Abstract
The inability of the yeast Saccharomyces cerevisiae to ferment xylose effectively under anaerobic conditions is a major barrier to economical production of lignocellulosic biofuels. Although genetic approaches have enabled engineering of S. cerevisiae to convert xylose efficiently into ethanol in defined lab medium, few strains are able to ferment xylose from lignocellulosic hydrolysates in the absence of oxygen. This limited xylose conversion is believed to result from small molecules generated during biomass pretreatment and hydrolysis, which induce cellular stress and impair metabolism. Here, we describe the development of a xylose-fermenting S. cerevisiae strain with tolerance to a range of pretreated and hydrolyzed lignocellulose, including Ammonia Fiber Expansion (AFEX)-pretreated corn stover hydrolysate (ACSH). We genetically engineered a hydrolysate-resistant yeast strain with bacterial xylose isomerase and then applied two separate stages of aerobic and anaerobic directed evolution. The emergent S. cerevisiae strain rapidly converted xylose from lab medium and ACSH to ethanol under strict anaerobic conditions. Metabolomic, genetic and biochemical analyses suggested that a missense mutation in GRE3, which was acquired during the anaerobic evolution, contributed toward improved xylose conversion by reducing intracellular production of xylitol, an inhibitor of xylose isomerase. These results validate our combinatorial approach, which utilized phenotypic strain selection, rational engineering and directed evolution for the generation of a robust S. cerevisiae strain with the ability to ferment xylose anaerobically from ACSH.
Introduction
As the world’s human population increases, so does the demand for energy. Renewable biofuels offer a route to replace a portion of the finite amounts of liquid petroleum and natural gas-based fuels. Although bioethanol produced from grain has been employed as a partial replacement for gasoline, this process is generally viewed as unsustainable [1]. An alternative to grain-based ethanol, which has been traditionally produced by microbial fermentation of starch sugars, is bioethanol generated from lignocellulosic (LC) sugars derived from renewable and sustainable plant feedstocks [2]. Despite well over two decades of research, microbial-based production of cellulosic ethanol at an industrial scale remains largely unpracticed throughout the world. Part of the reason for this is a number of molecular barriers that have profound impact on the metabolic catabolism of LC sugars, thereby limiting their efficient and cost-effective conversion into ethanol. In particular, the yeast Saccharomyces cerevisiae, in its native form, does not convert most LC sugars into ethanol due to insufficient biochemical activities and metabolic inhibition.
Although S. cerevisiae excels at fermenting glucose from cornstarch and sugar cane juice, the fermentation of pentose sugars from the hemicellulose component of lignocellulosic biomass is challenging. In particular, xylose, a pentose sugar and a major component of hemicellulose, composes 30–40% of total cell-wall carbohydrate in grasses and some woody biomass [3]. Conversion of xylose to ethanol is crucial to maximize the economic return from fuel production in excess of feedstock and production costs. However, native S. cerevisiae cannot efficiently ferment xylose, as most strains have either lost or downregulated the activities of xylose catabolism enzymes [4] and lack specific xylose transporters [5]. To overcome this deficiency, yeast have been engineered to express a minimal enzyme set from native xylose-metabolizing organisms that allow conversion of xylose into xylulose-5-phosphate (X5P), which can then be catabolized by the pentose phosphate pathway into ethanol. Specifically, engineering of S. cerevisiae to express xylose reductase (XR) and xylitol dehydrogenase (XDH), or xylose isomerase (XI) alone, has permitted the limited conversion of xylose into xylulose, which can then be phosphorylated to X5P by overexpression of native or exogenous xylulokinase (XK) (for reviews, see [6]–[8]). With additional rational engineering approaches, yeast strains with improved xylose fermentation in lab medium have been created (reviewed in [9]–[11]). Some of these approaches have been employed with varying degrees of success, including metabolic reengineering of S. cerevisiae strains through overexpression of native pentose phosphate pathway enzymes [12], [13], deletion of genes such as PHO13 to improve xylose metabolism [14], and heterologous expression of putative xylose transporters [5], [15]. Experimental directed evolution is another well-utilized means to improve desired phenotypic traits (reviewed in [16]). A combination of rational engineering followed by directed evolution on xylose-containing medium under aerobic [17], [18] or oxygen-limited [10], [12] conditions has generated yeast strains with increased anaerobic xylose consumption rates relative to their parental strains. Most recently, two sequential anaerobic selections of an XR-XDH engineered S. cerevisiae strain on xylose resulted in an evolved isolate with a significantly faster anaerobic consumption rate of xylose than its ancestor, although most of the xylose appears to be converted to xylitol and glycerol [19]. These approaches have allowed for effective consumption of xylose in innocuous and nutrient-rich lab medium; however, conversion of complex, LC-derived xylose from lignocellulosic hydrolysates into biofuels is much more challenging.
Before being deconstructed into fermentable sugars, plant biomass requires chemical, thermal and/or mechanical pretreatments that alter cellulose, hemicellulose and lignin organization, thereby allowing hydrolytic enzymes greater access to sugar polymers for faster rates of enzymatic hydrolysis. Numerous pretreatment methods have been developed and they include the use of dilute acid, bases and ionic liquids (IL) (reviewed in [20]). Although it significantly increases the rate and effectiveness of biomass deconstruction, lignocellulose pretreatment generates a number of common degradation products released from plant cell walls. These chemical compounds include hemicellulose-derived acetate and lignin-derived aromatic aldehydes that induce microbial stress by draining reducing cofactors, limiting ATP generation and causing cellular damage (reviewed in [21] and [22]). Additionally, each pretreatment process can generate its own set of dominant stress-inducing compounds, such as furans generated from dilute acid pretreatment [23], [24] and hydroxycinnamic acids in alkaline hydrogen peroxide (AHP) pretreatment [25]. In some cases, the pretreatment compound itself can be a major biological inhibitor, as is the case for the IL, 1-ethyl-3-methylimidazolium acetate ([C2mim][OAc]) [26]. Ammonia Fiber Expansion (AFEX) is a highly effective pretreatment for herbaceous biomass. In contrast to dilute acid, which degrades the hemicellulose, AFEX pretreatment retains hemicellulose as intact polymers [27] that can then be hydrolyzed into fermentable sugars for additional fuel production. However, AFEX pretreatment of corn stover generates diverse inhibitory small molecules, including phenolic amides, which impair xylose fermentation by S. cerevisiae [28], [29]. These effects are often compounded during xylose fermentation in the absence of oxygen, likely due to reduced ATP yield from pentose sugars compared to hexoses, combined with decreased energetic yield under anaerobic conditions. Given that ATP drives numerous detoxification processes [21], the cellular stress induced by these compounds has profound impacts on xylose fermentation. To cope with these diverse inhibitory compounds present in LC hydrolysates and their impacts on cellular physiology and metabolism, many researchers have opted to employ industrial or environmental S. cerevisiae strains with innate stress tolerant properties [12], [25], [30]–[32]. A combination of directed engineering and evolution approaches with S. cerevisiae for xylose metabolism in the presence of defined spruce [30] or AHP [25] hydrolysate inhibitors or undefined raw spruce hydrolysates [12] have resulted in a small number of S. cerevisiae strains with improved xylose fermentation properties in lignocellulosic hydrolysates compared to their parental strains.
At present, little is known about how the inhibitors found in AFEX-pretreated corn stover hydrolysate (ACSH) impact xylose fermentation by S. cerevisiae, particularly under strict, industrially relevant, anaerobic conditions where ethanol production is maximized. As the foundation for investigating this knowledge gap, and the goal of determining what genetic factors are important for improving anaerobic xylose fermentation, we sought to develop and compare closely related S. cerevisiae strains with varying anaerobic xylose fermentation phenotypes. Previously, we identified a natural S. cerevisiae strain, GLBRCY0 (originally named as NRRL YB-210 [33]) with growth tolerance to individual LC-derived inhibitors [25] as well as ACSH at elevated temperature [34]. Despite engineering NRRL YB-210 (YB-210) with known XR-XDH genes, the strain fermented xylose in lab medium and ACSH at slow rates, even in the presence of limited oxygen. Here, we report the development of an engineered and evolved derivative of YB-210, which displayed robust cell growth in a variety of pretreated lignocellulosic hydrolysates (LCH) relative to other strains, with the ability to rapidly ferment xylose from lab medium and ACSH under completely anaerobic conditions. Combined genetic and metabolomic analyses indicate that the evolved strain, named GLBRCY128 (Y128), incurred a missense mutation in GRE3, which, combined with additional unknown mutations, allowed for faster anaerobic xylose consumption rates relative to its parent. Together, these results identify Y128 as a novel S. cerevisiae strain with evolved genetic traits for robust anaerobic fermentation of xylose from ACSH. They also illustrate how careful selection of genetic background can accelerate development of biocatalysts with the ability to ferment xylose anaerobically in an inhibitor-laden LCH.
Materials and Methods
AFEX pretreated corn stover hydrolysate (ACSH) preparation
Zea mays (Pioneer hybrid 36H56) corn stover from Field 570-C Arlington Research Station, University of Wisconsin was harvested in 2008 for use in 96-well plate phenotyping or in 2009 for use in anaerobic fermentation experiments. AFEX pretreatment of corn stover was performed as described previously [27]. AFEX pretreated corn stover was hydrolyzed at 6% or 9% glucan loading in a 1.5 L reaction volume in a 3 L Applikon fermenter (Applikon Biotechnology Inc. USA) with Spezyme CP (15 FPU/g glucan loading, DuPont Danisco), Multifect Xylanase (10% of Spezyme CP, DuPont Danisco), and Novovzyme 188 (64 pNPGU/g glucan, Sigma-Aldrich) at 50°C for 5 days. Tetracycline (40 mg/L) was used to prevent microbial contamination and pH 4.8 was maintained during the hydrolysis process. Biomass was added to the reaction mixture in 4 batches within 4 h from the start of hydrolysis to facilitate better mixing at 1000 rpm. After 120 h, the hydrolysis mixture was centrifuged (2500×g for 30 min.) and sterile filtered (0.22 µm pore size; Millipore Stericup). For 6% glucan loading ACSH, the final sugar concentrations were 53 g/L glucose and 21.7 g/L xylose. For 9% glucan loading ACSH, the final sugar concentrations were 80 g/L glucose and 36 g/L xylose. For ACSH used in bioreactor fermentations, hydrolysates were prepared as described previously [35] with one additional modification; prior to fermentation, the hydrolysate was adjusted to pH 5.0 and again filtered through a 0.2 µm filter to remove precipitates and to ensure sterility.
