Abstract
The development of pancreatic ductal adenocarcinoma (PDAC) is heavily influenced by local stromal tissues, or desmoplasia. Biomimetic hydrogels capable of mimicking tumor niches are particularly useful for discovering the role of independent matrix cues on cancer cell development. Here, we report a photo-curable and bio-orthogonal thiol-ene (i.e., cross-linked by mutually reactive norbornene and thiol groups via photoinitiation) hydrogel platform for studying the growth, morphogenesis, drug resistance, and cancer stem cell marker expression in PDAC cells cultured in 3D. The hydrogels were prepared from multi-arm poly(ethylene glycol)-norbornene cross-linked with protease sensitive peptide to permit cell-mediated matrix remodeling. Collagen 1 fibrils were incorporated into the covalent network while cytokines (e.g., EGF and TGF-β1) were supplemented in the culture media for controlling cell fate. We found that the presence of collagen 1 enhanced cell proliferation and Yes-associated protein (YAP) translocation to cell nuclei. Cytokines and collagen 1 synergistically up-regulated MT1-MMP expression and induced cell spreading, suggestive of epithelial-mesenchymal transition (EMT) in the encapsulated cells. Furthermore, PDAC cells cultured in 3D developed chemo-resistance even in the absence of collagen 1 and cytokines. This phenotype is likely a consequence of the enrichment of pancreatic cancer stem cells that expressed high levels of CD24, sonic hedgehog (SHH), and vascular endothelial growth factor (VEGF).
Keywords: Pancreatic ductal adenocarcinoma, hydrogel, chemo-resistance, pancreatic cancer stem cell, epithelial-mesenchymal transition
1. Introduction
Pancreatic cancer is the fourth leading cause of all cancer-related deaths in the US with more than 43,000 new cases and 37,000 deaths in 2012 [1]. Pancreatic ductal adenocarcinoma (PDAC) accounts for more than 80% of all pancreatic cancers, which are difficult to diagnose at an early stage [2]. The late diagnosis and poor prognosis in PDAC lead to limited treatment options and extremely low survival rate [3]. Chemotherapies are generally not effective because PDAC patients quickly develop chemo-resistance by mechanisms that remain elusive [4,5]. Increasing evidence has suggested that the clinical complications associated with PDAC cells are contributed by a subset of cancer cell populations, namely pancreatic cancer stem cells (PCSCs) [6]. Sharing many characteristics with normal stem cells, cancer stem cells not only self-renew, but also differentiate into mature cancer cells. In addition, these cancer stem cells are prone to metastasize and to resist drug-induced apoptosis [7-9]. Although only 1-5% of total PDAC cell population is characterized as PCSCs [10], this cell population is enriched after drug treatment. PCSCs are characterized by the expression of surface markers CD24, CD44, CD133, and epithelial specific antigen (ESA) [6]. These cells also express high levels of sonic hedgehog (SHH), which has been associated with the resistance of PDAC cells to drug (e.g., gemcitabine) treatment [6,11].
While most of the in vitro PDAC and PCSC studies were conducted in conventional two-dimensional (2D) tissue culture plastics (TCP), increasing evidence has suggested that cells behave in a more pathophysiological relevant manner when cultured in a three-dimensional (3D) niche [12-16]. 3D matrices derived from animal-based products (e.g., Matrigel, collagen gel, etc.) often possess ill-defined compositions and weak mechanical properties. Furthermore, it may be difficult to study the influence of a specific matrix cue on PDAC cell fate using animal models. We hypothesized that a semi-synthetic microenvironment capable of mimicking aspects of pancreatic desmoplasia (i.e., malignant stromal tissues containing high amount of collagen 1, myofibroblastic pancreatic stellate cells, and immune cells) could be used to study how changes in matrix compositions affect PDAC cell behaviors. We have previously shown that hydrogels prepared from norbornene-functionalized 4-arm poly(ethylene glycol) (e.g., PEG4NB) and protease-sensitive peptides supported the growth and morphogenesis of PDAC cells (PANC-1) in 3D [13]. We have also shown that the susceptibility of PANC-1 cells to a peptide drug (NYQQN) in 3D culture is matrix stiffness-dependent [14]. Although these studies shed light on the utility of PEG-based hydrogels on PDAC research in vitro, we have not evaluated the synergistic influences of various extracellular matrix (ECM) cues presenting in the desmoplasia on the growth, epithelial-mesenchymal transition (EMT), and drug resistance in PDAC cells cultured in 3D.
To bridge this knowledge gap and to reveal the potential enrichment of PCSCs in 3D culture, we systematically investigate the effects of cell culture platforms on PDAC cell fate processes. We used COLO-357 cells in this study as this cell type is highly sensitive to cytokine (e.g., TGF-β1) and chemotherapeutic treatment (e.g., gemcitabine). To study the influence of matrix cues on PDAC cell behaviors, we utilized a modular synthetic hydrogel platform prepared from a light-mediated orthogonal thiol-norbornene photochemistry [13,14,17]. We evaluated the cytocompatibility of this desmoplasia-mimetic hydrogel system on in situ encapsulation of PDAC cells. We also examined the synergistic influence of matrix components (e.g., collagen 1 and cytokines) on PDAC cell proliferation and EMT in 3D. Finally, we studied the impact of culture context on chemo-resistance and enrichment of PCSC-like cells in 3D.
2. Materials and Methods
2.1. Materials
4-arm poly(ethylene glycol)-amine (20 kDa) was purchased from JenKem Technology USA. Reagents and chemicals for peptide synthesis were were acquired from Anaspec or Chempep. Bovine type I collagen was purchased from Amsbio. AlamarBlue reagents were purchased from AbD Serotec. Live/Dead staining kit for mammalian cells and DAPI stain were obtained from Invitrogen. Gemcitabine was purchased from TSZ CHEM. YAP rabbit mAb, E-cadherin rabbit mAb, vimentin rabbit mAb, anti-rabbit IgG, anti-mouse IgG HRP-linked, and Alexa Fluor® 488-labeled anti-mouse IgG F(ab’)2 antibodies were obtained from Cell Signaling Technology. hVEGF ELISA kit was purchased from PeproTech. HPLC grade acetonitrile and water were acquired from Fisher Scientific and VWR International, respectively. All other chemicals were purchased from Sigma-Aldrich unless noted otherwise.
2.2. Macromer, photoinitiator, and peptide syntheses
PEG-tetra-norbornene (PEG4NB) was synthesized according to an established protocol [18]. Briefly, 4-arm PEG-NH2 was dried in a vacuum oven overnight and dissolved dimethylformamide (DMF). 5-norbornene-2-carboxylic acid (5 eq. of amine group on PEG) was activated by 2-(1h-benzotriazol-1-yl)-1,1,3,3-tetramethyluronoium hexafluorphosphate (HBTU, 5.5 eq.) and hydroxybenzotriazole (HOBt, 5.5 eq.) in DMF for 3 min at room temperature. Next, N,N-diisopropylethylamine (DIEA, 6 eq.) was added into the solution and stirred for 5 min. The 4-arm PEG-NH2 solution was added to the activated norbornene acid solution. All reactions were performed under nitrogen. After 12 h reaction, the product was filtered and precipitated in cold ethyl ether, dried in a vacuum desiccator, and dialyzed against ddH2O for 3 days. The degree of functionalization (80–90%) was determined using 1H NMR (Avance III 500, Brüker) (Fig. S1). Photoinitiator lithium arylphosphinate (LAP) was synthesized according to a published protocol without modification [19].
MT1-MMP sensitive linker (KCGPLGLYAGCK) was synthesized using Fmoc-Rink-Amide HBMA resin in a microwave-assisted peptide synthesizer (CEM Discover SPS) following standard HOBt/HBTU coupling chemistry. Peptide cleavage was also performed in the microwave peptide synthesizer (38 °C, 20 W, 30 min) using a cleavage cocktail containing 95% trifluoroacetic acid (TFA), 2.5% water, and 2.5% triisopropylsilane (TIS) in the presence of 5% (w/v) phenol. Crude peptide was precipitated in cold ethyl ether, dried overnight in a vacuum desiccator, purified using HPLC (PerkinElmer Flexar System), and characterized by mass spectrometry (Agilent Technologies) (Fig. S2). Purified peptides were lyophilized and stored in −20 °C. The concentration of thiol groups on purified cysteine-containing peptides was quantified using Ellman’s reagent (Pierce).
