Abstract
Previous studies indicate that a subgroup of suicide completers has low cortical brain expression levels of TrkB-T1, a TrkB gene transcript that is highly expressed in astrocytes. Epigenetic modifications, including methylation changes in the TrkB promoter, partially explain TrkB-T1 low expression levels in brain tissue from suicide completers. The aim of this study was to investigate whether methylation changes in other regions of the TrkB gene could also contribute to the significant downregulation of the TrkB-T1 transcript observed in the brain of suicide completers. Methylation levels were assessed on BA8/9 from suicide completers expressing low TrkB-T1 transcript levels and controls, using custom-made Agilent arrays tiling the whole TrkB gene. After statistical correction for multiple testing, five probes located in the TrkB-T1 3′UTR region were found hypermethylated in the frontal cortex of suicide completers. These results were validated for four CpGs spanning a 150 bp sequence by cloning and Sanger sequencing bisulfite treated DNA. We found a significant correlation between the methylation level at these four CpGs and TrkB-T1 expression in BA8/9. Site-specific hypermethylation on this 3′UTR sequence induced decreased luciferase activity in reporter gene cell assays. Site-specific differential methylation in the TrkB-T1 3′UTR region associates with functional changes in TrkB-T1 expression and may play a significant role in the important decrease of cortical TrkB-T1 expression observed among suicide completers.
Keywords: suicide, epigenetics, DNA methylation, BA8/9, TrkB-T1, astrocytes, luciferase assay
Introduction
Suicide is a major public health problem responsible for 10 to 20 deaths per 100 000 people.1 Whereas suicidal behavior is multifactorial and results from the interaction of several different elements,2 epigenetic factors seem to play an important role, for instance, by modifying stress response to environmental factors.3,4 Accordingly, recent studies have shown differential histone modifications and DNA methylation in brain regions of suicide completers as compared with controls.5,6 These epigenetic changes, especially DNA methylation, have been associated with environmental stressors7 and correlated with altered gene expression.8
Previous studies point to a change in the level of TrkB expression in suicide completers.9-11 The TrkB gene, located on chromosome 9, encodes three receptors of neurotrophins, characterized by specific 3′UTR sequences. TrkB gene products are involved in neurotransmission, calcium release, synaptic plasticity, and cell survival,12 processes whose altered functions have been implicated in the neurobiology of suicide. The shortest TrkB transcript encodes a protein without a tyrosine kinase domain, which is involved in the release of intracellular calcium in astrocytes.13 Downregulation of this truncated TrkB transcript, in coordination with a network of several other astrocytic genes,14 has been reported in several brain regions of suicide completers.9 Interestingly, convergent evidence from studies investigating gene expression patterns15 to studies focusing on cell morphology,16 has been increasingly suggesting that astrocyte dysregulation plays an important role in depression and suicide.
We have shown in the past that a subgroup of suicide completers has very low expression of the TrkB-T1 transcript in the frontal cortex. Studies of DNA methylation in the TrkB promoter sequence in cortical tissue from that brain region, and of microRNA binding in the 3′UTR sequence suggested that these marks could be partially responsible for the regulation of TrkB-T1 expression.9,10 However, DNA methylation in promoter sequence and the functional effect of microRNA binding described in the 3′UTR sequence, whether individually or combined, seem not to be sufficient to explain the large and specific decrease of TrkB-T1 transcript levels associated with suicide. Other mechanisms could be involved in that transcript specific regulation.
In this study, we focused on the frontal cortex, where the difference in gene expression between controls and suicide completers is the most important. Furthermore, this brain area is involved in executive function, working memory and decision making,17 which are known to be impaired in patients with suicidal ideation.18
Most methylation studies conducted to date have focused on the investigation of DNA methylation differences in the promoter regions of genes of interest. However, growing evidence has indicated that gene body, as well as intergenic methylation associates with differential gene function regulation.19 Thus, this study aimed to investigate whether other TrkB gene regions may be differentially methylated and contribute to the significant downregulation of TrkB-T1 reported in the frontal cortex of suicide completers.
A custom-built microarray followed by a validation step allowed us to find a differentially methylated region in the TrkB gene between controls and suicide completers and determine if differential methylation could be correlated with TrkB-T1 mRNA expression. We assessed whether an A/G polymorphism (rs1624327), located in the DMR and able to generate a CpG site, could be associated with suicide or could drive TrkB-T1 expression through a CpG site formation. Finally, performing a patch methylation luciferase assay, we analyzed the functional effect of differential DNA methylation of the TrkB-T1 3′UTR sequence on TrkB-T1 expression.
Results
Microarray analysis
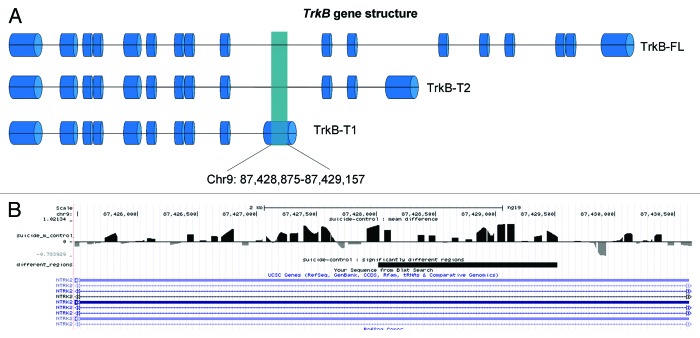
Following meDIP precipitation, samples were hybridized in custom-designed arrays, tiling the full-length of the TrkB gene in a total sample of 24 individuals, including 11 suicide completers having low TrkB-T1 expression levels and 13 controls not selected for TrkB-T1 levels. After correction for multiple testing, we found 5 consecutive probes differentially methylated (Fig. 1A; Table 1). These probes were located in a 500-bp range within the TrkB-T1 3′UTR (Chr9: 87.428.875–87.429.157). The five differentially methylated regions were hypermethylated in suicide completers in a log2FC ranging from 0.8 to 1.0. The length of DNA characterized as differentially methylated and captured by the probes was estimated to be approximately 1.2 kb (Fig. 1B), representing 25% of TrkB-T1 3′UTR sequence.
