Short abstract
Silent information regulator 2 (Sir2) proteins, or sirtuins, are protein deacetylases dependent on nicotine adenine dinucleotide (NAD) and regulate transcriptional repression, recombination, the cell-division cycle, microtubule organization, and cellular responses to DNA-damaging agents.
Abstract
Silent information regulator 2 (Sir2) proteins, or sirtuins, are protein deacetylases dependent on nicotine adenine dinucleotide (NAD) and are found in organisms ranging from bacteria to humans. In eukaryotes, sirtuins regulate transcriptional repression, recombination, the cell-division cycle, microtubule organization, and cellular responses to DNA-damaging agents. Sirtuins have also been implicated in regulating the molecular mechanisms of aging. The Sir2 catalytic domain, which is shared among all sirtuins, consists of two distinct domains that bind NAD and the acetyl-lysine substrate, respectively. In addition to the catalytic domain, eukaryotic sirtuins contain variable amino- and carboxy-terminal extensions that regulate their subcellular localizations and catalytic activity.
Gene organization and evolutionary history
Protein acetylation regulates a wide variety of cellular functions, including the recognition of DNA by proteins, protein-protein interactions, and protein stability (reviewed in [1]). Post-translational modification of proteins at lysine residues by reversible acetylation is catalyzed by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs), which act on both histone and non-histone substrates despite their names. HDACs are grouped into three classes on the basis of their homology to yeast transcriptional repressors. Class I and class II HDACs, which share significant similarity to each other in their catalytic cores, are homologs of the yeast deacetylases Rpd3p and Hda1p, respectively (reviewed in [2,3]). The class III HDACs are homologous to the yeast transcriptional repressor Sir2p and have no sequence similarity to class I and II HDACs; these Sir2 proteins, also called sirtuins, are the focus of this article.
The founding member of class III HDACs, Saccharomyces cerevisiae Sir2p, functions in transcriptional repression at the telomeres [4,5], the silent mating-type loci [6-8], and ribosomal DNA loci [9,10]. Sir2p has been implicated in the repair of double-strand DNA breaks, cell-cycle progression, and chromosomal stability in yeast and plays a pivotal role in the molecular mechanisms of aging in both S. cerevisiae and Caenorhabditis elegans [11-15].
Sirtuins have been found in bacteria to eukaryotes [16,17]. The hallmark of the family is a domain of approximately 260 amino acids that has a high degree of sequence similarity in all sirtuins. The family is divided into five classes (I-IV and U) on the basis of a phylogenetic analysis of 60 sirtuins from a wide array of organisms [17] (Figure 1). Class I and class IV are further divided into three and two subgroups, respectively. The U-class sirtuins are found only in Gram-positive bacteria [17]. The S. cerevisiae genome encodes five sirtuins, Sir2p and four additional proteins termed 'homologs of sir two' (Hst1p-Hst4p) [11] (Figure 1). The human genome encodes seven sirtuins, with representatives from classes I-IV (Table 1) [17].
Figure 1.
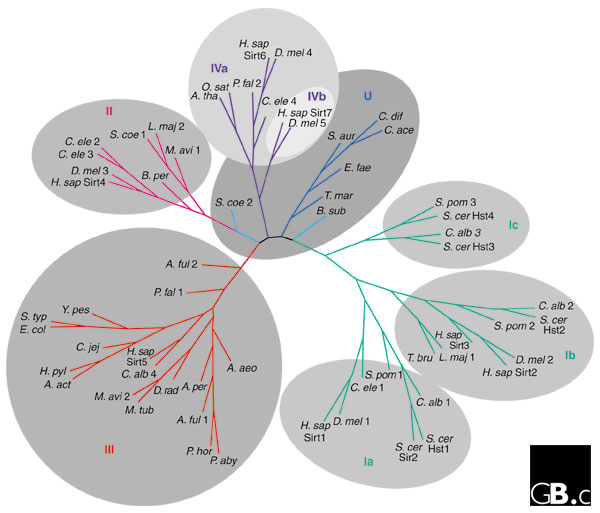
An unrooted tree diagram derived from phylogenetic analysis of the conserved domains of 60 sirtuin sequences from all sirtuin classes. Classes I, II, III, IV, and U and subdivisions of classes I and IV are indicated. Organism abbreviations: A. act, Actinobacillus actinomycetemcomitans; A. aeo, Aquifex aeolicus; A. ful, Archaeoglobus fulgidus; A. per, Aeropyrum pernix; A. tha, Arabidopsis thaliana; B. per, Bordetella pertussis; B. sub, Bacillus subtilis; C. ace, Clostridium acetabutylicum; C. alb, Candida albicans; C. dif, Clostridium difficile; C. ele, Caenorhabditis elegans; C. jej, Campylobacter jejuni; D. mel, Drosophila melanogaster; D. rad, Deinococcus radiodurans; E. col, Escherichia coli; E. fae, Enterococcus faecalis; H. sap, Homo sapiens; H. pyl, Helicobacter pylori; L. maj, Leishmania major; M. avi, Mycobacterium avium; M. tub, Mycobacterium tuberculosis; O. sat, Oryza sativa; P. aby, Pyrococcus abyssi; P. fal, Plasmodium falciparum; P. hor, Pyrococcus horikoshii; S. aur, Staphylococcus aureus; S. coe, Streptomyces coelicolor; S. pom, Schizosaccharomyces pombe, S. typ, Salmonella typhimurium; S. cer, Saccharomyces cerevisiae; T. bru, Trypanosoma brucei; T. mar, Thermotoga maritima; Y. pes, Yersinia pestis. Modified from [17].
Table 1.
Classification and chromosomal locations of human sirtuin genes
| Orthologs | |||||
| Name | Class | Chromosomal location | OMIM ID | Yeast | Mouse |
| Sirt1 | Ia | 10q21.3 | 604479 | Sir2, Hst1 | Sir2β |
| Sirt2 | Ib | 19q13.2 | 604480 | Hst2 | Sir2l2 |
| Sirt3 | Ib | 11p15.5 | 604481 | Hst2 | Sir2l3 |
| Sirt4 | II | 12q24.31 | 604482 | Sirt4 | |
| Sirt5 | III | 6p23 | 604483 | Sirt5 | |
| Sirt6 | IVa | 19p13.3 | 606211 | Sirt6 | |
| Sirt7 | IVb | 17q25.3 | 606212 | Sirt7 | |
Mouse orthologs of human Sirt4-7 have not been characterized but are found in sequence databases. OMIM IDs are from the Online Mendelian Inheritance in Man repository at the National Center for Biotechnology Information (NCBI) [76].