2.3. Collagen-FAM conjugation
To prepare fluorescently labeled collagen 1 for visualizing collagen distribution in hydrogels, bovine type 1 collagen was diluted in PBS at a working concentration of 2 mg/mL. 5(6)-Carboxyfluorescein succinimidyl ester (5(6)-FAM-SE, from Anaspec) was dissolved in dimethyl sulfoxide (DMSO) and diluted 100-fold to yield a working concentration of 1 mg/mL. pH of the mixture was adjusted to 7.5 using 7.5% (w/v) of sodium bicarbonate aqueous solution. The mixture was protected from light and incubated on ice for 2 h. The product was used without further purification.
2.4. Cell culture and encapsulation
COLO-357 cells were maintained in high glucose Dulbecco’s modified eagle medium (DMEM, HyClone) containing 10% of fetal bovine serum (FBS) (Gibco) and penicillin streptomycin (Gibco, 50 U/mL penicillin, 50 U/mL streptomycin). Prior to cell encapsulation, pre-polymer solutions containing PEG4NB, dithiol cross-linker (DTT or MT1-MMP-sensitive peptide), and 1 mM of LAP were prepared according to the hydrogel formulations shown in Table S1. COLO-357 cells were trypsinized and suspended in a prepolymer solution at 2×106 cells/mL. 25 μL of pre-polymer solution was placed in a 1 mL disposable syringe with a cut-open tip. The sample was exposed to 365 nm light at 5 mW/cm2 for 2 min (Fig. 1A-D).
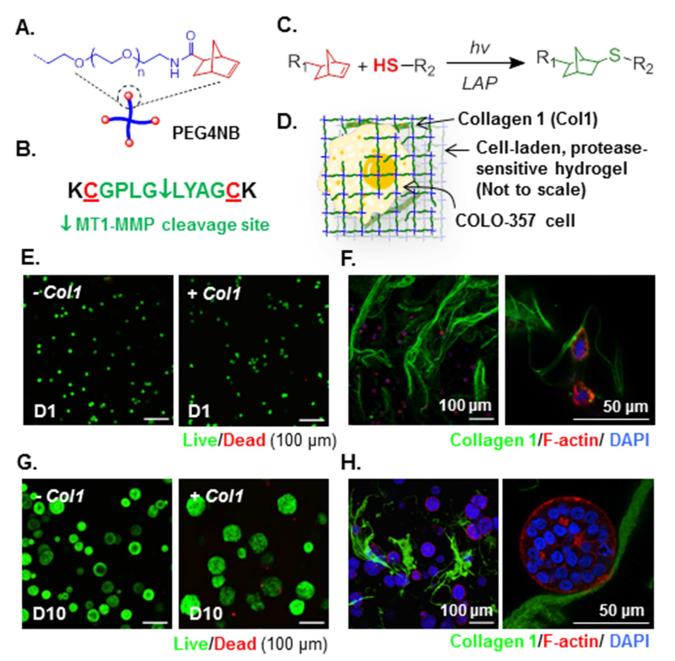
Figure 1.
(A) Schematic of amide-linked PEG-tetra-norbornene (PEG4NB). (B) Bis-cysteine bearing MT1-MMP sensitive peptide cross-linker. (C) Light-mediated thiol-ene reaction (R1: PEG, R2: MT1-MMP sensitive linker). (D) Schematic of a COLO-357 cell encapsulated in the desmoplasia-mimetic hydrogel network. (E) Day 1 Live/dead images of encapsulated cells (Z-stack: 100 μm, live: green, dead: red). (F) Fluorescence-staining of encapsulated cells and entrapped Col1 (two magnifications) at Day 1 post-encapsulation. (G) Day 10 Live/dead staining images of encapsulated cells (Z-stack: 100 μm, live: green, dead: red). (H) Fluorescence-staining of encapsulated cells and entrapped Col1 (two magnifications) at Day 10 post-encapsulation.
2.5. Characterization of cell-laden hydrogel
Shear moduli of the cell-laden hydrogels were measured on a digital rheometer (Bohlin CVO 100). To prepare flat gels for rheomety analysis, cell-laden hydrogels were prepared between two glass slides separated by 1 mm thick spacers. Circular discs were punched out from the hydrogel slab using a biopsy punch (diameter 8 mm). Shear moduli of the cell-laden hydrogels were measured at 1- and 10-day post-encapsulation. To visualize cell morphology and collagen distribution, cell-laden hydrogels were collected 1- and 10-day post-encapsulation and fixed with 4% paraformaldehyde solution for 20 min at RT, followed by washing and permeabilization with 1 mg/mL sapponin. The hydrogels were incubated with 100 nM rhodamine phalloidin (Cytoskeleton Inc.) solution for 2 h at RT and counter-stained with DAPI. Confocal images of the stained samples were obtained using an Olympus Fluoview FV100 laser scanning microscope. To identify proliferating cells, 10 nM of EdU reagent (Click-IT® EdU staining kit) was added into the culture media 9 days post-encapsulation, followed by incubation at 37 °C for 24 hr. The cell-laden gels were collected and rinsed with PBS twice, fixed with 4% paraformaldehyde solution for 20 min, and permeabilized with 0.5% Triton X-100 for 20 min. The gels were washed with PBS twice and immersed in EdU reaction cocktail prepared following manufacturer’s instruction. The gels were counter-stained with DAPI for 1 h and the stained gels were imaged by confocal microscopy.
Cell viability in 2D was determined by MTT assay. COLO-357 cells seeded in 96-well plate at 10,000 cells/well were cultured for 1 day, followed by treatment of cells with gemcitabine-containing media (10−1~105 nM) for 4 days (media change every 2 days). After the incubation, the old media were removed and each well was rinsed with PBS. Then, 10 μL of 5 mg/mL MTT solution was diluted in 100 μL of culture medium and added to the cells for 4 h. The purple formazan crystal formed was dissolved by dimethyl sulfoxide, followed by quantifying the absorbance at 540 nm. Cell morphology was observed using Live/Dead staining and confocal imaging. Cell-laden hydrogels were incubated in Live/Dead staining solution for 1 h at room temperature with gentle shaking. Z-stack images (100 μm thick, 10 μm per slice) of at least four random fields were acquired by using the confocal microscope. Initial cell viability was obtained by counting live and dead cells from 12 randomly selected z-stack live/dead images. The live cell number was divided by total cell count to obtain initial cell viability. Cell cluster diameters were measured 10-day post-encapsulation. For non-spherical clusters, the distance along the longest axis in a cluster was measured instead of diameter.
2.6. RNA isolation, reverse transcription PCR and real-time PCR
For RNA extraction, gels were collected in DNase/RNase-free microtubes, flash frozen with liquid nitrogen, and stored at −80 °C until use. RNA extraction was performed using a combination of guanidinium thiocyanate-phenol-chloroform extraction [20] and NucleoSpin RNA II kit (Clontech). In brief, frozen gels were homogenized in 900 μL of QIAzol (Qiagen) as the samples thaw and incubated at room temperature for 5 min. The samples were filtered through NucleoSpin Filters to clear lysates before the addition of 180 μL of 1-bromo-3-chloropropane (BCP, Sigma). The mixtures were vortexed for 15 s and incubated at room temperature for 3 min before centrifuging at 13800 g for 15 min at 4 °C for aqueous/organic phase separation. Subsequently, the colorless aqueous layers (~600 μL) were transferred to clean DNase/RNase-free microtubes, followed by the addition of equal volume of RNase-free 70% ethanol, and vortexed for 20 s. The mixtures were then added to NucleoSpin RNA columns, and RNA isolation was performed by following the manufacturer’s protocol. The final total RNA products were eluted in 30 μL of DNase/RNase-free water, and the yield was quantified by UV spectrometry. Aliquots of RNA samples were stored at −80 °C until use. In a reverse transcription reaction, 250-500 ng of the isolated total RNA were converted into single-stranded cDNA using the PrimeScript RT reagent kit (Clontech). Quantitative real-time PCR was performed by using SYBR Premix Ex Taq II kit (Clontech) on an Applied Biosystems 7500 fast real-time PCR machine as described in the manufacturer’s protocol. The samples were run at 95 °C for 30 s, followed by 45 cycles of 95 °C for 5 s, 55 °C for 30 s, and 72 °C for 30 s. The amplification/SYBR signals were detected at the end of each cycle when 30 s at 72 °C was completed. The relative quantity of each gene was normalized to the GAPDH internal control and evaluated by using the 2−ΔΔCT method. Forward and reverse primers were listed in Table S2.
