Abstract
Background
Vitiligo is a depigmenting disorder resulting from loss of functional melanocytes in the skin. NPY plays an important role in induction of immune response by acting on a variety of immune cells. NPY synthesis and release is governed by IL1B. Moreover, genetic variability in IL1B is reported to be associated with elevated NPY levels.
Objectives
Aim of the present study was to explore NPY promoter −399T/C (rs16147) and exon2 +1128T/C (rs16139) polymorphisms as well as IL1B promoter −511C/T (rs16944) polymorphism and to correlate IL1B transcript levels with vitiligo.
Methods
PCR-RFLP method was used to genotype NPY -399T/C SNP in 454 patients and 1226 controls; +1128T/C SNP in 575 patients and 1279 controls and IL1B −511C/T SNP in 448 patients and 785 controls from Gujarat. IL1B transcript levels in blood were also assessed in 105 controls and 95 patients using real-time PCR.
Results
Genotype and allele frequencies for NPY −399T/C, +1128T/C and IL1B −511C/T SNPs differed significantly (p<0.0001, p<0.0001; p = 0.0161, p = 0.0035 and p<0.0001, p<0.0001) between patients and controls. ‘TC’ haplotype containing minor alleles of NPY polymorphisms was significantly higher in patients and increased the risk of vitiligo by 2.3 fold (p<0.0001). Transcript levels of IL1B were significantly higher, in patients compared to controls (p = 0.0029), in patients with active than stable vitiligo (p = 0.015), also in female patients than male patients (p = 0.026). Genotype-phenotype correlation showed moderate association of IL1B -511C/T polymorphism with higher IL1B transcript levels. Trend analysis revealed significant difference between patients and controls for IL1B transcript levels with respect to different genotypes.
Conclusion
Our results suggest that NPY −399T/C, +1128T/C and IL1B −511C/T polymorphisms are associated with vitiligo and IL1B −511C/T SNP influences its transcript levels leading to increased risk for vitiligo in Gujarat population. Up-regulation of IL1B transcript in patients advocates its possible role in autoimmune pathogenesis of vitiligo.
Introduction
Vitiligo is an acquired hypomelanotic disorder characterized by circumscribed depigmented macules in the skin resulting from the loss of functional melanocytes from the cutaneous epidermis. It is frequently associated with positive family history [1]–[2]. The most accepted view in vitiligo pathogenesis is the interaction between genetic and non-genetic factors that influence melanocyte survival and function [3]–[4]. Neurochemical mediators secreted by the nerve endings such as acetylcholine and catecholamines might lead to the destruction of melanocytes [4], [5].
Neuropeptide-Y (NPY) is a 36-amino acid peptide which is able to induce melanocyte dendricity and participates in the regulation of cell substrate adhesion, cell motility and shape [6]. Out of five receptors of NPY, Y1 receptor is present on immune cells [7]. Interleukin-1B (IL1B) is a key regulator of the body's inflammatory responses which exerts numerous biological effects and its defects result in various diseases [8]. IL1B induces biosynthesis and release of NPY which in turn induces the catecholamine release [9]. In order to understand the possible mechanism of action of NPY in vitiligo pathogenesis, neuro-immune-cutaneous system may be considered. NPY is one of the most abundant neuropeptides in the central and peripheral nervous system. Under physiological conditions, NPY is secreted either by sympathetic postganglionic nerve fibers or by activated macrophages and binds to Y1 receptors expressed on B cells, T cells, dendritic cells and macrophages. Interestingly, the NPY system has emerged to have an important role in the induction of a number of immune responses by acting on a variety of immune cells [10]. The possible roles of NPY include immune cell distribution, production of cytokines by T helper cells and release of inflammatory mediators from macrophages [11].
Abnormal release of catecholamines from autonomic nerve endings may play an etiological role in the onset and development of vitiligo by an overproduction of toxic radicals in the microenvironment of melanocytes [5]. Sympathetically-derived NPY is preferentially released, which functions to homeostatically regulate norepinephrine release. Moreover, altered balance of neuropeptides in vitiliginous skin supports the role of nervous system in the pathogenesis of vitiligo [12]. The levels of NPY in the tissue fluids from skin lesions were significantly higher than those from unaffected skin in segmental vitiligo [13]. Thus, NPY might play a critical role in the pathogenesis of vitiligo, via neuro-immune mechanism on the melanocytes. Furthermore, immunological and inflammatory processes, endogenous mediators, and cell and tissue injury can trigger interleukin-1 (IL1) production [14]. IL1B is the predominant form of IL1 being produced and secreted by activated macrophages, monocytes, dendritic cells, natural killer (NK) cells and B cells [15], [16], thereby potentially contributing to the autoimmune pathogenesis of vitiligo via inflammosome formation.
The human NPY gene has two well known single nucleotide polymorphisms (SNPs) i.e., in promoter region, −399 T/C (rs16147) and a non-synonymous polymorphism in exon 2, +1128 T/C (rs16139). Sommer et al. [17] suggested that promoter region variation at −399 T/C results in elevated expression of NPY. The 1498 bp long IL1B gene consists of seven exons and six introns, which encodes for a 269 amino acid protein. IL1B also plays an important role during inflammation, acute phase reaction and apoptosis. IL1B −511C/T (rs16944) polymorphism has been reported to be involved in several autoimmune diseases [18].
The present study was designed to explore the role of NPY in vitiligo pathogenesis by addressing its non-synonymous (+1128 T/C) and promoter (−399 T/C) polymorphisms. In addition, the study investigated IL1B −511C/T promoter polymorphism and its transcript level changes in a case-control approach.
Materials and Methods
Study subjects
The study group included 575 vitiligo patients (256 males and 319 females) who referred to S.S.G. Hospital, Vadodara and Civil Hospital, Ahmedabad, Gujarat, India. A total of 1279 unaffected individuals (552 males and 727 females) were included in this study (Table S1). None of the healthy individuals had any evidence of vitiligo and any other disease. The importance of the study was explained to all participants and written consent was obtained from all patients and controls before performing the studies. The study plan and consent forms were approved by the Institutional ethical committee for human research (IECHR), Faculty of Science, The Maharaja Sayajirao University of Baroda, Vadodara, Gujarat, India.
The diagnosis of vitiligo by dermatologists was clinically based on characteristic skin depigmentation with typical localization and white colour on the skin lesions under Woods lamp and confirmed as two different types: generalized vitiligo (including acrofacial vitiligo and vitiligo universalis) (GV) and localized vitiligo (LV). The patients with active vitiligo had existing lesions spreading and/or new lesions had appeared within the previous 6 months, whereas patients with no increase in lesion size or number in last six months were considered as stable vitiligo.
Genotyping of NPY structural and promoter polymorphisms
Genomic DNA was extracted from whole blood using Qiagen DNA extraction kit (Qiagen, USA). PCR-RFLP method was used to genotype +1128 T/C and −399 T/C polymorphisms of NPY gene. Partial gene sequence containing NPY +1128 T/C SNP was amplified using the forward 5′-ATTGGGGGTCGCGTGTGGTAG-3′ and reverse 5′-GTCCTGCCCTGGGATAGA GCG-3′ primers. Partial gene sequence containing -399 T/C SNP was amplified using the forward 5′-TTCCTACTCCGGCACCCAGTGAG-3′ and reverse 5′-GGGCTTTTATGGAGC TTCCTCGC-3′ primers. The amplified products for +1128 T/C and −399 T/C polymorphisms were 418 bp and 402 bp amplicons respectively. Restriction enzymes BseNI and AluI (Fermentas, Vilnius, Lithuania) were used for detection of NPY +1128 T/C and −399 T/C SNPs respectively (Figs. S1A & B).
Genotyping of IL1B promoter polymorphism
The IL1B −511C/T SNP was genotyped using forward 5′-GTTTAGGAATCTTCCCACTT-3′ and reverse 5′-TGGCATTGATCTGGTTCATC-3′ primers which yielded a 305 bp amplicon. Restriction enzyme Bsu36I (New England Biolabs Inc., UK) was used and it cuts the IL1B amplicon at the ancestral allele ‘T’ to give 113 bp and 192 bp products but the polymorphic C allele remains uncut (Fig. S1C).