Characteristic structural features
The sirtuins have a catalytic domain, unique to this family, characterized by its requirement for nicotine adenine dinucleotide (NAD) as a cofactor [18]. The structures of four sirtuins (Archaeoglobus fulgidus Sir2-Af1 and Sir2-Af2, human Sirt2, and yeast Hst2) have been obtained at atomic resolution, and a number of common features are emerging [18-21]. All four are organized in two bilobed globular domains: a small domain with two distinct modules and a large domain (Figure 2).
Figure 2.
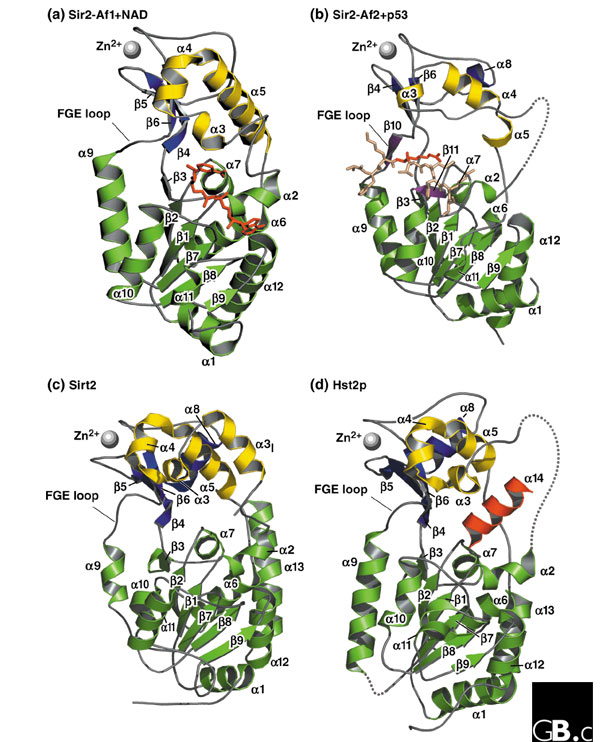
Three-dimensional high-resolution crystal structures of four sirtuin proteins. The zinc-binding module is shown at the top left in each panel, the helical modules of the small domain are lighter and at the top right, and the large Rossmann-fold domain is in the lower half. Each α helix and β strand is labeled to facilitate comparisons. (a) Sir2-Af1 complexed with NAD (in stick representation; PDB accession number: 1ICI) [20]. (b) Sir2-Af2 complexed with acetylated p53 peptide (in stick notation, with acetyl-lysine darker). Two β strands (β10 and β11) are shown that might mediate the binding of the substrate peptide (PDB accession number: IMA3) [21]. An acetylated peptide, such as p53, may be bound through the formation of an enzyme-substrate β sheet, in which the substrate β strand is sandwiched between the β11 strand within the Rossmann fold and a β10 strand within the FGE loop, named for its highly conserved FGExL motif [21]. (c) Human Sirt2 (catalytic core; PDB accession number: 1J8F) [18]. (d) Full-length yeast Hst2p with the carboxy-terminal α14 helix interacting with the NAD-binding pocket (PDB accession number: 1Q14) [19]. Structural coordinates were taken from the Protein Data Bank and models were drawn with PYMOL [77].
The large domain
The large domain contains an inverted classical open α/β Rossmann-fold structure, which is commonly found in proteins that bind oxidized or reduced NAD or NADP [18,20,21] (Figure 2). This domain consists of six parallel β strands (β1-β3 and β7-β9) that form a central β sheet, and eight α helices (α1, α2, α6, α7, and α9-α12) that pack against the β sheet (Figure 2). Sirt2 and the yeast Hst2p each have an additional α helix in the crystal structure, α13, that packs against the outside of the large domain but is not found in Sir2-Af1 or Sir2-Af2. The most significant difference in the large domains is an insertion in the region of the α11 helix in Sirt2 and Hst2p that is absent from Sir2-Af1 and Sir2-Af2. This helix is located outside the catalytic pocket but on the same face; it could possibly have a role in the recognition of substrate or of other members of a macromolecular complex [18].
The small domain
The small domain has two structural modules that result from two insertions in the Rossmann fold of the large domain [18,20,21]. The first insertion consists of three α helices (α3, α4, and α5) that fold to form the helical module (Figure 2). This module has a pocket, lined with hydrophobic residues, that intersects the large groove between the small and large domains. The properties of the pocket suggest that it may be a class-specific protein-protein interaction domain, possibly recognizing specific residues in the substrate protein. Sequence alignments show that all class I sirtuins have the α3I, α3, α4, and α5 helices that form the hydrophobic pocket, but classes II, III, and IV sirtuins have deletions in this area, suggesting class-specific differences. Structural data from Sir2-Af1 bound to NAD, Sir2-Af2 bound to p53, and unbound human Sirt2 suggest that the helical module undergoes a conformational change upon binding of NAD, possibly to allow interaction with acetyl-lysine substrates [21].
The second insertion forms a zinc-binding module (see Figure 2), composed of antiparallel (β strands containing two Cys-X-X-Cys motifs (where X is any amino acid) separated by 15-20 amino acids that are involved in zinc coordination [18,20,21]. Replacing the cysteines with alanines abolishes enzymatic activity in vitro and the silencing activity in vivo at silent mating-type, telomeric, and rDNA loci [20,22]. Likewise, the presence of zinc is required for enzymatic activity, as the zinc chelator o-phenanthroline inhibits enzymatic activity [20]. The localization of zinc away from the NAD-binding pocket suggests that the zinc ion does not participate directly in catalysis. This is in contrast to class I and class II HDACs, in which zinc ions are part of the active site [20,23].
The NAD-binding pocket
The NAD-binding pocket, located within the large domain at the interface of the large and small domains, can be divided into three spatially distinct regions: the A site, where the adenine-ribose moiety of NAD is bound, the B site where the nicotinamide-ribose moiety is bound, and the C site, located deep in the NAD-binding pocket (see also Figure 3) [20]. The B and C sites are thought to be directly involved in catalysis. In the presence of an acetyl-lysine, NAD bound to the B site can undergo a conformational change, bringing the nicotinamide group in proximity to the C site, where it can be cleaved [20]. The ADP-ribose product of this reaction may then return to the B site, where deacetylation of the acetyl-lysine occurs. The organization of the NAD-binding pocket might explain how nicotinamide inhibits sirtuin activity. At high concentrations, free nicotinamide may occupy the C site, irrespective of whether any acetylated lysine is bound, and block the conformational change of NAD [24].
