Abstract
Microbiological data regarding KPC-producing Enterobacter spp. are scarce. In this study, 11 unique KPC-producing Enterobacter isolates were identified among 44 ertapenem-non-susceptible Enterobacter isolates collected between 2009 and 2013 at a hospital system in Western Pennsylvania. All cases were healthcare-associated and occurred in medically complex patients. While pulsed-field gel electrophoresis (PFGE) showed diverse restriction patterns overall, multilocus sequence typing (MLST) identified Enterobacter cloacae isolates with sequence types (STs) 93 and 171 from two hospitals each. The levels of carbapenem minimum inhibitory concentrations were highly variable. All isolates remained susceptible to colistin, tigecycline, and the majority to amikacin and doxycycline. A blaKPC-carrying IncN plasmid conferring trimethoprim-sulfamethoxazole resistance was identified in three of the isolates. Spread of blaKPC in Enterobacter spp. appears to be due to a combination of plasmid-mediated and clonal processes.
Keywords: Klebsiella pneumoniae carbapenemease, Enterobacter cloacae, Enterobacter aerogenes
1. Introduction
Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae has become endemic in many hospitals and long-term care facilities in the United States. The rate of carbapenem resistance has exceeded 10% among Klebsiella isolates causing healthcare-associated infections in U.S. hospitals as of 2010 (Sievert, et al., 2013). Most of this is presumed to be due to production of KPC (Kaiser, Castanheira, Jones, Tenover, & Lynfield, 2013). Enterobacter spp., also part of the family Enterobacteriaceae, ranks eighth among the most common pathogens causing healthcare-associated infections (Sievert, et al., 2013). While less common than in Klebsiella spp., the rate of carbapenem resistance in Enterobacter spp. has reached approximately 4% in U.S. hospitals in 2010 (Sievert, et al., 2013). However, microbiological data regarding KPC-producing Enterobacter spp. are still limited. Here, we report the microbiological characteristics of KPC-producing Enterobacter spp. collected at our hospitals between 2009 and 2013.
2. Materials and methods
2.1. Enterobacter clinical isolates
Enterobacter clinical isolates resistant to ertapenem or meropenem were collected from two clinical microbiology laboratories serving four hospitals in Pittsburgh, Pennsylvania between 2009 and 2013. The isolates were identified as Enterobacter cloacae or Enterobacter aerogenes using either MicroScan WalkAway (Siemens, Tarrytown, NY) or Vitek2 (bioMérieux, Durham, NC) automated instruments in the clinical microbiology laboratories. Only one isolate was collected per patient. De-identified medical records were provided to the investigators for review by a certified honest broker under approval from the University of Pittsburgh Institutional Review Board (PRO12060302).
2.2. Susceptibility testing and identification of antimicrobial resistance genes
The ertapenem-non-susceptible isolates were subjected to PCR for detection of the KPC gene blaKPC (Kim, et al., 2012). The PCR products were sequenced to determine the blaKPC allele. Antimicrobial susceptibility was determined by the broth microdilution method using Sensititre GN2XF (TREK Diagnostics, Cleveland, OH). For carbapenems, the agar dilution method was used to test a wider range of minimum inhibitory concentrations (MICs) (0.06 μg/ml to 256 μg/ml). The assays were conducted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines and their breakpoints (Clinical and Laboratory Standards Institute, 2012a, 2014). Potential ESBL genes blaCTX-M, blaSHV and blaTEM were sought by PCR (Kim, et al., 2012). Amplified products were sequenced to determine whether they represented ESBL genes or not. Plasmid-mediated AmpC β-lactamase genes were sought using previously described PCR primers, except for blaACT/MIR which originates from the E. cloacae chromosome (Perez-Perez & Hanson, 2002). Plasmid-mediated fluoroquinolone resistance genes qnrA, qnrB and qnrS were detected by PCR (Tian, et al., 2010).
2.3. Pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST)
PFGE was conducted to determine the genomic relatedness of the KPC-producing Enterobacter isolates using restriction enzyme XbaI (Kim, et al., 2012). Dendrograms were generated by the weighted pair group method with arithmetic mean using Bionumerics (Austin, TX).