Determination of IL1B and GAPDH mRNA expression
RNA extraction and cDNA synthesis
Total RNA from whole blood was isolated using Ribopure blood Kit (Ambion inc. Texas, USA) by following the manufacturer's protocol. RNA integrity was verified by 1.5% agarose gel electrophoresis, RNA purity and yield was determined spectrophotometrically at 260/280 nm. RNA was treated with DNase I (Ambion inc. Texas, USA) before cDNA synthesis to avoid DNA contamination. One microgram of total RNA was used to prepare cDNA using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania) according to the manufacturer's instructions in the MJ Research Thermal Cycler (Model PTC-200, Watertown, MA, USA).
Real-time PCR
The expression of IL1B and GAPDH transcripts were measured by real-time PCR using SYBR Green method and gene specific forward 5′-AGATGAAGTGCTCCTTCCAGG-3′ and reverse 5′-TGGTCGGAGATTCGTAGCTG-3′ primers (Eurofins, Bangalore, India). GAPDH was used as a reference gene and its expression was monitored using forward 5′-ATCCCATCACC ATCTTCCAGGA-3′ and reverse 5′-CAAATGAGCCCCAGCCTTCT-3′ primers (Table S2). Real-time PCR was performed in duplicates in 20 µl volume using LightCycler480 SYBR Green I Master (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer's instructions and carried out in the LightCycler480 Real-Time PCR (Roche Diagnostics GmbH, Mannheim, Germany). The thermal cycling conditions included an initial activation step at 95°C for 10 min, followed by 45 cycles of denaturation, annealing and amplification (95°C for 10 sec., 65°C for 20 sec., 72°C for 20 sec.). The fluorescence data collection was performed during the extension step. At the end of the amplification phase a melt curve analysis was carried out to check the specificity of the products formed. The PCR cycle at which PCR amplification begins its exponential phase and product fluorescence intensity finally rises above background and becomes visible was considered as the crossing point (CP) or cycle threshold (CT). The ΔCT or ΔCP value was determined as the difference between the cycle threshold of target gene (IL1B) and reference gene (GAPDH). The difference between the two ΔCP values (ΔCP Controls and ΔCP patients) was considered as ΔΔCP to obtain the value of fold expression (2−ΔΔCp).
Statistical analyses
The distribution of the genotypes and allele frequencies of NPY and IL1B polymorphisms for patients and control subjects were compared using chi-squared test with 3×2 and 2×2 contingency tables respectively using Prism 4 software (Graphpad software Inc; San Diego CA, USA, 2003). p-values less than 0.017 were considered as statistically significant due to Bonferroni's correction for multiple testing. Odds ratio (OR) with respective confidence interval (95% CI) for disease susceptibility was also calculated. Haplotype analysis was carried out using http://analysis.bio-x.cn/myAnalysis.php [19]. The linkage disequilibrium (LD) coefficients D′ = D/Dmax and r2-values for the pair of the most common alleles at each site were estimated using the Haploview programe version 4.1 [20]. Age of onset analysis and relative expression of IL1B in patient and control groups were plotted and analyzed by Mann-Whitney Wilcoxon test using Prism 4 software (Graphpad software Inc; San Diego CA, USA, 2003). Cochran-Armitage trend test was performed using SAS 9.2 software for analyzing the trend of IL1B transcript levels with respect to the genotype for each group individually. Further, ANOVA's trend test was used to compare the mean ΔCp values across the different genotype categories using SPSS version 20 software.
Results
Analysis of association between NPY exon 2 (+1128 T/C) and promoter (−399 T/C) polymorphisms and vitiligo
We investigated 575 patients and 1279 controls for NPY exon 2 (+1128 T/C) polymorphism and 454 patients and 1226 controls for NPY promoter (−399 T/C) polymorphism. The genotype and allele frequencies for exon 2 (+1128 T/C) and −399 T/C promoter polymorphisms differed significantly between patients and controls (p<0.0001, p = 0.0161 and p<0.0001, p = 0.0035 respectively) (Table 1). The patient population deviated from Hardy-Weinberg equilibrium (HWE), whereas the control population was found to be in HWE for +1128 T/C (p<0.0001 and p = 0.7747 respectively) and for −399 T/C polymorphisms (p<0.0001 and p = 0.0759 respectively) (Table 2). NPY exon 2 (+1128; T/C) and promoter (−399; T/C) polymorphisms when compared between generalized (GV) and localized (LV) vitiligo patients (Table 2), the observed genotype frequencies for these polymorphisms in GV and LV patients with respect to unaffected controls were found to be significantly different (p<0.0001, p = 0.0002 and p = 0.029, p<0.0001). The allelic frequencies also differed significantly for exon 2 (+1128; T/C) (p<0.0001; p = 0.029) and promoter (−399; T/C) polymorphisms (p = 0.029, p = 0.019) respectively for NPY gene in GV and LV patients as compared to controls (Table 2). The observed genotype frequencies for the two polymorphisms also differed in active (AV) and stable vitiligo (SV) patients with respect to unaffected controls (p = 0.0002, p<0.0001 and p<0.0001, p = 0.0002 respectively) (Table 3). The allelic frequencies for both the above polymorphisms differed significantly in the case of AV patients (p = 0.0007 and p<0.0001 respectively) compared to unaffected controls. The allelic frequencies of NPY promoter polymorphism also differed significantly in SV patients as compared to controls (p = 0.0073), but the allelic frequencies of NPY exon 2 (+1128; T/C) polymorphism did not differ in stable vitiligo patients as compared to controls (p = 0.075).
Table 1. Association studies for NPY gene exon 2 (+1128 T/C), promoter (−399 T/C) and IL1B gene promoter (−511 C/T) polymorphisms in Gujarat vitiligo patients and unaffected controls.
| SNP | Genotype or allele | Patients (freq.) | Control (Freq.) | p for Association | p for HWE | Odds ratio (95% CI) |
| Genotype | (n = 575) | (n = 1279) | ||||
| rs16139 NPY Exon 2 (+1128 T/C) | TT | 495 (0.86) | 1130 (0.88) | <0.0001a | 0.7747 (C) <0.0001 (P) | 0.7147 (0.5473–0.9333) |
| TC | 66 (0.11) | 145 (0.11) | ||||
| CC | 14 (0.03) | 4 (0.01) | ||||
| Allele | ||||||
| T | 1056 (0.92) | 2405 (0.94) | 0.0161b | |||
| C | 94 (0.08) | 153 (0.06) | ||||
| Genotype | (n = 454) | (n = 1226) | ||||
| rs16147 NPY Promoter (−399 T/C) | TT | 129 (0.28) | 347 (0.28) | <0.0001a | 0.0759 (C) <0.0001 (P) | 0.7945 (0.6820–0.9255) |
| TC | 162 (0.36) | 581 (0.47) | ||||
| CC | 163 (0.36) | 298 (0.24) | ||||
| Allele | ||||||
| T | 420 (0.46) | 1275 (0.52) | 0.0035b | |||
| C | 488 (0.54) | 1177 (0.48) | ||||
| Genotype | (n = 448) | (n = 785) | ||||
| rs16944 IL1B Promoter (−511 C/T) | CC | 234 (0.52) | 489 (0.62) | <0.0001a | 0.1243(C) 0.0002 (P) | 0.5919 (0.4905–0.7143) |
| CT | 156 (0.35) | 270 (0.34) | ||||
| TT | 58 (0.13) | 26 (0.03) | ||||
| Allele | ||||||
| C | 624 (0.70) | 1248 (0.79) | <0.0001b | |||
| T | 272 (0.30) | 322 (0.21) |
‘n’ represents number of Patients/ Controls, HWE refers to Hardy-Weinberg Equilibrium, (P) refers to vitiligo patients, (C) refers to healthy control individuals and CI represents Confidence Interval.
Vitiligo patients vs. Controls using the chi-squared test with 3×2 contingency table,
Vitiligo patients vs. Controls using the chi-squared test with 2×2 contingency table.
Odds ratio is based on allele frequency distribution. Statistical significance was considered at p value ≤0.017 due to Bonferroni's correction for multiple testing.
