Abstract
Gene loci on different chromosomes can preferentially colocalize in the cell nucleus. However, many of the mechanisms mediating this spatial proximity remain to be elucidated. The IgH locus on Chromosome 12 and the Myc locus on Chromosome 15 are a well-studied model for gene colocalization in murine B cells, where the two loci are positioned in close proximity at a higher than expected frequency. These gene loci are also partners in the chromosomal translocation that causes murine plasmacytoma and Burkitt’s lymphoma. Because both Chromosome 12 and Chromosome 15 carry nucleolar organizer regions (NORs) in the most commonly studied mouse strains, we hypothesized that NOR-mediated tethering of the IgH and Myc loci to shared nucleoli could serve as a mechanism to drive IgH:Myc colocalization. Using mouse strains that naturally carry nucleolar organizer regions (NORs) on different sets of chromosomes, we establish that IgH and Myc are positioned proximal to nucleoli in a NOR dependent manner and show that their joint association with nucleoli significantly increases the frequency of IgH and Myc pairing. Thus we demonstrate that simple nucleolar tethering can increase the colocalization frequency of genes on NOR-bearing chromosomes.
Keywords: IgH, Myc, nucleolus, nucleolar organizer region, nuclear architecture, chromatin organization, gene colocalization
Introduction
The genome is spatially organized within the eukaryotic nucleus with both individual loci and entire chromosomes probabilistically positioned relative to structural landmarks and to each other.1,2 Changes in nuclear organization are functionally linked to both development and disease,1,3 and defining the mechanisms governing how that organization is established, maintained and modulated remains an important goal. Chromosome size, gene density, and expression of certain proteins that localize to the nuclear envelope and nuclear lamina are correlated with the position of chromosomes within the nucleus, but the actual mechanism by which these factors drive chromosome position remains unclear.2,4-6 The position of a chromosome territory largely defines the position of individual genes on that chromosome but select loci can also loop out of their respective chromosome territory to associate with nuclear structures such as the periphery.1,7,8 While progress has been made toward understanding the mechanisms underlying gene positioning, much work remains to be done. For example, there is debate over the relative significance of specific DNA sequences vs. chromatin state in determining peripheral association.9-12 Multiple mechanisms have also been implicated in the pairing between gene loci from different chromosomes, with several different DNA binding proteins such as transcription factors, polymerase, and recombinases as well as long non-coding RNA (lncRNA) scaffolds all potentially playing a role.13-15
Although defining the precise molecular mechanisms that position loci or bring them together in 3D space remains a challenge to the field, multiple modes of tethering likely make significant contributions to overall nuclear organization. For example, while there is ambiguity surrounding the nature of the attachment between a DNA locus and the nuclear periphery as discussed above, there is little debate that some attachment exists. Most tethering studies have focused on the periphery, but nucleoplasmic bodies such as the nucleolus could also be self-organizing anchor points. Nucleoli are large membrane-free organelles that assemble directly on rDNA repeat sequences (rDNA) which are present on a select subset of chromosomes. They are the sites of rRNA (rRNA) expression and assembly, and also play a role in numerous other regulatory processes.16,17 The rDNA arrays are classically referred to as Nucleolar Organizer Regions (NORs) because of their role in nucleating nucleoli.18 Transcription of rDNA genes and assembly of ribosomal processing components at each active NOR begins in late anaphase, and in early G1 these nascent nucleoli fuse to form mature functional nucleoli with multiple NORs anchored together in each nucleolus.19 Chromosomal regions outside of NORs, including protein coding genes and centromeric regions on both NOR-bearing and non-NOR-bearing chromosomes, also associate with nucleoli making the nucleolus a potential mediator of inter-chromosomal interactions.20-24
The IgH and Myc genes, which are the respective breakpoints in a recurrent chromosomal translocation that causes Burkitt lymphoma and murine plasmacytoma, are on murine Chromosome 12 (Chr 12) and Chromosome 15 (Chr 15), respectively. Parada et al.25 found that Chr 12 and Chr 15 pair more frequently in B-cells, the cell type in which these cancers emerge, than do other chromosomes. Moreover, the IgH and Myc gene loci were found in proximity to one another in the nucleoplasm of mature B-cells26,27 and to localize in shared transcription factories following stimulation.26 However, the mechanisms responsible for the steady-state positioning of Chr 12 and Chr 15 and the IgH and Myc loci in B-cells have not been defined.
We observed that previous studies relied on BALB/c and C57BL/6 (C57) mice, where both Chr 12 and Chr 15 carry NORs.25,26,28-32 We hypothesized that tethering Chr 12 and Chr 15 to the same nucleolus via their NORs could explain both the chromosome proximity and IgH:Myc colocalization frequency. An alternate possibility was that this colocalization was driven by factors unrelated to NOR-tethering. For example, in human cells, IgH and Myc also pair more frequently than would be expected for random genes,33 but while human IgH is on a NOR bearing chromosome, Myc is not.
To directly test the degree to which nucleolar tethering affects IgH and Myc colocalization frequency, we took advantage of the fact that mouse NORs reside on different chromosomes in a strain specific manner.28,29,34 We quantitated the positions of IgH and Myc gene loci and Chr 12 and Chr 15 territories in B-cells from C57 mice and in B-cells from mouse strains that lack a NOR on either Chr 12 or Chr 15. Importantly, by comparing the different mouse strains we can measure the outcome of effectively removing a NOR from a single chromosome. The results validated our hypothesis: IgH and Myc loci on NOR-bearing chromosomes were significantly closer to the nucleolus than when the same genes resided on non NOR-bearing chromosomes. Moreover, IgH and Myc were physically closer to one another in C57 B cells, where both genes are on NOR-bearing chromosomes, as compared with the strains where only one of the genes was on a NOR-bearing chromosome. Thus, we define nucleolar tethering via NORs as a simple mechanism to promote pairing between genes from different chromosomes.