For the 8 E. cloacae isolates, the sequence types (STs) were determined by MLST (Miyoshi-Akiyama, Hayakawa, Ohmagari, Shimojima, & Kirikae, 2013). Novel STs were registered through the database (pubmlst.org/ecloacae/).
2.4. Transfer of blaKPC-encoding plasmids
E. coli TOP10 transformants harboring blaKPC-carrying plasmids were obtained from each clinical isolate by electroporation as described previously (Kim, et al., 2012). The transformants were selected on lysogenic agar plates containing 50 μg/ml of ampicillin or 0.5 μg/ml of ertapenem. The presence of blaKPC was confirmed by PCR.
2.5. Characterization of blaKPC-encoding plasmids
The sizes of blaKPC-encoding plasmids were estimated from the E. coli TOP10 transformants using the S1 nuclease PFGE method (Bueno, Francisco, O’Hara, de Oliveira Garcia, & Doi, 2013). Replicon typing of the plasmids was conducted as described by Carattoli et al. (Carattoli, et al., 2005). Susceptibility of the transformants to ertapenem, tetracycline, gentamicin, amikacin, trimethoprim-sulfamethoxazole and nalidixic acid was tested by the standard disk diffusion method (Clinical and Laboratory Standards Institute, 2012b) to confirm reduced susceptibility to ertapenem and identify co-resistance to non-β-lactams conferred by the blaKPC-carrying plasmids.
3. Results
3.1. Identification of KPC-producing Enterobacter spp
A total of 4,687 unique Enterobacter isolates were identified at two clinical microbiology laboratories serving four hospitals in Pittsburgh, Pennsylvania between 2009 and 2013. Of them, 127 were non-susceptible to ertapenem or meropenem. Forty-four of them were available for testing in the research laboratory, all of which had been reported as non-susceptible to ertapenem in the clinical microbiology laboratories. Among them, 11 unique KPC-producing Enterobacter isolates were identified by PCR. Eight cases were due to E. cloacae, and the remaining 3 cases were due to E. aerogenes. The clinical features of the 11 cases are summarized in Table 1. All affected patients had substantial comorbidity, and had been in hospital for a median of 24 days (range, 0 to 200) before the first KPC-producing Enterobacter spp. isolate was identified. Six patients were deemed to be infected, and the remainder colonized, by the organism. The sources included blood (3), urine (3), post-operative drain (2), sputum (1), bronchoalveolar lavage (1), and cerebrospinal fluid (1). The antimicrobial therapy given was highly variable, both for the empiric and definitive phases. Three patients expired during the hospitalization, three were discharged to another healthcare setting (long-term acute care hospital, skilled nursing facility or hospice) and five were discharged home (Table 1).
Table 1. Clinical features of 11 KPC–producing Enterobacter infections.
| Patient | Age | Year | Comorbid conditions |
Admitted from |
Reason for admission |
Interval from admission to positive culture, days |
Culture site | Infection or colonization |
Empiric therapy | Definitive therapy | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 34 | 2009 | Multiple sclerosis, lacerated liver |
Hospital | Liver transplant | 56 | Post-operative drainage |
Likely colonization |
Tigecycline, metronidazole |
Meropenem, amikacin |
Died |
| 2 | 69 | 2011 | COPD, bladder cancer |
Home | Perforated gallbladder |
12 | Post-operative drainage |
Infection | Meropenem | Tigecycline | Discharged to LTACH |
| 3 | 32 | 2011 | Acute myelogenous leukemia |
Hospital | Encephalopathy | 36 | Blood | Colonization | Meropenem | Not applicable | Discharged to home hospice |
| 4 | 57 | 2011 | Peripheral vascular disease |
Home | Incarcerated umbilical hernia |
42 | Sputum | Likely colonization |
None | Meropenem, colistin |
Discharged to home |
| 5 | 58 | 2011 | Heart transplant | SNF | Hematochezia | 200 | Blood | Infection | Doripenem, colistin |
Doripenem, colistin |
Died |
| 6 | 57 | 2011 | Diabetes, peripheral vascular disease |
SNF | Arm abscess | 21 | Blood | Infection | Meropenem | Amikacin | Discharged to home |
| 7 | 21 | 2012 | Meningioma, complicated by pseudoaneurysm |
Hospital | Mental status change |
24 | Cerebrospinal fluid |
Infection | Cefepime, tobramycin |
Not applicable | Died |
| 8 | 35 | 2012 | Hereditary pancreatitis, esophagectomy |
Hospital | Revision of fundoplication conduit |
14 | Urine | Colonization | None | None | Discharged to home |
| 9 | 61 | 2013 | Quadriplegia | Home | Urinary tract infection |
0 | Urine | Infection | Ciprofloxacin | Amikacin, Fosfomycin |
Discharged to home |
| 10 | 61 | 2013 | Alcohol abuse | Home | Encephalitis | 164 | Urine | Infection | Trimethoprim- sulfamethoxazole |
Trimethoprim- sulfamethoxazole |
Discharged to SNF |
| 11 | 55 | 2013 | Chronic pancreatitis, cirrhosis, diabetes |
Home | Pneumonia | 21 | Bronchoalveolar lavage |
Colonization | None | None | Discharged to home |
COPD, chronic obstructive pulmonary disease; LTACH, long-term acute care hospital; SNF, skilled nursing facility.