2.7. Western blot
Total protein from the encapsulated cells was extracted by homogenizing cell-laden hydrogels using a pellet mixer and lysed with RIPA buffer containing halt protease inhibitor (10 μg/mL), 5 mM EDTA, and 20 μM of phenylmethylsulfonyl fluoride (PMSF). Protein extract was cleared by centrifugation at 8,000 rpm for 10 min at 4 °C and concentrated using a centrifugal filter unit (VWR) at 12,000 rpm for 20 min at 4 °C. Total protein concentration was determined by micro-BCA protein quantification kit (Pierce), separated by SDS-PAGE, and transferred to PVDF transfer membrane using a Trans-Blot Turbo Transfer System (BioRad). The blots were blocked overnight with 5% nonfat milk in TBS solution (PBS containing 0.05% Tween 20) at 4 °C, followed by incubation in primary antibody (rabbit anti-β-vimentin 1:400, rabbit anti-E-cadherin 1:400, or mouse anti-β-actin 1:450) and HRP-conjugated secondary antibody (Anti-rabbit IgG, Anti-mouse IgG, 1:800) for 1 h each at room temperature. The blots were washed with TBS solution for 1 h and treated with a chemiluminescence detection kit (SuperSignal West Pico Detection Kit, Thermo Scientific). Immunoblotting images were obtained using a chemiluminescence imaging system (LAS3000, Fuji Film).
2.8. Immunofluorescence staining and confocal microscopy
YAP and CD24 expression in the encapsulated cells was visualized by immunofluorescence staining and confocal microscopy. Cell-laden hydrogels were fixed in 4% paraformaldehyde at room temperature for 20 min with gentle shaking. Samples were then rinsed with PBS and the encapsulated cells were permeabilized with 1 mg/mL of saponin in PBS at room temperature for 20 min with gentle shaking. The samples were then washed with PBS and blocked with 1% BSA solution overnight at 4 °C, followed by overnight incubation with rabbit anti-YAP (1:100) or mouse-anti-CD24 (Santa Cruz Biotech, 1:100) at 4 °C. After 3 washes with PBS, the samples were incubated with 1:100 diluted goat anti-Rabbit IgG (H+L)-FITC (from KPL) or Alexa Fluor® 488-labeled goat anti-mouse IgG F(ab’)2 overnight at 4 °C. The samples were rinsed with PBS and counter-stained with DAPI for 1 h. The cells were visualized and imaged by confocal microscopy.
2.9. Gemcitabine resistance, cell viability, and apoptosis assays
To evaluate the responsiveness of COLO-357 cells to gemcitabine treatment, cells were seeded into a 96-well plate at 5~40×103 cells/well and incubated for one day. 1 μM of gemcitabine was added to the cells for 4 days with media change every 2 days. The metabolic activity of drug-treated cells was measured by AlamarBlue assay with 2 h incubation time. To assess drug resistance of cells in 3D, the encapsulated cells were treated with 1 μM of gemcitabine on day-10 post-encapsulation for 4 additional days. Metabolic activity and morphology of the cells were assessed by AlamarBlue reagent and Live/dead staining, respectively. Cell apoptosis was assessed by caspase 3/7 activity assay using cells recovered from the cell-laden hydrogels. Gels were eroded with 1 mg/mL of α-chymotrypsin for 15 min to liberate cell spheroids, which were dissociated into single cells using 1× trypsin solution. The harvested cells were re-suspended in PBS and transferred to a 96-well plate (2×104 cells/well). 100 μL of Caspase-Glo® reagent (Promega) was added to the cells for 1 h and the luminescence was measured by a microplate reader (Synergy HT, BioTek).
2.10. Flow cytometry
Single cells harvested from 2D culture or hydrogels (see section 2.9) were fixed by 4% paraformaldehyde for 20 min before the permeabilization by 1 mg/mL of saponin for 20 min. After washed by PBS, cells were blocked with1% BSA for 1 hr, and immuno-stained by mouse-anti-CD24 (Santa Cruz, 1:100) and rat-anti-CD44 (Thermo Scientific, 1:100) antibodies for 2 h. After three PBS washes, cells were incubated with Alexa Fluor® 488-labeled goat anti-mouse IgG F(ab’)2 (Cell signaling, 1:100) and PE conjugated goat anti-rat IgG-R (Santa Cruz, 1:100) for 30 min. After washing with PBS twice, the immuno-stained cells were analyzed by flow cytometry using BD FACSCalibur (BD Biosciences).
2.11 SHH and VEGF enzyme-linked immunosorbent assay (ELISA)
The secretion of SHH and VEGF from encapsulated cells was analyzed by using SHH and VEGF ELISA assay kits (obtained from Sigma-Aldrich and PeproTech, respectively) following manufacturers’ instructions.
2.12 Statistics
All experiments were conducted independently for at least three times, and the results were presented as mean ± SEM. Each condition or gel formulation used in each experiment contained at least three gel samples. Statistical significance was determined using one-way ANOVA test. Difference was considered statistically significant when p < 0.05.
3. Results
3.1. Desmoplasia-mimetic hydrogels for culturing PDAC cells in 3D
To prepare desmoplasia-mimetic hydrogels, poly(ethylene glycol)-tetra-norbornene (PEG4NB, Fig. 1A and Fig. S1) was cross-linked by a bis-cysteine terminated peptide substrate (sequence: KCGPLG*LYAGCK [21], *protease cleavage site. Fig. 1B and Fig. S2). A PEG-based macromer was preferred because of its hydrophilic, non-fouling, and cytocompatible property [22]. Further, the peptide cross-linker has high specificity for membrane type 1-matrix metalloproteinase (MT1-MMP). The use of this peptide linker permits cell invasion into neighboring matrix caused by membrane-tethered protease activity. Type 1 collagen (Col1), the major matrix component in the pancreatic desmoplasia [23-25], was physically entrapped in some hydrogels during light-induced orthogonal thiol-ene gelation and cell encapsulation (Fig. 1C-D). The entrapment of Col1 allows controlled cell-matrix binding independent of other matrix biophysical properties. The orthogonal thiol-ene hydrogels exhibited high cytocompatibility for in situ encapsulation of COLO-357 cells, a Smad4-positive PDAC cell line highly responsive to transforming growth factor-β (TGF-β1) [15,26]. High percentages of live cells (> 90%) were found in all live/dead staining images of cell-laden protease-sensitive hydrogels (Fig. 1E) and bio-inert hydrogels cross-linked by dithiothreitol (DTT) (Fig. S3). The incorporation of Col1 (FAM-labeled, green in Fig. 1F) in hydrogels promoted the formation of larger multi-cell clusters 10-day post-encapsulation (Fig. 1G and Fig. S4A). The increased cell cluster size may be a result of enhanced cell-Col1 interactions (Fig. 1H).