Table 2. Association studies for NPY gene exon 2 (+1128 T/C), promoter (−399 T/C) and IL1B gene promoter (−511 C/T) polymorphisms in generalized and localized vitiligo patients from Gujarat.
| SNP | Genotype or allele | Generalized Vitiligo Patients (Freq.) | Localized Vitiligo Patients (Freq.) | Controls (Freq.) | p for Association | p for HWE | Odds ratio (95% CI) |
| Genotype | (n = 335) | (n = 240) | (n = 1279) | ||||
| rs16139 NPY Exon 2 (+1128 T/C) | TT | 270 (0.81) | 225 (0.94) | 1130 (0.88) | <0.0001a 0.0002b | 0.001 (GV) <0.0001 (LV) 0.7747 (C) | 0.5074 (0.3774–0.6749) (GV) 0.6479 (0.3980–1.055) (LV) |
| TC | 55 (0.16) | 11 (0.04) | 145 (0.11) | ||||
| CC | 10 (0.03) | 4 (0.02) | 4 (0.01) | ||||
| Allele | |||||||
| T | 595 (0.89) | 461 (0.96) | 2405 (0.94) | <0.0001c | |||
| C | 75 (0.11) | 19 (0.04) | 153 (0.06) | 0.078d | |||
| Genotype | (n = 264) | (n = 190) | (n = 1226) | ||||
| rs16147 NPY Promoter (−399 T/C) | TT | 68 (0.26) | 61 (0.32) | 347 (0.28) | 0.029a <0.0001b | 0.012 (GV) <0.0001 (LV) 0.0759 (C) | 0.8114 (0.6721–0.9797) (GV) 0.7715 (0.6210–0.9584) (LV) |
| TC | 111 (0.42) | 51 (0.27) | 581 (0.47) | ||||
| CC | 85 (0.32) | 78 (0.41) | 298 (0.24) | ||||
| Allele | |||||||
| T | 247 (0.47) | 173 (0.46) | 1275 (0.52) | 0.029c | |||
| C | 281 (0.53) | 207 (0.54) | 1177 (0.48) | 0.019d | |||
| Genotype | (n = 261) | (n = 187) | (n = 785) | ||||
| rs16944 IL1B Promoter (−511 C/T) | CC | 125 (0.48) | 109 (0.58) | 489 (0.62) | <0.0001a 0.0003b | 0.007 (GV) 0.0145 (LV) 0.1243 (C) | 0.5116 (0.4109–0.6370) (GV) 0.7368 (0.5671–0.9573) (LV) |
| CT | 97 (0.37) | 59 (0.32) | 270 (0.34) | ||||
| TT | 39 (0.15) | 19 (0.10) | 26 (0.03) | ||||
| Allele | |||||||
| C | 347 (0.66) | 277 (0.74) | 1248 (0.79) | <0.0001c | |||
| T | 175 (0.34) | 97 (0.26) | 322 (0.21) | 0.0218d |
‘n’ represents number of Patients/ Controls, HWE refers to Hardy-Weinberg Equilibrium, (GV) refers to Generalized Vitiligo, (LV) refers to Localized Vitiligo, (C) refers to Controls and CI refers to Confidence Interval.
Generalized Vitiligo vs. Controls using the chi-squared test with 3×2 contingency table,
Localized Vitiligo vs. Controls using the chi-squared test with 3×2 contingency table,
Generalized Vitiligo vs. Controls using the chi-squared test with 2×2 contingency table,
Localized Vitiligo vs. Controls using the chi-squared test with 2×2 contingency table.
Odds ratio is based on allele frequency distribution. Statistical significance was considered at p value ≤0.017 due to Bonferroni's correction for multiple testing.
Table 3. Association studies for NPY gene exon 2 (+1128 T/C), promoter (−399 T/C) and IL1B gene promoter (−511 C/T) polymorphisms in active and stable vitiligo patients from Gujarat.
| SNP | Genotype or allele | Active Vitiligo Patients (Freq.) | Stable Vitiligo Patients (Freq.) | Controls (Freq.) | p for Association | p for HWE | Odds ratio (95% CI) |
| Genotype | (n = 756) | (n = 394) | (n = 1279) | ||||
| rs16139 NPY Exon 2 (+1128 T/C) | TT | 314 (0.83) | 181(0.94) | 1130 (0.88) | 0.0002a <0.0001b | 0.0064 (AV) <0.0001 (SV) 0.7747 (C) | 0.6044 (0.451–0.8099) (AV) 1.0757 (0.679–1.7041) (SV) |
| TC | 56 (0.15) | 10 (0.04) | 145 (0.11) | ||||
| CC | 8 (0.02) | 6 (0.02) | 4 (0.01) | ||||
| Allele | |||||||
| T | 684 (0.90) | 372 (0.94) | 2405 (0.94) | 0.0007c | |||
| C | 72 (0.10) | 22 (0.06) | 153 (0.06) | 0.075d | |||
| Genotype | (n = 285) | (n = 169) | (n = 1226) | ||||
| rs16147 NPY Promoter (−399 T/C) | TT | 56 (0.20) | 73 (0.43) | 347 (0.28) | <0.0001a 0.0002b | 0.0003 (AV) 0.0001 (SV) 0.0759 (C) | 0.5717 (0.4744–0.689) (AV) 1.3711 (1.0879–1.7281) (SV) |
| TC | 106 (0.37) | 56 (0.33) | 581 (0.47) | ||||
| CC | 123 (0.43) | 40 (0.24) | 298 (0.24) | ||||
| Allele | |||||||
| T | 218 (0.38) | 202 (0.59) | 1275 (0.52) | <0.0001c | |||
| C | 352 (0.62) | 136 (0.41) | 1177 (0.48) | 0.0073d | |||
| Genotype | (n = 259) | (n = 189) | (n = 785) | ||||
| rs16944 IL1B Promoter (−511 C/T) | CC | 119 (0.46) | 115 (0.61) | 489 (0.62) | <0.0001a 0.08b | 0.0001 (AV) 0.07 (SV) 0.1243 (C) | 0.4454 (0.3586–0.5532) (AV) 0.9314 (0.7085–1.2244) (SV) |
| CT | 90 (0.34) | 66 (0.35) | 270 (0.34) | ||||
| TT | 50 (0.20) | 8 (0.04) | 26 (0.03) | ||||
| Allele | |||||||
| C | 328 (0.63) | 296 (0.78) | 1248 (0.79) | <0.0001c | |||
| T | 190 (0.37) | 82 (0.22) | 322 (0.21) | 0.06d |
‘n’ represents number of Patients/ Controls, HWE refers to Hardy-Weinberg Equilibrium, (AV) refers to Active Vitiligo, (SV) refers to Stable Vitiligo, (C) refers to Controls and CI refers to Confidence Interval.
Active Vitiligo vs. Controls using the chi-squared test with 3×2 contingency table,
Stable Vitiligo vs. Controls using the chi-squared test with 3×2 contingency table,
Active Vitiligo vs. Controls using the chi-squared test with 2×2 contingency table,
Stable Vitiligo vs. Controls using the chi-squared test with 2×2 contingency table.
Odds ratio is based on allele frequency distribution. Statistical significance was considered at p value ≤0.017 due to Bonferroni's correction for multiple testing.
Linkage disequilibrium and haplotype analysis
The LD analysis revealed that the two polymorphisms investigated in the NPY gene were in low LD association (+1128 T/C: −399 T/C; D′ = 0.260, r2 = 0.004) (Table 4). Estimated frequencies of haplotypes differed significantly between vitiligo patients and controls (global p-value <0.0001). However, the TC haplotype was more frequently observed in vitiligo patients and increased the risk of vitiligo by 2.3-fold [p<0.0001; odds ratio (OR): 2.312; 95% confidence interval (CI): (1.790–2.985)] (Table 4).
Table 4. Distribution of haplotypes frequencies for NPY gene structural and promoter polymorphisms (1128 T/C and −399 T/C) among vitiligo patients and controls.
| Haplotype (1128 T/C and −399 T/C) | Vitiligo Patients (Freq. %) (n = 575) | Controls (Freq. %) (n = 1279) | p for Association | p (global) | Odds ratio (95% CI) |
| CC | 1.31 (0.004) | 44.94 (0.060) | <0.0001 | <0.0001 | 0.056 (0.010–0.322) |
| CT | 14.69 (0.040) | 25.06 (0.034) | 0.5690 | 1.210 (0.627–2.332) | |
| TC | 199.69 (0.549) | 257.06 (0.345) | <0.0001 | 2.312 (1.790–2.985) | |
| TT | 148.31 (0.407) | 418.94 (0.561) | <0.0001 | 0.537 (0.416–0.692) |
‘n’ represents number of Patients/ Controls and CI represents Confidence Interval (frequency <0.03 in both control & case has been dropped and was ignored in analysis).