3.2. Antimicrobial susceptibility
A wide range of carbapenem MICs were observed among the KPC-producing Enterobacter isolates (Table 2). Ertapenem MICs ranged between 0.25 and 128 μg/ml, and a similar range was observed for the other carbapenems tested as well. Overall, 7, 3 and 1 isolates were susceptible to doripenem, meropenem, imipenem and ertapenem by the agar dilution method, respectively. As expected, most isolates were resistant to cephalosporins and β-lactam/β-lactamase inhibitor combinations. Among the non-β-lactam agents tested, all were susceptible to tigecycline and colistin. All but one isolates were susceptible to amikacin, whereas susceptibility to gentamicin, tobramycin, trimethoprim-sulfamethoxazole and ciprofloxacin was variable. Of note, 7 isolates were susceptible to doxycycline.
Table 2. Antimicrobial susceptibility and plasmid characteristics of 11 KPC–producing Enterobacter isolates.
| # | Year | Species | Source | Minimum inhibitory concentrations (MICs; μg/ml) | KPC type |
ESBL | blaKPC plasmid | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETP | IPM | MEM | DOR | TIM | TZP | FEP | CTX | CAZ | ATM | AMK | GEN | TOB | SXT | CIP | DOX | TGC | CST | Size (Kb) |
Inc type |
Co- resistan ce |
||||||
| 1 | 2009 | E. cloacae | Post- operative drainage |
128 | 32 | 64 | 64 | >128/2 | >64/4 | >16 | >32 | >16 | >16 | ≤4 | >8 | >8 | >4/76 | >2 | ≤2 | 0.5 | 0.25 | KPC- 3 |
SHV-5 | 90 | N | None |
| 2 | 2011 | E. cloacae | Post- operative drainage |
32 | 8 | 16 | 8 | >128/2 | >64/4 | 8 | 16 | 16 | >16 | ≤ | >8 | 2 | ≤0.5/9.5 | ≤0.25 | ≤2 | ≤0.25 | ≤0.25 | KPC- 2 |
None | 30 | FIB | None |
| 3 | 2011 | E. cloacae | Blood | 64 | 16 | 32 | 16 | >128/2 | >64/4 | >16 | >32 | >16 | >16 | ≤4 | >8 | >8 | >4/76 | ≤0.25 | ≤2 | ≤0.25 | 1 | KPC- 3 |
CTX- M-15 |
90 | N | GEN, SXT |
| 4 | 2011 |
E.
aerogenes |
Sputum | 4 | 2 | 2 | 2 | 64/2 | 16/4 | 16 | >32 | 16 | >16 | ≤4 | >8 | 8 | >4/76 | 2 | 8 | ≤0.25 | 1 | KPC- 3 |
CTX- M-15 |
90 | N | SXT |
| 5 | 2011 |
E.