Alkaline hydrogen peroxide pretreated hydrolysates
Pioneer hybrid 36H56 corn stover described above and switchgrass (Panicum virgatum cv. Cave-In-Rock) described elsewhere [36] were milled (Circ-U-Flow model 18-7-300, Schutte-Buffalo Hammermill, LLC) to pass through a 5 mm screen. AHP pretreatment was performed as reported previously [37] at a hydrogen peroxide loading of 0.125 g H2O2/g biomass in an incubator with shaking at 150 rpm at 30°C for 24 h with periodic pH readjustment to 11.5 during pretreatment using 5 N NaOH. For switchgrass, pretreatment was conducted at biomass concentration of 20% w/w. For corn stover, pretreatment was conducted at biomass concentration of 15% w/w. Following pretreatment, the whole slurry was adjusted to pH 4.8 using 72% H2SO4. Accelerase 1000 (Novozymes A/S), Multifect xylanase, and Multifect pectinase (DuPont Danisco) were used at the protein ratio of 0.62∶0.24∶0.14 with a total protein loading of 30 mg/g glucan as assayed by the Bradford Assay. Hydrolysis was performed at 50°C with shaking speed of 180 rpm for 24 h. After enzymatic hydrolysis, the whole slurry was centrifuged at 18,000×g for 30 min. The supernatant was used as undetoxified raw hydrolysate or for detoxified hydrolysate, activated carbon (Fisher Scientific #05-690A) was mixed with undetoxified hydrolysate at 5% concentration (5 g activated carbon with 100 mL hydrolysate) and incubated at 50°C for 1 h in an unbaffled shake flask at 150 rpm. After centrifugation at 18000×g for 30 min, the supernatant was used as the detoxified hydrolysate. All hydrolysates were filter-sterilized (0.22 µm pore size; Millipore Stericup). Final sugar concentrations for AHP hydrolysates were 30 g/L glucose and 20 g/L xylose for raw AHP corn stover hydrolysate, 35 g/L glucose and 23 g/L xylose for detoxified AHP CSH, and 27 g/L glucose and 19 g/L xylose for both raw and detoxified AHP SGH.
Dilute acid pretreated lignocellulosic hydrolysates
An industrial collaborator provided two different versions of dilute acid pretreated wheat straw that was hydrolyzed using a proprietary blend of cellulase enzymes at pH 5.0 and 50°C. Both hydrolysates were diluted 4∶5 in sterile water supplemented with 10 g/L yeast extract and 20 g/L peptone.
Ionic liquid pretreated switchgrass hydrolysate (IL-SGH)
Switchgrass was pretreated with [C2mim][OAc] (1-ethyl-3-methylimidazolium acetate) at 15% solids loading as described elsewhere [38]. IL-pretreated switchgrass was hydrolyzed with CTec2 (54 mg/g glucan) and HTec2 (6 mg/g glucan) enzymes (Novozyme) for 72 h in a 2 L IKA bioreactor. [C2mim][OAc]-pretreated SGH was generated at Advanced Biofuels Process Demonstration Unit (batch ABPDU 110201S02). Final sugar concentrations in the IL-SGH were 41 g/L glucose and 10 g/L xylose.
Lab media
Standard undefined yeast lab medium was prepared as previous described [39]. Briefly, liquid and plate-based medium contained 10 g/L yeast extract and 20 g/L peptone (YP), and various sugar concentrations (X = 20 g/L xylose, D = 20 g/L dextrose or glucose, DX = 60 g/L glucose and 30 g/L xylose). Where indicated, hydrolysates were supplemented with 10 g/L yeast extract and 20 g/L peptone. For bioreactor experiments, this YPDX medium also contained 50 mM potassium phosphate, pH 5.0.
Saccharomyces cerevisiae strains, 96-well growth assays and hierarchical clustering
Native S. cerevisiae strains used in this study (see Table S1) were obtained from Dr. Cletus Kurtzmann (USDA ARS, Peoria, IL), National Collection of Yeast Cultures (Norwich, UK), and Dr. Justin Fay (Washington University, Saint Louis, MO). Aerobic growth assays in microtiter plates were performed as previously described [40] [25] [34], with one exception; 10 µL of saturated culture was inoculated into 190 µL of YPD or a single type of pretreated lignocellulosic hydrolysate. Cell growth was measured by optical density at 595 nm every 10 min for 24 h in Tecan F500 or M1000 multimode plate readers with an interior temperature of 30°C. Background-subtracted cell density readings for each strain were analyzed by the program GCAT [25]. Normalized specific growth rates for each strain from three independent biological replicates in pretreated hydrolysates were normalized to their average growth rate in YPD alone, and then ranked ordered from 1 to 117 (including control strains – Y389, BY4741, CEN.PK113-5D and CEN.PK2-1D in duplicate) for highest average specific growth to lowest, respectively. For all strains with no detectable specific growth rates, strains were assigned a rank of 117. Strain ranks in each medium condition were hierarchically clustered and displayed with Spotfire (TIBCO).
DNA constructs and strain engineering
Genotypes of engineered strains used in this study are described in Table 1 . Construction of the GLBRCY73 strain has been described elsewhere [40]. The expression cassette containing TAL1 from S. cerevisiae S288c (ScTAL1), xylA from Clostridium phytofermentans ISDg (CpxylA) and XYL3 from Scheffersomyces stipitis CBS 6054 (SsXYL3) was generated in a similar manner with some modifications. Codon-optimized versions of each gene were synthesized (GeneArt, Life Technologies) and inserted via homologous recombination in the following promoter-open reading frame-terminator combinations in order from 5′ to 3′: ScPGK1 promoter-ScTAL1-ScTDH3 terminator, ScTDH3 promoter-CpxylA-ScTEF2 terminator, ScTEF2 promoter-SsXYL3-ScCYC1 terminator. This cassette, which also contains a loxP-KanMX-loxP selection marker [41], was flanked by ScHO sequences [42] for targeted recombination at the genomic HO locus. The complete CpxylA cassette was amplified by standard polymerase chain reaction (PCR) using primers that anneal to the 5′ ends of the HO flanking sequences, gel purified and transformed into the NRRL YB-210 strain. Following selection on YPD plates containing 200 µg/mL Geneticin (Life Technologies), verification of cassette insertion was determined by PCR using combinations of primers that anneal outside of the HO flanking sequence and specific to synthesized DNA cassette. The engineered YB-210 diploid strain was then subjected to sporulation and tetrad dissection. One spore, which was derived from a tetrad with three other inviable spores, was confirmed for a single MATa mating type and subsequently named GLBRCY22-3 (Y22-3). LoxP-KanMX-loxP marker rescues from Y22-3, Y127 and Y128 were carried out by expression of Cre recombinase as published elsewhere [41] to generate the respective Y36, Y132 and Y133 strains. Deletion of engineered CpxylA and ScTAL1, and endogenous ScGRE3 were performed by integration of PCR product using a loxP-KanMX-loxP cassette [41] into the marker-rescued versions of GLBRCY36, GLBRCY132 and GLBRCY133 strains. Sanger sequencing of PCR products and DNA plasmids was performed by the University of Wisconsin-Madison Biotechnology Center.
Table 1. Engineered and evolved S. cerevisiae strains used in this study.