Analysis of association between IL1B −511 C/T promoter polymorphism and vitiligo
The genotype and allele frequencies for −511 C/T promoter polymorphism of IL1B differed significantly between vitiligo patients and controls (p<0.0001; p<0.0001 respectively). The patient population was deviated whereas the control population was found to be in HWE for this polymorphism (p = 0.0002 and p = 0.1243 respectively). This study has 90% statistical power for the effect size 0.1 to detect association of the investigated polymorphisms at p<0.05. Association studies for IL1B promoter (−511; C/T) polymorphism was compared in GV and LV patients from Gujarat (Table 2). The observed genotype and allelic frequencies differed significantly for this polymorphism between GV and LV patients as compared to healthy controls (p<0.0001, p = 0.0003 and p<0.0001, p = 0.0218 respectively). Similar studies were performed to compare the AV and LV patients (Table 3). Both genotype and allelic frequencies for IL1B promoter polymorphism differed significantly between AV patients and controls (p<0.0001 and p<0.0001 respectively). However the genotype as well as allelic frequencies for SV patients did not differ as compared to controls (p = 0.08 and p = 0.06 respectively).
Relative gene expression of IL1B in vitiligo patients and controls
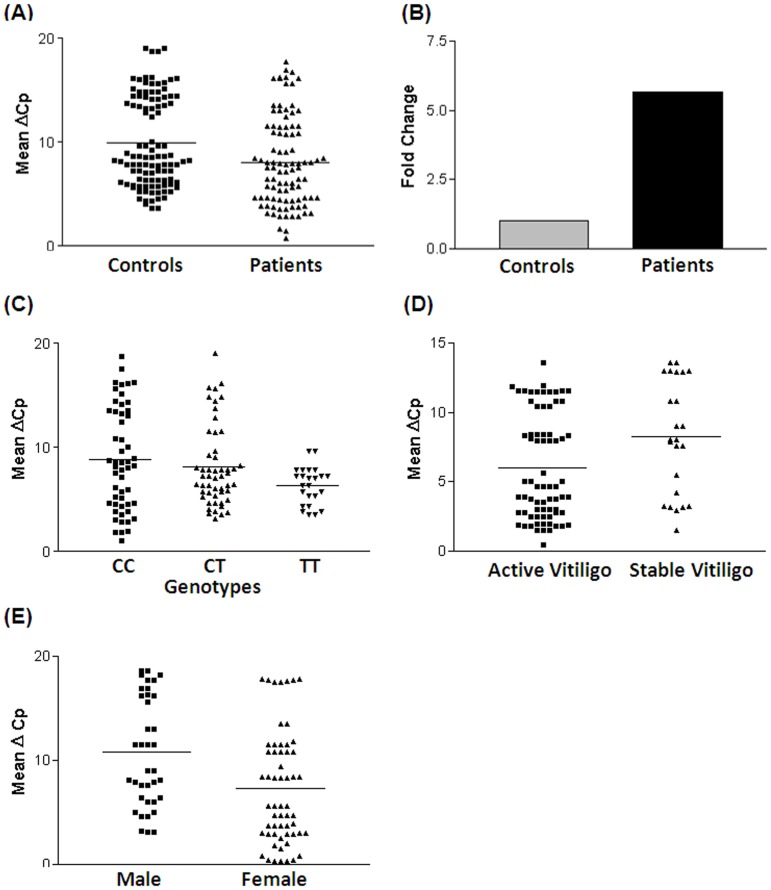
The IL1B transcript levels were compared in 95 vitiligo patients and 105 age matched unaffected controls after normalization with GAPDH transcript levels. The IL1B transcript levels in vitiligo patients were significantly higher than in controls (p = 0.003) as suggested by mean ΔCp values (Fig. 1A). The 2−ΔΔCp analysis showed approximately 6 fold higher expression of IL1B transcript in patients as compared to controls (Fig. 1B).
Figure 1. Relative gene expression of IL1Β in controls and vitiligo patients.
(A) Expression of IL1Β transcripts in 105 (42 male and 63 female) controls, 95 (37 male and 58 female) vitiligo patients, as suggested by Mean ΔCp. Vitiligo patients showed significantly increased mRNA levels of IL1Β as compared to controls (Mean ΔCp±SEM: 9.730±0.5521 vs 7.187±0.6169; p = 0.003). (B) Expression fold change of IL1Β transcripts in 95 vitiligo patients against controls 105 showed 5.83 fold higher expression as determined by 2−ΔΔCp method. (C) Expression of IL1Β transcripts with respect to IL1Β C/T (rs16944) promoter polymorphism as suggested by Mean ΔCp. Individuals with TT genotype showed significantly increased mRNA levels of IL1Β as compared to individuals having CC genotype (Mean ΔCp±SEM: 6.358±0.3659 vs 8.829±0.6770; p = 0.019) and CT genotype (Mean ΔCp±SEM: 6.358±0.3659 vs 8.136±0.5390; p = 0.036). There was no significant difference in the expression of IL1Β between individuals with CC genotype as compared to individuals with CT genotype (Mean ΔCp±SEM: 8.829±0.6770 vs 8.136±0.5390; p = 0.426). Cochran-Armitage trend analysis revealed significant difference between patients and controls (Odds ratio = 0.574, Chi-square = 28.67, p = 8.588e-08) for IL1B transcript levels with respect to different genotype categories. ANOVA's trend test confirmed a significant decrease in the mean ΔCp values for patients (p = 0.035), as compared to controls (p = 0.154). (D) Expression of IL1Β transcripts with respect to activity of the disease in 71 (28 male and 43 female) patients with active vitiligo and 24 (10 male and 14 female) patients with stable vitiligo, as suggested by Mean ΔCp. Active vitiligo patients showed significantly increased mRNA levels of IL1Β as compared to stable vitiligo patients (Mean ΔCp±SEM: 4.720±0.7307 vs 8.285±1.289; p = 0.015). (E) Expression of IL1Β transcripts with respect to gender differences in 37 male patients and 58 female patients with vitiligo, as suggested by Mean ΔCp. Female patients with vitiligo showed significantly increased mRNA levels of IL1Β as compared to male vitiligo patients (Mean ΔCp±SEM: 10.84±1.203 vs 6.808±1.255; p = 0.026).
Correlation of IL1B transcript levels with −511 C/T promoter polymorphism
Expression of IL1Β with respect to IL1Β C/T (rs16944) promoter polymorphism revealed that individuals with TT genotype showed significantly increased IL1Β mRNA levels as compared to individuals having CC (p = 0.019) and CT genotypes (p = 0.036). There was no significant difference in the expression of IL1Β between individuals with CC genotype as compared to individuals with CT genotype (p = 0.426) (Fig. 1C). Further, we performed Cochran-Armitage trend test for analyzing the trend of IL1B transcript levels with respect to −511 C/T promoter polymorphism genotypes for each group individually. We found significant difference in IL1B transcript levels between vitiligo patients and controls (Odds ratio = 0.574, Chi-square = 28.67, p = 8.588e-08) with respect to different genotypes. Moreover, ANOVA's trend test was used to see the change in mean ΔCp values across the different −511 C/T promoter genotypes. The analysis revealed a significant decrease in the mean ΔCp values for patients (p = 0.035), as compared to controls (p = 0.154).
Effect of IL1B expression on disease progression
The transcript levels of IL1B were analyzed with respect to progression of disease in 71 patients with active vitiligo and 24 patients with stable vitiligo. As suggested by mean ΔCp, AV patients showed significant increase in IL1B mRNA levels as compared to SV patients (p = 0.015, Fig. 1D). The elevated levels of IL1B transcript in patients with AV suggest that IL1B may be involved in progression of the disease.
Gender based analysis of IL1B expression
Expression of IL1B transcripts was analyzed with respect to gender differences in 37 male patients and 58 female patients with vitiligo. The female patients showed significant increase in IL1B mRNA levels as compared to male vitiligo patients (p = 0.026, Fig. 1E). However, we did not find any significant difference in IL1B mRNA levels between AV and SV patients based on gender distribution (data not shown).