Figure 3.
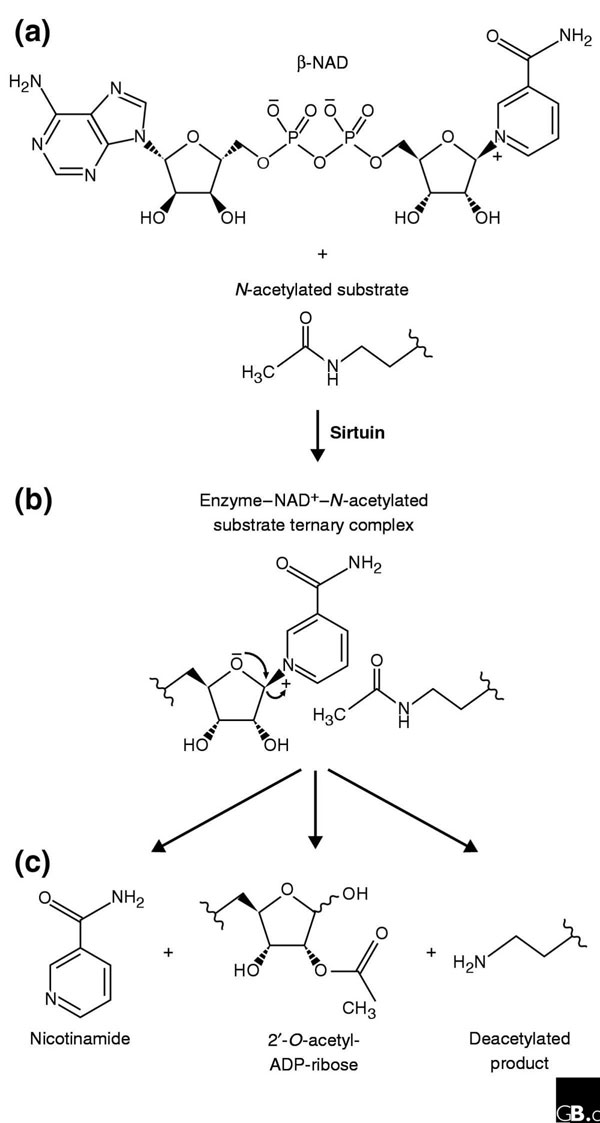
The enzymatic activity of sirtuins. (a) The components necessary for sirtuin-mediated deacetylase activity are the sirtuin, β-NAD, and the N-acetylated substrate. (b) The components form a tertiary complex and, during the enzymatic reaction, the nicotinamide is expelled from bound NAD to generate an oxocarbonium-like transition state in which the carbonyl oxygen of the acetyl group attacks the C1 carbon of ADP. After alkylamidate and cyclic intermediates and possibly protonation of the amine leaving group (not shown), the products (c) are formed: the deacetylated protein, 2'-O-acetyl-ADP-ribose, and nicotinamide. The 2'-O-acetyl-ADP-ribose is released into solution, where it equilibrates with 3'-O-acetyl-ADP-ribose. Adapted with modifications from [78].
The large groove
A large groove is formed at the interface between the large and small domains and runs perpendicular to the long axis formed by the two domains (Figure 2). On the basis of mutagenesis studies, a role in substrate recognition and catalysis has been proposed for this groove [18,20]. Analysis of the crystal structure of A. fulgidus Sir2-Af2 complexed with a P53 peptide (Figure 2b) demonstrates that the peptide substrate lies in the large groove [21]. The binding of an acetylated peptide, such as p53, may occur through the formation of an enzyme-substrate β sheet, in which the substrate β strand is sandwiched between the β11 strand within the Rossmann fold and a β10 strand within the FGE loop, named for its highly conserved FGExL motif [21]. A high degree of conservation between sirtuins is found in the residues implicated in substrate peptide binding, specifically in the region of the FGE loop [21]. The predicted β10 and β11 strands of Sir2-Af2 are absent from the solved structures of Sirt2, Sir2-Af1, and Hst2p, however. These observations suggest that these two regions become more ordered and form the enzyme-substrate β sheet upon substrate binding, as seen in the Sir2-Af2 p53 structure [21].
Amino- and carboxy-terminal extensions
The solved structures of full-length A. fulgidus Sir2-Af1, A. fulgidus Sir2-Af2 and human Sirt2 correspond primarily to the catalytic domain found within sirtuins. Yeast Hst2 and human Sirt2 have large amino- and carboxy-terminal extensions (not shown for Sirt2 in Figure 2c) that are likely to play a role in the regulation of enzymatic activity. In support of this model, studies have indicated that different modes of silencing are affected by various mutations outside the catalytic core [19]. In addition, the structure of full-length yeast Hst2p indicates that the carboxy-terminal extension folds into the NAD-binding pocket and that the amino-terminal extension may occlude the substrate binding cleft [19]. The carboxy-terminal α14 helix of yeast Hst2p interacts extensively with residues within the large groove between the large and small catalytic domains of the protein, as well as with residues in these domains. In the apo (unbound) form of Hst2p the α14 helix probably partially occludes the cofactor NAD-binding site and the loop following β1 is disordered. NAD binding then promotes dissociation of the α14 helix and ordering of the loop after β1, to facilitate enzymatic activity. Likewise, the human Hst2p homolog Sirt3 is enzymatically inactive as a full-length protein and becomes catalytically active after proteolytic cleavage of its amino terminus following import in the mitochondrial matrix [25].
Localization and function
Sirtuin proteins have been found in a wide variety of subcellular locations. Human Sirt1 localizes to the nucleus, as does yeast Sir2p [26]; Sirt1 appears to repress transcription in the nucleus by various different mechanisms. Human Sirt2 and Sirt3 localize to extra-nuclear compartments, much like yeast Hst2p: Sirt2 is found in the cytosol [27,28], whereas Sirt3 is found primarily in the mitochondria [25,29] (see Figure 4).
Figure 4.