| Strain name | Genotype | Reference |
| GLBRCY73 | NRRL YB-210 MATa/α HOΔ::SsXYL1-SsXYL2-SsXYL3-loxP-KanMX-loxP, aerobically evolved on YPX | [40] |
| GLBRCY22-3 | NRRL YB-210 MATa spore HOΔ::ScTAL1-CpxylA-SsXYL3-loxP-KanMX-loxP | This study |
| GLBRCY127 | GLBRCY22-3 MATa, aerobically evolved isolate on YPDX | This study |
| GLBRCY128 | GLBRCY127 MATa, anaerobically evolved isolate on YPDX | This study |
| GLBRCY36 | GLBRCY22-3 with loxP-KanMX-loxP marker excised by Cre | This study |
| GLBRCY132 | GLBRCY127 with loxP-KanMX-loxP marker excised by Cre | This study |
| GLBRCY133 | GLBRCY128 with loxP-KanMX-loxP marker excised by Cre | This study |
| GLBRCY132 xylAΔ | GLBRCY132 xylAΔ::loxP-KanMX-loxP | This study |
| GLBRCY132 tal1Δ | GLBRCY132 synthetic tal1Δ::loxP-KanMX-loxP | This study |
| GLBRCY133 xylAΔ-A | GLBRCY133 xylAΔ::loxP-KanMX-loxP transformant A | This study |
| GLBRCY133 xylAΔ-B | GLBRCY133 xylAΔ::loxP-KanMX-loxP transformant B | This study |
| GLBRCY36 gre3Δ | GLBRCY36 gre3Δ::loxP-KanMX-loxP | This study |
| GLBRCY132 gre3Δ | GLBRCY132 gre3Δ::loxP-KanMX-loxP | This study |
| GLBRCY133 gre3Δ | GLBRCY132 gre3Δ::loxP-KanMX-loxP | This study |
Quantitative RT-PCR
GLBRCY128 and GLBRCY128 xylAΔ strains were aerobically cultured at 30°C in baffled shake flasks containing YPD media. When the culture reached log phase growth (OD600 = 0.8), 7 mL of cell culture was then harvested by centrifugation at 3,000×g for 3 min, the supernatant was decanted and then cell pellets were flash frozen on dry ice/ethanol. Frozen cell pellets were resuspended and then vortexed in 0.8 mL phenol, pH 4.3 and 0.8 mL lysis buffer (10 mM Tris-Cl, pH 7.4, 10 mM EDTA, 0.5% SDS). Cell lysates were incubated at 65°C for 30 min and then centrifuged at 20,000×g at 4°C. The aqueous phase was then transferred to a new 2 mL centrifuge tube and further extracted with two additional rounds of phenol and then chloroform. The final extracted aqueous phase was then transferred to RNase-free minicentrifuge tubes and RNA precipitated by addition of 0.1 volumes of 0.3 M NaOAc, pH 5.2 and 2.5 volumes of 100% ethanol at −20°C for 1 h. Precipitated RNA was pelleted by centrifugation at 20,000×g for 30 min at 4°C. The RNA pellet was washed with 2 mL 70% ethanol, dried in a Speed-Vac, and dissolved in 0.2 mL RNase-free TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). RNA was further purified by RNeasy Mini Kit (Qiagen) according to the manufacturer protocol. cDNA synthesis from 10 µg total RNA was performed with Superscript III reverse transcriptase (Life Technologies) according to manufacturer protocol. Generated cDNA was purified and concentrated with a PCR Minelute Purification kit (Qiagen) into 12 ml elution buffer according to manufacturer protocol. For quantitative reverse-transcriptase polymerase chain reaction (qPCR) of xylA, 10 ng of cDNA was mixed with 500 nM SynCpXylA FWD (5′-GGTGGATGCTAGGTTGTCTTT-3′) and 250 nM SynCpXylA REV (5′-CACGCCTTCTTGCTCAAATAAC-3′), or ScERV25qPCRfor and ScERV25 FWD (5′-GTCGCGGATATTCACTCAGATG-3′) and ScERV25 REV (5′-CCTGCAAAGTCCCTCTTTCTAC-3′) primers and SYBR JumpStart Taq Ready Mix with Rox internal standard (Sigma-Aldrich) according to manufacturer protocol. Relative quantities of CpxylA and ScERV25 RNA were determined using an ABI7500 Real-Time PCR instrument (Applied Biosystems). Relative concentrations of CpxylA and ScERV25 transcripts were determined by ΔCt method.
Directed Evolution
Cell density measurements were determined from OD600 measurements from the cultures diluted 1∶10 in 1 cm path length cuvettes by a Beckman Coulter DU720 spectrophotometer. For aerobic adaptation, GLBRCY22-3 was inoculated at OD600 = 0.05–0.1 in 250 mL YP medium containing 2% xylose and 0.1% glucose. The first 15 transfers took place over 53 days with serial 1∶10 dilutions in fresh medium occurring every 3–4 days. After transfer 15, the adaptation culture was diluted every 2–3 days over the course of 44 additional days, ending after transfer 34. For anaerobic adaptation, GLBRCY127 was inoculated at OD600 = 0.05–0.1 in a flask containing 50 mL YP medium with 2% xylose, 0.1% glucose and 50 µg/mL Geneticin, and then placed in an anaerobic chamber. For the first two anaerobic transfers, the medium also contained 40 µg/L ergosterol (Sigma-Aldrich) and 4 g/L Tween-80 (Sigma-Aldrich). Anaerobic cultures were maintained in suspension using a stir bar and magnetic stir plate, and passaged every 7 days during the first 5 transfers. After the 5th transfer, the culture was passaged every 3–4 days with the final 14th transfer finishing 66 days after the start of the anaerobic adaptation. Xylose concentrations in the medium at the end of each transfer cycle were measure by YSI 2700 Select instrument. At the end of each adaptation, the culture at 1∶10,000 dilution was spread onto multiple YPD-Geneticin plates and incubated at 30°C for 48 h. Single clonal isolates were picked and evaluated for growth in YPX medium either aerobically or anaerobically in an anaerobic chamber as described previously [25], [40].
Aerobic and anaerobic fermentations
Aerobic and anaerobic batch fermentations in 3 L bioreactors (Applikon Biotechnology) were conducted using 2.1 L of ACSH or 1.7 L of YPX or YPDX with 50 mM potassium phosphate medium. Vessels were sparged in the headspace with N2 (150 sccm) for anaerobic fermentation or in the medium with air (200 sccm) for aerobic fermentation. Inocula were prepared from single colonies grown in YPD-Geneticin medium aerobically for ∼9 h, and then were diluted in ACSH or YPD medium with Geneticin at an initial OD600 = 0.1 (approximately 0.08–0.1 g DCW/L), and then grown aerobically for approximately 20 h. Starter cultures were then inoculated at a starting OD600 = 0.1 in bioreactor vessels maintained at 30°C and pH 5.0 with NaOH or HCl, and stirred at 500 rpm. For aerobic and anaerobic YPX growth assays, inoculum cultures were started from single colonies grown in YPD-Geneticin medium overnight and then diluted to OD600 = 0.1 in YPX (no Tween-80 or ergosterol) at the start of the assay. Yeast cultures were grown in culture tubes containing 5–7.5 mL of medium shaken at 30°C, or in 30 mL of medium stirred in flasks placed in an anaerobic chamber (Coy) purged with hydrogen. For anaerobic experiments, bioreactors containing YPX, YPDX or ACSH were sparged with N2 into the medium for at least 2 h. Flasks containing YPX were placed in the anaerobic chamber for at least 2 h prior to inoculation. Cell density measurements were determined by OD600 measurements from cultures diluted 1∶10 or 1∶25 in water. All OD600 measurements were blanked against uninoculated medium diluted in the same manner. Dry cell weight (DCW) was determined by vacuum filtering 50 mL of culture at 4 time points onto pre-weighed filters, washing with water and microwaving on 10% power for 15 minutes. Filtered cells were additionally dried by desiccant for 2–3 days and then weighed. DCW values in grams per 50 mL of culture included subtraction of the filter weight alone. Correlations between g DCW/L and OD600 concentrations were calculated to provide cell density measurements based on cell mass/L. Medium glucose and xylose concentrations from aerobic tube and anaerobic flask experiments were determined by YSI instrument. Extracellular glucose, xylose, ethanol, glycerol and xylitol concentrations from aerobic and anaerobic bioreactor experiments were determined by high performance liquid chromatography (HPLC) and refractive index detection (RID) as published elsewhere [35].
Quantification of intracellular pentose metabolic intermediates
To quantify intracellular metabolites, 5–10 mL of cell culture was rapidly removed from bioreactors with a 20 mL sterile syringe and 4 mL aliquots were applied to a filtration manifold unit (Hoefer FH 225 V) outfitted with sterile 25 mm nylon filters (Whatman; Nylon; 0.45 µm pore size), and the cells captured on the filters under vacuum. To reduce the background associated with metabolites present in ACSH, the cells were then rapidly washed with 5 mL of synthetic hydrolysate media [35] at pH 5.0 lacking amino acids and replacing 9% sorbitol in place of 6% glucose and 3% xylose. The filters were then removed and rapidly placed in 15 mL conical tubes containing ice-cold extraction buffer ([43]; acetonitrile-methanol-water, 40∶40∶20, 0.1% formic acid) and flash frozen in a dry ice ethanol bath.
The concentration of xylulose-5-phosphate was determined using reverse phase ion pairing HPLC [44] and electrospray ionization tandem mass spectrometry (ESI-MS/MS). Reagents and nonlabeled reference compounds were from Sigma Aldrich Co. (Saint Louis, Missouri, USA). Compounds were separated on an Ascentis HSS-T3 C18 column, 150×2.0 mm, 1.8 µm particle size (Waters Acquity). The mobile phase A consisted of 92.5∶7.5 water:methanol, 10 mM tributylamine, and 15 mM acetic acid, and mobile phase B was isopropyl alcohol. Xylulose-5-phosphate, whose peak overlapped with ribulose-5-phosphate, was quantified by integrating the portion of the partially resolved peak that was clearly attributable to X5P.
For quantifying intracellular xylose, xylulose, and xylitol, 20 µL aliquots of metabolite extract were transferred to 2.5 mL centrifuge tubes along with 20 µL of a solution containing 100 µM U-13C5-xylitol (Sigma-Aldrich), 100 µM U-13C5-xylose (Sigma-Aldrich) and 50 µM U-13C5 xylulose. U-13C5 xylulose was obtained by enzymatic conversion of U-13C5-xylose by immobilized xylose isomerase (Sigma-Aldrich). Samples were then evaporated to dryness under reduced pressure in a rotary evaporator (Savant SPD131A) with cryogenic cold trap for 3–4 hours. Dried samples were incubated with 30 µL 2% methoxyamine hydrochloride in anhydrous pyridine at 60°C for 45 min, and then derivatized at 60°C for an additional 30 min with 70 µL N-methyl,N-(trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane (Fisher Scientific). Derivatized samples were then analyzed by gas chromatography coupled with mass spectrometry (GC-MS) on an Agilent 5975 MSD with a Combi PALl autosampler (CTC analytics), and a 6890A GC oven equipped with a 30 m×0.25 mm ID×0.25 µm film HP5-MS capillary column. The inlet was held at 250°C and operated in split mode with a ratio of 10∶1 with a helium carrier gas flow rate maintained at 1.2 mL/min. The oven temperature was held at 125°C for 47 min then increased linearly at 40°C/min to a final temperature of 300°C.
The mass spectrometer was operated in SIM mode divided into time segments so that only the relevant masses were monitored over the times when each target compound eluted, allowing optimal dwell times of 100–150 ms while still recording at least 20 points over the width of a peak. SIM masses were selected corresponding to fragments (M+–15) containing all 5 13C atoms for the labeled internal standards of xylose, xylulose (m/z 457) and xylitol (m/z 427) to allow detection without interference from the isotopic masses of the coeluting natural abundance compounds. The naturally-occurring 12C compounds were measured by peak areas of the corresponding 12C (xylose, m/z 452, xylitol, m/z 422) ions except for xylulose, which was monitored with a much more abundant m/z 263 ion that was found to be free of interference from the labeled internal standard.