Figure 1. Differential DNA methylation in frontal cortex of suicide completers. (A) TrkB gene structure. (B) Differentially methylated region show through UCSC genome browser with the Track reporting the methylation level and differences between control and suicide.
Table 1. Probes differentially methylated between control and suicide completers.
| Chromosome | Start | End | cpg level | Methylation level | P value | Q value | Differential | Gene name | Gene strand | tss distance | More methylated in |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 87428875 | 87428928 | 0.1331 | 0.7637 | 0.0018 | 0.0029 | 1.0213 | NTRK2 | 1 | –143326 | Suicide |
| 9 | 87429019 | 87429078 | 0.1313 | 0.7735 | 0.0077 | 0.0029 | 0.8204 | NTRK2 | 1 | –143473 | Suicide |
| 9 | 87429029 | 87429084 | 0.1300 | 0.7735 | 0.0038 | 0.0029 | 0.9259 | NTRK2 | 1 | –143481 | Suicide |
| 9 | 87429105 | 87429156 | 0.1292 | 0.7296 | 0.0017 | 0.0029 | 0.9471 | NTRK2 | 1 | –143555 | Suicide |
| 9 | 87429107 | 87429157 | 0.1292 | 0.7296 | 0.0061 | 0.0029 | 0.8241 | NTRK2 | 1 | –143557 | Suicide |
Criteria used to select the differential probes are the following: P values: < 0.01; Q values < 0.05; CpG level > 0.1, Methylation level > 0.6 ; Log2 Fold Change: < -0.8 and > 0.8.
The TrkB-T1 transcript was distinguished from the other TrkB transcripts by its 3′ untranslated region. The differential methylation between control and suicide groups in the region Chr9: 87.428.875–87.429.157, which is specifically transcribed in TrkB-T1 mRNA, suggests that this genomic region plays a key role in the regulation of TrkB-T1 transcription/expression in the frontal cortex.
Validation of the DMR corresponding to TrkB-T1 3′UTR sequence
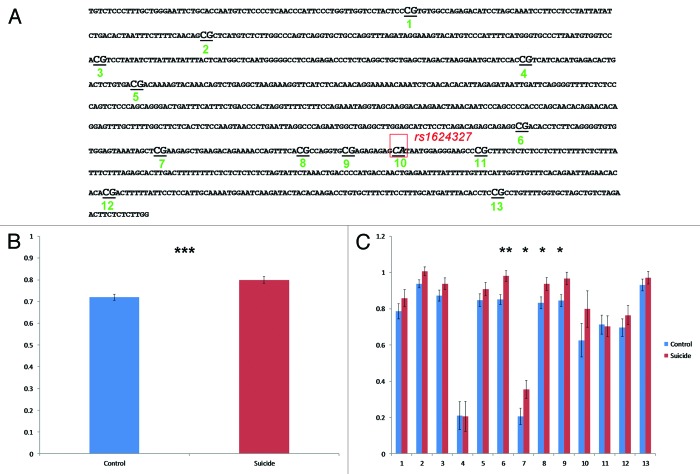
To validate the results observed with the array, we focused on the 1 kb region encompassing the five probes that were differentially methylated between groups. This region contains 13 CpGs (Fig. 2A). We initially cloned and then Sanger sequenced PCR products of bisulfite treated DNA. Twenty clones for each subject and for each of the 3 PCR fragments covering the 1 kb long, differentially methylated region were sequenced. We found significant overall hypermethylation in TrkB DNA sequence (chr9:87.428.550–87.429.575) suicide completers (0.800 ± 0.015) when compared with controls (0.720 ± 0.014) (f = 14.848; df = 169 ; P = 1.67 × 10−4) (Fig. 2B).
Figure 2. Validation of hypermethylation in the DNA sequence coding for TrkB-T1 3′UTR sequence in the frontal cortex of suicide completers. (A) Schematic representation of the 1 kb sequence encompassing the five probes found significantly hyper methylated in frontal cortex of suicide completers. The rs1624327 is indicated in red. The CpG islands are numbered from 1 to 13 including the rs1624327 in position number 10. (B) Overall methylation in the whole region of interest in controls and suicide completers. (C) Site specific methylation in suicide completers compared with controls.
When looking at site-specific CpG sites, we found a group of four CpGs (CpG6 to CpG9), spanning 150 bp, that were differentially methylated between suicide completers and controls (Fig. 2C; Table 2; Fig. S1). These data suggest that the significant difference in methylation found across this 1 kb region was driven by these four CpG.
Table 2. Descriptive statistics of the differential methylation levels in CpG6, 7, 8, and 9 in the region differentially methylated between control and suicide.
| Mean | Std. Error | Mean | Std. Error | |||
|---|---|---|---|---|---|---|
| CpG | Control | Suicide | F | Sig | ||
| 6 | 0.851 | 0.026 | 0.981 | 0.029 | 11.082 | 0.003 |
| 7 | 0.207 | 0.045 | 0.355 | 0.049 | 4.960 | 0.036 |
| 8 | 0.834 | 0.032 | 0.937 | 0.035 | 4.505 | 0.045 |
| 9 | 0.846 | 0.032 | 0.967 | 0.035 | 6.454 | 0.018 |
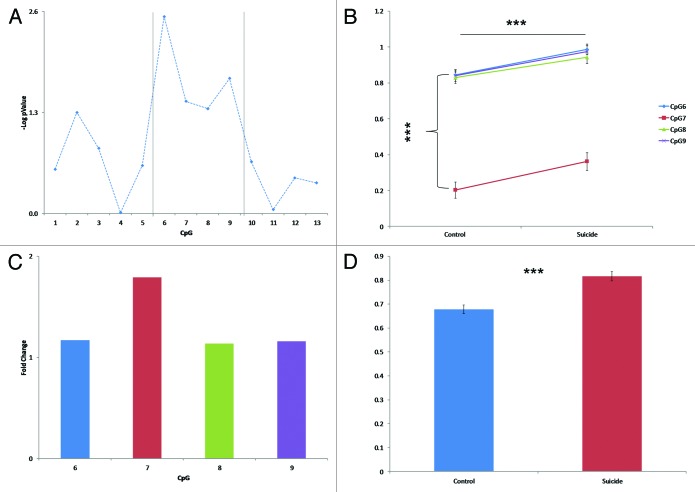
To determine whether these CpG sites could be considered as a cluster with regard to the differential methylation observed between suicide completers and controls, we compared the –logP values (Fig. 3A) of these four CpGs to the values for the other CpGs. Using ANOVA, we found a significant differences in the –logP values between CpG regions (f = 4.198; df = 2; P = 0.047). The CpG6–9 region has a higher –logP value (1.764 ± 0.269) compared with the CpG1 to 5 region (0.665 ± 0.208) (P = 0.026) and to the CpG 10 to 14 region (0.389 ± 0.127) (P = 0.026).