3.2. Effect of collagen 1 on PDAC cell proliferation, Hippo pathway inhibition, and gel property
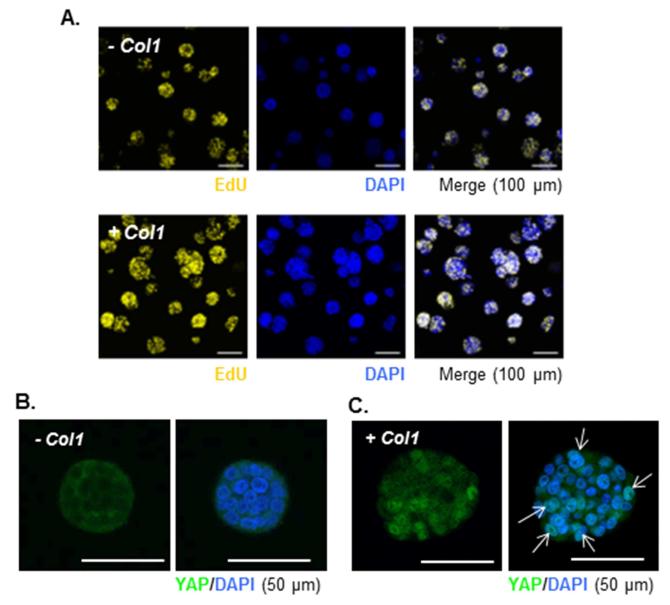
Recent studies have revealed the negative effect of Hippo pathway activation on cell proliferation [27-29]. In the current study, we found that only about 44% of the encapsulated cells were actively proliferating (EdU+) in the absence of Col1 in the gels, whereas more than 60% of the cells were EdU+ in the presence of Col1 (Fig. 2A, Table S3). The enhancement in cell proliferation in the presence of Col1 was also confirmed by total cell counts following gel erosion and cell recovery using exogenous chymotrypsin treatment (Fig. S4B) [30]. AlamarBlue assay results showed that metabolic activity of the encapsulated cells was higher in the presence of Col1, but in both systems (−Col1 and +Col1) cell metabolic activity reached maximum after 7 days of in vitro culture (Fig. S4C), suggesting that the cells reached homeostasis within the 3D hydrogels. Although cell metabolic activity dropped slightly from day 7 to day 10, no significant cell death was observed in live/dead staining (data not shown). The presence of Col1 also caused translocation of yes-associated protein (YAP) from cytoplasm (Fig. 2B) to cell nuclei (Fig. 2C), suggesting the activation (i.e., dephosphorylation) of YAP and inhibition of Hippo pathway in the encapsulated COLO-357 cells. To understand the influence of increased cell cluster size on mechanical property of the cell-laden thiol-ene hydrogels, we measured elastic moduli of the hydrogels over time. Initially, both hydrogels (−Col1 and +Col1) showed similar elastic moduli (G’ ~ 2 kPa) 1-day post-encapsulation. In the presence of Col1, however, gel modulus degreased significantly after 10 days of culture (Fig. S6). This result could potentially be attributed to increased cellular activity, which enhanced peptide linker cleavage and consequently reduced bulk gel mechanical properties.
Figure 2.
(A) Click-IT® EdU staining of proliferating COLO-357 cells in hydrogels without or with Col1. (B and C) Immunofluorescence images of cell clusters encapsulated in hydrogels formed without/with Col1 (B: −Col1, C: + Col1) 10-day post-encapsulation (YAP: green, nuclei: blue). Arrows indicate co-localization of YAP and cell nuclei.
3.3. Effect of cytokines on PDAC cell morphology in 3D
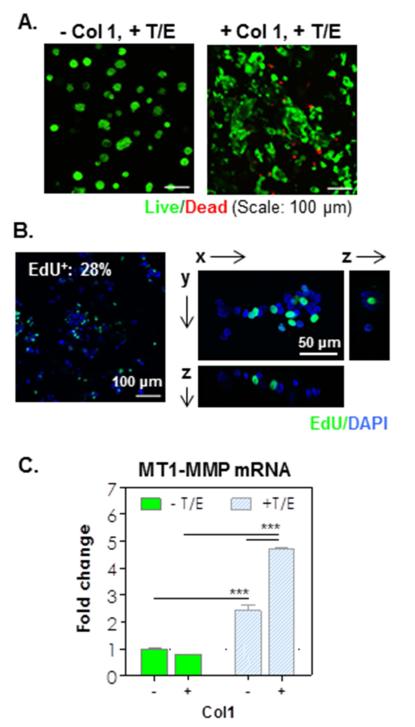
The progression of PDAC has been associated with autocrine and/or paracrine effects of cytokines, such as TGF-β1 and EGF [15,31]. To evaluate the potential effects of these cytokines on PDAC cell fate in 3D, COLO-357 cells were encapsulated in MT1-MMP sensitive hydrogels in the absence or presence of Col1. Cell-laden hydrogels were maintained in culture media containing recombinant TGF-β1 (0.5 nM) and EGF (1 nM). Live/dead staining and confocal microscopy images showed that COLO-357 cells formed compact cell clusters in −Col1 hydrogels after 10 days (Fig. 3A, left); however, these cell clusters appeared to be smaller than those formed in the same type of gels (−Col1) without cytokine treatment (Fig. 1G, left). Interestingly, in +Col1 hydrogels treated with TGF-β1 and EGF, the shape of the cell clusters became highly irregular (Fig. 3A, right), suggesting the enhancement in either cancer cell proliferation or spreading, or both. The addition of cytokines also increased metabolic activity of the encapsulated cells in both types of hydrogels (Fig. S7, compared with Fig. S4C) and the enhancement did not taper off after 10 days of culture. To further elucidate the cause of these irregular cell shapes, we stained the encapsulated cells with Click-iT® EdU Alexa Fluor® 488 staining kit. Results showed that the spreading/invading cell phenotype in the presence of both cytokines and Col1 was not due to increased cell proliferation because the number of EdU+ cells decreased markedly (~24 ± 1%, Fig. 3B, Table S3) when compared with the results shown in Fig. 2A where no cytokines were added. Furthermore, staining images revealed that EdU+ cells were randomly scattered in any given cell cluster after cytokine treatment (Fig. 3B right), whereas only the peripheral cells of the clusters were stained EdU+ in the absence of cytokines (Fig. S5).
Figure 3.
(A) Live/dead images of encapsulated cells in hydrogel formed without/with Col1 and in presence of cytokines (TGF-β1/EGF) 10-day post-encapsulation (Z-stack: 100 μm, live: green, dead: red). (B) Click-IT EdU staining images (left: Z-stack; right: single slice; EdU+: green, DAPI: blue). (C) MT1-MMP mRNA expression in the encapsulated cells (1-fold: mRNA levels in cells cultured in 2D. *** p<0.0001).
Since the thiol-ene hydrogels used in this study were cross-linked by a peptide linker highly sensitive to MT1-MMP, the only route by which these cells invaded into their neighboring matrix is through increased MT1-MMP expression and matrix cleavage. Therefore, we examined MT1-MMP mRNA level in the encapsulated cells using quantitative real-time PCR. Results showed that MT1-MMP mRNA level was significantly increased in cells treated with cytokines (Fig. 3C), suggesting that EGF and TGF-β1 are necessary and sufficient in up-regulating the expression of MT1-MMP in cells encapsulated in these hydrogels. On the other hand, Col1 by itself did not increase MT1-MMP mRNA level but showed synergistic effect with EGF and TGF-β1 in up-regulating MT1-MMP expression (Fig. 3C).
3.4. Effect of collagen and cytokines on EMT in PDAC cells cultured in 3D
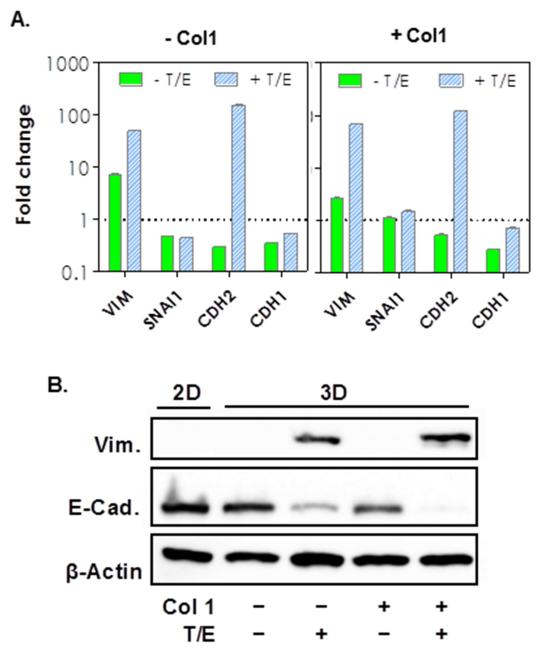
We reasoned that the increased MT1-MMP expression in the encapsulated cells has a positive correlation with the degree of epithelial-mesenchymal transition (EMT). To verify this, we evaluated the expression of markers specific to epithelial and mesenchymal tissues (Fig. 4A). We found that COLO-357 cells cultured in 3D hydrogel, when compared with 2D culture (level set as 1-fold), expressed a higher level of vimentin (VIM). Treatment with cytokines increased the expression of mesenchymal markers, VIM and N-cadherin (CDH2), but did not change the expression of Snail-1 (SNAI1) and an epithelial maker, E-cadherin (CDH1). The influence of Col1 was only apparent in the expression of SNAI1. We also examined selected EMT marker expressions in the protein level using Western Blot (Fig. 4B). While VIM protein expression was un-detectable without cytokine treatment (regardless of 2D or 3D culture), its expression was up-regulated when cytokines were added in the culture media. Increased VIM protein expression was accompanied with a decrease in the levels of an epithelial marker, E-cadherin (E-Cad), and the addition of Col1 potentiated this trend.