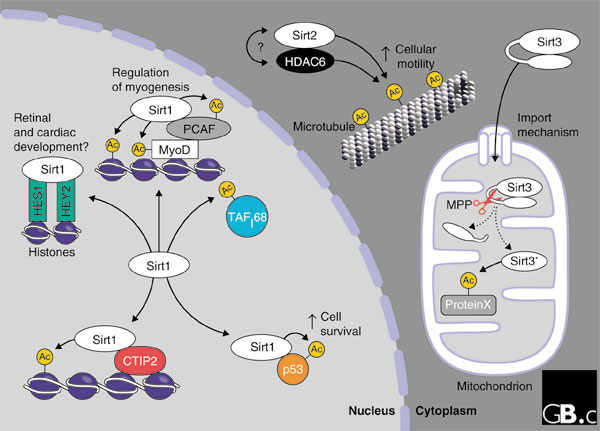
Localization and functions of the human sirtuins Sirt1, Sirt2 and Sirt3. Sirt1 is found in the nucleus; it associates with several partners and targets for deacetylation, as discussed in the text. Sirt2 is found in the cytoplasm bound to the microtubule network, where it forms a complex with the histone deacetylase HDAC6. Both proteins can deacetylate α-tubulin. Sirt3 is imported into the mitochondrial matrix and proteolytically processed by the peptidase MPP, leading to its enzymatic activation (asterisk). Ac, acetyl moiety; ProteinX, an unknown protein substrate.
A key function of sirtuins is their regulation of transcriptional repression, mediated through binding of a complex containing sirtuins and other proteins. The silencing function of the yeast Sir2p is mediated by different multiprotein complexes at different genomic sites. Silencing at the telomeres and at the mating-type loci is mediated by a protein complex consisting of Sir2p and the structurally unrelated proteins Sir3p and Sir4p. Sequence-specific DNA-binding proteins mediate the initial recruitment of Sir protein complexes to the telomeres and mating-type loci. At those loci, Sir3p and Sir4p blanket the underlying nucleosomes by interacting with hypoacetylated tails of histones H3 and H4, and Sir2p interacts with Sir3p and Sir4p (reviewed in [30]). Sir2p-dependent silencing of ribosomal DNA (rDNA) is mediated by the 'regulator of nucleolar silencing and telophase exit' (RENT) complex, containing Sir2p, Net1p, and a telophase-regulating phosphatase, Cdc14p, which is released in late metaphase [31,32]. At rDNA loci, Sir2p can silence transcription of RNA polymerase II-dependent marker genes inserted within the rDNA array and also suppresses homologous recombination among the tandemly repeated rDNA copies [9,33,34].
Enzyme mechanism
Initial enzymatic experiments with sirtuins, carried out in the bacterium Salmonella typhimurium, revealed only their activity as ADP-ribosyltransferases, not protein deacetylases. During the biosynthesis of cobalamin (also known as vitamin B12), the CobT enzyme in Salmonella catalyzes the transfer of phosphoribose from nicotinic acid mononucleotide (NaMN) to dimethylbenzimidazole (DMB) to form DMB-5'-ribosyl-phosphate [35,36]. In the absence of CobT, the Sir2-like protein CobB can partially compensate in the catalysis of this reaction [37]. In later studies, recombinant Sir2 proteins from both bacteria and humans were shown to have NAD-dependent ADP-ribosyltransferase activity in vitro [16,38].
Although the enzymatic function initially associated with sirtuins was ADP-ribosyltransferase activity, NAD-dependent histone-deacetylase activity was subsequently shown to be the primary enzymatic activity of Sir2p and other sirtuins [39-41]. The deacetylation reaction generates three products: acetyl-ADP-ribose, nicotinamide, and a deacetylated peptide substrate (Figure 3). The ratio of these products is 1:1:1, consistent with the model that hydrolysis of one NAD to acetyl-ADP-ribose and nicotinamide occurs for each acetyl group removed, and that deacetylation requires an enzyme-ADP-ribose intermediate (Figure 3) [42]. The demonstration that the ribosyltransferase and NAD-cleavage activities are both dependent on an acetylated substrate confirms the fundamental link between the two activities [38,39,42-44]. The recent observation that O-acetyl-ADP-ribose delays oocyte maturation and cell division in blastomere-stage embryos suggests that this compound might be a bona fide second messenger linked to the enzymatic activity of sirtuins [43-45].
It remains likely that there is a physiological role for the ADP-ribosyltransferase activity of individual sirtuins. A recently identified Trypanosoma brucei class Ib sirtuin, TbSIR2RP1, exerts both histone-deacetylase and robust ADP-ribosyltransferase activity on histones H2A and H2B [46]. García-Salcedo et al. [46] have suggested that the activity of TbSIR2RP1 and extent of chromatin ADP ribosylation correlates with the sensitivity of trypanosomes to agents that damage DNA.
It is not clear whether all five classes of sirtuins have deacetylase activity. For instance, only class I human sirtuins (Sirt1, Sirt2, and Sirt3) have robust enzymatic activity on a peptide corresponding to the amino-terminal tail of histone H4 [47]. Sirt5, a class III sirtuin, has low but detectable activity in comparison with the class I sirtuins [47]. Interestingly, the class III sirtuin from A. fulgidus, Sir2-Af1, also has low activity on a histone peptide but significantly stronger activity on an acetylated bovine serum albumin substrate, suggesting that the level of deacetylase activity is substrate-specific [20,21].
Phenotypic screens using yeast strains that have either URA3 or TRP1 inserted within Sir2p-silenced loci have led to the identification of α-substituted β-naphthol compounds, sirtinol and splitomicin, which inhibit the enzymatic activity of Sir2 proteins [48,49]. Sirtinol inhibits recombinant Sir2p and human Sirt2 in vitro and silencing at the telomeres, silent mating-type, and rDNA loci in vivo. Sirtinol was recently used to define further the role of mammalian Sir2 proteins in the regulation of muscle gene expression and differentiation in response to alterations in the ratio of concentrations of NAD+ and NADH [50].
Other inhibitors have been developed that capitalize on the dependence of the sirtuins' enzymatic activity on NAD. A non-hydrolyzable NAD molecule, carba-NAD, inhibits sirtuin activity, consistent with the requirement for NAD cleavage during the enzymatic reaction [42]. It is unlikely, however, that this inhibitor will prove useful in vivo because it cannot permeate cells and because of its potential to affect other cellular NAD-dependent enzymatic activities or biological pathways [48]. Nicotinamide, a byproduct of the NAD-dependent deacetylation reaction, inhibits sirtuins both in vitro and in vivo [42,51,52].