Instrument operation, data collection, and calculation of results were conducted by Agilent MassHunter for GC software VB.07.00 and Mass Hunter Workstation Quantitative Analysis v.B.06.00. Results were calculated from relative peak areas of analytes to their corresponding internal standards interpolated with a calibration curve of relative natural standard/13C internal standard peak areas versus relative standard/13C internal standard concentrations.
In vitro xylose reductase activity assays
In vitro xylose reductase (XR) activities were performed as previously described [45]–[47] with minor modifications. Y132, Y132 gre3Δ, Y133 and Y133 gre3Δ strains were grown in YPD medium to OD600 = 0.8 and then cells from 45 mL of culture were harvested, washed with 10 mL 0.85% NaCl and re-suspended in an equal volume of breaking buffer (50 mM potassium phosphate, pH 7.4, 1 mM β-mercaptoethanol and 1x Thermo HALT protease inhibitor cocktail). Resuspended cells were then transferred to a glass tube with glass beads and vortexed. The resulting material was centrifuged at 4°C and the clarified cell lysate used immediately for activity assays. A Tecan M1000 microplate reader was used to measure absorbance at 340 nm in technical triplicates. Protein concentrations from cell extracts were determined using a modified Bradford assay [48] with bovine serum albumin as the standard. Units of enzyme activity were normalized to total protein extract concentration and averaged from two independent biological samples. Specific XR activities were measured in range of 2–40 mU activity/mg total protein in the presence of NADPH co-factor.
Results and Discussion
Phenotypic screening identifies natural S. cerevisiae strains tolerant to pretreated lignocellulosic hydrolysates
The primary goal of this study was to create a strain of S. cerevisiae that can effectively ferment xylose anaerobically from AFEX-pretreated lignocellulosic biomass. We first evaluated two well-characterized laboratory strains, BY4741 [49] and CEN.PK2 [50] as potential starting points of this research by evaluating their growth abilities in lignocellulosic hydrolysates (LCHs) generated from a variety of established pretreatments and feedstocks (see Materials and Methods). Although both strains reached saturated cell density within 8 h after inoculation in YPD medium, they grew at substantially slower rates and reached low cell densities in the pretreated LCHs compared to standard medium (Figs. S1A and B), even though glucose concentrations were significantly higher in LCHs. These results suggest that the BY4741 and CEN.PK2 lab strains would not be able to generate sufficient cell biomass for rapid fermentation of inhibitor-laden hydrolysates. Thus, we sought an alternative strain background with robust cell growth in multiple LCHs in hopes of utilizing a strain with sufficient tolerance to LCH inhibitors. To find such strain, we performed comprehensive phenotyping of a collection of publicly available wild and domesticated S. cerevisiae strains obtained from a variety of locations and environments (Supplemental Strain Table 1, [51]–[53]) for cell growth in multiple pretreated LCHs. Individual strains were inoculated into 96-well plates containing 6% or 9% glucan-loading AFEX pretreated corn stover (ACS), raw or detoxified AHP pretreated corn stover (CS) or switchgrass (SG), [C2mim][OAc]-pretreated SG (IL SGH), or two different proprietary dilute acid pretreated biomass materials, supplemented with yeast extract and peptone (YP). Cell densities were continuously measured for 24–48 h, from which specific growth rates for each strain in every medium condition were calculated and normalized relative to their growth rate in YPD medium.
The collection of strains displayed wide ranges of aerobic growth rates in the various hydrolysate conditions (Figs. S2–11 and Table S1). Supplementation of YP to 6% glucan-loading ACSH (Figs. S2 and S6) improved growth, suggesting that growth defects in hydrolysates were due to lack of specific nutrients or additional nutrients allowed strains to overcome the effect of toxins. Not surprisingly, detoxification of AHP CSH and SGH (Figs. S4, S5, S7 and S8) significantly improved overall rates compared to raw AHP hydrolysates, while growth rates in 9% glucan-loading ACSH, which contained higher concentrations of sugars and inhibitory compounds [34], were lower than in 6% glucan medium (Figs. S2 and S3). In AHP CSH (Fig. S4) and the dilute acid hydrolysates (Figs. S9 and S10), many strains did not achieve enough cell growth to calculate exponential growth rates, suggesting that these hydrolysates contained higher inhibitor concentrations, different combinations of inhibitors, or both, compared to the other hydrolysates. Direct growth-rate comparisons between different hydrolysate pretreatments were not made since the hydrolysates were not standardized for glucan-loading or feedstock source (e.g., AHP SGH vs. IL SGH).
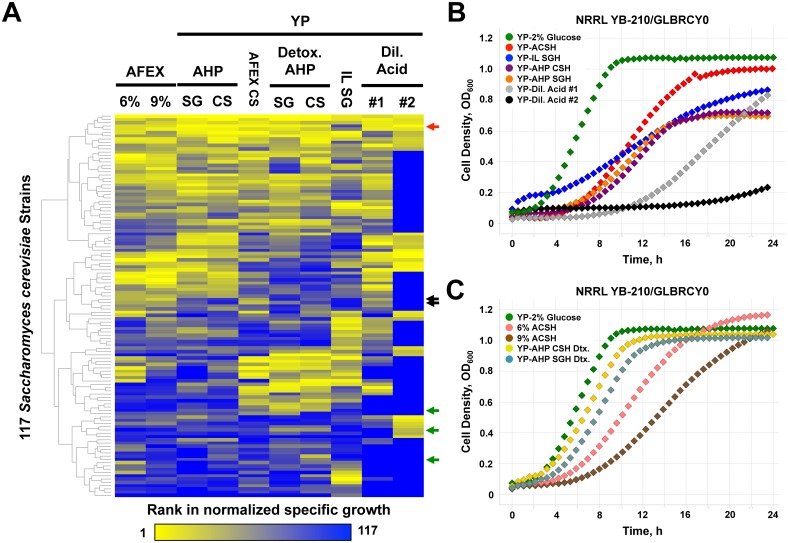
To glean insights from the large datasets we amassed, we used hierarchical clustering to organize strains based on their rank in growth rate for each hydrolysate relative their growth rate rank in YPD medium (the reference media), and according to similarities in phenotypes across strains and conditions ( Fig. 1A ). Clustering the hydrolysates based on the distribution of growth phenotypes showed that AHP and ACS hydrolysates grouped together, regardless of plant feedstock, which is consistent with the fact that these LCHs are generated by alkaline-based pretreatments. AHP and AFEX pretreatments have been shown to produce lower levels of inhibitory furans commonly generated from acid dehydration of hexose sugars, particularly furfural, and this absence may be a significant driver in the clustering of alkaline hydrolysates from acid hydrolysates [23]–[25], [29]. In addition, growth in IL and dilute acid-pretreated LCHs differed significantly from growth in other hydrolysates, with most strains unable to grow in dilute acid pretreated LCH #2. One study suggested that the predominant inhibitor in IL-derived LCHs is residual IL itself [26], which likely drives the unique phenotypic profile of yeast strain growth. It is unclear, due to restrictions on proprietary information, how the acid pretreated LCH #1 and #2 differ. These results also suggest that the driving difference between strain profiles is the pretreatment method, with lesser impact of the type of biomass used. Upon further inspection of individual strain performance, we found the BY4741 lab strain and CEN.PK2 ( Fig. 1A , green arrows) clustered with a group of strains that generally grew slowly in most hydrolysates; this group also included many sake-producing strains, several bread-making strains, and strains isolated from natural fermentations. One cluster contains 11 strains with high growth-rate ranks across most hydrolysate conditions. These include isolates from soil (DBVPG1373), fermentation (Y9, Y12), lab (FL100), banana (YB-210), oak (YPS1009), clinical (YJM440, YJM653, YJM978, YJM981) and unknown (NCYC361) sources. Given our interest in understanding the genetic determinants of xylose fermentation from AFEX-pretreated LCHs, we focused on YB-210 ([33], also referred to as GLBRCY0 [40]), that displayed broad tolerance across the hydrolysates tested but ranked highest in growth for alkaline pretreatments. YB-210 grew robustly in AFEX, AHP and dilute acid pretreated LCHs, and less robustly in IL pretreated LCH ( Figs. 1B and C ) under aerobic conditions. Furthermore, YB-210 displays higher tolerance to elevated temperature [34] and inhibitors found in AHP-pretreated LCHs [25]. Therefore, the YB-210 strain background was selected for metabolic engineering and evolution of anaerobic xylose fermentation. As an added benefit of this work, other researchers can utilize this phenotypic dataset to select from publically available strains tailored for tolerance to their pretreated biomass of interest (e.g., YPS163 or YPS1000 for dilute acid pretreated wheat straw).
Figure 1. Phenotypic screening of wild and domesticated S. cerevisiae strains identifies NRRL YB-210 with tolerance to hydrolysates made from a variety of pretreated lignocellulose.
In (A), 117 S. cerevisiae strains (including some in duplicate) were cultured in 96-well plates and monitored for changes cell density and growth rates calculated as described in Materials and Methods. All strains in each condition were then ranked from 1 (highest growth rate in yellow) to 117 (lowest growth rate, or no growth, in blue) and hierarchically clustered. Arrows indicate clustered rows for BY4741 (green), CEN.PK2 (black) in duplicate microtiter wells, and NRRL YB-210/GLBRCY0 (red). Representative growth data for the YB-210/GLBRCY0 strain in the indicated media from Fig. 2A are plotted (B–C). CS, corn stover; SG, switchgrass; YP; Yeast Extract and Peptone supplementation, 6%; 6% glucan loading ACSH, 9%; 9% glucan loading ACSH, Dtx.; Detoxified.