Figure 3. (A) Plot of –logP values for the 13 CpGs group in the TrkB-T1 3′UTR DNA sequence (B) Representation of the paired comparison of methylation levels on individuals CpG6, 7, 8, and 9 between control and suicide. (C) Histogram of the individual fold changes in methylation on CpG6, 7, 8, and 9 in suicide group compared with control. (D) Histogram of the average level of methylation in the CpG6, 7, 8, and 9 in suicide and control group.
Comparison of methylation levels in the 150 bp fragment containing the CpG sites 6 to 9 between suicides and controls showed a significant difference (P = 4.3 × 10−6) (Fig. 3B and D), which is characterized by a significant hypermethylation in suicide completers (0.817 ± 0.020) as compared with controls (0.679 ± 0.018) (Fig. 3D). The fold change in average methylation levels in the suicide group as compared with controls was: (1) CpG6: FC = 1.17; (2) CpG7: FC = 1.79; (3) CpG8: FC = 1.14; and (4) CpG9: FC = 1.16 (Fig. 3C). Interestingly, however, in both groups, the CpG7 had lower methylation levels than the other CpGs (Fig. 3B; Table S1).
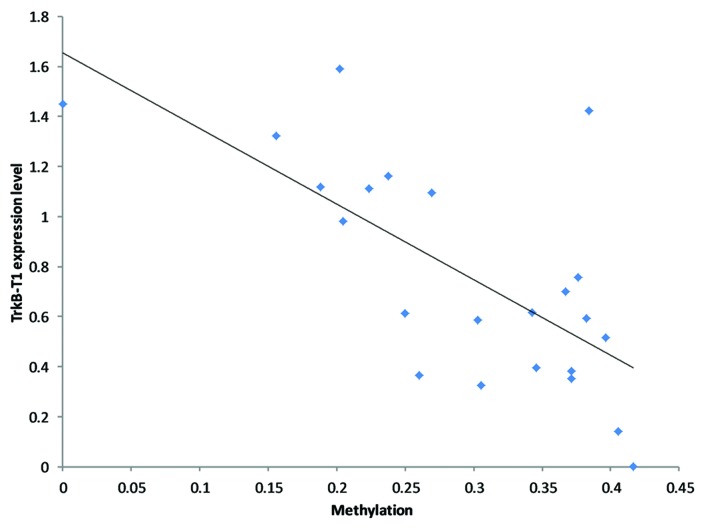
We then determined if the methylation levels were associated with TrkB-T1 expression levels. Using a linear regression model, we found significant correlation between the methylation levels and TrkB-T1 expression (P = 5.7 × 10−4) (Fig. 4). These data suggested that TrkB-T1 decrease in BA8/9 of suicide completers is associated with methylation levels in CpG6 to 9 in our sequence of interest.
Figure 4. Linear regression analysis of the TrkB-T1 expression depending on DNA methylation levels on CpG6 to 9, corrected for age and PMI.
Relationship between TrkB-T1 expression levels and the polymorphism at the rs1624327 locus
One of the CpG sites in the differentially methylated region contains the SNP rs1624327, an A/G substitution (minor allele frequency A = 0.247) that is the tenth CpG of the sequence investigated in the validation studies (Fig. 2A). This A/G polymorphism significantly changes methylation levels, as homozygous AA would not have any methylation at this locus; heterozygous would have a maximum of 50%, and homozygous GG could present up to 100% of methylation. We found, in average, a 17% difference in methylation between the control (0.623 ± 0.092) and the suicide groups (0.799 ± 0.100), with high coefficients of variation being, respectively, 53.2% and 41.7% (Fig. 3C). In fact, with the exception of one control subject, the methylation levels directly reflected the genotype (Fig. S2A). With average methylation levels at: (1) 0.94 ± 0.04 for the G/G patients, (2) 0.54 ± 0.05 for G/A patients, and (3) 0 for the A/A patients. We found significant correlation between genotype and methylation level (R = 0.923 P = 1.4 × 10−10). Consequently, we genotyped the rs1624327 locus in our initial sample of 24 patients (13 controls and 11 suicide completers), as well as in an extended sample of 61 patients (28 controls and 33 suicide completers with low TrkB-T1 expression) (Fig. S2B; Table S2). The genotypic distributions are presented in Tables 3A and C. We found no significant difference in the allelic distribution between controls and suicides, whether in the initial cohort (χ 2 = 0.16 and P = 0.69; Table 3B) or in the extended cohort (χ 2 = 0.09 P = 0.77; Table 3D). Although the methylation level at the rs1624327 is driven by the presence of a G allele at this locus, the absence of significant association between the allele at the rs1624327 and suicide suggests that the allele at the rs1624327 is not involved in the regulation of TrkB-T1 expression, as the results of bisulfite sequencing were suggestive, in spite of high variances in each group.