aerogenes |
Blood | 2 | 1 | 1 | 0.5 | >128/2 | 32/4 | 4 | 32 | >16 | >16 | 16 | >8 | >8 | 1/19 | >2 | ≤2 | ≤0.25 | ≤0.25 | KPC- 3 |
SHV- 154 |
190 | NT | None |
| 6 | 2011 | E. cloacae | Blood | 2 | 1 | 0.5 | 0.25 | >128/2 | 64/4 | 8 | 16 | >16 | >16 | 8 | >8 | >8 | >4/76 | >2 | 8 | 0.5 | ≤0.25 | KPC- 2 |
SHV- 12 |
40 | NT | None |
| 7 | 2012 | E. cloacae | Cerebrospi nal fluid |
4 | 2 | 2 | 1 | >128/2 | >64/4 | >16 | >32 | >16 | >16 | ≤4 | >8 | >8 | >4/76 | >2 | >16 | 2 | 1 | KPC- 2 |
CTX- M-15 |
90 | N | SXT |
| 8 | 2012 | E. cloacae | Urine | 2 | 2 | 1 | 0.5 | >128/2 | 16/4 | <2 | 2 | 2 | 8 | ≤4 | ≤1 | ≤1 | ≤0.5/9.5 | ≤0.25 | ≤2 | ≤0.25 | 1 | KPC- 3 |
None | 40 | NT | None |
| 9 | 2013 |
E.
aerogenes |
Urine | 4 | 2 | 1 | 1 | >128/2 | 64/4 | 8 | 16 | >16 | >16 | 32 | >8 | >8 | 2/38 | >2 | 4 | ≤0.25 | 0.5 | KPC- 3 |
SHV- 12 |
120 | NT | None |
| 10 | 2013 | E. cloacae | Urine | 0.25 | 0.5 | 0.25 | 0.12 | >128/2 | 16/4 | ≤2 | 8 | 16 | >16 | ≤4 | 8 | 8 | ≤0.5/9.5 | ≤0.25 | ≤2 | ≤0.25 | 0.5 | KPC- 2 |
SHV-5 | 160 | NT | None |
| 11 | 2013 | E. cloacae | Bronchoalv eolar lavage |
4 | 2 | 1 | 0.5 | >128/2 | >64/4 | 8 | 32 | >16 | >16 | ≤4 | ≤1 | ≤1 | <0. 5/9 . 5 | >2 | 8 | 0.5 | ≤0.25 | KPC- 2 |
SHV-5 | 40 | NT | None |
ETP, ertapenem; IPM, imipenem; MEM, meropenem; DOR, doripenem; TIM, ticarcillin-clavulanic acid; TZP, piperacillin-tazobactam; FEP, cefepime; CTX, cefotaxime; CAZ, ceftazidime, ATM, aztreonam; AMK, amikacin; GEN, gentamicin; TOB, tobramycin; SXT, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; DOX, doxycycline; TGC, tigecycline; CST, colistin; NT, non-typeable. The carbapenems were tested by the agar dilution method. The remainder was tested by the broth dilution method. Bold numbers indicate MICs in the non-susceptible or susceptible-dose dependent range. Plasmid sizes were estimated by S1 nuclease PFGE.
3.4. blaKPC alleles, ESBL, plasmid-mediated AmpC and Qnr genes
Five and 6 isolates possessed blaKPC-2 and blaKPC-3, respectively, distributed in both E. cloacae and E. aerogenes. Nine of the 11 isolates co-produced ESBL. Six isolates had blaSHV-5, blaSHV-12 or blaSHV-154, and 3 isolates harbored blaCTX-M-15. Therefore, like in the case of K. pneumoniae (Endimiani, et al., 2009) and unlike in the case of Escherichia coli (Kim, et al., 2012), co-production of ESBL appeared to be a common phenomenon in KPC-producing Enterobacter spp. In addition, 8 isolates had blaTEM-1, which encodes a non-ESBL, broad-spectrum β-lactamase. No plasmid-mediated AmpC genes were detected. Three and 2 isolates possessed plasmid-mediated fluoroquinolone resistance genes qnrA and qnrB, respectively. However, the blaKPC-harboring transformants were negative for the qnr genes, consistent with their full susceptibility to nalidixic acid.