Two-stage directed evolution using YB-210 harboring xylose isomerase, transaldolase and xylulokinase permitted anaerobic xylose fermentation
After phenotypic observations revealed its stress tolerant properties, the YB-210 was engineered for xylose metabolism by insertion of an expression cassette containing the XYL1 (xylose reductase, XR), XYL2 (xylitol dehydrogenase, XDH) and XYL3 (xylulokinase) genes from Sch. stipitis [40], and then aerobically adapted on xylose. One evolved isolate (GLBRCY73, or Y73) that displayed improved xylose consumption rates in both lab media (Fig. S12A) and AHP SG hydrolysate (ASGH) was selected for further study [25]. We first examined the ability of Y73 to ferment xylose under controlled anaerobic conditions in N2-sparged bioreactors containing YPDX in lab medium (Fig. S12B) or ACSH (Fig. S12C). Although the Y73 strain aerobically consumed ∼40% of the xylose in 48 h, it anaerobically fermented <20% and <5% of the xylose in YPDX and ACSH, respectively, in the same time period. These results indicated that the XR-XDH engineered strain is severely impaired for anaerobic fermentation of xylose, particularly in ACSH, relative to aerobic culturing and suggested that this engineered strain would not be useful for our goals of better understanding anaerobic xylose fermentation. Attempts to adapt Y73 for anaerobic growth on xylose yielded no improved clones.
Similar to our observations with the GLBRCY73 strain, reduced anaerobic xylose consumption rates from other S. cerevisiae strains expressing Sch. stipitis XR-XDH enzymes have been reported. This limitation is likely due to redox cofactor imbalance. Heterologous engineering of Sch. stipitis XYL1, which primarily utilizes NADPH as its reducing cofactor, and XYL2, which uses NAD+ as its oxidizing cofactor, introduces non-regenerative cycles in S. cerevisiae that are rapidly imbalanced in the absence of oxygen [4]. A possible alternative to circumvent this problem is to utilize xylose isomerase (XI) [54]–[57], which catalyzes the conversion of xylose into xylulose without cofactors, in place of XYL1 and XYL2. We therefore re-engineered the diploid YB-210 strain with an expression cassette containing the ScTDH3 promoter upstream of the Clostridium phytofermentans xylose isomerase (CpxylA), which has been shown to confer anaerobic xylose fermentation onto S. cerevisiae after additional genetic modifications [54]. Our cassette also included ScTAL1, a pentose phosphate pathway transaldolase enzyme that can improve xylose metabolism when overexpressed [58], [59], and SsXYL3 driven by the ScPGK1 and ScTEF2 promoters, respectively. Finally, to simplify future genomic resequencing of evolved descendants and to rapidly uncover beneficial recessive traits during directed evolution, we isolated one haploid spore, named GLBRCY22-3 (Y22-3), which maintained the TAL1-xylA-XYL3 gene expression cassette.
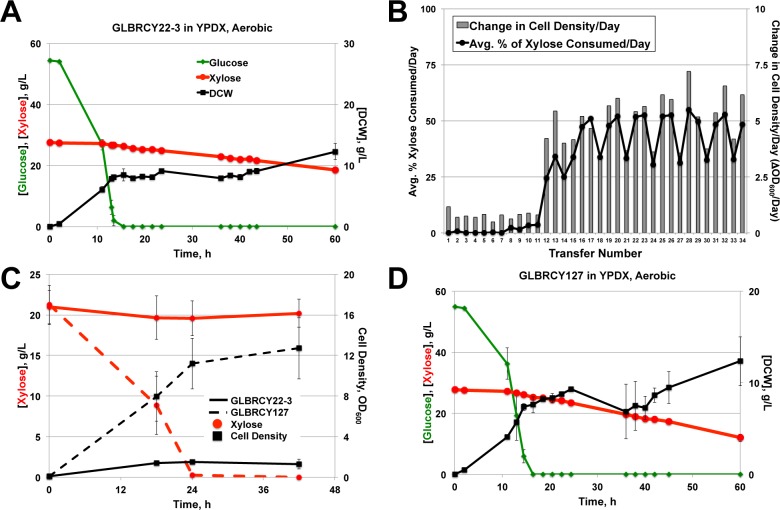
To assess whether the engineered Y22-3 strain could metabolize xylose, Y22-3 was cultured aerobically in bioreactors with YPDX medium. Concentrations of extracellular glucose, xylose and dry cell weights were measured over the course of the fermentation ( Fig. 2A ). The Y22-3 strain consumed less than half of the xylose within 64 h, which was significantly less than the Y73 strain. Thus, the Y22-3 strain was subjected to aerobic batch selection in YP medium containing 0.1% glucose and 2% xylose and without exogenous mutagens. Anaerobic batch selection of Y22-3 in YP medium containing 0.1% glucose and 2% xylose was also performed without observing appreciable cell growth and was therefore abandoned, while adaptation in ACSH was not performed because high glucose concentrations (60 g/L in 6% glucan loading ACSH) and diauxic xylose consumption would prevent selection for improved growth on xylose present at 30 g/L. For the first seven transfer cycles in YP-0.1% glucose and 2% xylose, each of which took place over 3–4 day periods, the culture grew at rates of ∼1 generation per day with limited xylose consumption from the medium ( Fig. 2B ), suggesting that most of the growth was on glucose. Over the 8th to 11th transfers, slightly greater xylose consumption was observed, but without substantially faster cell growth rates. By the 12th transfer and beyond, the culture adapted to xylose, consuming all of the sugars within the 2–4 day passaging cycle and reaching saturated growth. After the 34th transfer (∼115 generations), the culture was plated and single clones were screened for growth on xylose-containing medium. One clone, GLBRCY127 (Y127), displayed rapid aerobic growth in YPX by 96-well plate assay (data not shown), and was evaluated for aerobic xylose consumption in culture tubes containing YPX medium ( Fig. 2C ) or bioreactors containing YPDX medium ( Fig. 2D ). Compared to the Y22-3 strain, the evolved Y127 isolate displayed 15 to 17-fold faster absolute and specific xylose consumption rates than the Y22-3 parent in aerobic YPDX medium ( Figs. 2A, D and Table 2 ). These results indicate that the Y127 isolate evolved from Y22-3 with properties allowing faster aerobic xylose consumption.
Figure 2. The GLBRCY127 strain developed by directed engineering with xylose isomerase coupled with batch evolution can rapidly consume xylose aerobically.
Average sugar consumption and cell growth of unevolved GLBRCY22-3 strain engineered with ScTAL1, CpxylA and SsXYL3 cultured in bioreactors containing YPDX media and sparged with air from biological duplicates is shown (A). Indicated components were quantified from media samples at times from initial inoculation. In (B), the average percentage of xylose consumed and change in cell density per day are plotted for each transfer during the adaption of the Y22-3 strain in YP media containing 0.1% glucose and 2% xylose. The pattern of lower % of xylose consumed and change in cell density per day during every third transfer is due to reaching saturated growth prior to transfer. Average extracellular xylose concentrations and cell density measurements from parental Y22-3 and evolved Y127 strains grown aerobically in culture tubes with YPX media from three independent biological replicates are plotted in (C). In (D), evolved isolate Y127 was cultured in the same conditions as in (A), and samples measurements taken in an identical manner.
Table 2. Fermentation kinetic profiles for engineered and evolved S. cerevisiae strains.
| Media | Aerobic YPDX | Anaerobic YPDX | Anaerobic ACSH | ||||||
| Strain | Y73 | Y22-3 | Y127 | Y73 | Y127 | Y128 | Y73 | Y127 | Y128 |
| Absolute xyloseconsumption rate1 | 0.47±0.02 | 0.17±0.003 | 0.31±0.01 | 0.30±0.02 | 0.094±0.031 | 1.68±0.06 | 0.28±0.01 | 0.04±0.03 | 0.52±0.01 |
| Specific xyloseconsumption rate2 | 0.039±0.001 | 0.019±0.000 | 0.036±0.01 | 0.066±0.010 | 0.016±0.007 | 0.27±0.06 | 0.059±0.01 | 0.013±0.01 | 0.18±0.02 |
| % of theoretical ethanolyield for consumedsugars3 | ND | ND | ND | 80.1±1.4 | 86.0±2.5 | 87.5±1.1 | 72.1±10.4 | 78.9±14.3 | 77.2±6.4 |
| % of theoretical ethanolyield for consumedxylose4 | ND | ND | ND | 9.8±5.2 | ND* | 86.2±15.6 | 20.9±18.9 | ND* | 71.5±7.0 |
| Yx/glc 5 | ND | ND | ND | 0.08±0.00 | 0.09±0.00 | 0.11±0.02 | 0.066±0.012 | 0.045±0.004 | 0.05±0.01 |
| Yglycerol/glc 6 | ND | ND | ND | 0.10±0.00 | 0.07±0.00 | 0.08±0.01 | 0.051±0.002 | 0.038±0.006 | 0.04±0.00 |
ND, Not Determined for aerobic conditions; ND*, Not Determined – no ethanol produced.
In g xylose consumed/L/h.
In g xylose consumed/g DCW/h.
Calculated from the maximum ethanol concentration produced divided by the consumed xylose concentration at that time.
Calculated from the ethanol concentration produced between two time points after glucose depletion.
Yield of g DCW/g glucose consumed calculated at or near the time of glucose depletion and prior to xylose consumption. No cell growth was observed during xylose consumption.
Yield of g glycerol/g glucose consumed calculated at or near the time of glucose depletion and prior to xylose consumption.