Discussion
Melanocytes are neural crest derived cells with an embryological link to the nervous system [21]. Neurochemical hypothesis of vitiligo pathogenesis is based on certain abnormalities observed in depigmented skin such as perturbed acetylcholine esterase activity, neuropeptide distribution and catecholamine metabolism [22]. Previously, we reported decrease in acetylcholine esterase activity in vitiligo patients as compared to controls suggesting the involvement of neural factors in pathogenesis of vitiligo [23].
In the present study, we have investigated structural and promoter polymorphisms of NPY which are reported to have strong association with higher levels of NPY. We found that Leu7Pro (exon 2 +1128 T/C) polymorphism as well as (−399 T/C) promoter polymorphism of NPY were significantly associated with vitiligo susceptibility. Moreover, the observed genotype and allele frequencies for these polymorphisms in GV and LV patients were found to be significantly different as compared to controls (Table 2, Table 3). The polymorphisms were also found to be in association with active and stable vitiligo as compared to controls (Table 3). Exon 2 +1128 T/C (Leu7Pro) SNP is located in the signal peptide region which influences the intracellular processing of preproNPY peptide, resulting in altered processing of the prohormone and storage or kinetics of NPY release [24]. The Leu7Pro polymorphism has shown to confer susceptibility to development of diabetic nephropathy in Swedish female type 1 diabetes patients [25]. Moreover, individuals with Leu7/Pro7 genotype have an average 42% maximal increase in plasma NPY as compared to Leu7/Leu7 individuals [24]. The promoter (−399 T/C) polymorphism of NPY was suspected to be responsible for inter-individual variation in resiliency to stress, a risk factor for many diseases [26]. Moreover, elevated serum levels of NPY were detected in patients with asthma and systemic lupus erythematosus suggesting its role in acute inflammatory diseases [27]–[28]. Reactivity against NPY antibody was found to be higher in vitiligo patients in lesional and marginal areas during immunoreactivity studies with a neuronal marker suggesting that NPY may be involved in vitiligo pathogenesis [29]. Caixia et al. [30] reported increase in NPY levels from skin lesion and plasma of vitiligo patients compared to unaffected controls, suggesting the involvement of NPY in vitiligo pathogenesis.
The catecholamine secretion by chromaffin cells is regulated by neuropeptides and cytokines that are co-released with catecholamines or are present in circulating blood [31]–[33]. Further experimental evidences suggested that the role of IL1B is not limited to catecholamine release from chromaffin cells, but is also involved in neuropeptide biosynthesis [34]–[35]. Joana et al. [9] also showed that IL1B plays a critical role in biosynthesis and release of NPY which in turn induces the catecholamine release. L-tyrosine serves as a precursor to melanin pigment, catecholamines and thyroid hormones [36] and L-dihydroxyphenylalanine (L-DOPA) serve as substrates and intermediates of melanogenesis [37]. Depending on genetic makeup of the individual and environmental factors, L-tyrosine and L-DOPA can act as inducers or modifiers of melanogenesis and melanocytic phenotype [38]. It has been suggested that nitrification of L-tyrosine, and other byproducts of melanogenesis, may serve both as a protective means for the cell survival and as an inducer of apoptosis [38]. Moreover, L-DOPA stimulates nitric oxide synthetase [39], and nitrate levels which can serve as markers of nitrosative stress in vitiligo [40]. Such mechanism might play a role in neurodegenerative diseases including vitiligo [39], [41], [42]. In addition, L-tyrosine and L-DOPA both are positive regulators of melanocyte-stimulating hormone (MSH) receptor activity [43], [44]. A dual hormonal control of color change is regulated by MSH and melanin-concentrating hormone (MCH) [45]. Melanin-concentrating hormone receptor 1 (MCHR1) expression has been detected in human melanocytes. Importantly, MCHR1 has been identified as an autoantigen in vitiligo patients [46]. It has been shown that MCH stimulation in cultured human melanocytes, reduced α-MSH-induced increase in cAMP production. Moreover, the melanogenic actions of α-MSH were inhibited by MCH [47]. Therefore, the MCH/MCHR1 system may regulate skin pigmentation through modifications of melanocortin signaling.
It is worth noting that in addition to neuropeptide Y, the neuroendocrine modulators controlling the activity of the hypothalamus-pituitary-adrenal axis are expressed in the skin including CRH, urocortin, and POMC, with its products ACTH, alpha-MSH, and beta-endorphin. These neuroendocrine modulators are produced in the skin under stress [48]–[50]. Therefore, it has been suggested that skin neuroendocrine system communicates with itself and with the systemic level through humoral and neural pathways to induce vascular, immune, or pigmentary changes, to directly buffer noxious agents or neutralize the elicited local reactions [48]. Recently, Slominski et al. [50] has suggested that expression of skin cells producing proinflammatory CRH and anti-inflammatory POMC derived peptides along with CRFR-1α is environmentally regulated, and their dysfunction can lead to skin and systemic diseases that worsen with stress. Environmentally stressed skin can activate both the central and local HPA axis through either sensory nerves or humoral factors to turn on homeostatic responses counteracting cutaneous and systemic environmental damage [50], [51].
There are reports that IL1B evokes catecholamine synthesis by activating the enzyme tyrosine hydroxylase (TH) by Ser40 phosphorylation through different intracellular pathways such as protein kinase C (PKC), protein kinase A (PKA), and mitogen-activated protein kinase (MAPK) [23], [52], [53]. Increased catecholamines inhibits mitochondrial calcium uptake which results in generation of free radicals [54]. Moreover, IL1B is a potent activator of superoxide dismutase 2 (SOD2) in different tissues and cell types [55]; which could lead to increased H2O2 production as observed in vitiligo patients. Interestingly, our recent study suggests increased levels of SOD2 transcripts and SOD2 activity in vitiligo patients which might be one of the consequences of increased IL1B [56].
In the present study, we show that −511C/T promoter polymorphism of IL1B gene is associated with vitiligo in Gujarat population (Table 1). Moreover, higher frequency of TT and CT genotypes in patients than that of controls indicating the profound effect of allele ‘T’ with susceptibility to vitiligo. The observed genotype and allelic frequencies differed significantly for this polymorphism between generalized and localized vitiligo patients with respect to healthy controls. The genotype and allele frequencies differed significantly in active and stable vitiligo patients, suggesting association of this polymorphism with the disease progression (Table 3). Interestingly, the mRNA levels of IL1B in patients were found to be significantly higher than controls suggesting the possible involvement of IL1B in vitiligo pathogenesis (Fig. 1A). The −511 T allele is reported to increase the IL1B mRNA levels [57], [58]. Further, genotype-phenotype correlation analysis revealed that individuals with TT genotype showed significantly increased mRNA levels of IL1Β as compared to individuals having CC genotype (p = 0.019) and CT genotype (p = 0.036), which shows disease-independent effect of rs16944 on IL1B mRNA level. However, there was no significant difference in the expression of IL1Β between individuals with CC genotype as compared to individuals with CT genotype (p = 0.426) (Fig. 1C). Furthermore, Cochran-Armitage trend analysis demonstrated significant increase in IL1B transcript levels in vitiligo patients with respect to the different genotypes compared to those of controls (p = 8.588e-08). Moreover, ANOVA's trend test confirmed and revealed a significant decrease in the mean ΔCp values for patients (p = 0.035), as compared to controls (p = 0.154). Our results showed that the genotype and allele frequencies for -511C/T promoter polymorphism of IL1B differed significantly in active and stable vitiligo patients (Table 3), suggesting association of this polymorphism with the disease progression. Moreover frequency of the susceptible allele −511 T is significantly lesser in stable cases as compared to active cases of the disease which might be one of the reasons to have effect of this SNP with the active form of the disease. In addition, our results also demonstrated increased IL1B mRNA levels in active vitiligo as compared to stable cases (Fig. 1D), further confirming the important role of IL1B in disease progression. Higher IL1B mRNA levels are seen in female patients compared to male patients (Fig. 1E). However, we did not find any significant difference between active and stable vitiligo patients based on gender distribution (data not shown).