Figure 4.
(A) mRNA expression levels of epithelial and mesenchymal markers in the absence or presence of Col1 (1-fold: mRNA levels in cells cultured in 2D). (B) Western blot analysis of EMT markers (Vim: vimentin. E-Cad: E-cadherin) in COLO-357 cells cultured in various conditions as indicated (Col1: Collagen 1. T: TGFβ1. E: EGF).
3.5. Chemo-resistance of PDAC cells in 3D
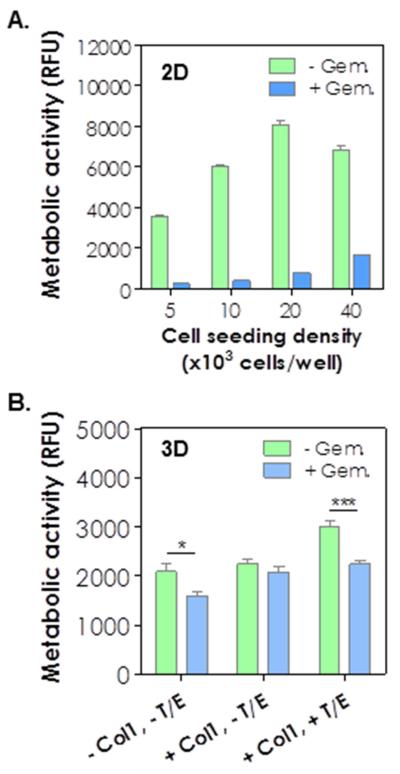
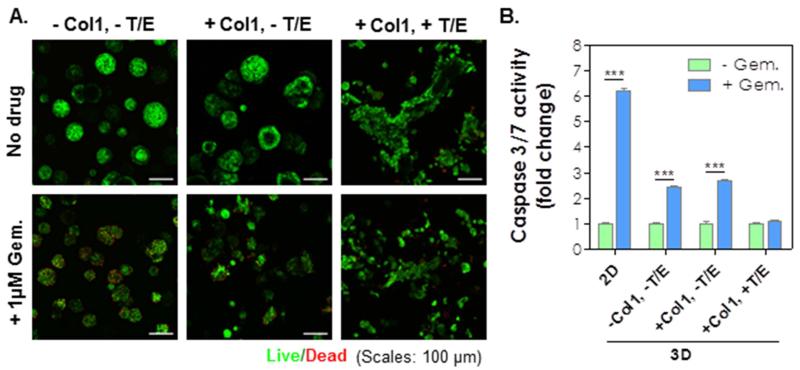
PDAC cells are prone to develop chemo-resistance by mechanisms that are still elusive. We found that COLO-357 cells cultured on 2D TCP are sensitive to gemcitabine (Gem), a chemotherapeutic agent commonly used to treat PDAC [3], with a LD50 of 6×10−3 μM (Fig. S8). We further evaluated whether increasing cell-cell interaction confers chemoresistance on COLO-357 cells cultured in 2D. As shown in Fig. 5A, cells cultured at various cell densities on 2D TCP did not alter their susceptibility to gemcitabine treatment as 1 μM Gem caused significant cell death regardless of cell seeding density. To investigate whether culture platform (i.e., 2D TCP or 3D desmoplasia-mimetic hydrogels) played a role in the development of chemo-resistance in PDAC cells, COLO-357 cells were encapsulated in hydrogels and cultured in the presence of 1 μM of Gem. Surprisingly, under Gem treatment, only slight reduction (< 20%) of cell metabolic activity (Fig. 5B) and limited cell death (mostly on the surface of cell clusters, Fig. 6A) were observed in cells cultured in 3D hydrogels. While Gem treatment induced slight reduction in cell cluster size, it did not alter the spreading cell phenotype caused by EGF/TGF-β1 (Fig. 6A). The influence of Gem on COLO-357 cells was also verified via caspase activity assay (Fig. 6B). There was a 6-fold increase in the degree of cell apoptosis in 2D culture with Gem. By contrast, Gem treatment induced less than 3-fold increase in caspase activity when the cells were cultured in 3D and without cytokine treatment. No enhancement in caspase activity was detected when the cells were maintained in 3D hydrogels incorporated with Col1 and cytokines.
Figure 5.
(A) Metabolic activity of COLO-357 cells with or without 1 μM gemcitabine treatment (4 days) at various seeding densities in 2D TCP. (B) Metabolic activity of COLO-357 cells encapsulated in hydrogels with indicated conditions (n = 3; *, p < 0.05; ***, p < 0.001).
Figure 6.
(A) Live/dead images of COLO-357 cells encapsulated in hydrogels with indicated conditions. (B) Caspase-3/7 activities of COLO-357 cells cultured in indicated conditions with or without 1 μM gemcitabine treatment. Caspase activity without gemcitabine treatment at respective group was set as 1-fold (n = 3, *** p < 0.001).