Results
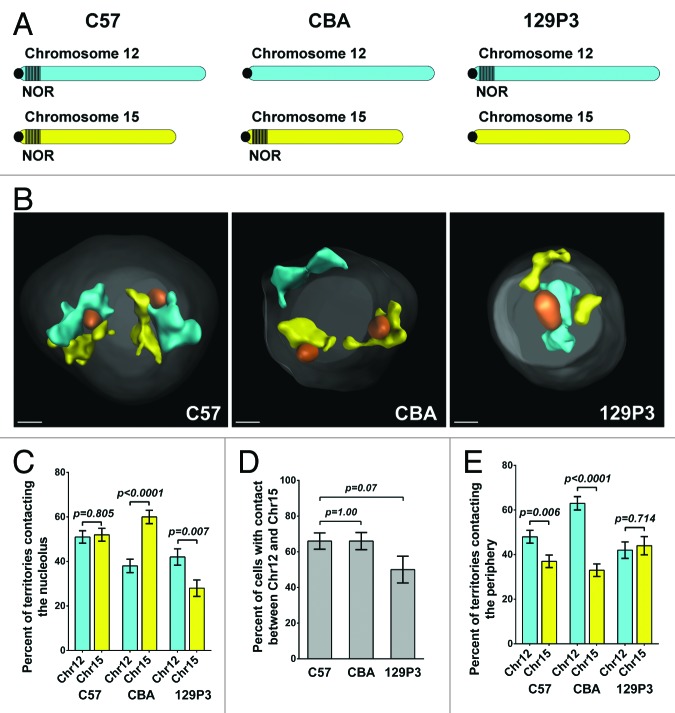
We first asked if the presence of a NOR influences the position of chromosomes relative to the nucleolus. C57 mice have NORs on both Chr 12 and Chr 15, while CBA/CaJ (CBA) mice have a NOR on Chr 15 but not on Chr 12, and 129P3/J (129P3) mice have a NOR on Chr 12 but not on Chr 15 (Fig. 1A).28-32 We isolated mature resting B-cells from the spleens of C57, CBA, and 129P3 mice and performed DNA immunofluorescence (immunoFISH) using whole chromosome paints of Chr 12 and Chr 15 along with an antibody to label nucleoli (Fig. 1B; Fig. S1). As predicted, in B-cells from C57 mice both Chr 12 and Chr 15 contacted the nucleolus at an equivalent frequency whereas in B-cells from CBA mice, Chr 12 contacted nucleoli significantly less frequently than Chr 15 (P < 0.0001) and in 129P3 B-cells the converse was true: Chr 15 contacted nucleoli significantly less frequently than Chr 12 (P = 0.007) (Fig. 1C). We note that because there are slight strain-specific differences in the nuclear diameter and number of nucleoli per nucleus (Table S1), we limited our analysis to comparisons within a given strain.
Figure 1. The presence of a NOR modulates Chr 12 and Chr 15 territory position. (A) Idiograms depicting the presence or absence of NORs on Chr 12 and Chr 15 in the different mouse strains used in this study. (B) 3D reconstruction and rendering of C57, CBA, and 129P3 B-cell chromosome paint images. Original images are shown in Figure S1. Chr 12, cyan; Chr 15, yellow; nucleoli, orange; DAPI, gray. Scale bar = 1 µm. (C) Percentage of Chr 12 and Chr 15 territories contacting nucleoli in each strain. (D) Percentage of cells with at least one contact between Chr 12 and Chr 15 in each strain. (E) Percentage of Chr 12 and Ch15 territories contacting the nuclear periphery in each strain.
We also asked whether nucleolar tethering affected inter-chromosomal associations. We quantified the percentage of cells in which Chr 12 and Chr 15 territories contacted each other but found no significant difference in Chr 12:Chr 15 pairing among the strains (Fig. 1D). These results suggest that although Chr 12 and Chr 15 positioning relative to the nucleolus is NOR dependent, other mechanisms in addition to nucleolar tethering are involved in their relative positioning.
We next determined whether nucleolar tethering affects the frequency with which chromosomes associate with other nuclear compartments by quantitating contact between the chromosome territories and the nuclear periphery (Fig. 1E, note that a territory may contact both the nucleolus and the periphery). In C57 B-cells, Chr 12 territories contacted the periphery more frequently than Chr 15 territories (P = 0.006). In CBA B-cells, where Chr 12 lacks a NOR, Chr 12 territories also contacted the periphery more frequently than Chr 15 territories (P < 0.0001). Moreover, the ratio of peripherally localized Chr 12 to peripherally localized Chr 15 was significantly higher in CBA cells than in C57 cells (1.91 vs 1.30; P = 0.02 (χ2 test)). These observations suggest that in C57 cells, nucleolar tethering restricts the frequency with which Chr 12 localizes near the periphery. Conversely, in 129P3 cells, Chr 15 contacted the periphery in equal proportion to Chr 12, demonstrating that the presence of a NOR also restricts the frequency with which Chr 15 localizes near the periphery (Fig. 1E). Together these data support the hypothesis that nucleolar tethering via NORs constrains the intra-nuclear position of both Chr 12 and Chr 15.