A recent study found a number of compounds that increase the enzymatic activity of sirtuins [53], one of which, resveratrol, activates both yeast Sir2p and human Sirt1 in vitro and in vivo. Plant polyphenols such as resveratrol - which is found in grapes and red wine - have been associated with health benefits such as cardioprotection, neuroprotection, and cancer suppression [54-56]. Sirtuins have been implicated in the regulation of cellular and organismal aging in several model organisms (see Frontiers); regulation of sirtuins by polyphenols may provide a functional link between the effects of plant products such as resveratrol on health and longevity and the regulation of aging [53].
Substrates and functions
The functions and substrates have been studied most for human Sirt1, Sirt2 and Sirt3 and their closest yeast homologs Sir2, Hst1 and Hst2; little is known about the functions of Sirt4, Sirt5, Sirt6, and Sirt7.
Sirt 1
TAFI68, a transcription factor necessary for regulation of the RNA polymerase I transcriptional complex, was the first substrate to be identified for Sirt1 [57]; the fact that S. cerevisiae Sir2p also regulates rDNA suggests that Sir2p and Sirt1 may have similar functions. Deacetylation of TAFI68 by Sir2α - the mouse ortholog of human Sirt1 - inhibits transcriptional initiation in vitro [57]. Sirt1 is reported to associate physically with the human basic helix-loop-helix (bHLH) repressor proteins hHES1 and hHEY2 [58] (Figure 4); a similar interaction is also found in Drosophila, in which the bHLH repressor proteins Hairy and Deadpan recruit a sirtuin protein [59]. The Hairy-related bHLH proteins function as transcriptional repressors and play important roles in diverse aspects of metazoan development. Sirt1 has also been shown to form a complex with the histone acetyltransferase PCAF and the muscle transcription factor MyoD, and it deacetylates both proteins [50]. Although transcriptional regulation by Sirt1 through TAFI68, bHLH, MyoD and PCAF, and p53 (see below) identify non-histone targets for the sirtuins, Sirt1 might also regulate histone acetylation directly through its interaction with sequence-specific DNA binding factors, such as COUP-TF-interacting proteins 1 and 2 (CTIP1 and CTIP2) or a MyoD/PCAF complex [50,60]. These findings implicate Sirt1 as a transcriptional repressor that functions through deacetylation of histones and non-histone proteins.
Sirt1 deacetylase activity has also been implicated in the repair of DNA damage, through its ability to deacetylate the tumor suppressor p53 [26,52], a sequence-specific transcription factor that regulates processes such as the cell cycle, cell death, and DNA repair, in response to a variety of stress signals. The p53 protein is acetylated at two lysine residues, Lys32O and Lys382, in response to DNA damage, leading to its activation [61-63]. Acetylation by the acetyltransferase P3OO positively regulates p53 activity, and deacetylation by HDAC1 and Sirt1 negatively regulates its activity [26,63]. Sirt1-mediated deacetylation of p53 suppresses the induction of apoptosis and prolongs cellular survival in response to DNA damage [52]. Both Sirt1 and p53 can be localized in promyelocytic leukemia (PML) bodies [51], subnuclear structures that are altered or disrupted in certain tumors and in response to various different cellular stresses [64]. These studies [26,51,52,61-63] suggest that Sirt1 deacetylates the p53 tumor suppressor protein to dampen apoptotic and cellular senescence pathways.
Two studies [65,66] have used gene-targeted mutagenesis experiments in mice to examine the consequences of expressing a mutant Sirt1 protein lacking part of the catalytic domain or of deleting the ortholog of the Sirt1 gene completely. McBurney et al. [65] showed that animals homozygous for a null allele of sir2α are born at only half the expected frequency, suggesting prenatal lethality; homozygous embryos and pups are smaller than their wild-type and heterozygous littermates and have developmental defects of the eyes, lungs, and pancreas. In an outbred background, the Sir2α-null animals often survive to adulthood, but both sexes are sterile because of a failure to ovulate in females and inefficient spermatogenesis in males [65]. Unexpectedly, there is no defect in gene silencing in Sir2α-null animals, nor in Sir2α-null homozygous embryonic stem cells, suggesting either that Sirt1 orthologs have a different role in mammals from that in S. cerevisiae or that its role in gene silencing is confined to a small subset of mammalian genes [65,67]. In the second study [66], mice were generated that lacked Sir2a or expressed a mutant Sir2a protein lacking part of the catalytic domain. Both types of Sir2α mutant mice were smaller than their wild-type and heterozygous littermates and had developmental defects in the retina and heart; most died postnatally [66]. Sir2α-deficient thymocytes showed an increase in p53 hyperacetylation in response to ionizing radiation, followed by apoptosis [66]. The phenotype of the Sir2α-null mice suggests that the Sirt1 protein is essential for normal embryogenesis and for normal reproduction in both sexes in mammals. Further study of the function of Sirt1 orthologs at the molecular level will help to unravel the origins of these developmental defects.
Sirt2
The human Sirt2 protein is similar in sequence to yeast Hst2p and both proteins are located in the cytoplasm [27,28] (Figure 4), where Hst2p affects chromatin silencing through an unknown mechanism [27]. Sirt2 colocalizes with the microtubule network and deacetylates Lys40 of α-tubulin [47]. The same residue of α-tubulin is also deacetylated by HDAC6, a class II HDAC, and deacetylation by HDAC6 leads to changes in cellular motility [68]. Sirt2 and HDAC6 are found along microtubules and can be co-immunoprecipitated with each other, suggesting that the two proteins coordinately regulate the level of tubulin acetylation [47]. Recent evidence shows that Sirt2 is upregulated before mitosis and suggests a role for this protein in cell-cycle regulation [69]. Sirt2 is rapidly degraded after mitosis, and cells overexpressing mutant forms of Sirt2 show a delay in exit from mitosis. Whether Sirt2-dependent regulation of α-tubulin acetylation is related to cell-cycle arrest remains to be determined.
A role for Sirt2 in cancer pathogenesis was recently demonstrated using a proteomic approach [70]. The Sirt2 gene, which is located at chromosome 19q13.2, lies within a region that is frequently deleted in human gliomas, and levels of Sirt2 mRNA and Sirt2 protein expression are severely reduced in a large fraction of human glioma cell lines [70]. Ectopic expression of Sirt2 in these cell lines suppressed colony formation and modified the microtubule network. These results indicate that Sirt2 may act as a tumor suppressor and may function to control the cell cycle by acetylation of α-tubulin.
