates are considerably higher than centromere detachment from the microtubule. This telomere study indicates that all tethers are not equal, and highlights the state and magnitude of the tether as critical parameters moving forward, as they can readily be incorporated in the model.
Figure 6. Scatter plots of loci positions, for (A) different locations within the chromosome arm and (B) telomere sites. (A) is adapted from reference 26. In a chromosome arm, the two extreme tethering points are the centromere and the telomere. Behavior of the telomere is fundamentally different from that of the centromere, implying different dynamics governing each site.
Finally, the model predicts that tethers, defined as any physical means that restricts motion, throughout the chromosome in living cells will influence motion of the adjacent loci. The homogeneous depiction we choose here, borrowed from polymer physics, may not be able to capture the wealth of tethering behavior that can occur in the nucleus. Tethering in polymers arise from entanglements and covalent cross-links between chains. In the chromosome they arise from microtubule-dependent tethering of the centromere to a spindle pole body, or interaction of the telomere with sites on the nuclear envelop. Secondary tethers may arise from protein-protein interactions between DNA binding proteins, entrapment in ring-like molecules, plectonemic structures, strand entanglement, binding of chromosomal components such as condensin, cohesin, or topoisomerase, or accumulation of replication or repair factories. These tethers, through their consequence on DNA dynamics, may be secondary drivers of gene activity and or gene silencing. In this sense, different parts of the chain can respond differently to the same binding complexes. This feature predicts that a given binding protein may have different consequences depending on the particular location of the target gene within the nucleus. As a consequence, predictive models need to include heterogeneous behavior of the chain properties and spatially dependent tethering dynamics when the experimental information becomes available.
Interaction heat maps
Chromosome interactions are captured by recording the average distances between different sites in a given chromosome. Figure 7A shows a typical distribution of bead positions for our 4-chromosomes configuration. Figure 7B shows the corresponding contact map. Here the distances have been normalized, so that 1 is the maximum separation between any two beads in a given numerical simulation. Dark red represents beads “in contact” and blue are beads that in average don’t come into contact. By plotting bead separation in this fashion the contact map captures both the intra-chain dynamics (main diagonal squares) and the inter-chain interactions (off-diagonal squares). This contact map illustrates that different chains rarely come into contact, (blue on the heatmaps), while within each chain there is a great deal of interaction. These heatmaps show in a compact form all the information contained in the mean-square distance discussed above.
Figure 7B shows that chains 1 and 4, with corresponding telomere sites are closer to the centromere (Fig. 7A), exhibit a more uniform distribution of bead separation compared with the other two chains. The main differences of these two chains, compared with chains 2 and 3 are (1) interactions of these chains with the boundary wall are greater than in other chains. And (2) they have smaller territories, so that their range of motion is more restricted. These data suggest that territories location and size influence the interactions dynamics of different DNA regions.
If the locations of the telomere tethers are decreased to two (Figs. 7C-D) there is a marked effect on chain interactions. While chains 1–2 and 3–4 share the same tether sites, chains 3 and 4 (purple- green, Fig. 7C) come into contact at greater frequency relative to chains 1 and 2 (blue-red, Fig. 7D). The position of the tether site dictates the shape of the territory in which a chain resides. This in turn, dictates the nature of the interactions of beads within that chain as well as the interactions with other chains. Since a polymer behaves like a spring whose restoring force is given by entropy, itself determined by the number of available configurations of the polymer chain, larger territories result in larger restoring forces. This causes the beads to increase their interactions in response to its own motion and the motion of other chains.
The polymer model of chromosome interactions has implications for DNA transactions such as DNA repair and chromosomal translocations. Increased DNA motility is observed upon DNA damage, but the precise driving mechanism has been unclear. The increased motion is a physical consequence following the release of internal constraints within a chain. Likewise, the incidence of specific translocations in natural human populations most likely reflects preferred chromosome territories.
Parameter Study
Previously we have shown that tethers affect the chain dynamics, through variations in the spring constant and MSD data.26 In addition, from the mean-squared distance and contact maps, we showed that the position of the tethers have a great influence on the nature of the chromosome interactions.26 Here we investigate other model parameters that might also affect those dynamics and interactions.