Table 3. Genotypic and allelic distributions at the rs1624327 for the initial and extended cohorts of controls and suicide completers.
| A | Initial cohort | ||
| Genotype | G/G | A/G | A/A |
| Control | 7 | 4 | 2 |
| Suicide | 7 | 3 | 1 |
| B | Initial cohort | ||
| Allele | G | A | |
| Control | 18 | 8 | |
| Suicide | 17 | 5 | |
| C | Extended cohort | ||
| Genotype | G/G | A/G | A/A |
| Control | 16 | 10 | 2 |
| Suicide | 20 | 11 | 2 |
| D | Extended cohort | ||
| Allele | G | A | |
| Control | 42 | 14 | |
| Suicide | 51 | 15 | |
(A) Genotype at the rs1624327 for the initial cohort of 13 controls and 11 suicides with low-TrkB-T1 expression. (B) Allelic frequency at the rs1624327 for the initial cohort of 13 controls and 11 suicides with low TrkB-T1 expression. (C) Genotype at the rs1624327 for the extended cohort of 28 controls and 33 suicides with low TrkB-T1 expression. (D) Allelic frequency at the rs1624327 for the extended cohort of 28 controls and 33 suicides with low TrkB-T1 expression.
Functional validation of differential methylation of TrkB-T1 3′UTR sequence
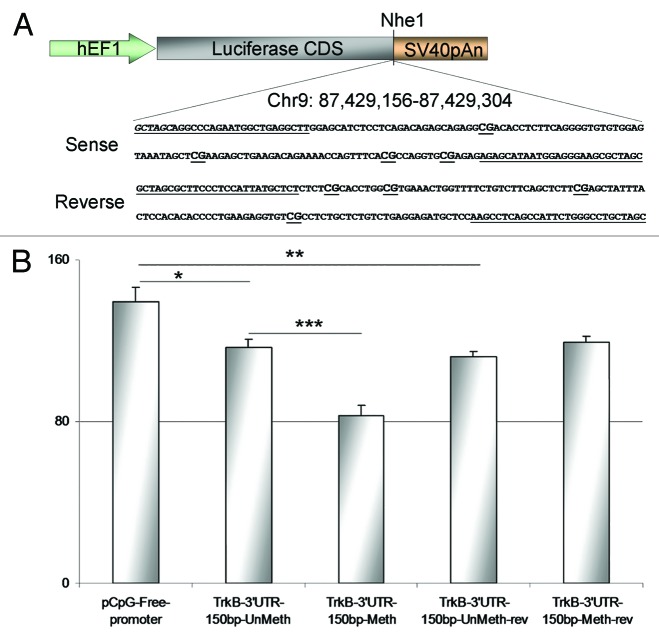
To assess the functional effect of site specific differential methylation in TrkB-T1 3′UTR sequence, a 150 bp fragment containing the CpG6, 7, 8, and 9 was cloned in a pCpGfree vector, 3′ to the luciferase coding sequence, in sense and antisense orientation (Fig. 5A). These constructs (methylated or unmethylated) were transfected into HEK293 and luciferase activity was compared among cells containing native vector, unmethylated or methylated vector.
Figure 5. Functional analysis of site-specific methylation (A) Schematic representation of pCpGfreepromoter lucia vector with 150 bp TrkB-T1 3′UTR DNA sequence inserted 3′ to the luciferase coding sequences before an SV40 polyAn signal. Primers are underlined. CpG island are underlined and labeled in bold. (B) Luciferase activities in HEK293 cells, 24 h after transfection
Twenty-four hours after transfection into HEK293 cells, we found significant differences in luciferase activity among conditions (f = 23.965; df = 20; P = 1.37 × 10−6) (Fig. 5B). Luciferase activity in cells transfected with unmethylated 150 bp TrkB-T1 sequences cloned in sense and antisense orientations were significantly lower (15.9% for sense orientation [P = 0.033] and 19.4% for antisense orientation [P = 0.002]) than luciferase activity in cells transfected with empty vector. A 29.1% decrease of luciferase activity was found in cells transfected with the methylated 150 bp in sense orientation compared with the luciferase activity from cells transfected with the unmethylated constructs (P = 9.08 × 10−5). No significant difference in luciferase activities were found between cells transfected either with: (1) unmethylated sense construct; (2) unmethylated antisense construct, and (3) methylated antisense construct (f = 1873; df = 13; P = 0.199).
These data suggest that the 150 bp sequence containing the significantly differentially methylated CpGs has an inhibitory effect on TrkB-T1 expression. They also suggest that methylation on CpG6, 7, 8, and 9 contributes to TrkB-T1 reduced expression in HEK cells. These results show that methylation has an effect on luciferase activity only when the sequence is cloned in a sense orientation.
Discussion
Previous studies had shown differences in DNA methylation9 and histone modifications5 in the TrkB promoter and microRNA binding10 in TrkB-T1 3′UTR sequences between controls and suicide completers. However, the results from these previous studies suggested that additional mechanisms were necessary to explain the extreme decrease in TrkB-T1 expression observed in frontal cortical areas of suicide completers.
In this study, using a custom designed microarray, we analyzed full methylation patterns of the TrkB gene in suicide completers with significant TrkB-T1 downregulation in the frontal cortex. Our results point to hypermethylation in the 3′UTR region of the TrkB-T1 transcript, in the frontal cortex of suicide completers. We validated these results by bisulfite sequencing and found significant differences in methylation levels at four CpG sites spanning a 150 bp sequence. Interestingly, methylation levels at these sites correlate with TrkB-T1 expression levels in the frontal cortex. A fifth CpG site located at 10 bp downstream of the 150 bp sequence is generated by an A/G polymorphism (rs1624327) that is responsible for high variation in methylation levels at this site. After genotyping a cohort of 28 controls and 33 suicide completers, we found no significant association between that SNP and suicide. Finally, a functional in vitro assay indicated that DNA methylation on the four CpG sites located in the 150 bp sequence coding for TrkB-T1 3′UTR sequence leads to a 30% decrease of luciferase activity. All together, the results of the present study demonstrate that TrkB-T1 expression is partially regulated by DNA methylation in the 3′UTR region. Furthermore, compared with other correlation studies5,9 and functional analyses,10 the effect of methylation on TrkB-T1 3′UTR DNA sequence, described here, suggests that methylation in this DNA region may be among the main causes of TrkB-T1 decrease in suicide completers.