Sirt3
Human Sirt3 is primarily located in the mitochondrial matrix [25,29] (Figure 4), and its mitochondrial import is mediated by an amphipathic α helix at its amino terminus. The protein, whether endogenous or overexpressed, is proteolytically clipped at its amino terminus in vivo, resulting in the removal of the first 100 amino acids. This clipping can be recapitulated in vitro with purified mitochondrial-matrix processing peptidase (MPP) [25]. Interestingly, the unprocessed protein is enzymatically inactive in vitro and becomes enzymatically active after proteolytic processing by MPP [25]. Negative regulation of the enzymatic activity of a sirtuin by its amino terminus is also observed in the case of another class Ib sirtuin, yeast Hst2p. This protein may form a homotrimer in solution, such that the amino-terminal methionine of each molecule interacts with the active site in another Hst2p molecule within the quaternary structure [19].
Although Sirt3 shows robust histone deacetylase activity on a histone H4 peptide in vitro, the absence of histones in mitochondria suggest that non-histone proteins are its primary target [25,29]. Mouse Sirt3, also called Sir2L3, lacks the amino-terminal extension that mediates the mitochondrial targeting of human Sirt3 and is located in cytoplasmic vesicles [71]. Finally, a small fraction of cells overexpressing Sirt3 shows reproducible nuclear staining (B.J.N. and E.V., unpublished observations); the relevance of this observation is unclear, but it could indicate selective targeting of Sirt3 to different compartments under different physiological conditions.
Frontiers
Much excitement has been generated by the recent observations that sirtuin proteins might play a significant role in the genetic control of aging. In S. cerevisiae, lifespan is shortened by a null mutation in SIR2 and is extended by the presence of an extra copy of SIR2 [12,72]. Loss of Sir2p leads to a derepression of silencing at the rDNA locus, which increases recombination between rDNA repeats and results in the accumulation of extrachromosomal rDNA circles. High numbers of these circles in older mother cells promote senescence by an undefined mechanism, possibly through the titration of necessary factors away from other promoters [73]. Likewise, the C. elegans Sir2p homolog, Sir-2.1, mediates dauer-larva formation and regulates lifespan [15]. The dauer larva represents a specialized survival form of the worm; the molecular mechanisms by which Sir-2.1 controls dauer formation remain to be elucidated, however.
It has been speculated that the metabolic rate of the cell may be important in the regulation of the function of various sirtuins, given their dependency on NAD for enzymatic activity. This idea is further supported by evidence that NAD metabolism directly participates in controlling the aging process [12,74,75]. Translating these ideas to mouse and human sirtuins could give novel insights into the regulation of mammalian lifespan. In mammals, Sirt1 could be a key part of the decision of a cell whether to live or die in response to DNA damage. As the cell ages and NAD levels become lower, the resulting reduced activity of Sirt1 could drive the decision to cell senescence or apoptosis rather than cellular survival. Likewise, lowered activity of Sirt2 via reduced levels of NAD could alter the rate of cell division through cell-cycle regulation. The localization pattern of Sirt3 in the mitochondrial matrix and its dependence on NAD suggest a possible function in the regulation of cellular metabolism as a sensor for intramitochondrial NAD levels. Further work is required to determine the endogenous target of this deacetylase in the mitochondrial compartment, however. Genetic deletion of the genes encoding different sirtuins in mice will facilitate studies of their roles in mammalian aging.
In addition to a connection between metabolism and sirtuin activity via NAD, the formation of O-acetyl-ADP-ribose as an enzymatic byproduct represents another promising area of investigation. Microinjection of O-acetyl-ADP-ribose delays or blocks oocyte maturation and cell division in blastomeres [45]. A similar effect is observed after microinjection of low levels of active yeast Hst2 or human Sirt2 enzyme, but not with a catalytically impaired mutant, indicating that the enzymatic activity is essential for the observed effects.
Human Sirt4, Sirt5, Sirt6, and Sirt7 show low or undetectable enzymatic activity on histone H4 peptide [47]. This could reflect differing substrate specificities or different requirements for cofactors, a field of investigation that is likely to yield interesting insights in the future. Finally, an increased understanding of the relationship between the structure and function of sirtuin proteins will be important in designing specific inhibitors and exploring their potential therapeutic value in a variety of pathological conditions.
Acknowledgments
Acknowledgements
We thank John Carroll and Jack Hull for graphics, Stephen Ordway and Gary Howard for editorial assistance, and Sarah Sande for manuscript preparation. Our work is supported by the NIH and the J. David Gladstone Institutes.