We next asked if the intra-nuclear positioning of IgH and Myc gene loci is also NOR dependent. The IgH gene locus is approximately 110 MB from the NOR on Chr 12 and Myc is approximately 60 MB from the NOR on Chr 15, thus a priori one cannot assume these genes would be proximal to the nucleolus. We performed multi-color DNA immunoFISH to simultaneously label IgH and Myc gene loci and nucleoli in B-cells from each of the three strains (Fig. 2B). We then measured the 3D distance from each locus to the edge of the closest nucleolus (Fig. 2C; Table S2). Computer simulations of random gene placement were performed within nuclei modeled using measured parameters specific to each strain (Table S3). In C57 cells, both IgH and Myc were significantly closer to nucleoli than would be expected if they were randomly positioned (median distances 0.35 µm and 0.55 µm, respectively, vs 1.31 µm). By contrast, in cells from CBA mice, IgH was significantly further from the nucleolus than Myc (median distance 1.00 µm vs 0.60 µm, P < 0.0001) and in cells from 129P3 mice, Myc was significantly further from the nucleolus than IgH (median distance 1.15 µm vs 0.70 µm, P < 0.0001) (Fig. 2C; Table S2). Given the striking difference in nucleolar proximity for genes on NOR-bearing chromosomes as compared with those that are not, the data strongly support our hypothesis that nucleolar tethering constrains the position of IgH and Myc gene loci.
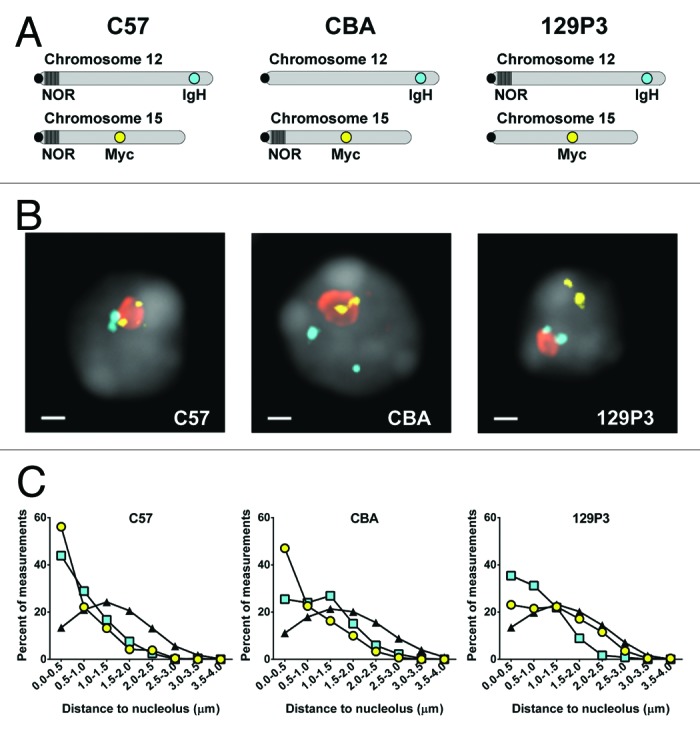
Figure 2.IgH and Myc genes are closer to nucleoli when on a NOR-bearing chromosome. (A) Idiograms depicting the position of the IgH and Myc gene loci as well as the presence or absence of NORs in the different mouse strains used in this study. (B) DNA immuno-FISH images of C57, CBA, and 129P3 B-cells. (C) Quantitation of the distance between gene loci and the closest nucleolus in C57, CBA, and 129P3 B-cells: IgH, cyan squares; Myc, yellow circles; nucleoli, orange; DAPI, gray. Scale bar = 1 µm. Also shown in (C) is the distance between simulated random genes and nucleoli, black triangles.
We noted that the distributions of distances between nucleoli and the “untethered” genes (i.e., the IgH in CBA cells and the Myc gene in 129P3 cells) approached, but did not fully overlap the simulated random distribution (Fig. 2C). For these simulations, we assumed the nucleus was topologically uniform and thus each simulated gene had an equal probability of being positioned anywhere within the nuclear volume. However, in vivo, nuclear topology constrains gene position, for example dense heterochromatin or nuclear bodies may physically occlude a portion of the nuclear volume.35-37 We therefore asked if introducing topological constraints to the simulations would alter the distribution of distances between the simulated genes and nucleoli. The simulations were modified to introduce an excluded volume, i.e., nuclear space that is inaccessible to the simulated loci (Fig. S2A).The distances between the nucleolus and simulated genes in these modified simulations more closely mirrored the measured distances between the nucleolus and the untethered genes in CBA and 129P3 cells, respectively (Fig. S2B; Table S2), implying that no gene is truly randomly positioned within the nucleus.
Many of the tethered IgH and Myc alleles are localized on the surface of, or within nucleoli. Because the nucleolus is surrounded by a rim of heterochromatin and many rDNA repeats are heterochromatized,37 we wondered whether nucleolar association inhibited IgH or Myc transcription. We therefore performed primary transcript RNA immunoFISH and found that both IgH and Myc transcripts were readily detected within nucleoli (Fig. S3). These data directly demonstrate that these genes are expressed while in association with the nucleolar compartment.