3.6. Enrichment of pancreatic cancer stem cell markers in 3D culture
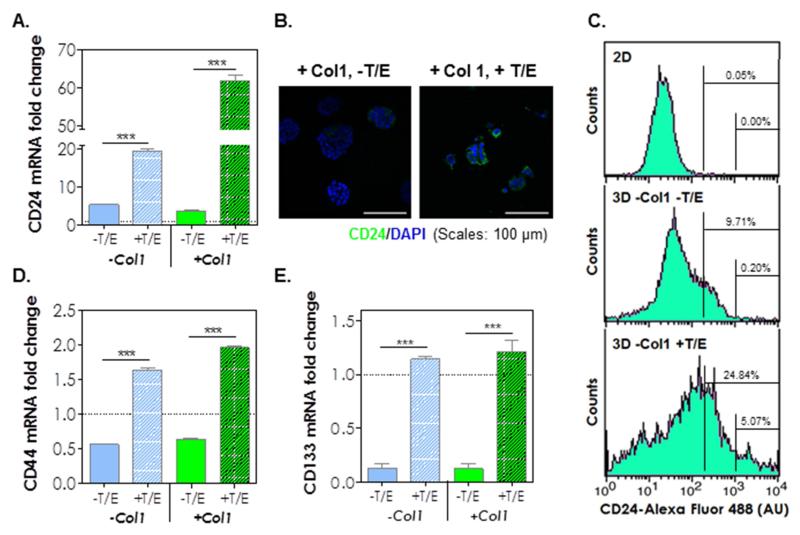
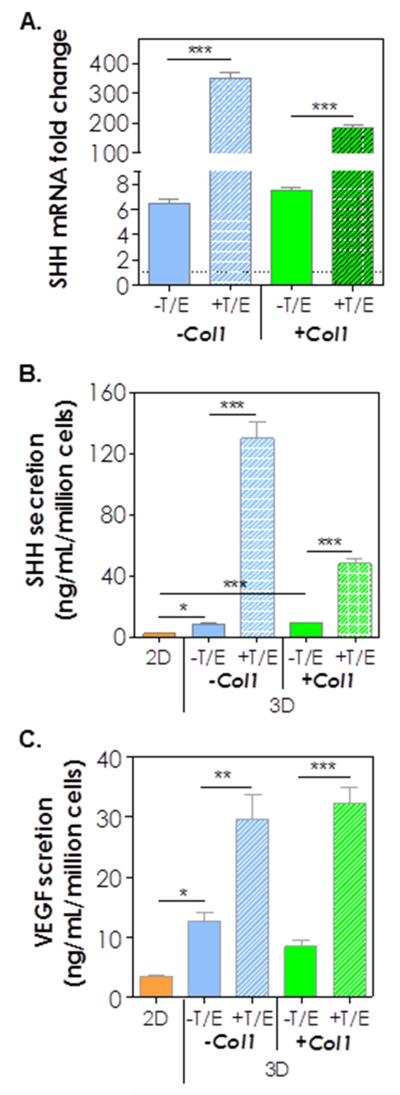
Because cancer stem cells (CSC) are highly resistant to chemotherapy, we speculated that most of the cells killed in the gemcitabine experiment above were mature cancer cells whereas the cells that survived gemcitabine treatment were CSC-like cells. To evaluate this possibility, we examined the expression of markers associated with pancreatic cancer stem cells (PCSCs), including CD24, CD44, CD133, sonic hedgehog (SHH), and vascular endothelial growth factor (VEGF) in COLO-357 cells encapsulated in desmoplasia-mimetic hydrogels (Fig. 7). Strikingly, the expression of CD24 mRNA was increased 6-fold simply by culturing cells in 3D and the treatment of encapsulated cells with cytokines (TGF-β1 and EGF) further increased CD24 expression to 20-fold (Fig. 7A). While Col1 by itself did not alter the expression of CD24, it synergistically increased CD24 expression with cytokines (+T/E), evidenced by a 61-fold increase when both components were added to the culture system. The enhancement in CD24 expression was also evidenced by immunofluorescence staining using an anti-CD24 antibody (Fig. 7B). Flow cytometry results support the observation of CD24 up-regulation in 3D hydrogels (Fig. 7C). Compared with cells cultured in 2D, a higher percentage of cells were CD24+, and the addition of cytokines further increased CD24+ cell population (Fig. 7C) in 3D. We also found that CD44 (Fig. 7D) and CD133 (PROM1, Fig. 7E) were down regulated by 3D culture but their expressions were restored and slightly up-regulated by the addition of cytokines. SHH, another PCSC marker, was up-regulated at both mRNA (Fig. 8A) and protein (Fig. 8B) levels. Specifically, when compared with 2D culture, COLO-357 cells in 3D (without cytokine treatment) exhibited 6.5-fold (−Col1) to 7.5-fold (+Col1) increase in SHH mRNA expression and 3.7-fold (−Col1) to 4.4-fold (+Col1) increase in protein secretion. Strikingly, cytokine treatment led to 54-fold up-regulation of SHH mRNA expression (Fig. 8A) and 16-fold increase in protein secretion when the cells were cultured in hydrogels without Col1. Interestingly, the addition of Col1 somewhat attenuated SHH mRNA expression and protein secretion in the presence of cytokines (i.e., 25-fold increase in mRNA and 5-fold increase in protein level. Fig. 8A and 8B). The amounts of VEGF secretion from COLO-357 cells were also significantly increased when the cells were cultured in 3D, especially in the presence of TGF-β1 and EGF (Fig. 8C).
Figure 7.
(A) CD24 expressions at the mRNA level. Expression levels were compared with the level in cells cultured in 2D (level in 2D = 1-fold; n = 3; ***, p < 0.001). (B) Immunofluorescence images of CD24 (CD24: green; nuclei: blue). (C) Flow cytometry analysis of CD24 expression. (D) CD44 and (E) CD133 expressions at the mRNA levels. (level in 2D = 1-fold; n = 3; ***, p < 0.001).
Figure 8.
(A) SHH mRNA expression levels at 10 days post-encapsulation (the level in 10-day 2D cultured cells were set as 1-fold; n = 3; ***, p < 0.001). Secretion of: (B) SHH and (C) VEGF from cells cultured on 2D TCP or 3D hydrogels at day 10 post-encapsulation (n = 3. *, **, *** = p < 0.05, 0.01, 0.001).
4. Discussion
Three-dimensional cell culture matrices capable of deciphering multiple extracellular cues, including, but not limited to, matrix stiffness, ECM proteins, and cytokines can facilitate the understanding of cancer cell progression and its response to drug treatment. While Matrigel and collagen gels are commercially available and commonly used as 3D matrices for studying cancer cell biology in vitro, the properties of these animal-derived matrices are difficult to control (e.g., weak stiffness, batch-dependent components). Consequently, the use of animal-derived matrices for in vitro cell study is sub-optimal if one wishes to understand the influence of a specific matrix cue on call fate process. On the contrary, hydrogels cross-linked by orthogonal thiol-ene photochemistry, such as the system reported here, is appropriate and relevant for studying cancer cell biology in vitro and ex vivo. As reported here, we adjusted the content of PEG4NB macromer in thiol-ene hydrogels so that the gel shear moduli were at around 2 kPa (Young’s modulus ~6 kPa. Fig. S6), which is within the stiffness range of solid pancreatic tumor tissue [32]. These PEG-based hydrogels were hydrolytically stable but were sensitive to protease mediated matrix cleavage. Specifically, the gels were cross-linked by peptide linkers sensitive to MT1-MMP, a protease highly expressed on cell surface of many PDAC cells [33,34]. This feature allowed us to observe cell invasion into the neighboring matrix due to up-regulation of MT1-MMP. To further control cell behaviors, Col1 fibril and cytokines (TGF-β1 and EGF) were added independently in gel matrix and culture medium, respectively. This design permitted the manipulation of cancer cell behaviors in an orthogonally controlled manner, which decouples the influence of various ECM cues. Although the presence of other proteases in the cell culture media may cause nonspecific cleavage of the peptide cross-linker (or any peptide/protein incorporated in any hydrogel), the evaluation of this effect is outside of the scope of the current study.
We found that Col1 enhanced the proliferation and the formation of larger cell clusters of encapsulated COLO-357 cells (Fig. 1). The observation is likely a result of enhanced cell-collagen binding and integrin activation because Col1 contains many integrin-binding sites (e.g., GFOGER, GLPGER, GASGER) [35-37]. It is worth noting that we did not extract numerical data from confocal images about cell-Col1 interactions because such data would be semi-quantitative at best. Furthermore, after 10-day post-encapsulation it was impossible to distinguish individual cells in any given cell cluster unless images with higher magnification (e.g., Fig. 1H, right) were taken. Practically it would require an unreasonable amount of time on confocal image acquisitions and analysis. The observation of the translocation of YAP to cell nuclei in the +Col1 hydrogels (Fig. 2C) suggests a possible regulatory role of Col1 in inhibiting the Hippo pathway. To the best of our knowledge, the inhibitory regulation of Col1 in the Hippo pathway has not been reported previously and the results shown here warrant future investigation. The EdU staining results confirmed that cytokines not only caused growth inhibition in 3D, but also attenuated the growth promoting effect of Col1 (Fig. 3B). In fact, the addition of cytokines and Col1 induced significant EMT in PDAC cells, evidenced by increased cell invasion into the neighboring matrix (Fig. 3A) and increased MT1-MMP expression from the encapsulated cells (Fig. 3C). These results suggest that the presence of EGF and TGF-β1 stimulates EMT in COLO-357 cells encapsulated in desmoplasia-mimetic hydrogels and synergistically promotes cell spreading with Col1 via MT1-MMP dependent pathway. Recent studies using collagen gels have concluded that collagen promotes the expression of MT1-MMP and regulates the expression of EMT markers [25,38]. Our results, however, showed that the up-regulation of MT1-MMP and mesenchymal markers (e.g., vimentin) was not determined by collagen, but by culturing cells in 3D (Fig. 4). These results reaffirm the unique utility of synthetic biomimetic matrices on studying cancer cell biology in 3D.