Methylation in the TrkB gene
A previous study by our group found that histone modification and site-specific differential methylation in the TrkB promoter contribute to regulate TrkB-T1 expression.5,9 While the current study did not find evidence of differential methylation in the promoter region, our current results are not discordant from our previous findings. The microarray methodology used in this study integrates methylation levels from several CpG sites,20 whereas the findings reported previously on the TrkB promoter were observed using bisulfite sequencing and, thus, had single base resolution but interrogated a very short sequence. In the array study, each probe covered a 600 bp sequence,20,21 and this method is particularly powerful to detect signals coming from a cluster of differentially methylated CpGs, as was the case for the 3′UTR findings reported here. Together, our results suggest that TrkB-T1 transcription is regulated by multiple factors: DNA methylation and histone modification in the promoter region and DNA methylation and microRNA binding in 3′UTR sequence.
Function of methylation in 3′UTR sequences
Although DNA methylation is a relatively stable epigenetic modification,22 it has primarily been investigated in the promoter regions because of its repressive function on gene expression. Consequently, the functional effect of DNA methylation on other regulatory regions, such as in 3′UTR regions, as well as in other non-coding and coding regions remain still poorly understood, including their functional consequence on gene function. A growing number of studies, however, have recently investigated the role of DNA methylation in the 3′UTR on the regulation of gene expression.23-26
One of the first studies showing differential methylation in the 3′UTR region reported that hypermethylation of the cell cycle inhibitor gene p15INK4b interferes with its expression in primary lymphomas.23 Hypomethylation of CCDN1 3′UTR DNA region, concomitant with an increased expression of the gene, was observed in many neuroblastoma cases.24 Similarly, methylation of Lhx8 DNA 3′UTR region was inversely correlated with the protein expression during different phases of oogenesis in the mouse.26 Finally, hypermethylation in 3′UTR sequence of the PDX1 and OTX1 genes were positively correlated with gene expression in colorectal cancers.25 Whereas promoter methylation has extensively been investigated and correlated with gene expression, our study, together with those cited above, have suggested that methylation in the 3′UTR could regulate the levels of RNA and/or proteins. These findings are in line with the idea that methylation changes in regions other than the promoter can lead to transcript specific regulation. In consequence, TrkB-T1 regulation may result from combined mechanisms involving different epigenetic mechanisms, including promoter DNA methylation, histone modifications, and microRNA binding. In addition, the current study suggests a potential role for methylation of the 3′UTR, specifically targeting regulation of the TrkB-T1 transcript.
Transcript-specific regulation and methylation in the 3′UTR position
Several studies, including ours, have shown that methylation of the 3′UTR has regulatory effects on gene expression. However, the mechanisms by which methylation in the 3′UTR change gene expression are not well understood. One possibility is through impacting regulation by the CTCF factor, which is part of a regulatory complex that binds intragenic regions on insulator sequences.27 A recent study showed that CTCF contributes to regulate alternative splicing. CTCF allows the insertion of exon five of the CD45 gene and promotes the elongation of a weakly expressed RNA,28 whereas DNA methylation alters DNA binding of the CTCF factor. Interestingly, the 150 bp sequence of TrkB-T1 3′UTR sequence has been picked up by a CTCF ChIP-seq experiment in human cell lines.29,30 Furthermore, other epigenetic modifications, such as H3K36me3,30 which has been suggested to be an epigenetic mark contributing to alternative splicing regulation,31 have been found in this region. Characterizing CTCF binding sites and mapping H3K36me3 epigenetic marks on TrkB in the frontal cortex of suicide completers could highlight the mechanism by which DNA methylation on TrkB-T1 3′UTR sequence leads to a specific decrease of TrkB-T1 transcript in context of suicide.
Methylation, vulnerability factors to suicidal behavior, and astrocytic dysfunction
DNA methylation of the TrkB-T1 3′UTR sequence in the brain from suicide completers might be related to changes in the maintenance of synapses32 and/or to inflammatory mechanisms. There is substantial evidence suggesting that suicidal risk is related to molecular deregulations associated with early life events.4 Results of studies investigating gene expression profiles related to suicidal risk, notably in the context of childhood abuse,3,8 suggest that early-life adversity increases vulnerability to suicidal behavior by altering stress response later on in life, through modulation of genes that belong to HPA axis.33,34 Furthermore, stressful events and depressive disorders were shown to alter expression of genes that normally promote cell survival.35-37 Depression and suicidal behaviors were linked to inflammatory processes, as an increased level of kynurenine was found in the CSF of patients diagnosed with MDD and past history of suicide attempts compared with control.38 In addition, our group has found a hypertrophy of astrocytes, cells that act as modulator of the immune response in the CNS,39 in the BA24 of suicide completers.16 Finally, TrkB is part of an astrocytic gene network that has been reported as downregulated in the brain of suicide completers.14 Consequently, differential DNA methylation in the TrkB gene investigated in the current study could be related either to: (1) the molecular changes that translate stress responses; (2) inflammatory processes, which specifically involve astrocytes; or (3) a decrease of astrocytes or a decrease of astrocyte functioning linked to suicidal behaviors. Further studies need to be performed in order to support these hypotheses.
To conclude, the current study, based on the downregulation of the TrkB-T1 transcript observed in brain of suicide completers, focused on the functional effect of methylation of DNA in the TrkB-T1 3′UTR. We found that DNA methylation in the TrkB-T1 3′UTR sequence has an effect on TrkB-T1 expression. Compared with previous findings, DNA methylation in this genomic region seems to be the most important mechanism regulating TrkB-T1 expression in the brain of a subgroup of suicide completers. Further studies need to be performed to investigate: (1) the mechanisms, molecular partners, and interactions by which methylation of 3′UTR DNA sequence affects TrkB-T1 expression and (2) the consequences of TrkB gene methylation on cell functioning and network properties, especially on morphology and astrocytic activation. This study reinforces the idea that astrocytic dysfunction occurs in suicide and depressive pathophysiology, potentially related to impairments in cell survival and communication and/or dysregulation leading to inflammatory processes.