We next asked if nucleolar tethering promotes close pairing of IgH and Myc loci in murine B-cells. We measured and analyzed all pairwise distances between IgH and Myc alleles in B-cells from the three mouse strains. We then compared the medians of all pairwise inter-allele distances and found that IgH and Myc loci in C57 cells (median inter-locus distance 2.00 µm, n = 450) were significantly closer to each other than in CBA cells (median inter-locus distance 2.20 µm, n = 518, P < 0.0001) but the differences were not significant when comparing C57 to 129P3 cells (median inter-locus distance 2.10 µm, n = 469) (Fig. S4A and C; Table S4). However, the nuclei in 129P3 B-cells are smaller than those from C57 and CBA (Table S1), so it was possible that the smaller nuclear diameter itself increased the probability of close gene pairing. In fact, the distribution of randomly simulated IgH:Myc distances is influenced by nuclear topology, similar to gene-to-nucleolus distances (Fig. S4E). Therefore, all inter-allele distances were normalized as a percentage of nuclear diameter. When median distances are calculated from normalized pairwise IgH:Myc distances, median inter-locus distance was significantly smaller in C57 cells (38.6% of the nuclear diameter) as compared with both CBA (41.51% of the nuclear diameter, P = 0.001) and 129P3 cells (40.82% of the nuclear diameter, P = 0.011) (Fig. S4B and D; Table S4). This subtle but significant difference between the median pair distance in cells from C57 and the other strains is likely influenced by the fact that Chr 12 and Chr 15 pair at similar frequencies in all of the strains (see above).
We then compared the frequency of close IgH:Myc pairs, defined as loci within 1 µm of each other, among the strains. We observed a significantly higher proportion of close IgH:Myc pairs in cells from C57 mice (11% of pairs) as compared with cells from either CBA (5% of pairs, P = 0.0005) or 129P3 (7% of pairs, P = 0.025) mice, or from random simulations (4% of pairs, P < 0.0001) (Fig. 3A). We also performed the same analysis with the inter-allele distances normalized to nuclear diameter and found that the fraction of close IgH:Myc pairs, defined as alleles separated by less than 20% of the nuclear diameter, remained significantly higher in C57 cells as compared with either CBA or 129P3 cells (P = 0.004, P = 0.006, respectively, Fig. 3A). Together these results support the hypothesis that nucleolar tethering of Chr 12 and Chr 15 increases the likelihood of close pairing between IgH and Myc.
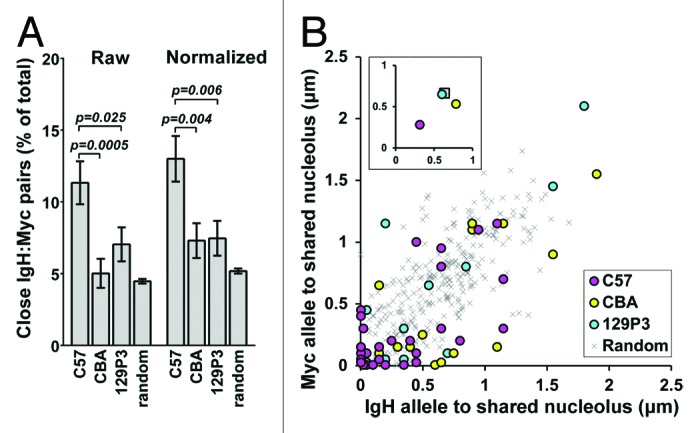
Figure 3. Nucleolar tethering mediates IgH and Myc gene pairing. (A) Percentage of total pairwise IgH:Myc measurements with genes positioned ≤1 µm apart (Raw) or ≤20% of the nuclear diameter apart (normalized). (B) For close IgH:Myc gene pairs, the distance from the IgH allele (x axis) and Myc allele (y axis) to the closest shared nucleolus is plotted. C57, magenta; CBA, yellow; 129P3, cyan; random simulations, gray x. Inset: Average position for each strain. Coloration as above but random gene pairs are represented as a gray square.
Our previous analyses considered the distance between each allele and its closest nucleolus (Fig. 2C) or the distance between IgH and Myc alleles without regard to nucleoli (Fig. 3A; Fig. S4A-D). However, if nucleolar tethering of Chr 12 and Chr 15 has a role in mediating proximity between IgH and Myc alleles, both alleles of close IgH:Myc gene pairs should be positioned adjacent to the same nucleolus even when more than one nucleolus is present. We therefore measured the distance from each allele of an IgH:Myc gene pair to the closest common nucleolus in all cells with two nucleoli. As predicted, closely paired (<1 µm) IgH and Myc alleles were most frequently found adjacent to the same nucleolus in cells from C57 mice (Fig. 3B). The closely paired alleles in cells from CBA and 129P3, by contrast, scattered further from nucleoli (Fig. 3B). We interpret these results to suggest that nucleolar tethering of both IgH and Myc promotes gene colocalization but tethering of a single gene does not. As expected, IgH and Myc alleles more than 1 µm apart from each other are not closely associated with a shared nucleolus in cells from any of the strains (Fig. S5). In summary, there are nearly twice as many close IgH:Myc gene pairs in C57 cells as compared with CBA and 129P3 cells and those gene pairs in C57 cells are positioned at shared nucleoli whereas the close pairs that are present in CBA and 129P3 cells are not.