Rigidity of the confining wall
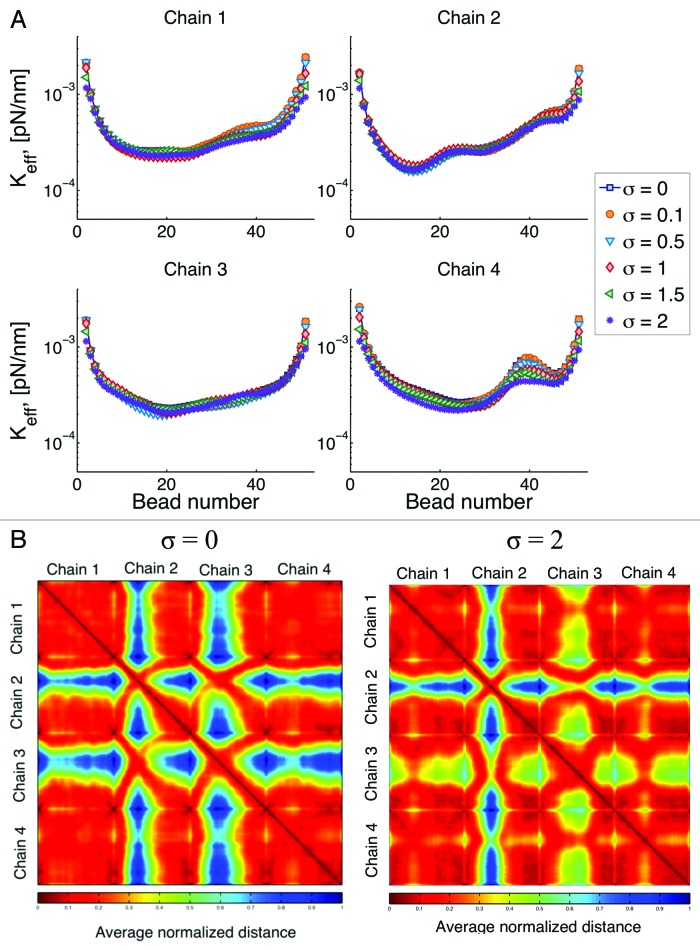
Instead of a hard wall confinement, where beads do not leave the circular domain, we assume a “soft-wall” where beads can leave the domain with a given probability. The probability of leaving the domain is governed by a normal distribution (Gaussian) with mean in the boundary and a given standard deviation. In Figure 8A, we show results of varying this standard deviation, σ.
Figure 8. (A) Effective spring constant for different type of interactions with the confining wall. After a bead leaves the simulation domain (circle of radius 1 micron) the probability of coming back to the confining boundary is Gaussian, with standard deviation σ. So that if σ = 0 the boundary is a “hard-wall”, and as σ increases the “softness” of the wall increases. (B) Interaction heat maps for hard and soft walls.
In general, softer walls reduce the stiffness of the chain by allowing beads to explore a greater space. We note that the marked increased in the effective spring constant observed around bead 40 in chain 4, is reduced as the wall becomes softer and the behavior along the chain becomes more homogeneous. This is a consequence of the chain being able to explore more configurations.
The biological consequence is that explored volume and chain heterogeneity are influenced by wall stiffness. The softer the wall the more space explored and the chain behaves more homogeneously. Whether the mechanical stiffening observed in cancer cells results in a commensurate change in chromosome motion remains to be tested.
However, it is known that mutations in nuclear lamins (underlying the nuclear envelop) lead to Progeria syndrome that has been linked to nuclear mechanics.34
This change in the stiffness of the chains, results in changes in the interaction among chains as shown in Figure 8B. As the chains behave like “softer” springs, they tend to interact more with other chains, represented in the figures by a reduction of the “blue” regions in the contact maps. Moreover, although the addition of soft-wall boundary conditions is a global change, changes in the motion of the chains are not uniform. For instance, chain 4 exhibits the largest change and the magnitude of this change varies for different regions within the chain.