3.5. Clonality of the clinical isolates
MLST for E. cloacae isolates showed 6 STs, with 4 isolates sharing 2 of the STs (ST93 and ST171). Three of the STs were novel, and were assigned ST252, 253 and 254. None of the identified STs belonged to major clonal complexes that are known to date.
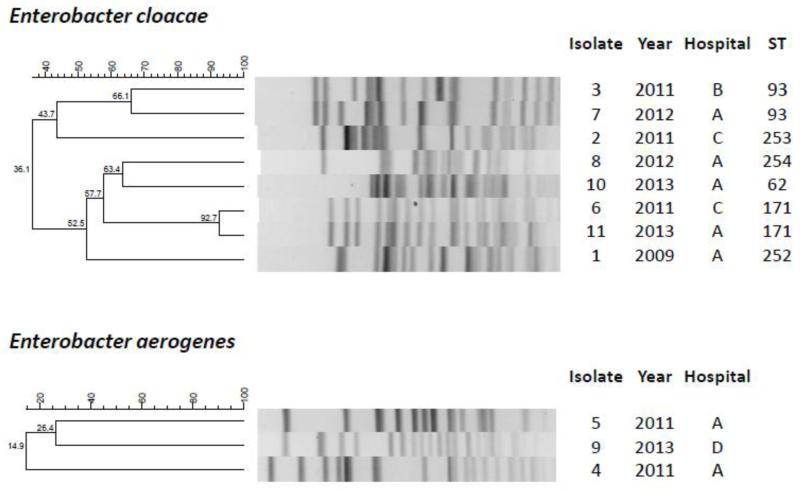
PFGE showed diverse restriction profiles overall (Figure). The 2 E. cloacae ST171 isolates (isolates 6 and 11) shared 92.7% identity, and the 2 E. cloacae ST93 isolates (isolates 3 and 7) shared 66.1% identity. The ST171 cases occurred at 2 hospitals 18 months apart, and the ST93 cases occurred at 2 hospitals 10 months apart. These cases were considered epidemiologically unrelated based on review of the hospitalization history. The level of clonal diversity based on PFGE was greater than that observed in a polyclonal outbreak of KPC-producing E. cloacae that occurred among 16 patients (6 identified by clinical cultures and 10 identified by rectal surveillance cultures) at a Canadian hospital in 2011 (Haraoui, et al., 2013).
Figure.
PFGE profiles and STs of the KPC-producing Enterobacter isolates.
3.6. Characterization of blaKPC-carrying plasmids
The sizes of the blaKPC-encoding plasmids were estimated to be between 30 kb to 190 kb by S1 nuclease PFGE (Table 2). The incompatibility groups could be determined for 5 of the 11 plasmids. The most common was IncN, accounting for 4 plasmids appearing in both species. Their size was approximately 90kb, and 3 of them shared an identical restriction pattern (isolates 3, 4 and 7; data not shown). Another plasmid belonged to IncFIB, but the remaining plasmids were non-typeable. Plasmid-mediated co-resistance to non-β-lactam agents was relatively uncommon, with only 3 plasmids, all IncN, conferring resistance to trimethoprim-sulfamethoxazole and one plasmid also to gentamicin.
4. Discussion
KPC-producing Enterobacteriaceae have become endemic at health care institutions in many parts of the world. Production of KPC is most commonly identified in K. pneumoniae, and its detection in non-K. pneumoniae species remains relatively rare (Munoz-Price, et al., 2013). We here studied the microbiologic characteristics of KPC-producing Enterobacter isolates, which were collected over five years within our hospital system. Carbapenem non-susceptibility rate was 2.7% during this period. Of the 44 unique ertapenem-non-susceptible isolates available for workup, only 11 were found to be positive for blaKPC (25%). Unlike in K. pneumoniae, Enterobacter spp. may become resistant to carbapenems in the absence of acquired carbapenemase by means of porin disruption and derepressed production of chromosomal AmpC (Doumith, Ellington, Livermore, & Woodford, 2009). This may account for the relative low prevalence of blaKPC observed among ertapenem-non-susceptible Enterobacter isolates in our study.