Discussion
The data presented here support the hypothesis that nucleolar tethering is sufficient to promote pairing between the IgH and Myc gene loci. We demonstrated that the position of Chr 12 and Chr 15 relative to nucleoli and the nuclear periphery is NOR-dependent. Moreover, IgH and Myc loci on NOR-bearing chromosomes were significantly closer to the nucleolus than the same genes were in the mouse strains where they were on non NOR-bearing chromosomes. Strikingly, when both IgH and Myc were on NOR-bearing chromosomes, they were more closely paired than in cells from the mouse strains where only one of the genes was on a NOR-bearing chromosome (Fig. 4). This simple mechanism explains the previous reports of non-random Chr 12 and 15 proximity and IgH:Myc gene association, which relied on cells from strains that carry NORs on both Chr 12 and Chr 15.25-27 Moreover, it explains how IgH and Myc loci are positioned prior to B-cell stimulation such that they can then rapidly engage shared transcription machinery.26
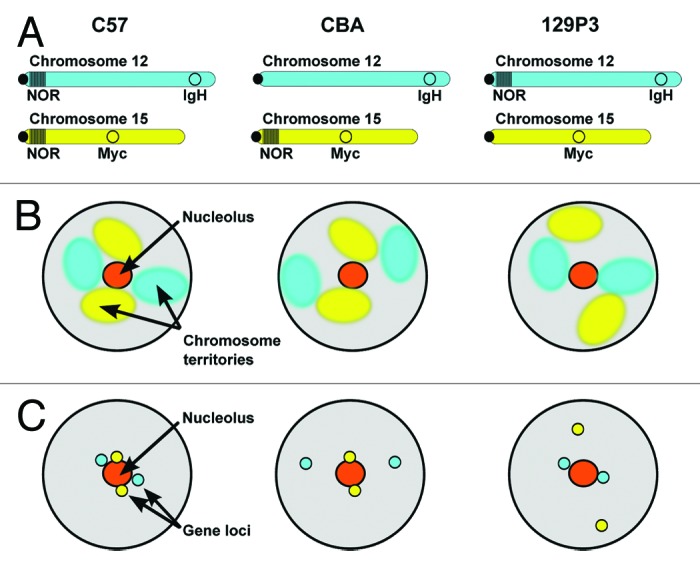
Figure 4. Nucleolar tethering via NORs is a mechanism that contributes to nuclear organization and promotes gene pairing. (A) Idiograms depicting Chr12 and Chr 15, the position of the IgH and Myc gene loci as well as the presence or absence of NORs in the different mouse strains used in this study. (B) Schematic diagram summarizing the effects on chromosome territory position when a NOR on Chr 12 or Chr 15 is absent. (C) Schematic diagram summarizing the effects on IgH and Myc gene position when a NOR on Chr 12 or Chr 15 is absent.
Tethering chromosomes to shared nucleoli is likely a general and tissue type-independent mechanism contributing to 3D nuclear organization. For example, in Arabidopsis, only NOR-bearing chromosomes co-associate with non-random frequencies during interphase.38 Further, in their study of chromosome pairing in B-cells and hepatocytes from C57 mice, Parada et al.25 observed that although the frequency with which non NOR-bearing Chr 5 and Chr 6 were proximal to each other was tissue type-dependent (pairing in 69% of hepatocytes vs 33% of B-cells), the percentage of cells in which Chr 12 and Chr 15 were close to one another was cell-type independent (pairing in 50% of B-cells and 55% of hepatocytes). This observation is completely consistent with the nucleolar tethering mechanism proposed here, but also highlights the possibility that chromosomes which are paired via NOR-based tethers may be more resistant to tissue type-specific chromosome organization than their non NOR-bearing counterparts. According to this model, individual NOR-bearing chromosomes would still be free to adopt tissue-specific positions because nucleolar position is reset after each cell division,19,39 but the position of NOR-bearing chromosomes relative to each other would be buffered by tethering to shared nucleoli.
We observed that IgH and Myc position relative to the nucleolus was more strongly influenced by the presence of a NOR than was the positioning of their respective chromosomes (Fig. 1C vs. Fig. 2C). Similarly, the presence or absence of a NOR did not affect the frequency of Chr 12:Chr 15 pairing but did affect IgH:Myc colocalization frequency (Fig. 1D vs. Fig. 3A). It is unclear whether this enhanced effect on gene positioning reflects local chromosome folding or represents a superimposed gene-specific mechanism. Future studies aimed at elucidating the mechanisms controlling gene position within a chromosome territory will likely help answer this question. It is also worth noting that individual genes, even on NOR-bearing chromosomes, may be more susceptible to tissue type-specific repositioning than the chromosomes themselves as discussed above. The IgH locus, for example, is positioned at the periphery in non B-cells40 but the fraction of nucleoplasmic Chr 12 territories remains relatively unchanged in multiple tissue types.25
Sequences from non NOR-bearing chromosomes can also closely associate with nucleoli. Pericentromeric heterochromatin has long been known to localize adjacent to nucleoli20,37 and more recent genomics studies on purified nucleoli identified contiguous regions of sequence bound to nucleoli which were termed Nucleolar Associated Domains (NADs).22,23 Many NADs are neither proximal to NORs in primary sequence nor even on NOR-bearing chromosomes, suggesting that broadly employed mechanisms beyond NOR-based tethering can also promote nucleolar localization. Interestingly, NADs substantially overlap genomic regions that associate with the nuclear lamina and regions that associate with the lamina during one interphase often associate with nucleoli in the next interphase, i.e., following cell division.12,22,23 This exchange between positioning at the nucleolus and the lamina may in part explain why Chr 12 and Chr 15 increased peripheral association in CBA and 129P3 cells.
At the level of individual loci, nucleolar positioning can be stable, even for loci on chromosomes without a NOR. For example, the murine Zac1 gene on Chromosome 10 preferentially localizes to the nucleoplasm and closely associates with nucleoli.21 Also, human Myc, which is on non-NOR-bearing Chr 8 localizes near nucleoli in tissue culture cells.24 Moreover, human IgH is on NOR-bearing Chr 14 and as alluded to earlier, IgH and Myc also colocalize at a higher than expected frequency in human cells.33 Therefore, colocalization of IgH and Myc via nucleolar association may be a mechanism promoting their pairing in human cells as well. In this model though, IgH would be tethered via a NOR but Myc would localize via an alternative mechanism. While the mechanisms responsible for NOR-independent gene localization are largely unknown, it is interesting to note that the CTCF protein binds the abundant nucleolar protein, nucleophosmin. Moreover, the Myc promoter has well described CTCF binding sites suggesting a possible localization mechanism.41,42 Additionally, nucleolin, another highly abundant nucleolar protein, binds a G-quadruplex structure also found in the Myc promoter.43 However, our data from 129P3 cells demonstrate that in contrast to the human locus, murine Myc requires a NOR to preferentially associate with nucleoli even though it also harbors CTCF binding sites and G-quadruplex forming sequences.