Material and Methods
Subjects
This study was approved by the Douglas Institute Research Ethics Board, and all participating families signed consent forms. Brain tissue was obtained from the Quebec Suicide Brain Bank (QSBB, www.douglasrecherche.qc.ca/suicide). The methylation arrays and validations were performed on a sample of 24 patients, which includes 11 suicides with low TrkB-T1 expression levels (mean age is 37 ± 13 and mean PMI is 26 ± 6), and 13 healthy controls not selected for TrkB-T1 levels (mean age is 33 ± 11 and mean PMI is 29 ± 6). Genotyping of the rs1624327 variant was performed on a total sample of 61 subjects including the 24 individuals that were part of the array study and 22 additional low TrkB-T1 expressors, as well as 15 healthy controls. The mean age of the 33 suicide completers was 39 ± 11 and the average PMI was 49 ± 26. The mean age of the 28 controls was 42 ± 18 and the mean PMI was 32 ± 17. All subjects were male and of French Canadian origin, a homogeneous population with a well-defined founder effect. All subjects died suddenly and could not have undergone any resuscitation procedures or other type of medical intervention, and thus did not experience prolonged agonal states.
Toxicological analyses at the time of death are systematically performed and past history of substance abuse and dependence are investigated. Among the suicide completers, 45% had a history of major depressive disorder (n = 15), 16% were diagnosed with depressive disorder not otherwise specified (n = 5), and 9% (n = 3) had substance dependence. One suicide completer was diagnosed with bipolar disorder and another with a psychotic disorder. Eight suicide completers (24%) did not have evidence of psychopathology and all controls were psychiatrically normal.
Diagnostic procedures
Diagnoses were obtained by means of psychological autopsies performed by trained clinicians using the SCID-I40 with an informant best-acquainted with the deceased, as described elsewhere.41 Briefly, this is a structured diagnostic procedure to elicit diagnostic information by means of proxy-based interviews complemented with medical and coroner records, followed by a consensus diagnosis reached by a panel of clinicians42,43 using DSM-IV criteria.44
DNA extraction
The DNA was extracted from 20 mg of brain tissue using the DNA extraction kit (Qiagen) and the procedure provided by the manufacturer.
Methylated DNA immunoprecipitation, labeling, and hybridization
Methylated DNA was extracted following an adaptation of a methylated DNA immunoprecipitation (meDIP) method developed45 using 5′ methylcytosine antibody bound to sepharose beads. Input, unmethylated, and methylated fractions were purified by phenol-chloroform and precipitated in ethanol. Labeling, hybridization, and data extraction were performed following the manufacturer’s (Agilent Technologies) instructions.
Every subject was hybridized on a separate microarray. Microarrays were scanned (High-Resolution C Scanner; Agilent Technologies), and data were extracted using commercial software (Feature Extraction; Agilent Technologies).
Microarray
A custom-designed 4 × 44K promoter tiling array was used for this study (Agilent Technologies). We tiled a 625 000 bp region centered at the transcription start site of TrkB gene. All regions tiled contain one probe approximately every 100 bp. Extracted microarray intensities were processed and analyzed using the R software environment for statistical computing.46 Data were corrected for multiple tests (FDR = 0.05).47
Bisulfite treatment
The DNA sample were bisulfite treated using the following kit EpiTect Bisulfite Kit (Qiagen) and the protocol defined by the manufacturer.
Primer design and PCR
Primers (shown in Table 4) were design by methyl primer express software to generate three PCR products which covers a 1 kb region of TrkB-T1 3′UTR DNA sequence
Table 4. Primers used for to generate PCR products of TrkB-T1 3′UTR from bisulfite converted DNA and the constructs in pCpGfreepromoter lucia vector.
| Primer | Sequence | Length |
|---|---|---|
| TrkB-T1 3′UTRup1 | TGTTTTTTTT TGTTGGGAAT T | 261 bp |
| TrkB-T1 3′UTRdw1 | ACCCCCATTA AACCATAAAT AA | |
| TrkB-T1 3′UTRup2 | TTTATGGGTG TTTTTAATGT GG | 249 bp |
| TrkB-T1 3′UTRdw2 | AAAAAAAAAA ACCCCTAAAT CAA | |
| TrkB-T1 3′UTRup3 | AGGTTTAGAA TGGTTGAGGT TT | 432 bp |
| TrkB-T1 3′UTRdw3 | ATCAAATTCA AACCAAAAAA AA | |
| TrkB-T13’UTRNHE1 up | GCTAGCAGGC CCAGAATGGC TGAGGCTT | 150 bp |
| TrkB-T13’UTRNHE1 dw | GCTAGCGGCT TCCCTCCATT ATGCTCT |
Cloning
PCR product from bisulfite-treated DNA were cloned into a topo4 TA cloning vector and transformed into the Top10 bacterial strain. Plasmidic DNA from 20 to 30 individual colonies were extracted and sequenced by the Genewiz company according to the guidelines provided by the manufacturer. FASTA sequences and chromatograms were aligned and analyzed using Bioedit version 7.0.9.0 and Geneious version 5.5.2 software
Statistical analysis
Methylation data were analyzed using linear mixed models as implemented in SPSS 18. The best fit model was chosen comparing AIC and AICC scores. CpG were considered as repetitive elements and groups defined by the status control or suicide. The results were corrected for PMI and age as covariates when applicable. P values related to differences in site specific methylation between control and suicide were log transformed when required.
Allelic distributions were assessed by using Chi Square test. Comparisons of TrkB-T1 expression levels in controls and suicide group were also performed using a linear mixed model The graphic representation (Fig. 4) of the partial correlation between methylation of TrkB-T1 3′UTR sequence and TrkB-T1 expression was obtained performing a linear regression under SPSS and translating coordinates with a constant term in X and Y axes in order to get positives values for those two variables. Dot plots of TrkB-T1 expression were performed using GraphPad Prism 4 software.