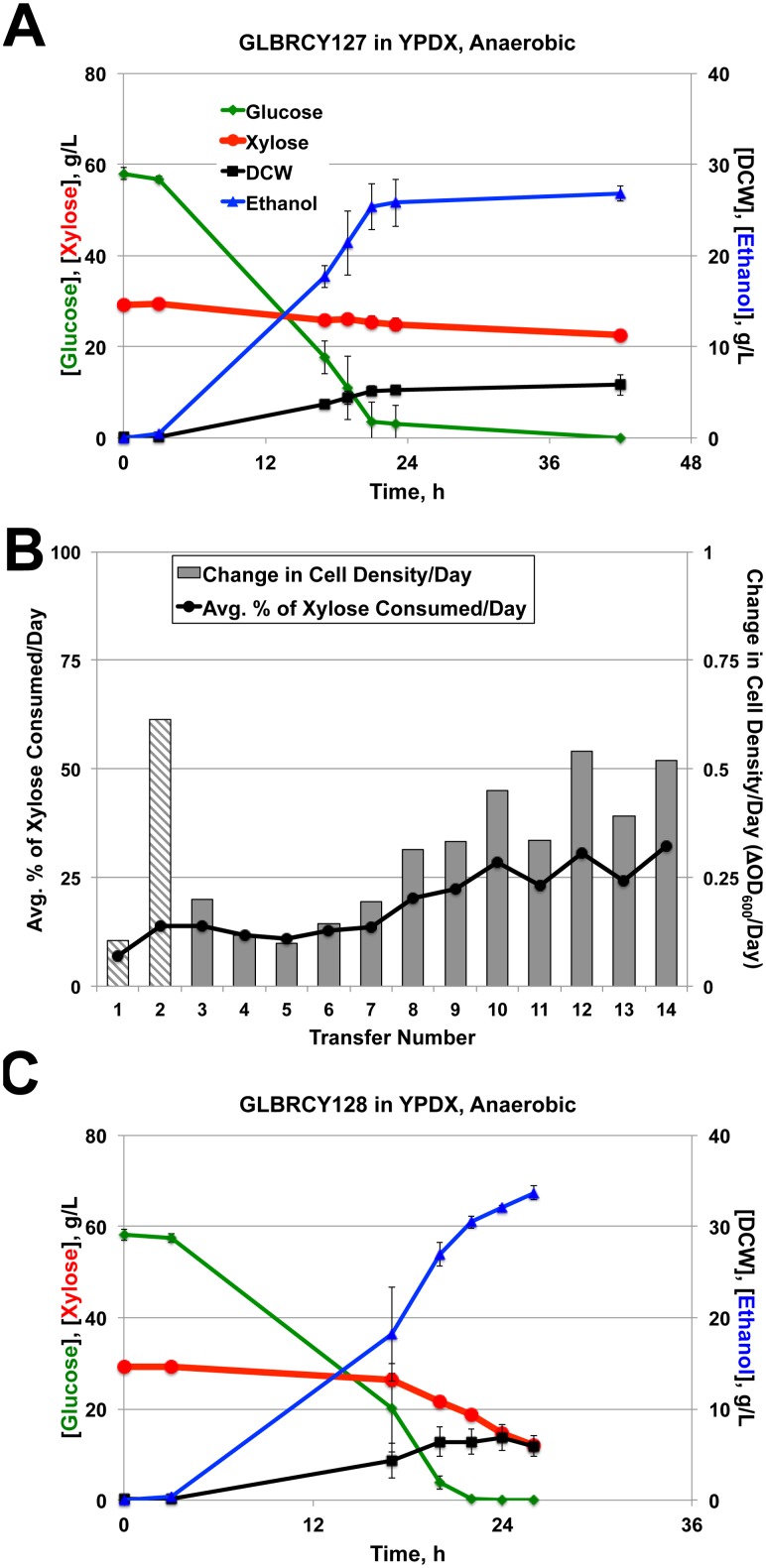
We next assessed the ability of the Y127 strain to ferment xylose anaerobically in bioreactors sparged with N2. Similar to the XR-XDH engineered Y73 strain, the aerobically evolved Y127 strain displayed limited xylose fermentation from YPDX medium, consuming less than 30% of the total xylose within 42 h, and did not appear to convert the small amount of consumed xylose into ethanol ( Fig. 3A ). This suggested that, like Y73, the Y127 strain was not capable of effectively fermenting xylose in the absence of oxygen. In an attempt to overcome this barrier, we performed a second round of batch selection of the Y127 strain cultured in YP medium containing 0.1% glucose and 2% xylose under completely anaerobic conditions ( Fig. 3B ). During the first two transfers, 40 µg/L ergosterol and 4 g/L Tween-80 were added to support anaerobic growth, but then omitted for all successive transfers. For the first six transfers, the cell population doubled approximately twice per week. After the 6th transfer, the culture began to grow faster and consumed a greater percentage of the total xylose per day. After reaching 33 generations at the 10th transfer, the culture appeared to plateau in anaerobic growth and xylose consumption rate. After the 14th transfer (∼47 generations), the culture was plated and colonies were screened for fastest growth rate in YPX medium by 96-well plate assay. One clone (GLBRCY128, Y128) displaying rapid anaerobic growth on xylose (data not shown) was then evaluated in bioreactors containing YPDX medium sparged with N2. In contrast to Y127 ( Fig. 3A ), the Y128 strain rapidly fermented xylose in the absence of oxygen, during which time the extracellular ethanol concentration increased ( Fig. 3C ). Consistent with this result, Y128 exhibited higher absolute and specific xylose consumption rates in anaerobic YPDX medium than the Y127 strain ( Table 2 ). These results indicate that the two-stage directed evolution yielded Y127 and Y128 strains with enhanced aerobic and anaerobic xylose metabolism, respectively.
Figure 3. Second stage anaerobic adaptation on xylose enabled rapid xylose fermentation by evolved GLBRCY128 isolate.
Average fermentation kinetic profiles of the GLBRCY127 strain cultured in bioreactors containing YPDX media and sparged with nitrogen from biological duplicates are shown (A). Average concentrations with standard deviations of indicated compounds were quantified from media samples at times from initial inoculation. In (B), the percentage of xylose consumed and change in cell density per day is plotted for each transfer during the anaerobic adaptation of Y127 in YP media containing 0.1% glucose and 2% xylose. In the first two transfers (hatched bars), Tween-80 and ergosterol were added to the media. In (C), evolved isolate Y128 was cultured in biological duplicate under the same conditions as in (A), and samples measurements taken in an identical manner.
The evolved Y128 strain can anaerobically convert xylose into ethanol faster than the parental strains
While the YPDX media matched the predominant sugar concentrations found in 6% glucan loading ACSH, the inclusion of 60 g/L glucose clouded our ability to directly compare the abilities of our engineered and evolved strains in the anaerobic conversion of xylose into ethanol. Therefore, we performed anaerobic fermentations with Y22-3, Y127 and Y128 strains in bioreactors containing YP media with 20 g/L xylose, and quantified xylose consumption and ethanol production from the media over time. For Y128, there was production of ethanol and cell biomass with simultaneous depletion of xylose from the media ( Fig. 4 ). In contrast, both Y22-3 and Y127 strains produced less than 1 g/L ethanol ( Table 3 ) and consumed less than 1 g/L xylose ( Fig. 4 ) by the end of fermentation. Importantly, these differences in xylose consumption and ethanol production showed that Y128 consumed xylose significantly faster and produced ethanol at a higher yield and titer than Y22-3 and Y127 ( Table 3 ). These results further support that the evolved Y128 strain displays marked improvement in anaerobic xylose fermentation compared to its parental strains.
Figure 4. GLBRCY128 can anaerobically ferment xylose from YPX medium into ethanol faster than its parental strains.
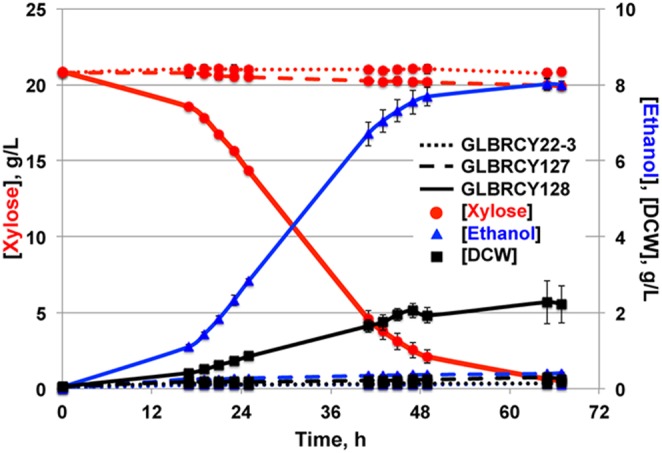
Fermentation kinetic profiles comparing Y22-3, Y127 and Y128 strains cultured anaerobically in bioreactors containing YPX medium and sparged with nitrogen are shown. Average concentrations with standard deviations of xylose, ethanol and DCW concentrations in the media were calculated from independent biological triplicates.
Table 3. Comparison of anaerobic fermentation kinetics for Y22-3, Y127 and Y128 in YPX medium.
| Strain | Y22-3 | Y127 | Y128 |
| Absolute xylose consumption rate1 | ND | 0.101±0.004 | 0.596±0.028 |
| Specific xylose consumption rate2 | ND | 0.515±0.009 | 0.587±0.038 |
| % of theoretical ethanol yield for consumed xylose3 | ND* | ND* | 77.7±0.6 |
| % of theoretical ethanol yield from total xylose4 | 1.2±0.8 | 3.9±0.2 | 74.7±0.5 |
| Avg. final ethanol titer5 | 0.11±0.03 | 0.42±0.02 | 8.0±0.1 |
ND, Not Determined – low xylose consumption; ND*, Not Determined – no ethanol produced and low xylose consumption.
In g xylose consumed/L/h.
In g xylose consumed/g DCW/h.
Calculated from the maximum ethanol concentration produced divided by the consumed xylose concentration at that time.
Calculated from the maximum ethanol concentration produced divided by the starting xylose concentration.
In g ethanol/L.