Expression of IgH and Myc is necessary for B-cell differentiation. Notably, we showed that despite being surrounded by a rim of heterochromatin,37 the nucleolar environment of murine B-cells is permissive to the Pol II transcription of both IgH and Myc (Fig. S3). Consistent with this observation, there is indirect evidence for perinucleolar Pol II transcription in tissue culture cells.21,24 Additionally, a recent study posited that CyclinD is upregulated in mantle cell lymphoma because it becomes localized to nucleoli following a characteristic translocation with IgH.44 Moreover, the enzyme activation-induced cytidine deaminase (AID), which catalyzes class switch recombination at immunoglobulin loci and is involved in IgH:Myc translocations has been detected in nucleoli.45,46 Taken together these data suggest that localization of genes, including IgH and Myc, to nucleoli does not interfere with their function. Nonetheless, the genes found in NADs tend to be repressed,22,23 thus it is not clear what role the nucleolar environment plays in modulating active and repressive chromatin states.
Although NOR based tethering is certainly not the only mechanism mediating gene colocalization, it may play a central role in organizing chromosomes and consequently genes within the nucleoplasm. The position of Chr 12 and Chr 15 as well as the IgH and Myc genes is significantly affected by the presence or absence of a NOR in the different mouse strains. Likewise, we expect nucleolar tethering to at least partly control the intra-nuclear position of the other NOR-bearing chromosomes and the genes that reside on them. The physical characteristics of the nucleolus are ideally suited for this function: The nucleolus, like the nuclear periphery, remains intact and relatively immobile throughout interphase, breaks down at mitosis and reassembles at G1, conferring both stability and plasticity to its structure. Because nucleolar formation begins in anaphase, subsequent NOR fusion could be a first seed driving interphase chromatin organization after mitosis.19,47 Further, nucleoli self-assemble directly on NORs and the features that are necessary and sufficient for nucleolar assembly are known.48 The concept that tethering multiple gene loci to a single structure increases their colocalization frequency is paralleled by the cell’s utilization of scaffolding proteins and compartmentalization to increase local protein concentration. In fact the nucleolus was recently used as a scaffold to artificially drive assembly of a higher order protein complex.49 A key challenge will be deciphering how cell type-dependent differences in nuclear diameter, chromatin organization and nuclear body topology35,36 as well as the number of nucleoli per cell50,51 contribute to cell-type specific nuclear organization .
Finally, we note that our use of unaltered primary cells in these experiments eliminates the possibility of artifacts due to genomic manipulations or pleotropic drugs. However, the mouse strains we used are not isogenic, which limits our ability to assess the functional consequences of the altered nuclear organization, for example, to ask if there are differences in translocation frequency between the strains. Nonetheless, by using the novel method of comparing gene and chromosome positions in mouse strains that have a different arrangement of NORs we are able to directly assess the contribution of nucleolar tethering to nuclear organization. We are optimistic that the general approach of capitalizing on the intrinsic variability of NOR arrangement among mouse strains, perhaps coupled with cross-breeding strategies and integration of ectopic neo-NORs,48 will lead to new insights regarding the role of nucleoli in nuclear organization and function.
Materials and Methods
Cell isolation and preparation
C57BL/6J (Stock # 000664), CBA/CaJ (Stock # 000654) and 129P3/J (Stock # 000690) mice were acquired from Jackson Labs. Spleens were collected from 3–6 mo old mice and manually disrupted by maceration through a 70 µm nylon cell filter and passage through a 25G syringe. The cell suspension was centrifuged over a Histopaque-1083 (Sigma-Aldrich; 10831) cushion and mononuclear lymphocytes were collected. Cells were labeled with anti-mouse CD43 microbeads (Miltenyi Biotec; 130-049-801) and CD43- resting B-cells were collected as flow-through from a MACS MS or LS column (Miltenyi Biotec; 130-042-201, 130-042-401). Collected cells were resuspended at 2–4 × 106 cells per ml in RPMI 1640 (Life Technologies; 11875-093) supplemented with 10% FBS (Thermo Scientific; SH40007-13) and 55 μM 2-ME (Life Technologies; 21985-023) and allowed to rest for 1 hr at 37 °C, 5% CO2. After recovery, the cells were pipetted onto to poly-l-lysine coated slides and fixed with 3.7% formaldehyde in PBS. Following fixation, slides were washed in PBS, equilibrated in 20% glycerol in PBS, flash frozen in liquid nitrogen, and stored at -80 °C until needed.
Chromosome paints and DNA immunoFISH
Digoxigenin and Cy3 labeled whole chromosome paints for Chr 12 and Chr 15, respectively, were purchased from Chrombios (PM12DIG, PM15OR) and used according to manufacturer’s directions. Digoxigenin was detected with a FITC labeled mouse anti-DIG antibody (Sigma-Aldrich; F3523) and nucleoli were labeled using a rabbit anti-nucleolin primary antibody (Abcam; ab22758) and an Alexa 647 labeled secondary antibody (Life Technologies; A-31573).