Our previous studies suggest that susceptibility to generalized vitiligo involves a number of immune regulatory genes such as cytotoxic T-lymphocyte associated antigen-4 (CTLA4), human leukocyte antigen (HLA), interleukin-4 (IL4), tumor necrosis factor-α (TNFA), tumor necrosis factor-β (TNFB), interferon-gamma (IFNG), melanocyte proliferating gene 1 (MYG1), and NACHT leucine-rich-repeat protein 1 (NLRP1) [59]–[67]. For the first time we report an association between IL1B promoter polymorphism and vitiligo along with higher transcript levels of IL1B in vitiligo patients as compared to controls. NPY synthesis and release is also governed by IL1B [9]. Taken together, altered IL1B transcript levels due to genetic variability in IL1B might be associated with elevated NPY levels in patients with vitiligo. Furthermore, Joana et al. [9] suggested that IL1B induces biosynthesis and release of NPY which in turn induces chromaffin cells to secrete catecholamines which enter the blood flow and interacts with α-adrenergic receptors resulting in vasoconstriction. Repeated vasoconstriction of blood vessels leads to epidermal and dermal hypoxia with hyper production of toxic oxyradicals by different pathways which might lead to the destruction of melanocytes [5]. Recent animal studies, in-vitro cultures and clinical trials provide evidence that a causative role for IL1B as the primary agonist in the loss of beta-cell mass in type 2 diabetes. In vitro, IL1B mediated autoinflammatory process results in beta-cell death [27].
Wheway et al. [10] provided new insights into the role of NPY in innate immunity by showing NPY autocrine stimulation pathway on macrophages. NPY signaling on dendritic cells promotes increased antigen uptake and IL12 secretion while on T cells it promotes Th2 responses by increasing IL4 production. Hernanz et al. [68] reported that NPY enhanced the production of IL6 and TNFα in peripheral blood cells. In addition, our previous studies have shown high levels of serum IL4 and TNFα as well as increased mRNA levels in patients with vitiligo compared to controls [62], [63] suggesting that NPY might be involved in the cell-mediated as well as humoral immune mechanisms and plays a crucial role in melanocyte destruction. However, the exact role of NPY in vitiligo pathogenesis is yet to be elucidated. Similarly, higher IL1B levels can cause autoimmune responses against melanocytes, by initiating a cascade of reactions involving inflammosome formation and ultimately resulting in vitiligo. NLRP1 is a key regulator of the innate immune system, particularly in the skin and the NLRP1 inflammasome promotes caspase 1 dependent cleavage of IL1B precursor in to bioactive IL1B, resulting in IL1B secretion and downstream inflammatory responses. Interestingly, our recent study shows significant association of NLRP1 promoter variants with GV and NLRP1 overexpression in patients compared to controls; which might lead to increased IL1B production in vitiligo patients [67].
IL1B can act on lymphocytes by up regulating IL2 receptor expression, prolonging survival of T cells, enhancing antibody production by B cells and increasing B cell proliferation [69], [70]. IL1B also plays a crucial role in driving the differentiation of Th17 and Th1 cells, ultimately evoking immune response against melanocytes [71]. Our recent studies have shown decrease in regulatory T-cells, increase in CD8+ T-cells and anti-melanocyte antibody levels in GV patients which were positively correlated with disease onset and progression [72], [73].
In conclusion, our findings suggest that NPY exon 2 +1128 T/C, −399 T/C and IL1B −511 C/T promoter polymorphisms are significantly associated with vitiligo susceptibility, which might result in higher levels of IL1B thereby leading to autoimmune mediated responses in vitiligo. In addition, the study also emphasizes the influence of NPY and IL1B genes on disease progression for developing vitiligo.
Supporting Information
(A) PCR-RFLP analysis of NPY (rs16139 T/C) exon 2 polymorphism on 2.5% agarose gel electrophoresis: lanes: 1, 3 & 4 show heterozygous (TC) genotypes; lanes: 2, 5 & 6 show homozygous (TT) genotypes; lane: 7 shows 100 bp DNA ladder. (B) PCR-RFLP analysis of NPY (rs16147 T/C) promoter polymorphism on 10% polyacrylamide gel electrophoresis: lanes: 1 & 5 show homozygous (TT) genotypes; lanes: 2 & 6 show heterozygous (TC) genotypes; lane: 3 shows homozygous (CC) genotype; lane 4 shows 100 bp DNA ladder. (C) PCR-RFLP analysis of IL1Β (rs16944) promoter polymorphism on 2.5% agarose electrophoresis: lanes: 1 & 6 show homozygous (CC) genotypes; lanes: 2, 3 & 5 show heterozygous (CT) genotypes; lane: 4 shows homozygous (TT) genotype.
(TIF)
Demographic characteristics of vitiligo patients and unaffected controls.
(DOC)
Primers and restriction enzymes used for genotyping of NPY, IL1B SNPs and IL1B gene expression.
(DOC)
Acknowledgments
We thank all vitiligo patients and control subjects for their participation in this study. This work was supported by grants to RB (BMS/Adhoc/122/11-2012) ICMR, New Delhi, India; (GSBTM/MD/PROJECTS/SSA/453/2010-2011), GSBTM, Gandhinagar, Gujarat, India and (BT/PR9024/MED/12/332/2007) DBT, New Delhi, India. MSM thanks UGC and NCL thanks CSIR, for awarding JRF/ SRF. We thank Mrs. Sadhana Kannan, ACTREC, Mumbai, India for her kind help in trend analysis.
Funding Statement
This work was supported by grants to RB (BMS/Adhoc/122/11-2012) ICMR, New Delhi, India; (GSBTM/MD/PROJECTS/SSA/453/2010-2011), GSBTM, Gandhinagar, Gujarat, India and (BT/PR9024/MED/12/332/2007) DBT, New Delhi, India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Alkhateeb A, Fain P, Thody A, Bennett DC, Spritz RA (2003) Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res 16: 208–214. [DOI] [PubMed] [Google Scholar]
- 2. Shajil EM, Agrawal D, Vagadia K, Marfatia YS, Begum R, et al. (2006) Vitiligo: Clinical profiles in Vadodara, Gujarat. Ind J Dermatol 51: 100–104. [Google Scholar]
- 3. Glassman SJ (2011) Vitiligo: Reactive Oxygen Species and T-Cells. Clin Sci Lond 120: 99–120. [DOI] [PubMed] [Google Scholar]
- 4. Shajil EM, Chatterjee S, Agrawal D, Bagchi T, Begum R (2006) Vitiligo: pathomechanisms and genetic polymorphism of susceptible genes. Indian J Exp Biol 44: 526–539. [PubMed] [Google Scholar]
- 5. Morrone A, Picardo M, De Luca C, Terminali O, Passi S, et al. (1992) Catecholamines and vitiligo. Pigment Cell Res 5: 65–69. [DOI] [PubMed] [Google Scholar]
- 6. Rozengurt E (1995) Convergent Signalling in the action of integrins, neuropeptides, growth factors and oncogenes. Cancer Surv 24: 81–96. [PubMed] [Google Scholar]
- 7. Prod'homme T, Weber MS, Steinman L, Zamvil SS (2006) A neuropeptide-mediated inflammation, Y? Trends Immunol 27: 164–167. [DOI] [PubMed] [Google Scholar]
- 8.Schwanstecher M (2011) Diabetes-Perspectives in Drug Therapy, Handbook of Experimental Pharmacology. Springer 410p.
- 9. Joana RS, Ines MA, Ana Rita A, Mandes AF, Ferreira L, et al. (2009) Regulation of catecholamine release and tyrosine hydroxylase in human adrenal chromaffin cells by interleukin-1β: role of neuropeptide Y and nitric oxide. J Neurochem 109: 911–922. [DOI] [PubMed] [Google Scholar]
- 10. Wheway J, Mackay CR, Newton RA, Sainsbury A, Boey D, et al. (2005) A fundamental bimodal role for neuropeptide Y1 receptor in the immune system. J Exp Med 202: 1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bedoui S, Miyake S, Straub RH, Horsten SV, Yamamura T, et al. (2004) More sympathy for autoimmunity with neuropeptide Y? Trends Immunol 25: 508–512. [DOI] [PubMed] [Google Scholar]
- 12. Liu P, Bondesson L, Loentz W, Jonansson O (1996) The occurrence of cutaneous nerve endings and neuropeptides in vitiligo vulgaris: a case control study. Arch Dermatol Res 288: 670–675. [DOI] [PubMed] [Google Scholar]
- 13. Tu C, Zhao D, Lin X (2001) Levels of neuropeptide-Y in the plasma and skin tissue fluids of patients with vitiligo. J Dermatol Sci 27: 178–182. [DOI] [PubMed] [Google Scholar]
- 14.Mahé Y, Oppenheim J (1992) Interleukin 1. In: Roitt, IM and Delves, PJ, editors Encycloepedia of Immunology, Harcourt Brace Jovanovich, Sandiego, pp. 897–901.