There were several interesting findings in this work. Firstly, the levels of ertapenem MICs were highly variable, including one isolate in the susceptible range (isolate 10; 0.25 μg/ml by agar dilution). The ertapenem MIC for this isolate was initially reported as 4 μg/ml by an automated instrument in the microbiology laboratory. Despite the low ertapenem MIC, this isolate was repeatedly positive for blaKPC by PCR, and yielded a blaKPC-positive transformant with reduced ertapenem susceptibility. The remaining 10 isolates were resistant or intermediately resistant at least to ertapenem. It is well documented that KPC-producing Enterobacteriaceae may present with variable levels of carbapenem resistance (Miriagou, et al., 2010). While the number is relatively small, it appears that KPC-producing Enterobacter spp. testing susceptible to ertapenem under the current CLSI breakpoints is a rare event.
Secondly, the KPC-producing Enterobacter isolates were clonally diverse as defined by PFGE, whereas some of the E. cloacae isolates shared the same ST. While clonal outbreaks of KPC-producing Enterobacter isolates have been reported (Haraoui, et al., 2013; Qin, Yang, Hu, & Zhu, 2014), our findings suggested that the occurrence of KPC-producing Enterobacter spp. remains sporadic and generally polyclonal in the absence of a local outbreak, as was the case in our series. This is in contrast with the highly clonal and global dissemination observed in K. pneumoniae, where a specific MLST clone, defined as sequence type 258 (ST258), and its related STs account for the majority of KPC-producing isolates worldwide including the U.S. (Munoz-Price, et al., 2013). However, 2 of the 6 STs were identified in E. cloacae isolates from different hospitals in the system. Therefore, prospective surveillance and typing of KPC-producing E. cloacae with MLST may be helpful in detecting future expansion of certain STs at an early stage.
Finally, while many blaKPC-carrying plasmids remained non-typeable, four were identified as IncN. Three of them shared an identical size and restriction pattern and conferred resistance to trimethoprim-sulfamethoxazole. IncN is a common incompatibility group observed among blaKPC-encoding plasmids in K. pneumoniae (Gootz, et al., 2009). While resistance genes are generally sparse in fully characterized blaKPC-encoding IncN plasmids, a recently reported IncN plasmid identified in K. pneumoniae from New York City also conferred resistance to trimethoprim-sulfamethoxazole (Chen, et al., 2013). All 4 blaKPC-encoding IncN plasmids we previously identified in E. coli also conferred resistance to this agent (O’Hara, et al., 2014). Therefore, resistance to trimethoprim-sulfamethoxazole may be a hallmark of blaKPC-encoding IncN plasmids currently circulating across Enterobacteriaceae in the Northeast U.S.
In summary, KPC-producing isolates accounted for approximately a quarter of Enterobacter spp. identified as non-susceptible to ertapenem in hospitals in Pittsburgh, Pennsylvania. These isolates caused healthcare-associated infections in medically complex patients. The isolates were diverse in both E. cloacae and E. aerogenes with PFGE, but some E. cloacae isolates from different hospitals shared the same STs by MLST. Some of the blaKPC-encoding plasmids were shared between the two species, including a trimethoprim-sulfamethoxazole-resistant IncN plasmid. These findings suggested a combination of plasmid-mediated and clonal processes underpinning their spread.
Acknowledgments
The authors thank Lloyd Clarke for his assistance in the collection of clinical data presented here, and Dr. Tohru Miyoshi-Akiyama for assignments of new STs.
Funding
The research effort of Y.D. was supported by grants from the National Institutes of Health (R21AI107302 and R01AI104895).
Transparency declarations
Y.D. previously received a research grant from Merck.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The other authors have no potential conflicts of interest.