In addition to tumor cell growth, invasion, and EMT, we also demonstrated that our orthogonally cross-linked hydrogels are ideal for studying cancer cell drug resistance. We used gemcitabine, a clinically prescribed anti-cancer therapeutic, to study PDAC cell responses to chemotherapy. Many studies have shown that COLO-357 cells were highly susceptible to gemcitabine treatment in 2D culture [2,39,40]. Indeed, we found that, regardless of cell seeding density, COLO-357 cells cultured in 2D had very low tolerance to gemcitabine treatment (Fig. 5A, S8). On the other hand, the same cells encapsulated in 3D remained largely viable and metabolically active under gemcitabine treatment at a concentration (1 μM) that killed most cells in 2D. This indicates that 3D culture stimulates the development of drug resistance. Using collagen gels, Dangi-Garimella et al. previously concluded that collagen promotes gemcitabine resistance in PDAC cells [24]. In our study, however, PDAC cells cultured in 3D hydrogels developed drug resistance spontaneously, and the embedded Col1 did not render these cells more resistant to drug treatment (Fig. 5B, 6). These results suggest that cell-ECM interaction, 3D culture context, and the stimulation of EGF and TGF-β1 together contribute to the development of chemo-resistance in PDAC cells.
The behaviors of COLO-357 cells in desmoplasia-mimetic hydrogels suggest that these cells might have adapted to exhibit characteristics that are different from their counterpart when cultured on 2D TCP. We speculated that these altered cell behaviors (i.e., increased proliferation, EMT, and drug resistance) were caused by the enrichment of certain cell populations, such as pancreatic cancer stem cells (PCSCs). Indeed, when compared with 2D culture, we found that the encapsulated COLO-357 cells expressed higher levels of PCSC markers, especially CD24, SHH, and VEGF (Fig. 7, 8). The addition of TGF-β1/EGF further increased the expression of these markers, as well as CD44 and CD133. CD44 is already abundantly expressed in COLO-357 cells (Fig. S9). On the other hand, the expression of CD24 has been implicated in the progression of PDAC [41]. Moreover, the increased secretion of SHH and VEGF also indicated the enrichment of PCSC-like cells in 3D matrix. SHH has been associated with PCSCs and drug resistance to chemotherapy [6,10,11], and VEGF is an important marker during carcinogenesis and metastasis [42,43]. One of the potential explanations of the increase of SHH and VEGF expression in 3D hydrogel is hypoxia. Hypoxic conditions are known to occur in tumor tissues and to induce VEGF secretion from cancer cells. Recent studies have also shown that hypoxic conditions activate SHH expression and signaling [44,45]. While the cells encapsulated in our hydrogels were mostly alive, they might experience certain degree of hypoxia in the cross-linked hydrogel network. Although hypoxia is a common challenge facing 3D cell culture, regardless of the material (collagen, Matrigel, PEG, etc.), the use of PEG-based matrices with defined compositions permits the decoupling of various matrix cues on cell fate choice.
Through an autocrine and/or paracrine mechanism, the increase of SHH secretion might also contribute to gemcitabine resistance and activated EMT in the encapsulated COLO-357 cells. Prior studies have shown that CD24+, CD44+, CD133+, and SHH are selection markers for PCSCs [10,39]. Our results, however, showed that the expression levels of these markers were differentially regulated, depending on the cell culture platform (2D vs. 3D; ± Col1 or ± cytokines). While CD24 and SHH were significantly up-regulated in our 3D cell culture, CD44 and CD133 expressions were reduced in 3D (Fig. 7). It is possible that PCSCs express differential sets of markers depending on microenvironment stimuli, such as matrix stiffness and context (i.e., 2D vs. 3D). It is also likely that our 3D hydrogels provide a more pathophysiological-relevant microenvironment that facilitates the selection of distinct sub-populations of PCSC-like cells. The capability of our 3D hydrogel to up-regulate selective PCSC makers such as CD24, SHH, and VEGF in COLO-357 cells provides a unique and useful system for future PCSC research targeting specific signaling pathways.
5. Conclusion
We have established a desmoplasia-mimetic semi-synthetic hydrogel with orthogonally controlled biophysical and biochemical properties for PDAC study in vitro. Current data suggest that PDAC cell behaviors are regulated by a myriad of local microenvironment cues. In particular, we found that collagen 1 promoted cell proliferation via nuclear translocation of YAP and potential inhibition of the Hippo pathway. We also showed that the up-regulation of MT1-MMP and mesenchymal markers was possible in the absence of Col1. Furthermore, Col1 and cytokines synergistically promoted EMT in PDAC cells. The encapsulated COLO-357 cells exhibited a high gemcitabine resistance, likely due to the enrichment of PCSC-like cells in a 3D microenvironment. Utilizing this desmoplasia-mimetic hydrogel platform, future work will focus on mechanistic understanding of potential synergistic influence of matrix stiffness and integrin binding on activation/inhibition of Hippo pathway and on evaluating the effects of matrix conditions on PCSC drug resistance.
Supplementary Material
Acknowledgment
This work was supported in part by IUPUI Biomechanics and Biomaterials Research Center (BBRC), a Faculty Start-up fund from IUPUI Department of Biomedical Engineering, and NIH (R21EB013717).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- [2].Liu F, Gore AJ, Wilson JL, Korc M. DUSP1 is a novel target for enhancing pancreatic cancer cell sensitivity to gemcitabine. PLoS One. 2014;9:e84982. doi: 10.1371/journal.pone.0084982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Heinemann V, Reni M, Ychou M, Richel DJ, Macarulla T, Ducreux M. Tumour-stroma interactions in pancreatic ductal adenocarcinoma: rationale and current evidence for new therapeutic strategies. Cancer Treat Rev. 2014;40:118–28. doi: 10.1016/j.ctrv.2013.04.004. [DOI] [PubMed] [Google Scholar]
- [4].McCarroll JA, Naim S, Sharbeen G, Russia N, Lee J, Kavallaris M, et al. Role of pancreatic stellate cells in chemoresistance in pancreatic cancer. Front Physiol. 2014;5:141. doi: 10.3389/fphys.2014.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Huang FT, Zhuan-Sun YX, Zhuang YY, Wei SL, Tang J, Chen WB, et al. Inhibition of hedgehog signaling depresses self-renewal of pancreatic cancer stem cells and reverses chemoresistance. Int J Oncol. 2012;41:1707–14. doi: 10.3892/ijo.2012.1597. [DOI] [PubMed] [Google Scholar]
- [6].Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. J Clin Oncol. 2008;26:2806–12. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- [7].Maccalli C, Volonte A, Cimminiello C, Parmiani G. Immunology of cancer stem cells in solid tumours. A review. Eur J Cancer. 2014;50:649–55. doi: 10.1016/j.ejca.2013.11.014. [DOI] [PubMed] [Google Scholar]
- [8].Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW, et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16:268–80. doi: 10.1038/ncb2910. [DOI] [PubMed] [Google Scholar]
- [9].Fessler E, Dijkgraaf FE, De Sousa EMF, Medema JP. Cancer stem cell dynamics in tumor progression and metastasis: is the microenvironment to blame? Cancer Lett. 2013;341:97–104. doi: 10.1016/j.canlet.2012.10.015. [DOI] [PubMed] [Google Scholar]
- [10].Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- [11].Xu M, Li L, Liu Z, Jiao Z, Xu P, Kong X, et al. ABCB2 (TAP1) as the downstream target of SHH signaling enhances pancreatic ductal adenocarcinoma drug resistance. Cancer Lett. 2013;333:152–8. doi: 10.1016/j.canlet.2013.01.002. [DOI] [PubMed] [Google Scholar]
- [12].Ng MR, Brugge JS. A stiff blow from the stroma: collagen crosslinking drives tumor progression. Cancer Cell. 2009;16:455–7. doi: 10.1016/j.ccr.2009.11.013. [DOI] [PubMed] [Google Scholar]