References
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. A review of the regulation of acetylation and acetylase enzymes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. This paper and [3] focus on the function and regulation of class I and II HDACs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. See [2]. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. Yeast telomeres have an epigenetic effect on the transcriptional repression of nearby genes. [DOI] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. Yeast Sir3p and Sir4p are localized to telomeric regions and are required for telomeric localization and formation as well as for telomere-associated gene repression. [DOI] [PubMed] [Google Scholar]
- Klar AJ, Strathern JN, Hicks JB. A position-effect control for gene transposition: state of expression of yeast mating-type genes affects their ability to switch. Cell. 1981;25:517–524. doi: 10.1016/0092-8674(81)90070-2. An increase in expression of the HML and HMR silent mating-type loci increases the rate of switching of the two loci, indicating a role for expression in transposition. [DOI] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. The initial identification of the SIR genes and their role in repression of gene expression at the silent mating-type loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth KA, Tatchell K, Hall BD, Astell C, Smith M. A position effect in the control of transcription at yeast mating type loci. Nature. 1981;289:244–250. doi: 10.1038/289244a0. The finding that there is a region close to each HML and HMR cassette that is responsible for transcriptional repression of these loci. [DOI] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. This paper and [10] indicate that transcriptional silencing at the rDNA locus is mediated by a distinct mechanism for the telomeres and silent mating-type loci, yet rDNA silencing still requires Sir2p. [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. See [9]. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. Identification of four homologs of Sir2 genes and their involvement in silencing at Sir2-dependent loci and a role in cell-cycle progression, radiation resistance, and genomic stability. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. Activation of Sir2p by NAD is a feature found during lifespan extension mediated by caloric restriction. [DOI] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/S0092-8674(00)80772-2. This paper and [14] demonstrate that in response to DNA damage, telomeric components are targeted to sites of DNA damage. [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/S0092-8674(00)80773-4. See [13]. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. A duplication of Sir2.1 in C. elegans increases lifespan by 50% and functions upstream of daf-16 in the insulin-like signaling pathway. [DOI] [PubMed] [Google Scholar]
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. Identification of five human sirtuins and demonstration that Sirt2 (as well as Salmonella typhimurium CobB) contain mono-ADP-ribosyltransferase activity on bovine serum albumin. [DOI] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. Identification of two additional human sirtuins and classification of all sirtuins into five classes. [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol. 2001;8:621–625. doi: 10.1038/89668. This paper and [19-21] describe high-resolution crystal structures of sirtuin proteins, indicating interactions with NAD and acetylated substrates and of their amino- and carboxy-terminal extensions with the catalytic site. [DOI] [PubMed] [Google Scholar]
- Zhao K, Chai X, Clements A, Marmorstein R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat Struct Biol. 2003;10:864–871. doi: 10.1038/nsb978. See [18]. [DOI] [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/S0092-8674(01)00317-8. See [18]. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell. 2002;10:523–535. doi: 10.1016/S1097-2765(02)00628-7. See [18]. [DOI] [PubMed] [Google Scholar]
- Sherman JM, Stone EM, Freeman-Cook LL, Brachmann CB, Boeke JD, Pillus L. The conserved core of a human SIR2 homologue functions in yeast silencing. Mol Biol Cell. 1999;10:3045–3059. doi: 10.1091/mbc.10.9.3045. The catalytic cores of human Sirt2 can be functionally substituted in yeast Sir2p, suggesting an evolutionarily conserved mode of function across species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. The high-resolution crystal structure of a histone-deacetylase homolog, which emphasizes significant differences between class I and II HDACs and the later-crystallized class III HDACs. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. Demonstration that nicotinamide, an inhibitor of class III HDACs, can shorten yeast lifespan similarly to Sir2p mutants. A model is proposed for nicotinamide inhibition of sirtuins via binding within the C site of the NAD-binding pocket. [DOI] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. This paper and [29] demonstrate that human Sirt3 is a mitochondrially localized sirtuin with NAD-dependent deacetylase activity. Furthermore, Sirt3 is rendered active in vitro by proteolytic processing by the mitochondrial matrix protein MPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53deacetylase. Cell. 2001;107:149–159. doi: 10.1016/S0092-8674(01)00527-X. This paper and [51,52] demonstrate a role for Sirt1 in p53 deacetylation and modulation of the sensitivity of cells to the p53-dependent apoptotic response. [DOI] [PubMed] [Google Scholar]
- Perrod S, Cockell MM, Laroche T, Renauld H, Ducrest AL, Bonnard C, Gasser SM. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J. 2001;20:197–209. doi: 10.1093/emboj/20.1.197. Demonstration that yeast Hst2p is localized to the cytoplasm where, as shown in overexpression studies in a Sir2-deficient strain, it can negatively regulate telomeric silencing and positively regulate rDNA silencing by an unknown mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar G, Murnane JP. Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene. 1999;234:161–168. doi: 10.1016/S0378-1119(99)00162-6. The first cloning of a human sirtuin gene and identification of a novel, cytoplasmic, subcellular localization of a Sir2 protein. [DOI] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. See [25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ. Mechanisms of silencing in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1998;8:233–239. doi: 10.1016/S0959-437X(98)80146-9. A review focused on transcription silencing at heterochromatin regions by SIR proteins and its link with the nucleolus and aging. [DOI] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. This paper and [32] demonstrate that Net1p, Cdc14p and Sir2p are components of a complex involved in mitotic exit and associated with silencing at the rDNA locus, called the regulator of nucleolar silencing and telophase exit (RENT) complex. [DOI] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. See [31]. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. This paper and [34] show that Sir2p in yeast functions at the rDNA array and suppresses recombination in addition to its roles in transcriptional repression. [DOI] [PubMed] [Google Scholar]
- Fritze CE, Verschueren K, Strich R, Easton Esposito R. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. See [33]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzebiatowski JR, O'Toole GA, Escalante-Semerena JC. The cobT gene of Salmonella typhimurium encodes the NaMN: 5,6-dimethylbenzimidazole phosphoribosyltransferase responsible for the synthesis of N1-(5-phospho-alpha-D-ribosyl)-5,6-dimethylbenzimidazole, an intermediate in the synthesis of the nucleotide loop of cobalamin. J Bacteriol. 1994;176:3568–3575. doi: 10.1128/jb.176.12.3568-3575.1994. This paper and [36,37] describe the role of CobT in cobalamin biosynthesis and the ability of CobB to substitute for CobT activity in a CobT-deficient background. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzebiatowski JR, Escalante-Semerena JC. Purification and characterization of CobT, the nicotinate-mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase enzyme from Salmonella typhimurium LT2. J Biol Chem. 1997;272:17662–17667. doi: 10.1074/jbc.272.28.17662. See [35]. [DOI] [PubMed] [Google Scholar]