Persistence length
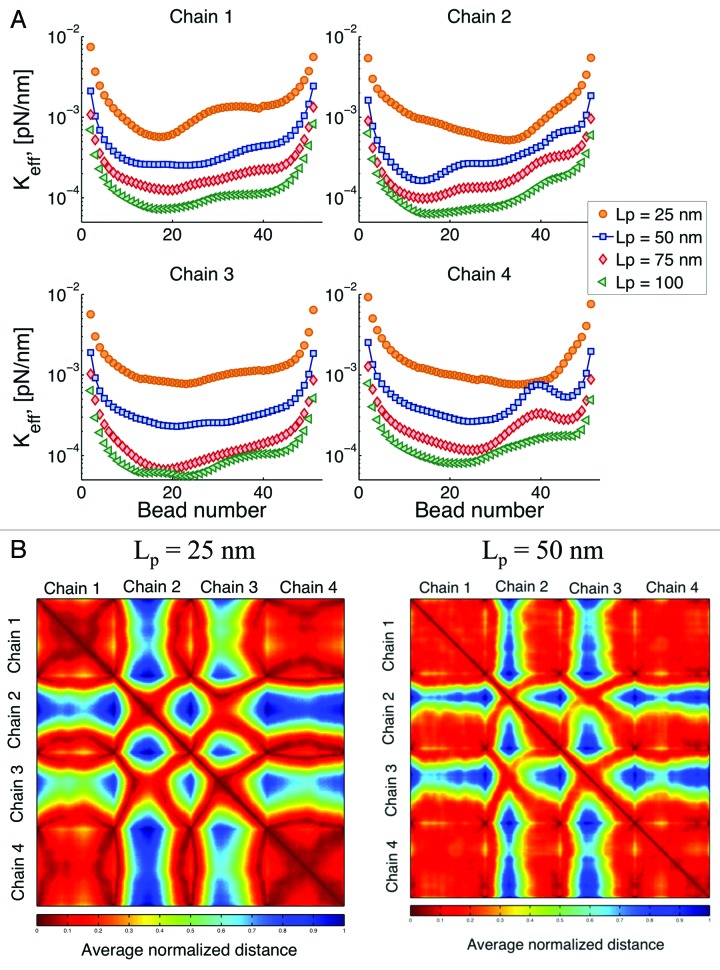
We vary the persistence length as shown in Figure 9A. In general, the flexibility of long polymers arises from the statistical fluctuations of segments larger than the persistence length. In addition, the number of unconstrained degrees of freedom governs the number of configurations. Smaller persistence length results in more degrees of freedom. This change is translated into a larger effective spring constant with decreasing persistence length, Lp.
Figure 9. (A) Effective spring constant after global increase of persistence length (Lp). From Eqn. (3) increasing Lp, results in a decrease of the “stiffness” of the chains. (B) Interaction heat maps for two different values of Lp. Reducing in persistence length results in an increase of the intra-chain interaction and a decrease of the inter-chain interactions.
From Eqn. (3), an increase in persistence length results in a decrease of the spring constant of the individual springs. However, if this was the only effect on the overall behavior, all the curves in Figure 8 should look similar with just a translation on the y-axis. On the contrary, we observe that not only the overall behavior is “shifted,” but the intra-chain dynamics are also modified. Since the confining circle remains the same for all simulations (1 μm), by varying the persistence length we are also changing the way confinement affects the chain behavior. Beads in chains that have a small Lp interact with the boundary more often than beads in chains that are stiffer. Roughly speaking, increasing the persistent length of a chain, decreases the influence the boundary has on its dynamics.
Figure 9B shows that when Lp is decreased the intra-chain interactions increase, as shown by larger dark red regions in the main diagonal squares. Conversely, the inter-chain contact decreases, as the blue regions off the main diagonal become larger. This indicates that as the chains become stiffer, there is a preferred contact of beads belonging to different chains than of beads within the same chain. This can be understood from an energetic point of view, as Lp increases “bending” becomes energetically unfavorable, resulting in fewer interactions of beads belonging to the same chain.