- [13].Raza A, Ki CS, Lin CC. The influence of matrix properties on growth and morphogenesis of human pancreatic ductal epithelial cells in 3D. Biomaterials. 2013;34:5117–27. doi: 10.1016/j.biomaterials.2013.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ki CS, Shih H, Lin CC. Effect of 3D matrix compositions on the efficacy of EGFR inhibition in pancreatic ductal adenocarcinoma cells. Biomacromolecules. 2013;14:3017–26. doi: 10.1021/bm4004496. [DOI] [PubMed] [Google Scholar]
- [15].Sempere LF, Gunn JR, Korc M. A novel 3-dimensional culture system uncovers growth stimulatory actions by TGF beta in pancreatic cancer cells. Cancer Biol Ther. 2011;12:198–207. doi: 10.4161/cbt.12.3.15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Miroshnikova YA, Jorgens DM, Spirio L, Auer M, Sarang-Sieminski AL, Weaver VM. Engineering strategies to recapitulate epithelial morphogenesis within synthetic three-dimensional extracellular matrix with tunable mechanical properties. Phys Biol. 2011;8:026013. doi: 10.1088/1478-3975/8/2/026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. A versatile synthetic extracellular matrix mimic via thiol-norbornene photopolymerization. Adv Mater. 2009;21:5005–10. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Raza A, Lin CC. The influence of matrix degradation and functionality on cell survival and morphogenesis in PEG-based hydrogels. Macromol Biosci. 2013;13:1048–58. doi: 10.1002/mabi.201300044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials. 2009;30:6702–7. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- [21].Patterson J, Hubbell JA. Enhanced proteolytic degradation of molecularly engineered PEG hydrogels in response to MMP-1 and MMP-2. Biomaterials. 2010;31:7836–45. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- [22].Tibbitt MW, Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng. 2009;103:655–63. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pandol S, Edderkaoui M, Gukovsky I, Lugea A, Gukovskaya A. Desmoplasia of pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol. 2009;7:S44–7. doi: 10.1016/j.cgh.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dangi-Garimella S, Krantz SB, Barron MR, Shields MA, Heiferman MJ, Grippo PJ, et al. Three-dimensional collagen I promotes gemcitabine resistance in pancreatic cancer through MT1-MMP-mediated expression of HMGA2. Cancer Res. 2011;71:1019–28. doi: 10.1158/0008-5472.CAN-10-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dangi-Garimella S, Strouch MJ, Grippo PJ, Bentrem DJ, Munshi HG. Collagen regulation of let-7 in pancreatic cancer involves TGF-beta 1-mediated membrane type 1-matrix metalloproteinase expression. Oncogene. 2011;30:1002–8. doi: 10.1038/onc.2010.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nicolas FJ, Hill CS. Attenuation of the TGF-beta-Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-beta-induced growth arrest. Oncogene. 2003;22:3698–711. doi: 10.1038/sj.onc.1206420. [DOI] [PubMed] [Google Scholar]
- [27].Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–92. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- [29].Diep CH, Zucker KM, Hostetter G, Watanabe A, Hu C, Munoz RM, et al. Down-regulation of yes associated protein 1 expression reduces cell proliferation and clonogenicity of pancreatic cancer cells. PLoS One. 2012;7:e32783. doi: 10.1371/journal.pone.0032783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lin CC, Raza A, Shih H. PEG hydrogels formed by thiol-ene photo-click chemistry and their effect on the formation and recovery of insulin-secreting cell spheroids. Biomaterials. 2011;32:9685–95. doi: 10.1016/j.biomaterials.2011.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Deharvengt S, Marmarelis M, Korc M. Concomitant targeting of EGF receptor, TGF-beta and Src points to a novel therapeutic approach in pancreatic cancer. PLoS One. 2012;7:e39684. doi: 10.1371/journal.pone.0039684. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [32].Arda K, Ciledag N, Aktas E, Aribas BK, Kose K. Quantitative assessment of normal soft-tissue elasticity using shear-wave ultrasound elastography. AJR Am J Roentgenol. 2011;197:532–6. doi: 10.2214/AJR.10.5449. [DOI] [PubMed] [Google Scholar]
- [33].Ottaviano AJ, Sun L, Ananthanarayanan V, Munshi HG. Extracellular matrix-mediated membrane-type 1 matrix metalloproteinase expression in pancreatic ductal cells is regulated by transforming growth factor-beta1. Cancer Res. 2006;66:7032–40. doi: 10.1158/0008-5472.CAN-05-4421. [DOI] [PubMed] [Google Scholar]
- [34].Bramhall SR, Neoptolemos JP, Stamp GW, Lemoine NR. Imbalance of expression of matrix metalloproteinases (MMPs) and tissue inhibitors of the matrix metalloproteinases (TIMPs) in human pancreatic carcinoma. J Pathol. 1997;182:347–55. doi: 10.1002/(SICI)1096-9896(199707)182:3<347::AID-PATH848>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- [35].Pugh N, Simpson AM, Smethurst PA, de Groot PG, Raynal N, Farndale RW. Synergism between platelet collagen receptors defined using receptor-specific collagen-mimetic peptide substrata in flowing blood. Blood. 2010;115:5069–79. doi: 10.1182/blood-2010-01-260778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Caswell CC, Barczyk M, Keene DR, Lukomska E, Gullberg DE, Lukomski S. Identification of the first prokaryotic collagen sequence motif that mediates binding to human collagen receptors, integrins alpha2beta1 and alpha11beta1. J Biol Chem. 2008;283:36168–75. doi: 10.1074/jbc.M806865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Siljander PR, Hamaia S, Peachey AR, Slatter DA, Smethurst PA, Ouwehand WH, et al. Integrin activation state determines selectivity for novel recognition sites in fibrillar collagens. J Biol Chem. 2004;279:47763–72. doi: 10.1074/jbc.M404685200. [DOI] [PubMed] [Google Scholar]
- [38].Shields MA, Dangi-Garimella S, Krantz SB, Bentrem DJ, Munshi HG. Pancreatic cancer cells respond to type I collagen by inducing snail expression to promote membrane type 1 matrix metalloproteinase-dependent collagen invasion. J Biol Chem. 2011;286:10495–504. doi: 10.1074/jbc.M110.195628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shah AN, Summy JM, Zhang J, Park SI, Parikh NU, Gallick GE. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol. 2007;14:3629–37. doi: 10.1245/s10434-007-9583-5. [DOI] [PubMed] [Google Scholar]
- [40].Banerjee S, Zhang Y, Ali S, Bhuiyan M, Wang Z, Chiao PJ, et al. Molecular evidence for increased antitumor activity of gemcitabine by genistein in vitro and in vivo using an orthotopic model of pancreatic cancer. Cancer Res. 2005;65:9064–72. doi: 10.1158/0008-5472.CAN-05-1330. [DOI] [PubMed] [Google Scholar]
- [41].Sagiv E, Kazanov D, Arber N. CD24 plays an important role in the carcinogenesis process of the pancreas. Biomed Pharmacother. 2006;60:280–4. doi: 10.1016/j.biopha.2006.06.006. [DOI] [PubMed] [Google Scholar]
- [42].Fantozzi A, Gruber DC, Pisarsky L, Heck C, Kunita A, Yilmaz M, et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res. 2014;74:1566–75. doi: 10.1158/0008-5472.CAN-13-1641. [DOI] [PubMed] [Google Scholar]
- [43].Liu W, Xu J, Wang M, Wang Q, Bi Y, Han M. Tumor-derived vascular endothelial growth factor (VEGF)-a facilitates tumor metastasis through the VEGF-VEGFR1 signaling pathway. Int J Oncol. 2011;39:1213–20. doi: 10.3892/ijo.2011.1138. [DOI] [PubMed] [Google Scholar]
- [44].Onishi H, Kai M, Odate S, Iwasaki H, Morifuji Y, Ogino T, et al. Hypoxia activates the hedgehog signaling pathway in a ligand-independent manner by upregulation of Smo transcription in pancreatic cancer. Cancer Sci. 2011;102:1144–50. doi: 10.1111/j.1349-7006.2011.01912.x. [DOI] [PubMed] [Google Scholar]
- [45].Spivak-Kroizman TR, Hostetter G, Posner R, Aziz M, Hu C, Demeure MJ, et al. Hypoxia triggers hedgehog-mediated tumor-stromal interactions in pancreatic cancer. Cancer Res. 2013;73:3235–47. doi: 10.1158/0008-5472.CAN-11-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.