- Tsang AW, Escalante-Semerena JC. cobB function is required for catabolism of propionate in Salmonella typhimurium LT2: evidence for existence of a substitute function for CobB within the 1,2-propanediol utilization (pdu) operon. J Bacteriol. 1996;178:7016–7019. doi: 10.1128/jb.178.23.7016-7019.1996. See [35]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. Sir2p can catalyze the addition of a phosphate from NAD onto itself as well as histones, suggesting Sir2p is a mono-ADP-ribosyltransferase. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. This paper and [40,41] demonstrate that Sir2p is an NAD-dependent histone deacetylase. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. See [39]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. See [39]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. The products of Sir2 NAD-dependent histone deacetylation are ADP-ribose, nicotinamide and deacetylated substrate; furthermore, a non-hydrolyzable NAD analog and nicotinamide can function as Sir2 inhibitors. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. This paper and [44] demonstrate that the ADP-ribose product of the Sir2 deacetylation reaction is 1-O-acetyl-ADP-ribose. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. See [43]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borra MT, O'Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J Biol Chem. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. When 1-O-acetyl-ADP-ribose is injected into oocytes, it causes a delay or block in oocyte maturation and in cell division in the blastomeres of the resulting embryo; the same effect can also be achieved by injection of active yeast Hst2 or human Sirt2 enzyme. [DOI] [PubMed] [Google Scholar]
- García-Salcedo JA, Gijon P, Nolan DP, Tebabi P, Pays E. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. EMBO J. 2003;22:5851–5862. doi: 10.1093/emboj/cdg553. A Sir2 protein from Trypanosoma brucei is the first sirtuin to be shown to have a physiological role associated with a robust ADP-ribosyltransferase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/S1097-2765(03)00038-8. Human Sirt2 is an NAD-dependent tubulin deacetylase localized on the microtubule network. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001;276:38837–38843. doi: 10.1074/jbc.M106779200. This paper and [49] identify inhibitors of sirtuins using high-throughput small-molecule screens in yeast that make use of the roles of Sir2p in variegated transcriptional repression. [DOI] [PubMed] [Google Scholar]
- Bedalov A, Gatbonton T, Irvine WP, Gottschling DE, Simon JA. Identification of a small molecule inhibitor of Sir2p. Proc Natl Acad Sci USA. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. See [48]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/S1097-2765(03)00226-0. Overexpression of Sirt1 produces retarded muscle differentiation, whereas reduced expression of Sirt1 results in premature muscle differentiation. Muscle differentiation via Sirt1 is regulated by the intracellular ratio of [NAD+]/[NADH]. [DOI] [PubMed] [Google Scholar]
- Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. See [26]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/S0092-8674(01)00524-4. See [26]. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. Resveratrol, a plant polyphenol and a component of red wine, is a potent activator of sirtuin proteins and mimics the lifespan extension seen in calorie-restricted organisms. [DOI] [PubMed] [Google Scholar]
- Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, Fong HH, Farnsworth NR, Kinghorn AD, Mehta RG, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. This paper and [55,56] characterize resveratrol as a natural compound produced in plants with positive health effects for a variety of diseases. [DOI] [PubMed] [Google Scholar]
- Middleton E, Jr, Kandaswami C, Theoharides TC. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol Rev. 2000;52:673–751. See [54]. [PubMed] [Google Scholar]
- Ferguson LR. Role of plant polyphenols in genomic stability. Mutat Res. 2001;475:89–111. doi: 10.1016/S0027-5107(01)00073-2. See [54]. [DOI] [PubMed] [Google Scholar]
- Muth V, Nadaud S, Grummt I, Voit R. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001;20:1353–1362. doi: 10.1093/emboj/20.6.1353. Identification of TAFI68 as a substrate for Sirt1 dependent deacetylation, resulting in repression of RNA polymerase I transcription in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata T, Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun. 2003;301:250–257. doi: 10.1016/S0006-291X(02)03020-6. This paper and [59] characterize an interaction between a Sir2 protein and bHLH transcriptional repressors and a role for this interaction in developmental processes. [DOI] [PubMed] [Google Scholar]
- Rosenberg MI, Parkhurst SM. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell. 2002;109:447–458. doi: 10.1016/S0092-8674(02)00732-8. See [58]. [DOI] [PubMed] [Google Scholar]
- Senawong T, Peterson VJ, Avram D, Shepherd DM, Frye RA, Minucci S, Leid M. Involvement of the histone deacetylase SIRT1 in COUP-TF-interacting protein 2-mediated transcriptional repression. J Biol Chem. 2003;278:43041–43050. doi: 10.1074/jbc.M307477200. Sirt1 interacts with the DNA-binding transcription factor CTIP2 to mediate histone deacetylation and transcriptional repression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. This paper and [62,63] describe the acetylation of p53 and how this increases its DNA-binding affinity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. See [61]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/S0092-8674(00)80521-8. See [61]. [DOI] [PubMed] [Google Scholar]
- Strudwick S, Borden KL. Finding a role for PML in APL pathogenesis: a critical assessment of potential PML activities. Leukemia. 2002;16:1906–1917. doi: 10.1038/sj.leu.2402724. A review of the current state of the molecular mechanisms of the involvement of promyelocytic leukemia bodies in the pathogenesis of acute promyelocytic leukemia. [DOI] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. This paper and [66,67] describe the generation of Sirt1 (Sir2α) knockout mice and a role for Sirt1 in developmental processes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. See [65]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Bieman M, Th'ng J, Lemieux M. The absence of SIR2alpha protein has no effect on global gene silencing in mouse embryonic stem cells. Mol Cancer Res. 2003;1:402–409. See [65]. [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. HDAC6 is associated with microtubules and functions to deacetylate lysine 40 of α-tubulin to regulate cellular motility. [DOI] [PubMed] [Google Scholar]
- Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. The enzymatic activity of Sirt2 is associated with cell-cycle arrest, and Sirt2 is dephophorylated by Cdc14b, leading to its ubiquitination and degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A, Oshimura M. Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun. 2003;309:558–566. doi: 10.1016/j.bbrc.2003.08.029. The Sirt2 gene is found in a chromosomal region frequently deleted in human gliomas. Ectopic expression of Sirt2 in one cell line demonstrates a dramatic alteration of microtubule network morphology. [DOI] [PubMed] [Google Scholar]
- Yang YH, Chen YH, Zhang CY, Nimmakayalu MA, Ward DC, Weissman S. Cloning and characterization of two mouse genes with homology to the yeast Sir2 gene. Genomics. 2000;69:355–369. doi: 10.1006/geno.2000.6360. Characterization of mouse Sirt2 and Sirt3, demonstrating that the subcellular localization of mouse Sirt2 is similar to that of its human homolog but that of Sirt3 is different. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. This paper and [73] show that Sir2p regulates lifespan in yeast by repressing recombination at the rDNA array, which in the absence of Sir2p increases the formation of extrachromosomal rDNA circles and a reduction in lifespan. An increased dosage of Sir2p results in a lifespan extension. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles - a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/S0092-8674(00)80493-6. See [72]. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. A review focused on the role of Sir2p in regulation of aging in the context of cellular metabolism. [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. Regulation of lifespan through caloric restriction works through Sir2p and requires NAD. [DOI] [PubMed] [Google Scholar]
- Online Mendelian Inheritance in Man http://www.ncbi.nlm.nih.gov/omim A database found at the National Center for Biotechnology Information (NCBI), authored and edited by Victor A. McCusick and colleagues (Johns Hopkins University), that catalogs human genes and genetic disorders.
- PyMOL http://pymol.sourceforge.net/ Software developed for viewing three-dimensional high-resolution crystal structure data.
- Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem Sci. 2003;28:41–48. doi: 10.1016/S0968-0004(02)00005-1. A review discussing the regulation of Sir2 protein activity by the regulation of metabolism. [DOI] [PubMed] [Google Scholar]