3D immunoFISH was performed as previously described.52 Briefly BACs covering IgH (clone CT7–34H6) and Myc (clone RP23–98D8) loci were nick translated in the presence of either DNP-11-dUTP (Perkin Elmer; NEL551001EA) or digoxigenin-11-dUTP (Roche; 11093088910) to generate 300–600 bp labeled fragments. Cells were denatured at 76–77 °C (CBA and 129P3) or 78–79 °C (C57) in 50% deionized formamide in 2× SSC and hybridized overnight at 37 °C with the labeled probes. Nucleoli were labeled as above. Loci were detected using unlabeled mouse anti-DIG and goat anti-DNP (Sigma-Aldrich; D8156, D9781) primary antibodies followed by Alexa 488 and Cy3 labeled secondary antibodies (Life Technologies; A-11055 or A-21202 Jackson ImmunoResearch; 715-165-150 or 705-165-003) or Alexa 488 labeled anti-DNP (Life Technologies; A-11097) along with the mouse anti-DIG primary and Cy3 labeled secondary antibodies. Coverslips were mounted with Prolong Gold antifade reagent with DAPI (Life Technologies; P36941).
Microscopy and image quantitation
3D images were collected using a 100× 1.4 NA objective on a DeltaVision microscope fitted with the appropriate fluorescence filters and the images were deconvolved using SoftWoRx (Applied Precision). The deconvolved images were rendered in 3D and analyzed using Imaris software (Bitplane). Only cells with good preservation of 3D structure (round nuclei with distinct DAPI-stained pericentromeric foci) and 1–3 nucleoli were used for quantitation. Chromosome territories were considered to touch the nucleolus or each other if 3 or more voxels had overlapping signal, and they were considered peripheral if at least 20% of the surface area abutted the edge of the DAPI signal. The distances between genes and nucleoli were determined by setting a spherical point in the center of the FISH signal for a given allele and then expanding the sphere’s radius until it touched the edge of the closest nucleolus. The distance between alleles was measured as the 3D linear distance between the centers of the respective FISH signals. For the data presented in Figure 3B and Figure S5, only cells with 2 nucleoli were included and the distance from a given allele to each nucleolus was measured as above. The closest common nucleolus was determined to be the nucleolus to which the sum of distances was lowest. A nucleolus automatically became the common nucleolus if one of the alleles was closely tethered to it (i.e., on the surface of or buried within). If both the IgH and Myc alleles were closely tethered to different nucleoli, the nucleolus with the Myc allele was chosen.
Statistics
Unless otherwise stated, median distances were compared using a Kruskal-Wallis test and Dunn’s post-test with Prism software (GraphPad) and where data were expressed as fraction of the population, a two-proportion Z test was applied.
Computer simulations
Simulations were performed using the Smoldyn software package.53 Nuclei were defined with the parameters listed in Table S3. Nuclei and nucleoli were modeled as spheres. Molecules with a diffusion coefficient of 0 were used to represent the nucleoli and genes. Their position was used as the centerpoint for calculating distances. For gene to nucleolus (edge) distances the nucleolar radius was subtracted from the point to point distance. The appropriate number of nuclei, 2 IgH alleles, and 2 Myc alleles were randomly positioned within the nucleus for each iteration and nucleoli were positioned so that their entire volume was within the nucleus.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
This work was funded by NIH grants CA146572, HL065440, and DK044746. We thank Jessica Hallow, Scott Mason, Agnes Telling, Michelle Thompson, and Lisa Yang for expert technical assistance and other members of the Groudine lab for their critical assessments of the data and manuscript.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/psb/article/36233
Glossary
Abbreviations:
- Chr
chromosome
- immunoFISH
immunofluorescence in situ hybridization
- NAD
nucleolar associated domain
- NOR
nucleolar organizing region
- rDNA
ribosomal DNA repeat sequences
- rRNA
ribosomal RNA
References
- 1.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–55. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meaburn KJ, Cabuy E, Bonne G, Levy N, Morris GE, Novelli G, Kill IR, Bridger JM. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell. 2007;6:139–53. doi: 10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- 5.Mewborn SK, Puckelwartz MJ, Abuisneineh F, Fahrenbach JP, Zhang Y, MacLeod H, Dellefave L, Pytel P, Selig S, Labno CM, et al. Altered chromosomal positioning, compaction, and gene expression with a lamin A/C gene mutation. PLoS One. 2010;5:e14342. doi: 10.1371/journal.pone.0014342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuleger N, Boyle S, Kelly DA, de Las Heras JI, Lazou V, Korfali N, Batrakou DG, Randles KN, Morris GE, Harrison DJ, et al. Specific nuclear envelope transmembrane proteins can promote the location of chromosomes to and from the nuclear periphery. Genome Biol. 2013;14:R14. doi: 10.1186/gb-2013-14-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol. 2002;159:753–63. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ragoczy T, Telling A, Sawado T, Groudine M, Kosak ST. A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res. 2003;11:513–25. doi: 10.1023/A:1024939130361. [DOI] [PubMed] [Google Scholar]
- 9.Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 2010;24:766–82. doi: 10.1101/gad.559610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–87. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 11.Demmerle J, Koch AJ, Holaska JM. Emerin and histone deacetylase 3 (HDAC3) cooperatively regulate expression and nuclear positions of MyoD, Myf5, and Pax7 genes during myogenesis. Chromosome Res. 2013;21:765–79. doi: 10.1007/s10577-013-9381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–92. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 13.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Curr Opin Genet Dev. 2010;20:127–33. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–64. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–6. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–85. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 18.McClintock B. The relation of a particular chromosomal element to the development of the nucleoli in Zea mays. Z Zellforsch Mikrosk Anat. 1934;21:294–326. doi: 10.1007/BF00374060. [DOI] [Google Scholar]