References
- Bueno MF, Francisco GR, O’Hara JA, de Oliveira Garcia D, Doi Y. Coproduction of 16S rRNA methyltransferase RmtD or RmtG with KPC-2 and CTX-M group extended-spectrum β-lactamases in Klebsiella pneumoniae. Antimicrobial agents and chemotherapy. 2013;57:2397–2400. doi: 10.1128/AAC.02108-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. Journal of microbiological methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Chen L, Chavda KD, Fraimow HS, Mediavilla JR, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrobial agents and chemotherapy. 2013;57:269–276. doi: 10.1128/AAC.01648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard - nineth edition (M07-A9) Wayne; PA: 2012a. [Google Scholar]
- Clinical and Laboratory Standards Institute . Performance standards for antimicrobial disk susceptibility tests; approved standard - eleventh edition (M02-A11) Wayne; PA: 2012b. [Google Scholar]
- Clinical and Laboratory Standards Institute . Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement (M100-S24) Wayne; PA: 2014. [Google Scholar]
- Doumith M, Ellington MJ, Livermore DM, Woodford N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. The Journal of antimicrobial chemotherapy. 2009;63:659–667. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, Oethinger M, Paterson DL, Adams MD, Jacobs MR, Diekema DJ, Hall GS, Jenkins SG, Rice LB, Tenover FC, Bonomo RA. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. The Journal of antimicrobial chemotherapy. 2009;63:427–437. doi: 10.1093/jac/dkn547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrobial agents and chemotherapy. 2009;53:1998–2004. doi: 10.1128/AAC.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraoui LP, Levesque S, Lefebvre B, Blanchette R, Tomkinson M, Mataseje L, Mulvey MR, Miller MA. Polyclonal outbreak of KPC-3-producing Enterobacter cloacae at a single hospital in Montreal, Quebec, Canada. Journal of clinical microbiology. 2013;51:2406–2408. doi: 10.1128/JCM.02480-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser RM, Castanheira M, Jones RN, Tenover F, Lynfield R. Trends in Klebsiella pneumoniae carbapenemase-positive K. pneumoniae in US hospitals: report from the 2007-2009 SENTRY Antimicrobial Surveillance Program. Diagnostic microbiology and infectious disease. 2013;76:356–360. doi: 10.1016/j.diagmicrobio.2013.03.032. [DOI] [PubMed] [Google Scholar]
- Kim YA, Qureshi ZA, Adams-Haduch JM, Park YS, Shutt KA, Doi Y. Features of infections due to Klebsiella pneumoniae carbapenemase-producing Escherichia coli: emergence of sequence type 131. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2012;55:224–231. doi: 10.1093/cid/cis387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miriagou V, Cornaglia G, Edelstein M, Galani I, Giske CG, Gniadkowski M, Malamou-Lada E, Martinez-Martinez L, Navarro F, Nordmann P, Peixe L, Pournaras S, Rossolini GM, Tsakris A, Vatopoulos A, Canton R. Acquired carbapenemases in Gram-negative bacterial pathogens: detection and surveillance issues. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2010;16:112–122. doi: 10.1111/j.1469-0691.2009.03116.x. [DOI] [PubMed] [Google Scholar]
- Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PloS one. 2013;8:e66358. doi: 10.1371/journal.pone.0066358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. The Lancet infectious diseases. 2013;13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara JA, Hu F, Ahn C, Nelson J, Rivera JI, Pasculle AW, Doi Y. Molecular epidemiology of KPC-producing Escherichia coli: Occurrence of ST131-fimH30 subclone harboring pKpQIL-like IncFIIk plasmid. Antimicrobial agents and chemotherapy. 2014 doi: 10.1128/AAC.02182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. Journal of clinical microbiology. 2002;40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Yang Y, Hu F, Zhu D. Hospital clonal dissemination of Enterobacter aerogenes producing carbapenemase KPC-2 in a Chinese teaching hospital. Journal of medical microbiology. 2014;63:222–228. doi: 10.1099/jmm.0.064865-0. [DOI] [PubMed] [Google Scholar]
- Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S, National Healthcare Safety Network, T. Participating NF. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infection control and hospital epidemiology: the official journal of the Society of Hospital Epidemiologists of America. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- Tian GB, Garcia J, Adams-Haduch JM, Evangelista JP, Destura RV, Wang HN, Doi Y. CTX-M as the predominant extended-spectrum β-lactamases among Enterobacteriaceae in Manila, Philippines. The Journal of antimicrobial chemotherapy. 2010;65:584–586. doi: 10.1093/jac/dkp480. [DOI] [PMC free article] [PubMed] [Google Scholar]