Stiffer and/or softer chains
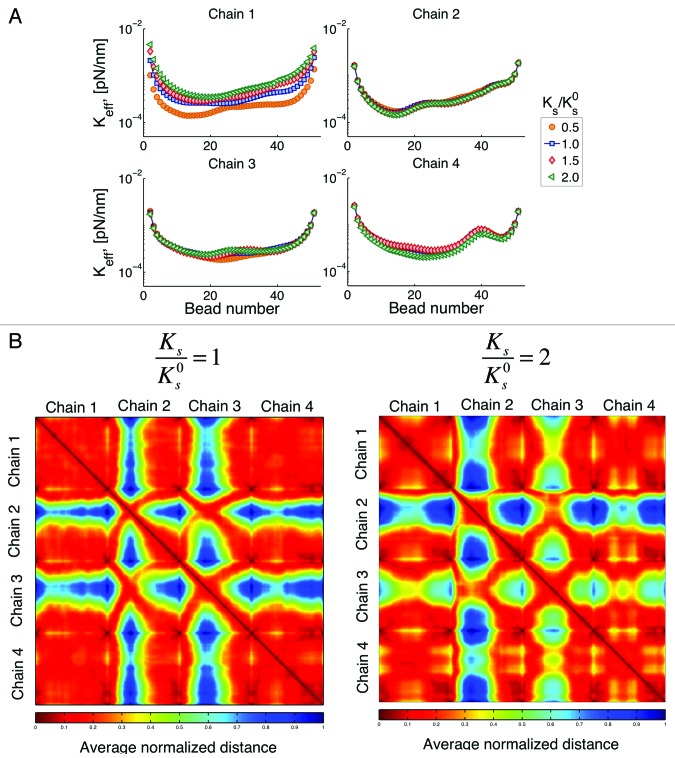
Our next question is whether changing the stiffness of an individual chain affects the behavior of other chains. One of such results is shown in Figure 10.
Figure 10. (A) Effective spring constant for variations of stiffness of chain 1. Ks is the spring constant for chain 1, is the spring constant for other chains obtained using Eqn. (3) and Lp = 50 nm. Changes of the properties of one chain affects the behavior of other chains through changes in their interactions. (B) Interaction heat maps for two different values of Ks. If chain 1 is stiffer, interaction of chain 2 with other chains is reduced, while interactions between chains 3 with other chains is increased. This figure shows how changes of the properties on a single chain, can affect the behavior of the whole system (global response to local changes). Similar behavior is observed if we choose to change the spring constant of any other chain.
Note that from Eqn. (3) this change on stiffness is related to changes in persistence length. However, unlike our previous point, here we modify the properties of a single chain. The question is whether variations on the properties of a single chain can affect the global behavior. When chain 1 is stiffer, its range of motion is reduced as expected. One will also expect that the available space left by the restriction of motion of chain 1 will result in an increased motion of other chains. Recall that chain 4 has the smallest territory, since it telomere site is the closest to the centromere. Figure 10A shows an increase in the motion of chain 4 that is larger relative to that of chains 2 and 3. That is, chain 4 the takes the most advantage of “available space” left by chain 1. In comparison, chain 2 (the one with the largest territory) is the one that is affected the least by changes in the properties of chain 1. These results point toward a global response, as it is not the chain closest to chain 1 the one that is affected the most.
We said before that an increase in motion does not necessarily translate into an increase of the interactions within a chain and among chains. This result is confirmed in Figure 10B, where even though chain 4 is the one with the largest increase in motion, it is chain 3 the one that shows the largest increase on interactions with other chains (less blue spots in the heatmap).
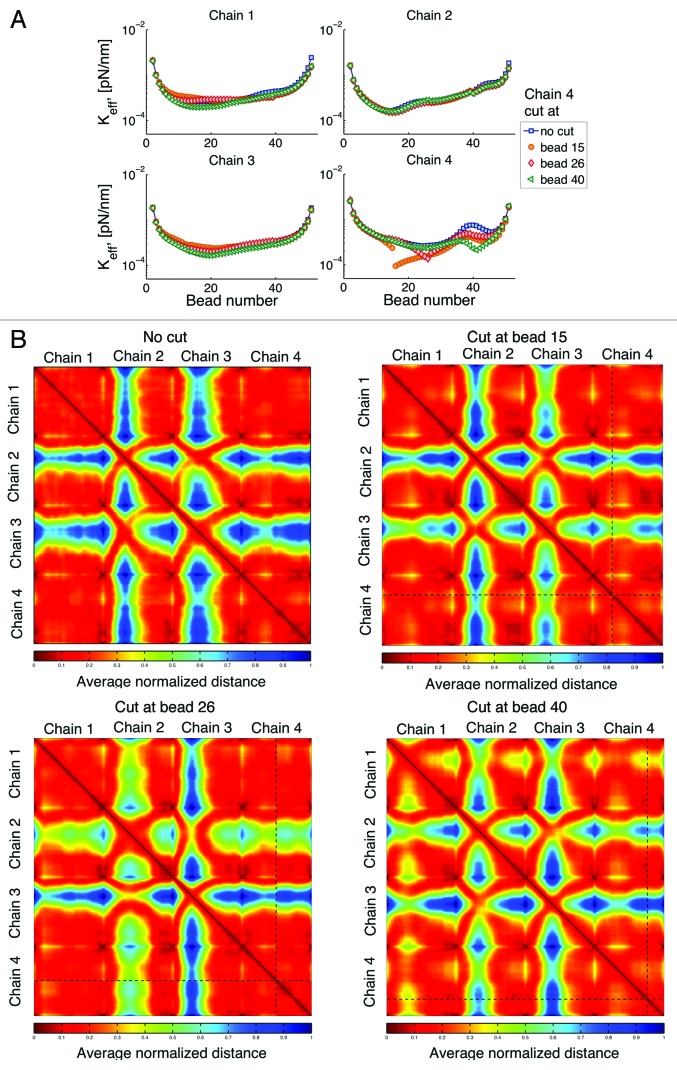
Single double strand DNA break: Cut one chain
In this numerical experiment we break chain 4 at one place. Figure 11A shows that chain breakage not only increases mobility (smaller spring constant) of the broken chain but also that of the uncut chains. The increase in motion is simply a consequence of the freedom afforded by release of the chain, and is supported with experimental observation.35,36 Interestingly, the exact location of the break plays a significant role in the resulting dynamics. Cutting the chain closer to the centromere (bead 15) results in a higher increase in mobility of the two cut segments. However, cutting the chain close to the telomere (bead 40) results in a higher increase in mobility of the uncut chains. Similarly, Figure 11B shows that interaction among chains after a cut, also depends on the location of the cut.