- 19.Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. 2011;2:189–94. doi: 10.4161/nucl.2.3.16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho C, Pereira HM, Ferreira J, Pina C, Mendonça D, Rosa AC, Carmo-Fonseca M. Chromosomal G-dark bands determine the spatial organization of centromeric heterochromatin in the nucleus. Mol Biol Cell. 2001;12:3563–72. doi: 10.1091/mbc.12.11.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Royo F, Paz N, Espinosa L, McQueen PG, Vellón L, Parada LA. Spatial link between nucleoli and expression of the Zac1 gene. Chromosoma. 2009;118:711–22. doi: 10.1007/s00412-009-0229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–48. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bártová E, Harnicarová A, Krejcí J, Strasák L, Kozubek S. Single-cell c-myc gene expression in relationship to nuclear domains. Chromosome Res. 2008;16:325–43. doi: 10.1007/s10577-007-1196-0. [DOI] [PubMed] [Google Scholar]
- 25.Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–6. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurihara Y, Suh DS, Suzuki H, Moriwaki K. Chromosomal locations of Ag-NORs and clusters of ribosomal DNA in laboratory strains of mice. Mamm Genome. 1994;5:225–8. doi: 10.1007/BF00360550. [DOI] [PubMed] [Google Scholar]
- 29.Dev VG, Tantravahi R, Miller DA, Miller OJ. Nucleolus organizers in Mus musculus subspecies and in the RAG mouse cell line. Genetics. 1977;86:389–98. [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson KR, Cook SA, Ward-Bailey P, Davisson MT. Genetic mapping of variable length rDNA segments to centromeric regions of mouse chromosomes 11, 12, 15, 16, and 18. Mamm Genome. 1993;4:49–52. doi: 10.1007/BF00364664. [DOI] [PubMed] [Google Scholar]
- 31.Romanova L, Korobova F, Noniashvilli E, Dyban A, Zatsepina O. High resolution mapping of ribosomal DNA in early mouse embryos by fluorescence in situ hybridization. Biol Reprod. 2006;74:807–15. doi: 10.1095/biolreprod.105.047340. [DOI] [PubMed] [Google Scholar]
- 32.Veĭko NN, Shubaeva NO, Malashenko AM, Beskova TB, Agapova RK, Liapunova NA. [Ribosomal genes in inbred mouse strains: interstrain and intrastrain variations of copy number and extent of methylation] Genetika. 2007;43:1226–38. [PubMed] [Google Scholar]
- 33.Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–91. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki H, Kurihara Y, Kanehisa T, Moriwaki K. Variation in the distribution of silver-staining nucleolar organizer regions on the chromosomes of the wild mouse, Mus musculus. Mol Biol Evol. 1990;7:271–82. doi: 10.1093/oxfordjournals.molbev.a040598. [DOI] [PubMed] [Google Scholar]
- 35.Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–3. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- 36.Pederson T. The nucleus introduced. Cold Spring Harb Perspect Biol. 2011;3:3. doi: 10.1101/cshperspect.a000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Politz JC, Scalzo D, Groudine M. Something silent this way forms: the functional organization of the repressive nuclear compartment. Annu Rev Cell Dev Biol. 2013;29:241–70. doi: 10.1146/annurev-cellbio-101512-122317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma. 2004;113:258–69. doi: 10.1007/s00412-004-0316-2. [DOI] [PubMed] [Google Scholar]
- 39.Cvacková Z, Masata M, Stanĕk D, Fidlerová H, Raska I. Chromatin position in human HepG2 cells: although being non-random, significantly changed in daughter cells. J Struct Biol. 2009;165:107–17. doi: 10.1016/j.jsb.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–62. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 41.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–8. doi: 10.1016/S1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 42.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–13. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.González V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J Biol Chem. 2009;284:23622–35. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allinne J, Pichugin A, Iarovaia O, Klibi M, Barat A, Zlotek-Zlotkiewicz E, Markozashvili D, Petrova N, Camara-Clayette V, Ioudinkova E, et al. Perinucleolar relocalization and nucleolin as crucial events in the transcriptional activation of key genes in mantle cell lymphoma. Blood. 2014;123:2044–53. doi: 10.1182/blood-2013-06-510511. [DOI] [PubMed] [Google Scholar]
- 45.Hu Y, Ericsson I, Torseth K, Methot SP, Sundheim O, Liabakk NB, Slupphaug G, Di Noia JM, Krokan HE, Kavli B. A combined nuclear and nucleolar localization motif in activation-induced cytidine deaminase (AID) controls immunoglobulin class switching. J Mol Biol. 2013;425:424–43. doi: 10.1016/j.jmb.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 46.Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–38. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert DM. Cell fate transitions and the replication timing decision point. J Cell Biol. 2010;191:899–903. doi: 10.1083/jcb.201007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grob A, Colleran C, McStay B. Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev. 2014;28:220–30. doi: 10.1101/gad.234591.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Liu Q, Yan Q, Shi L, Fang Y. Nucleolus-tethering system (NoTS) reveals that assembly of photobodies follows a self-organization model. Mol Biol Cell. 2014;25:1366–73. doi: 10.1091/mbc.E13-09-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shea JR, Jr., Leblond CP. Number of nucleoli in various cell types of the mouse. J Morphol. 1966;119:425–33. doi: 10.1002/jmor.1051190404. [DOI] [PubMed] [Google Scholar]
- 51.Berríos S, Koifman J, Fernández-Donoso R. Tissue and sex differences in the expression of nucleoli in mouse somatic cells. Eur J Morphol. 1992;30:297–303. [PubMed] [Google Scholar]
- 52.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–57. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrews SS, Addy NJ, Brent R, Arkin AP. Detailed simulations of cell biology with Smoldyn 2.1. PLoS Comput Biol. 2010;6:e1000705. doi: 10.1371/journal.pcbi.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.