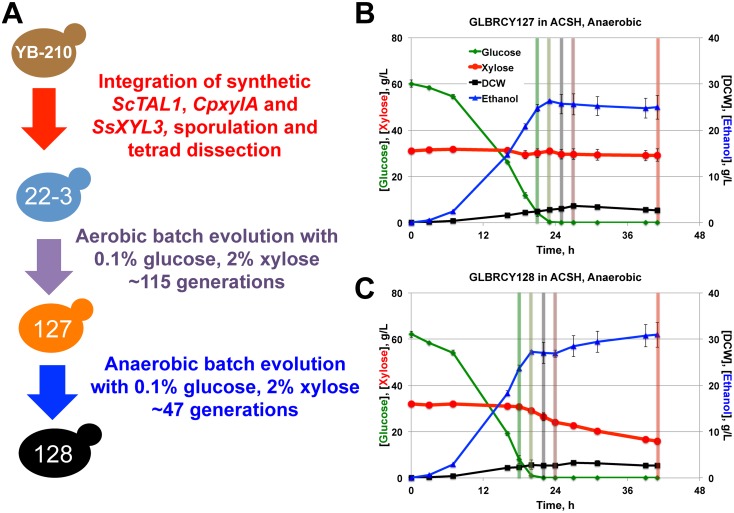
The evolved Y128 strain can anaerobically convert xylose from ACSH into ethanol
Within a relatively small number of generations (∼162 in total) without exogenous mutagens, our two-stage evolution yielded a xylA-engineered S. cerevisiae strain with the ability to consume xylose anaerobically in lab medium (summarized in Fig. 5A ). Although the Y128 genetic background originated from a hydrolysate-tolerant strain, Y128 could have lost stress-tolerance traits during the course of xylose evolution. If so, Y128 might not have been able to convert xylose in lignocellulosic hydrolysates into ethanol anaerobically, which was a chief goal of this research. Previous studies have shown that yeast strains able to ferment xylose rapidly in lab medium are severely impaired in LCHs [34]. Therefore, we assessed the abilities of the Y127 and Y128 strains to ferment sugars anaerobically from ACSH in bioreactors sparged with N2. Y22-3 was not used in this study, as it clearly does not significantly metabolize xylose aerobically ( Fig. 2 ) or anaerobically ( Fig. 4 ). Similar to our observations with YPDX lab medium, both Y127 and Y128 strains fermented glucose rapidly ( Figs. 5B and C ). However, the Y128 strain, but not Y127, consumed most of the xylose (∼50% within 44 hrs) once glucose was depleted from ACSH. Indeed the absolute and specific xylose consumption rates for Y128 were approximately 10-fold higher than Y127 ( Table 2 ). This also resulted in a higher yield in ethanol from xylose for Y128 compared to Y127 and Y73 ( Table 2 ). Importantly, the ethanol yield from glucose for Y128 was similar to Y127, suggesting that the anaerobic xylose evolution had little impact on the ability of Y128 to convert glucose into ethanol ( Table 2 ). Because Y128 ferments more xylose than Y127 anaerobically, this resulted in a higher ethanol titer for Y128 ( Fig. 5B and C ). Thus, despite the fact that evolution for xylose conversion occurred in lab medium lacking the inhibitors found in LCHs, the Y128 strain could effectively ferment xylose from an industrially relevant pretreated LCH in the absence of oxygen. This ability may be due in part to the innate hydrolysate-tolerant properties of the YB-210 genetic background.
Figure 5. GLBRCY128 can anaerobically ferment xylose from ACSH.
A diagram summarizing the engineering and evolution of the YB-210 strain into the evolved Y128 strain is provided in (A). Fermentation kinetic profiles of the Y127 (B) and Y128 (C) strains cultured in bioreactors containing ACSH and sparged with nitrogen from biological duplicates are shown. Average concentrations and standard deviations of indicated components were quantified from media samples at times from initial inoculation. Vertical colored bars indicate time points at which samples were taken for metabolomic analysis described in Fig. 7A–D.
Rapid xylose consumption by GLBRCY127 and Y128 is dependent upon xylA and TAL1 expression
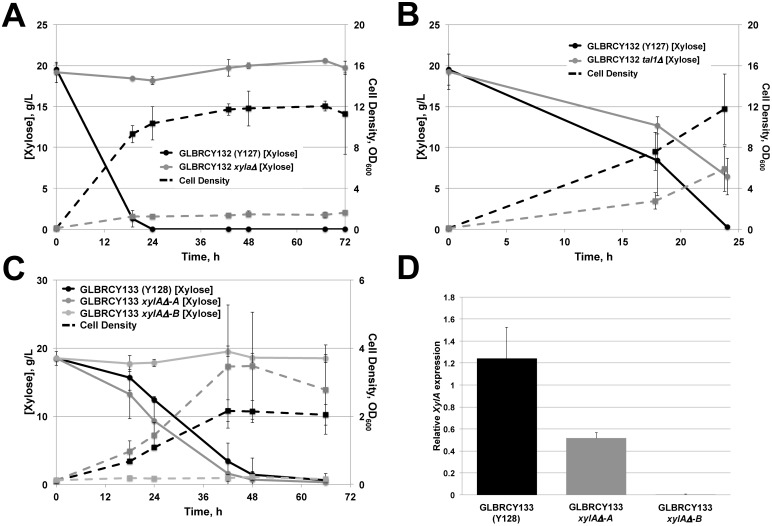
After clearly establishing the faster xylose consumption phenotypes of the Y127 and Y128 strains relative to the parental Y22-3 strain, we next wanted to better understand the potential genetic mechanisms by which these strains could have evolved. One obvious possibility would be through mutations in the engineered genes xylA, TAL1 and XKS1 that increase their expression or activities. However, when we sequenced the engineered gene cassette, no DNA sequence differences were identified. An alternative possibility is that the Y127 or Y128 strains obtained gain-of-function mutations in native genes that code for xylose metabolism enzymes, which are normally expressed at low levels or lack sufficient activities for rapid flux into the pentose phosphate pathway [4]. The S. cerevisiae genome contains a number of putative enzymes with xylose reductase activities, including Gre3p [60], [61], and xylitol dehydrogenases, including an ineffective XYL2 homolog [62] and XDH1, which is present only in some wild S. cerevisiae strains and confers detectable xylose consumption [63]. Thus, one possible model for the evolution of Y127 and Y128 is that genetic changes in one or more of these genes allowed for improved xylose consumption independent of engineered xylA and TAL1. We examined this possibility by first excising the loxP-KanMX-loxP selection marker from the engineered cassette from Y22-3, Y127 and Y128 to generate antibiotic-sensitive GLBRCY36, GLBRCY132 and GLBRCY133, respectively. We then deleted xylA from the engineered cassette of the Y127 marker rescued strain (Y132) and assessed its ability to consume xylose aerobically. Indeed, the Y132 xylA Δ strain was ablated of its ability to consume or produce cell biomass from xylose ( Fig. 6A ). In contrast, deletion of TAL1 from the engineered expression cassette, but not endogenous TAL1, in Y132 reduced the rate of xylose metabolism but did not impact the final amount of xylose consumed or the cell biomass produced from xylose ( Fig. 6B ), suggesting that the additional copy of engineered TAL1 was important for determining the rate of xylose consumption but was not essential. In addition, we confirmed two independent marker rescued Y128 (Y133) xylAΔ transformants that displayed separate xylose consumption phenotypes. Consistent with the Y132 xylAΔ strain, the Y133 xylAΔ-B strain could not consume xylose aerobically (data not shown) or anaerobically ( Fig. 6C ). Interestingly, the Y133 xylAΔ-A strain fermented xylose at a comparable rate to the Y133 strain. This suggested the possibility that the xylA gene was duplicated in cis, which could explain why the replacement of xylA with the KanMX deletion cassette could be verified by PCR. Indeed, when we compared xylA RNA expression in the two Y133 xylAΔ strains to the Y133 strain by qPCR, we found that Y133 xylAΔ-A expressed half as much xylA as Y133, whereas no xylA RNA was detected in the Y133 xylAΔ-B strain ( Fig. 6D ). Although this result does not rule out possible genetic changes in endogenous xylose metabolizing enzymes, our findings together suggest that the evolved xylose consumption phenotypes in Y127 and Y128 are dependent, at least in part, upon the engineered xylA and TAL1 genes.
Figure 6. The xylose consumption phenotypes of the evolved Y127 and Y128 strains are dependent upon CpxylA and ScTAL1.
Extracellular xylose concentrations (solid lines) and cell density (dashed lines) were measured by YSI instrument and OD600 readings, respectively, from cultures containing KanMX marker rescued versions of (A) GLBRCY127 (Y132) and GLBRCY132 xylAΔ or (B) Y132 and Y132 tal1Δ strains inoculated in aerobic YPX media. In (C), extracellular xylose concentrations (solid lines) and cell density (dashed lines) were measured as in (A, B) for KanMX marker rescued GLBRCY128 (Y133) and two independent GLBRCY133 xylAΔ strains inoculated in anaerobic YPX media. These selection marker-rescued Y128 strains were cultured in YPD media and total RNA isolated from a single time point. Expression of CpxylA was then quantified and normalized to ScERV25 RNA levels by qPCR. The bar graph in (D) displays the average values and standard deviations for CpxylA RNA from three independent biological replicates.