Figure 11. (A) Effective spring constant after cutting chain 4 at a given place. The total number of beads in a chain is 52. Breakage of a chain can affect the behavior of all other chains and these effects depend on the location of the breakage. (B) Interaction heat maps show an increase in chain interactions after breakage; however, where this increase occurs depends on the location of the breakage. Similar behavior is observed if other chains are cut.
Counter to intuition, “neighbor” chains are not necessarily the chains that have the greater magnitude of response. Rather, the response of other chains depends on the location of the telomere of the cut and uncut chains. This could be a regulated event, in that when one homolog is cut, the telomere of the homologous chromosome may be subject to regulation to tune interactions with homologous and non-homologous chromosomes.
Free telomere
We allow the telomere site of chain 2 to be detached from the wall and move freely in the domain. The resulting behavior in Figure 12 shows an increase in the motion of chain 2 (smaller spring constant). And, as with previous results, this change on the dynamics of chain 2 has a global effect on the other chains. However, the increase or decrease of the motion of a given chain is not uniform across the chain. For example, chain 1 shows a reduced motion for beads close to the centromere and an increase in motion for beads close to the telomere site. Similarly, the reduction of the motion around bead 40 observed in chain 4 is translated toward the middle of the chain.
Figure 12. Effective spring constant after the telomere site of chain 2 becomes untethered. Black symbols correspond to results where all the chains are tethered at both ends. Color symbols correspond to the resulting behavior for the free motion of the telomere site of chain 2. Motion at the telomere end in chain 2 is increased, while motion of other chains might increase or decrease depending of the location within the chain. Similar behavior is obtained when the telomere site of other chains is allowed to move freely.
Consistently with our previous numerical experiment, these data suggest that regulations of the dynamics of a telomere site can be exploited to regulate the motion and interaction dynamics of specific regions in other chains.
Conclusions
The polymer model described here provides a powerful tool that allows inquiring into non-trivial and experimentally testable consequences from studies of chain dynamics and modifications of simple model parameters. Without any assumptions of higher structural order or biochemical functions, other than modeling chromosomes as tethered polymer chains, we show how qualitatively predictions about experimental results can still be made.
Overall, it is not possible to modify a single chain without changing the global behavior of all the chains in the domain. This indicates that there exist several mechanisms in place that allow the cell to take advantage of the polymeric nature of chromosomes to perform remarkable tasks.
Further refinement of the model must include other species, e.g., proteins complexes; local changes in the chromosome chains, e.g., chromosome remodeling during transcription; and analysis of the effects the boundary conditions have in the chain dynamics.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We thank Dr M Gregory Forest (UNC-CH) for helpful discussions. This work was supported by NIH R37GM32238 to K.B.