Genotyping
Individuals were genotyped for the rs1624327 using a SNPassay (Cat. # 4351379) provided by Life Technologies. PCR data were analyzed using SDS software version 2.2.1.
Functional analysis
To assess the functional effect of methylation on TrkB-T1 expression, we cloned in 3′position of the luciferase coding sequence, a 150 bp DNA fragment encompassing CpG6 to CpG9, into the pCpGfreepromoter lucia vector in sense and antisense orientation using the primers (Table 4).
The construct was submitted to a 4-h treatment by Sss1 methylase (New England Biolabs) then purified.
This construct was co-transfected in HEK 293 cells with an equivalent amount of pGL3 control vector used for normalization. Luciferase activities were quantified using the dual luciferase reporter (Promega) kit 24 h after transfection in the cell medium and cellular extracts.
Statistical analyses were performed with SPSS 18 using ANOVA and post-hoc t test.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by NIH grant 8914. G.M. holds a fellowship from the American Foundation for Suicide Prevention.
References
- 1.World Health Organisation. Country Report. 2009
- 2.McGuffin P, Perroud N, Uher R, Butler A, Aitchison KJ, Craig I, Lewis C, Farmer A. The genetics of affective disorder and suicide. Eur Psychiatry. 2010;25:275–7. doi: 10.1016/j.eurpsy.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 3.McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turecki G, Ernst C, Jollant F, Labonté B, Mechawar N. The neurodevelopmental origins of suicidal behavior. Trends Neurosci. 2012;35:14–23. doi: 10.1016/j.tins.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Ernst C, Chen ES, Turecki G. Histone methylation and decreased expression of TrkB.T1 in orbital frontal cortex of suicide completers. Mol Psychiatry. 2009;14:830–2. doi: 10.1038/mp.2009.35. [DOI] [PubMed] [Google Scholar]
- 6.Labonté B, Suderman M, Maussion G, Lopez JP, Navarro-Sánchez L, Yerko V, Mechawar N, Szyf M, Meaney MJ, Turecki G. Genome-wide methylation changes in the brains of suicide completers. Am J Psychiatry. 2013;170:511–20. doi: 10.1176/appi.ajp.2012.12050627. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–40. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 8.Labonté B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, Bureau A, Mechawar N, Szyf M, Meaney MJ, et al. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. 2012;69:722–31. doi: 10.1001/archgenpsychiatry.2011.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst C, Deleva V, Deng X, Sequeira A, Pomarenski A, Klempan T, Ernst N, Quirion R, Gratton A, Szyf M, et al. Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Arch Gen Psychiatry. 2009;66:22–32. doi: 10.1001/archpsyc.66.1.22. [DOI] [PubMed] [Google Scholar]
- 10.Maussion G, Yang J, Yerko V, Barker P, Mechawar N, Ernst C, Turecki G. Regulation of a truncated form of tropomyosin-related kinase B (TrkB) by Hsa-miR-185* in frontal cortex of suicide completers. PLoS One. 2012;7:e39301. doi: 10.1371/journal.pone.0039301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey GN, Ren X, Rizavi HS, Conley RR, Roberts RC, Dwivedi Y. Brain-derived neurotrophic factor and tyrosine kinase B receptor signalling in post-mortem brain of teenage suicide victims. Int J Neuropsychopharmacol. 2008;11:1047–61. doi: 10.1017/S1461145708009000. [DOI] [PubMed] [Google Scholar]
- 12.Gupta VK, You Y, Gupta VB, Klistorner A, Graham SL. TrkB Receptor Signalling: Implications in Neurodegenerative, Psychiatric and Proliferative Disorders. Int J Mol Sci. 2013;14:10122–42. doi: 10.3390/ijms140510122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose CR, Blum R, Pichler B, Lepier A, Kafitz KW, Konnerth A. Truncated TrkB-T1 mediates neurotrophin-evoked calcium signalling in glia cells. Nature. 2003;426:74–8. doi: 10.1038/nature01983. [DOI] [PubMed] [Google Scholar]
- 14.Ernst C, Nagy C, Kim S, Yang JP, Deng X, Hellstrom IC, Choi KH, Gershenfeld H, Meaney MJ, Turecki G. Dysfunction of astrocyte connexins 30 and 43 in dorsal lateral prefrontal cortex of suicide completers. Biol Psychiatry. 2011;70:312–9. doi: 10.1016/j.biopsych.2011.03.038. [DOI] [PubMed] [Google Scholar]
- 15.Chandley MJ, Szebeni K, Szebeni A, Crawford J, Stockmeier CA, Turecki G, Miguel-Hidalgo JJ, Ordway GA. Gene expression deficits in pontine locus coeruleus astrocytes in men with major depressive disorder. J Psychiatry Neurosci. 2013;38:276–84. doi: 10.1503/jpn.120110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres-Platas SG, Hercher C, Davoli MA, Maussion G, Labonté B, Turecki G, Mechawar N. Astrocytic hypertrophy in anterior cingulate white matter of depressed suicides. Neuropsychopharmacology. 2011;36:2650–8. doi: 10.1038/npp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuster JM. The prefrontal cortex--an update: time is of the essence. Neuron. 2001;30:319–33. doi: 10.1016/S0896-6273(01)00285-9. [DOI] [PubMed] [Google Scholar]
- 18.Gorlyn M, Keilp JG, Oquendo MA, Burke AK, John Mann J. Iowa gambling task performance in currently depressed suicide attempters. Psychiatry Res. 2013;207:150–7. doi: 10.1016/j.psychres.2013.01.030. [DOI] [PubMed] [Google Scholar]
- 19.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lippman Z, Gendrel AV, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat Methods. 2005;2:219–24. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- 21.Oefner PJ, Hunicke-Smith SP, Chiang L, Dietrich F, Mulligan J, Davis RW. Efficient random subcloning of DNA sheared in a recirculating point-sink flow system. Nucleic Acids Res. 1996;24:3879–86. doi: 10.1093/nar/24.20.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malumbres M, Pérez de Castro I, Santos J, Fernández Piqueras J, Pellicer A. Hypermethylation of the cell cycle inhibitor p15INK4b 3′-untranslated region interferes with its transcriptional regulation in primary lymphomas. Oncogene. 1999;18:385–96. doi: 10.1038/sj.onc.1202299. [DOI] [PubMed] [Google Scholar]