- 15. Bendtzen K (1998) Interleukin–1, Interleukin-6 and tumor necrosis factor in infection, inflammation and immunity. Immunol Lett 19: 183–192. [DOI] [PubMed] [Google Scholar]
- 16. De Sanctis JB, Blanca I, Bianco NE (1997) Secretion of cytokines by natural killer cells primed with interleukin-2 and stimulated with different lipoproteins. Immunology 90: 526–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sommer W, Lidstrom J, Sun H, Passer D, Eskay R, et al. (2010) Human NPY Promoter Variation rs16147: T>C as a Moderator of Prefrontal NPY Gene Expression and Negative Affect. Hum Mutat 31: 1594–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alyssa K, Robert M, Vincent B, Christopher C, Gonzalez RD, et al. (2008) A Broad Analysis of IL1 Polymorphism and Rheumatoid Arthritis. Arthritis Rheum 58: 1947–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barrett J, Fry B, Maller J, Dally M (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 20. Shi Y, He L (2005) SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15: 97–98. [DOI] [PubMed] [Google Scholar]
- 21.Silver DL, Pavan WJ (2006) The Origin and Development of Neural Crest-Derived Melanocytes. In: Hearing VJ, Leong SPL, editors.From Melanocytes to Malanoma. Humana Press Inc. pp3–26. [Google Scholar]
- 22. Iyenger B (1989) Modulation of melanocytic activity by acetylcholine. Acta Anat (Basel) 136: 139–41. [DOI] [PubMed] [Google Scholar]
- 23. Shajil EM, Marfatia YS, Begum R (2006) Acetylcholine esterase levels in different clinical types of vitiligo in Baroda, Gujarat. Ind J Dermatol 51: 289–291. [Google Scholar]
- 24. Kallio J, Pesonen U, Kaipio K, Karvonen MK, Jaakkola U, et al. (2001) Altered intracellular processing and release of neuropeptide Y due to leucine 7 to proline 7 polymorphism in the signal peptide of preproneuropeptide Y in humans. FASEB J 15: 1242–1244. [PubMed] [Google Scholar]
- 25. Ma J, Nordman S, Möllsten A, Falhammer H, Brismar K, et al. (2007) Distribution of neuropeptide Y Leu7Pro polymorphism in patients with type 1 diabetes and diabetic nephropathy among Swedish and American populations. Eur J Endocrinol 157: 641–645. [DOI] [PubMed] [Google Scholar]
- 26. Zhou Z, Zhu G, Hariri AR, Enoch MA, Scott D, et al. (2008) Genetic variation in human NPY expression affects stress response and emotion. Nature 452: 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dinarello CA, Donath MY, Mandrup-Poulsen T (2010) Role of IL-1beta in type 2 diabetes. Curr Opin Endocrinol Diabetes Obes 17: 314–321. [DOI] [PubMed] [Google Scholar]
- 28. Solomon SS, Griffin WS, K O'Banion (2008) The role of interleukin-1 in neuroinflammation and Alzheimer disease: an evolving perspective. J Neuroinflam 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Al Abadie M, Senior H, Bleehen S, Gawkrodgen D (1994) Neuropeptide and neuronal marker studies in vitiligo. Brit J Dermatol 131: 160–165. [DOI] [PubMed] [Google Scholar]
- 30. Caixia T, Daming Z, Xiran L (2001) Levels of neuropeptide-Y in the plasma and skin tissue fluids of patients with vitiligo. J Dermatol Sci 27: 178–182. [DOI] [PubMed] [Google Scholar]
- 31. Nussdorfer G, Malendowicz L (1998) Role of VIP, PACAP, and related peptides in the regulation of the hypothalamo-pituitary-adrenal axis. Peptides 19: 1443–1467. [DOI] [PubMed] [Google Scholar]
- 32. Cavadas C, Silva A, Mosimann F, Cotrim M, Ribeiro C, et al. (2001) NPY regulates catecholamine secretion from human adrenal chromaffin cells. J Clin Endocrinol Metab 86: 5956–5963. [DOI] [PubMed] [Google Scholar]
- 33. Kobayashi H, Yanagita T, Yokoo H, Wada A (2003) Pathophysiological function of adrenomedullin and proadrenomedullin N-terminal peptides in adrenal chromaffin cells. Hypertens Res 26: S71–78. [DOI] [PubMed] [Google Scholar]
- 34. Ait-Ali D, Turquier V, Grumolato L, Yon L, Jourdain M, et al. (2004) The proinflammatory cytokines tumor necrosis factor-alpha and interleukin-1 stimulate neuropeptide gene transcription and secretion in adrenochromaffin cells via activation of extracellularly regulated kinase 1/2 and p38 protein kinases, and activator protein-1 transcription factors. Mol Endocrinol 18: 1721–1739. [DOI] [PubMed] [Google Scholar]
- 35. Cavadas C, Cefai D, Rosmaninho-Salgado J, Vieira MA, Moura E, et al. (2006) Deletion of the (NPY) Y1 receptor gene reveals a regulatory role of neuropeptide Y on catecholamine synthesis and secretion. Proc Natl Acad Sci USA 103: 10497–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yen PM (2001) Physiological and molecular basis of thyroid hormone action. Physiol Rev 81: 1097–1142. [DOI] [PubMed] [Google Scholar]
- 37. Simon JD, Peles D, Wakamatsu K, Ito S (2009) Current challenges in understanding melanogenesis: bridging chemistry, biological control, morphology and function. Pigment Cell Melanoma Res 22: 563–579. [DOI] [PubMed] [Google Scholar]
- 38. Slominski A, Zmijewski MA, Pawelek J (2012) L-tyrosine and L-dihydroxyphenyl alanine as hormone-like regulators of melanocyte functions. Pigment Cell Melanoma Res 25: 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pacher P, Beckman JS, Liaudet L (2007) Nitric oxide and peroxynitrite in health and disease. Physiol Rev 87: 315–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hazneci E, Karabulut AB, Ozturk C, Batcioglu K, Dogan G, et al. (2005) A comparative study of superoxide dismutase, catalase, and glutathione peroxidase activities and nitrate levels in vitiligo patients. Int J Dermatol 44: 636–640. [DOI] [PubMed] [Google Scholar]
- 41. Tsang AH, Chung KK (2009) Oxidative and nitrosative stress in Parkinson's disease. Biochim Biophys Acta 1792: 643–650. [DOI] [PubMed] [Google Scholar]
- 42. Schallreuter KU, Salem MM, Hasse S, Rokos H (2011) The redox – biochemistry of human hair pigmentation. Pigment Cell Melanoma Res 24: 51–62. [DOI] [PubMed] [Google Scholar]
- 43. McLane J, Osber M, Pawelek JM (1987) Phosphorylated isomers of L-dopa stimulate MSH binding capacity and responsiveness to MSH in cultured melanoma cells. Biochem Biophys Res Commun 145: 719–725. [DOI] [PubMed] [Google Scholar]
- 44. Slominski A, Jastreboff P, Pawelek J (1989) L-tyrosine stimulates induction of tyrosinase activity by MSH and reduces cooperative interactions between MSH receptors in hamster melanoma cells. Biosci Rep 9: 579–586. [DOI] [PubMed] [Google Scholar]
- 45. Slominski A, Tobin DJ, Shibahara S, Wortsman J (2004) Melanin pigmentation in mammalian skin and its hormonal regulation. Physiol Rev 84: 1155–1228. [DOI] [PubMed] [Google Scholar]
- 46. Kemp EH, Waterman EA, Hawes BE, O'Neill K, Gottumukkala RV, et al. (2002) The melanin- concentrating hormone receptor 1, a novel target of autoantibody responses in vitiligo. J Clin Invest 109: 923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hoogduijn MJ, Ancans J, Suzuki I, Estdale S, Thody AJ (2002) Melanin-concentrating hormone and its receptor are expressed and functional in human skin. Biochem Biophys Res Commun 296: 698–701. [DOI] [PubMed] [Google Scholar]
- 48. Slominski A, Wortsman J (2000) Neuroendocrinology of the skin. Endocr Rev 21: 457–487. [DOI] [PubMed] [Google Scholar]