References
- 1.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 3.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- 5.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 6.Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–16. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–8. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 8.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–91. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- 9.D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–27. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol. 2011;193:1167–80. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephens AD, Quammen CW, Chang B, Haase J, Taylor RM, 2nd, Bloom K. The spatial segregation of pericentric cohesin and condensin in the mitotic spindle. Mol Biol Cell. 2013;24:3909–19. doi: 10.1091/mbc.E13-06-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–22. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 13.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–58. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 14.Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–29. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 15.Martins RP, Finan JD, Guilak F, Lee DA. Mechanical regulation of nuclear structure and function. Annu Rev Biomed Eng. 2012;14:431–55. doi: 10.1146/annurev-bioeng-071910-124638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bustamante C, Bryant Z, Smith SB. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–7. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 17.Zlatanova J, Leuba SH. Stretching and imaging single DNA molecules and chromatin. J Muscle Res Cell Motil. 2002;23:377–95. doi: 10.1023/A:1023498120458. [DOI] [PubMed] [Google Scholar]
- 18.Leuba SH, Karymov MA, Tomschik M, Ramjit R, Smith P, Zlatanova J. Assembly of single chromatin fibers depends on the tension in the DNA molecule: magnetic tweezers study. Proc Natl Acad Sci U S A. 2003;100:495–500. doi: 10.1073/pnas.0136890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher JK, Ballenger M, O’Brien ET, Haase J, Superfine R, Bloom K. DNA relaxation dynamics as a probe for the intracellular environment. Proc Natl Acad Sci U S A. 2009;106:9250–5. doi: 10.1073/pnas.0812723106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song W, Gawel M, Dominska M, Greenwell PW, Hazkani-Covo E, Bloom K, Petes TD. Nonrandom distribution of interhomolog recombination events induced by breakage of a dicentric chromosome in Saccharomyces cerevisiae. Genetics. 2013;194:69–80. doi: 10.1534/genetics.113.150144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheutin T, Cavalli G. Polycomb silencing: from linear chromatin domains to 3D chromosome folding. Curr Opin Genet Dev. 2014;25:30–7. doi: 10.1016/j.gde.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 22.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamond AI, Sleeman JE. Nuclear substructure and dynamics. Curr Biol. 2003;13:R825–8. doi: 10.1016/j.cub.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Sleeman JE, Trinkle-Mulcahy L. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol. 2014;28:76–83. doi: 10.1016/j.ceb.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Underhill PT, Doyle PS. On the coarse-graining of polymers into bead-spring chains. J Non-Newt Fluid Mech. 2004;122:3–31. doi: 10.1016/j.jnnfm.2003.10.006. [DOI] [Google Scholar]
- 26.Verdaasdonk JS, Vasquez PA, Barry RM, Barry T, Goodwin S, Forest MG, Bloom K. Centromere tethering confines chromosome domains. Mol Cell. 2013;52:819–31. doi: 10.1016/j.molcel.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prakash JR. Rouse chains with excluded volume interactions in steady simple shear flow. Journal of Rheology. 2002;46:1353–80. doi: 10.1122/1.1514054. [DOI] [Google Scholar]
- 28.Berg HC. Random walks in biology. Princeton University Press, 1993. [Google Scholar]
- 29.Weber SC, Spakowitz AJ, Theriot JA. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys Rev Lett. 2010;104:238102. doi: 10.1103/PhysRevLett.104.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc Natl Acad Sci U S A. 2012;109:7338–43. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman A, Tseng Y, Wirtz D. Effect of length, topology, and concentration on the microviscosity and microheterogeneity of DNA solutions. J Mol Biol. 2002;323:199–215. doi: 10.1016/S0022-2836(02)00893-8. [DOI] [PubMed] [Google Scholar]
- 32.Doi M, Edwards S. The theory of polymer dynamics, 1986. Claredon, Oxford ISBN 0-19-852033-6 1986. [Google Scholar]
- 33.Münkel C, Eils R, Dietzel S, Zink D, Mehring C, Wedemann G, Cremer T, Langowski J. Compartmentalization of interphase chromosomes observed in simulation and experiment. J Mol Biol. 1999;285:1053–65. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- 34.Broedersz CP, Brangwynne CP. Nuclear mechanics: lamin webs and pathological blebs. Nucleus. 2013;4:156–9. doi: 10.4161/nucl.25019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol. 2012;14:502–9. doi: 10.1038/ncb2465. [DOI] [PubMed] [Google Scholar]
- 36.Miné-Hattab J, Rothstein R. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol. 2012;14:510–7. doi: 10.1038/ncb2472. [DOI] [PubMed] [Google Scholar]