- 24.Mayol G, Martín-Subero JI, Ríos J, Queiros A, Kulis M, Suñol M, Esteller M, Gómez S, Garcia I, de Torres C, et al. DNA hypomethylation affects cancer-related biological functions and genes relevant in neuroblastoma pathogenesis. PLoS One. 2012;7:e48401. doi: 10.1371/journal.pone.0048401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith JF, Mahmood S, Song F, Morrow A, Smiraglia D, Zhang X, Rajput A, Higgins MJ, Krumm A, Petrelli NJ, et al. Identification of DNA methylation in 3′ genomic regions that are associated with upregulation of gene expression in colorectal cancer. Epigenetics. 2007;2:161–72. doi: 10.4161/epi.2.3.4805. [DOI] [PubMed] [Google Scholar]
- 26.Zhang LJ, Pan B, Chen B, Zhang XF, Liang GJ, Feng YN, Wang LQ, Ma JM, Li L, Shen W. Expression and epigenetic dynamics of transcription regulator Lhx8 during mouse oogenesis. Gene. 2012;506:1–9. doi: 10.1016/j.gene.2012.06.093. [DOI] [PubMed] [Google Scholar]
- 27.Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–97. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 28.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–39. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee BK, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS, et al. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 2012;22:9–24. doi: 10.1101/gr.127597.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aanes H, Østrup O, Andersen IS, Moen LF, Mathavan S, Collas P, Alestrom P. Differential transcript isoform usage pre- and post-zygotic genome activation in zebrafish. BMC Genomics. 2013;14:331. doi: 10.1186/1471-2164-14-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartmann M, Brigadski T, Erdmann KS, Holtmann B, Sendtner M, Narz F, Lessmann V. Truncated TrkB receptor-induced outgrowth of dendritic filopodia involves the p75 neurotrophin receptor. J Cell Sci. 2004;117:5803–14. doi: 10.1242/jcs.01511. [DOI] [PubMed] [Google Scholar]
- 33.Labonte B, Yerko V, Gross J, Mechawar N, Meaney MJ, Szyf M, Turecki G. Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol Psychiatry. 2012;72:41–8. doi: 10.1016/j.biopsych.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 34.Wang SS, Kamphuis W, Huitinga I, Zhou JN, Swaab DF. Gene expression analysis in the human hypothalamus in depression by laser microdissection and real-time PCR: the presence of multiple receptor imbalances. Mol Psychiatry. 2008;13:786–99, 741. doi: 10.1038/mp.2008.38. [DOI] [PubMed] [Google Scholar]
- 35.McEwen BS. The ever-changing brain: cellular and molecular mechanisms for the effects of stressful experiences. Dev Neurobiol. 2012;72:878–90. doi: 10.1002/dneu.20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razzoli M, Domenici E, Carboni L, Rantamaki T, Lindholm J, Castrén E, Arban R. A role for BDNF/TrkB signaling in behavioral and physiological consequences of social defeat stress. Genes Brain Behav. 2011;10:424–33. doi: 10.1111/j.1601-183X.2011.00681.x. [DOI] [PubMed] [Google Scholar]
- 37.Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10:309–22. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- 38.Sublette ME, Galfalvy HC, Fuchs D, Lapidus M, Grunebaum MF, Oquendo MA, Mann JJ, Postolache TT. Plasma kynurenine levels are elevated in suicide attempters with major depressive disorder. Brain Behav Immun. 2011;25:1272–8. doi: 10.1016/j.bbi.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayo L, Quintana FJ, Weiner HL. The innate immune system in demyelinating disease. Immunol Rev. 2012;248:170–87. doi: 10.1111/j.1600-065X.2012.01135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spitzer RL, Williams JB, Gibbon M, First MB. The Structured Clinical Interview for DSM-III-R (SCID). I: History, rationale, and description. Arch Gen Psychiatry. 1992;49:624–9. doi: 10.1001/archpsyc.1992.01820080032005. [DOI] [PubMed] [Google Scholar]
- 41.Dumais A, Lesage AD, Alda M, Rouleau G, Dumont M, Chawky N, Roy M, Mann JJ, Benkelfat C, Turecki G. Risk factors for suicide completion in major depression: a case-control study of impulsive and aggressive behaviors in men. Am J Psychiatry. 2005;162:2116–24. doi: 10.1176/appi.ajp.162.11.2116. [DOI] [PubMed] [Google Scholar]
- 42.Lesage A, Séguin M, Guy A, Daigle F, Bayle MN, Chawky N, Tremblay N, Turecki G. Systematic services audit of consecutive suicides in New Brunswick: the case for coordinating specialist mental health and addiction services. Can J Psychiatry. 2008;53:671–8. doi: 10.1177/070674370805301006. [DOI] [PubMed] [Google Scholar]
- 43.Séguin M, Lesage A, Chawky N, Guy A, Daigle F, Girard G, Turecki G. Suicide cases in New Brunswick from April 2002 to May 2003: the importance of better recognizing substance and mood disorder comorbidity. Can J Psychiatry. 2006;51:581–6. doi: 10.1177/070674370605100906. [DOI] [PubMed] [Google Scholar]
- 44.American Psychiatric Association. American Psychiatric Association. Task Force on DSM-IV. Diagnostic and statistical manual of mental disorders: DSM-IV. Washington, DC: American Psychiatric Association, 1994. [Google Scholar]
- 45.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–53. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 46.Team RDCR. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria 2007. [Google Scholar]
- 47.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.