- 49. Slominski AT, Zmijewski MA, Skobowiat C, Zbytek B, Slominski RM, et al. (2012) Sensing the environment: regulation of local and global homeostasis by the skin's neuroendocrine system. Adv Anat Embryol Cell Biol 212: 1–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Slominski AT, Zmijewski MA, Zbytek B, Tobin DJ, Theoharides TC, et al. (2013) Key role of CRF in the skin stress response system. Endocr Rev Dec 34: 827–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Slominski A, Wortsman J, Luger T, Paus R, Solomon S (2000) Corticotropin releasing hormone and proopiomelanocortin involvement in the cutaneous response to stress. Physiol Rev 80: 979–1020. [DOI] [PubMed] [Google Scholar]
- 52. Morita K, Dohi T, Kitayama S, Koyama Y, Tsujimoto A (1987) Enhancement of stimulation-evoked catecholamine release from cultured bovine adrenal chromaffin cells by forskolin. J Neurochem 48: 243–247. [DOI] [PubMed] [Google Scholar]
- 53. Cox M, Parsons S (1997) Roles for protein kinase C and mitogen-activated protein kinase in nicotine-induced secretion from bovine adrenal chromaffin cells. J Neurochem 69: 1119–1130. [DOI] [PubMed] [Google Scholar]
- 54. Laddha NC, Dwivedi M, Mansuri MS, Gani AR, Ansarullah M, et al. (2013) Vitiligo: interplay between oxidative stress and immune system. Exp Dermatol 22: 245–250. [DOI] [PubMed] [Google Scholar]
- 55. Visner GA, Chesrown SE, Monnier J, Ryan US, Nick HS (1992) Regulation of manganese superoxide dismutase: IL-1 and TNF induction in pulmonary artery and microvascular endothelial cells. Biochem Biophys Res Commun 188: 453–462. [DOI] [PubMed] [Google Scholar]
- 56. Laddha NC, Dwivedi M, Gani AR, Shajil EM, Begum R (2013) Involvement of Superoxide Dismutase Isoenzymes and their genetic variants in progression and higher susceptibility towards Vitiligo. Free Rad Biol Med 65: 1110–1125. [DOI] [PubMed] [Google Scholar]
- 57. Hwang IR, Kodama T, Kikuchi S, Sakai K, Peterson LE, et al. (2002) Effect of interleukin 1 polymorphisms on gastric mucosal interleukin 1beta production in Helicobacter pylori infection. Gastroenterology 123: 1793–1803. [DOI] [PubMed] [Google Scholar]
- 58. Hulkkonen J, Laippala P, Hurme M (2000) A rare allele combination of the interleukin-1 gene complex is associated with high interleukin-1 beta plasma levels in healthy individuals. Eur Cytokine Network 11: 251–255. [PubMed] [Google Scholar]
- 59. Dwivedi M, Laddha NC, Imran M, Shah BJ, Begum R (2011) Cytotoxic T lymphocyte associated antigen-4 (CTLA-4) in isolated vitiligo: a genotype-phenotype correlation. Pigment Cell Melanoma Res 24: 737–740. [DOI] [PubMed] [Google Scholar]
- 60. Singh A, Sharma P, Kar HK, Sharma VK, Tembhre MK, et al. (2012) HLA alleles and amino acid signatures of the peptide binding pockets of HLA molecules in Vitiligo. J Invest Dermatol 132: 124–134. [DOI] [PubMed] [Google Scholar]
- 61. Birlea SA, Ahmad FJ, Uddin RM, Ahmad S, Begum R, et al. (2013) Association of Generalized Vitiligo with HLA Class II Loci in Patients from the Indian Subcontinent. J Invest Dermatol 133: 1369–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Imran M, Laddha NC, Dwivedi M, Mansuri MS, Singh J, et al. (2012) Interleukin-4 genetic variants correlate with its transcript and protein levels in vitiligo patients. Brit J Dermatol 167: 314–323. [DOI] [PubMed] [Google Scholar]
- 63.Laddha NC, Dwivedi M, Begum R (2012) Increased Tumor Necrosis Factor (TNF)-α and its promoter polymorphisms correlate with disease progression and higher susceptibility towards vitiligo. PLoS ONE 7: : 12 e52298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Laddha NC, Dwivedi M, Gani AR, Mansuri MS, Begum R (2013) Tumor Necrosis Factor B (TNFB) genetic variants and its increased expression are associated with vitiligo susceptibility. PLoS ONE 8: e81736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dwivedi M, Laddha NC, Shah K, Shah BJ, Begum R (2013) Involvement of Interferon-Gamma Genetic Variants and Intercellular Adhesion Molecule-1 in Disease Onset and Progression of Generalized Vitiligo. J Interferon Cytokine Res 33: 646–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dwivedi M, Laddha NC, Begum R (2013) Correlation of increased MYG1 expression and its promoter polymorphism with disease progression and higher susceptibility in vitiligo patients. J Dermatol Sci 71: 195–202. [DOI] [PubMed] [Google Scholar]
- 67. Dwivedi M, Laddha NC, Mansuri MS, Marfatia YS, Begum R (2013) Association of NLRP1 genetic variants and mRNA overexpression with generalized vitiligo and disease activity in a Gujarat population. Brit J Dermatol 169: 1114–1125. [DOI] [PubMed] [Google Scholar]
- 68. Hernanz A, Tato E, De la Fuente M, de Miguel E, Arnalich F (1996) Differential effects of gastrin-releasing peptide, neuropeptide Y, somatostatin and vasoactive intestinal peptide on interleukin-1 beta, interleukin-6 and tumor necrosis factor-alpha production by whole blood cells from healthy young and old subjects. J Neuroimmunol 71: 25–30. [DOI] [PubMed] [Google Scholar]
- 69. Ben-Sasson SZ, Hu-Li J, Quiel J, Cauchetaux S, Ratner M, et al. (2009) IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc Natl Acad Sci USA 106: 7119–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Maliszewski CR, Sato TA, Vanden Bos T, Waugh S, Dower SK, et al. (1990) Cytokine receptors and B cell functions. I. Recombinant soluble receptors specifically inhibit IL-1- and IL-4-induced B cell activities in vitro. J Immunol 144: 3028–3033. [PubMed] [Google Scholar]
- 71. Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, et al. (2009) Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 30: 576–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dwivedi M, Laddha NC, Arora P, Marfatia YS, Begum R (2013) Decreased regulatory T-Cells and CD4+/CD8+ ratio correlate with disease onset and progression in patients with generalized vitiligo. Pigment Cell Melanoma Res 26: 586–591. [DOI] [PubMed] [Google Scholar]
- 73. Laddha NC, Dwivedi M, Mansuri MS, Singh M, Gani AR, et al. (2014) Role of oxidative stress and autoimmunity in onset and progression of vitiligo. Exp Dermatol 23: 352–353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) PCR-RFLP analysis of NPY (rs16139 T/C) exon 2 polymorphism on 2.5% agarose gel electrophoresis: lanes: 1, 3 & 4 show heterozygous (TC) genotypes; lanes: 2, 5 & 6 show homozygous (TT) genotypes; lane: 7 shows 100 bp DNA ladder. (B) PCR-RFLP analysis of NPY (rs16147 T/C) promoter polymorphism on 10% polyacrylamide gel electrophoresis: lanes: 1 & 5 show homozygous (TT) genotypes; lanes: 2 & 6 show heterozygous (TC) genotypes; lane: 3 shows homozygous (CC) genotype; lane 4 shows 100 bp DNA ladder. (C) PCR-RFLP analysis of IL1Β (rs16944) promoter polymorphism on 2.5% agarose electrophoresis: lanes: 1 & 6 show homozygous (CC) genotypes; lanes: 2, 3 & 5 show heterozygous (CT) genotypes; lane: 4 shows homozygous (TT) genotype.
(TIF)
Demographic characteristics of vitiligo patients and unaffected controls.
(DOC)
Primers and restriction enzymes used for genotyping of NPY, IL1B SNPs and IL1B gene expression.
(DOC)