Y128 accumulates higher intracellular concentrations of xylose metabolic intermediates but little xylitol
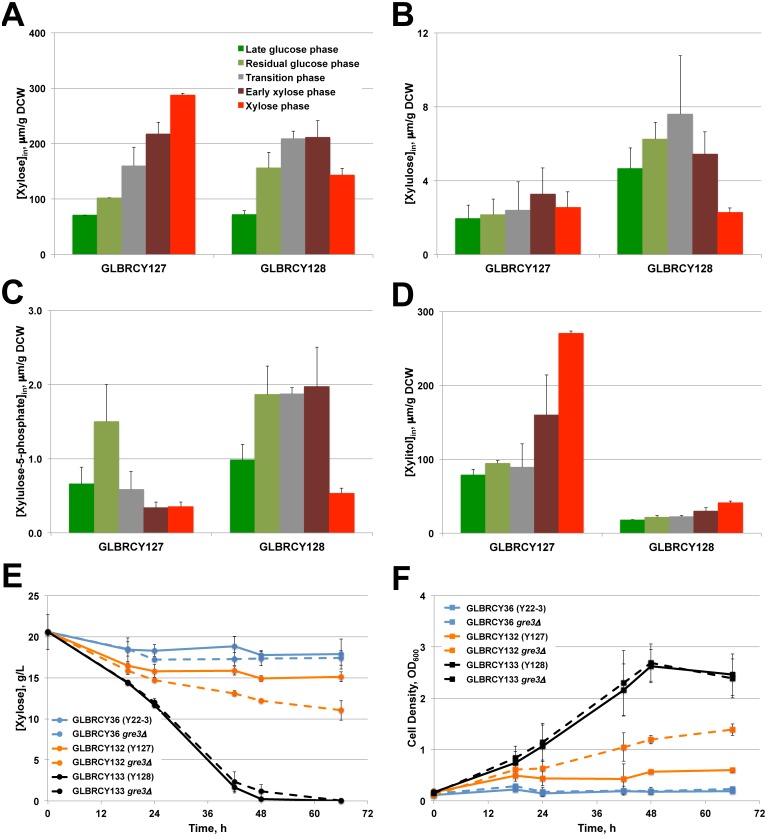
The data presented thus far suggest Y128 has an evolved ability to ferment xylose anaerobically in lab medium ( Figs. 3C and D ) and ACSH ( Fig. 5C ). To investigate how the evolved strains anaerobically ferment xylose, we analyzed the intracellular concentrations of xylose catabolism intermediates: xylose, xylitol, xylulose and xylulose-5-phosphate (X5P). During the anaerobic fermentation of ACSH by the Y127 and Y128 strains ( Figs. 5B and C ), cells were captured from the bioreactors using rapid vacuum filtration at five different time points spanning the glucose and xylose consumption phases; two samples were taken when glucose was detected in the hydrolysate (indicated by vertical lines in shades of green); one sample was taken during the transition to xylose after glucose was undetectable (grey vertical line), and two final samples were taken when xylose consumption (vertical lines in shades of red) was evident. Cell samples for Y127 and Y128 fermentations were taken at comparable sugar concentrations, whereas the last two samples were taken at the equivalent amount of time after the transition time point. During the course of the ACSH fermentation, the Y127 cells accumulated xylose ( Fig. 7A ), whereas xylulose, which is the product of isomerization of xylose by CpxylA, did not change ( Fig. 7B ). In contrast, Y128 peaked in intracellular xylose and xylulose levels during the transition phase, after which intracellular xylose decreased slightly at the end of the fermentation coincident with extracellular xylose depletion ( Figs. 7A and B ). The final metabolite of the engineered xylose metabolism pathway, X5P, peaked in intracellular concentration between the residual glucose and early xylose metabolism phases in Y128, whereas for Y127, X5P briefly peaked in the residual glucose phase, then decreased to low levels for the remainder of the fermentations ( Fig. 7C ). These patterns of intracellular xylose, xylulose and X5P accumulation, along with the simultaneous depletion of extracellular xylose and accumulation of ethanol ( Fig. 5C ), suggested the possibility that the xylose consumed by Y128 from ACSH was metabolized through the engineered xylA pathway at higher flux than in Y127.
Figure 7. Y128 has a mutation in GRE3 that reduces xylitol production and contributes towards anaerobic xylose fermentation.
Fermentation samples were taken at the indicated time points marked by vertical colored bars in Fig. 5B and C. Cells were then filter-captured, briefly washed and then intracellular metabolites extracted. Average concentrations of xylose (A), xylulose (B), xylulose-5-phosphate (C) and xylitol (D) from independent duplicate fermentations were determined by reverse phase ion pairing HPLC-ESI coupled with MS/MS or gas chromatography (see Materials and Methods). Average concentrations and standard deviations are based on two biological replicates. Y22-3, Y127 and Y128 strains with KanMX selection markers excised (Y36, Y132 and Y133, respectively) and with or without deletion of GRE3 were cultured under anaerobic conditions in YPX media. Samples were taken at the indicated time points to measure xylose concentrations (E) or cell density (F). Average values and standard deviations were calculated from biological triplicates.
The higher concentrations of catabolized pentose intermediates along with the faster xylose consumption rates ( Tables 2 and 3 ) suggest two possible biochemical mechanisms in the evolution of Y128. One possibility is that the apparent increase in xylose consumption from the hydrolysate is due solely to improved xylose transport, without further metabolic conversion of xylose to other products. However, the higher ethanol titer achieved by Y128 coincident with differences in intracellular concentrations of metabolized xylose intermediates supports alternate models in which the evolved Y128 strain has more active xylose catabolism or pentose phosphate enzymes, or both, relative to its Y127 parent, possibly allowing for greater flux of xylose to ethanol.
The accumulation of internalized xylose and unchanging levels of xylulose in Y127 during the ACSH fermentation, along with the paltry change in extracellular xylose levels, suggest a metabolic bottleneck in xylose isomerase activity. Xylitol is a known inhibitor of xylose isomerase [64], and it is know that deletion of GRE3, which encodes a reductase enzyme that can convert xylose into xylitol, in S. cerevisiae engineered with xylose isomerase improved xylose fermentation [17], [55], [57], [61], [65]. Therefore, we quantified and compared the intracellular levels of xylitol in Y127 and Y128 during ACSH fermentation. Strikingly, we observed a severe reduction in intracellular xylitol levels for Y128 at all time points compared to Y127, which accumulated xylitol over time ( Fig. 7D ). This suggested that one difference between the Y127 and Y128 could be an evolved mutation that alters GRE3 activity or expression. Thus, we sequenced the GRE3 open reading frames of the Y22-3, Y127 and Y128 strains. We found a single nucleotide polymorphism (SNP) in the Y128 strain producing a G-to-A mutation relative to both Y22-3 and Y127, which changed the alanine at amino acid residue 46 to threonine (A46T) in Gre3p. This residue is conserved in other S. cerevisiae strains, as well as in many other yeast species including Saccharomyces arboricola, S. kudriavzevii, Candida and Kluyveromyces, and resides within the aldo-ketoreductase catalytic domain, suggesting that the A46T substitution likely impairs Gre3p xylose reductase activity. Although others have rationally deleted GRE3 from strains to improve XI-mediated xylose metabolism prior to evolution, our observation of a spontaneous GRE3 mutation acquired from directed evolution of a xylA-engineered yeast strain confirms its importance in the xylose metabolism bottleneck.
Taken together, our results suggest that the gre3A46T mutation in Y128 may cause a partial or complete loss of Gre3p function, which in turn reduces xylitol production and thus minimizes inhibition of CpxylA. To further confirm the possibility that loss of Gre3p activity improves anaerobic xylose fermentation, we deleted GRE3 in the marker rescued Y22-3 (renamed Y36), Y127 (renamed Y132) and Y128 (renamed Y133) strains and compared their in vitro xylose reductase activities and abilities to ferment xylose in the absence of oxygen. First, xylose reductase activities from extracts generated from selection-marker rescued Y127 (Y132), Y132 gre3 Δ, marker rescued Y128 (Y133) and Y133 gre3 Δ cells were determined (Fig. S13). Extracts from the Y133 strain, which harbors the gre3A46T mutation, and gre3 Δ strains displayed similar decreased xylose reductase activity compared toY132, which contains wild-type GRE3. This result further supported our in vivo observations that strains containing the gre3A46T mutation behave biochemically similar to strains lacking GRE3. Finally, we found that the Y132 gre3Δ strain could anaerobically consume xylose ( Fig. 7E ) and grow faster than the parental Y132 strain ( Fig. 7F ), but not nearly as fast as Y133. This suggests that the Y128 strain contains mutations in addition to gre3A46 T that aid anaerobic xylose fermentation. Most importantly, there were no differences in the xylose consumption or growth rates between the Y133 and Y133 gre3Δ strains, which is consistent with the gre3A46 T mutation resulting in a loss of function. Finally, deletion of GRE3 in the Y36 parental strain had no effect, further indicating that loss of GRE3 function alone cannot confer anaerobic xylose fermentation. Together, these in vitro and in vivo results suggest that the Y128 strain evolved a loss-of-function mutation in GRE3, which along with other unknown mutations, contributed to improved xylose utilization by reducing the production of inhibitory xylitol.
Conclusions
Here, we report the development of an engineered haploid S. cerevisiae strain with the evolved ability to ferment xylose anaerobically in lab media and LCH. Although yeast strains with improved anaerobic xylose fermentation in lab media and pretreated LCHs have been reported, most are derived from polyploid industrial strains with robust tolerance traits [12], [17], [25], [30]–[32]. Although polyploidy can compensate for detrimental recessive alleles, the multiple gene copies present in these industrial strains make mapping and identifying the causal mutations that accelerate xylose conversion difficult. In contrast, we generated the haploid Y128 strain with the ability to rapidly ferment xylose anaerobically, even in the presence of ACSH inhibitors. Thus, haploid Y22-3 and its evolved Y127 and Y128 descendants are ideally suited for comparative analyses to identify gene sequences that contribute to xylose conversion in the presence and absence of oxygen, and to determine metabolomic and transcriptomic differences that underlie their respective phenotypes. As a proof of concept, we used metabolomic data and targeted gene sequencing to identify a loss-of-function mutation in GRE3, which we validated by strain reconstruction ( Fig. 7 ). Thus, these strains are exciting tools that will provide future opportunities for multi-omic dissection of the molecular bottlenecks to anaerobic xylose fermentation by S. cerevisiae under LCH inhibitor stress.
Supporting Information
Domesticated strains of S. cerevisiae grow poorly in lignocellulosic hydrolysates. Representative aerobic growth profiles of lab strains BY4741 (A) and CEN.PK2 (B) cultured in 96-well plates with hydrolysates made from various pretreated lignocellulose hydrolysates (see Materials and Methods) and supplemented with yeast extract and peptone (YP) are shown by plotting cell density (optical density at 595 nm) every 20 min for 24 h. ACSH; 6% glucan loading AFEX pretreated corn stover hydrolysate, AHP; Alkaline Hydrogen Peroxide pretreatment, IL; Ionic Liquid ([C2mim][OAc]) pretreated, Dil. Acid; Dilute Acid pretreated lignocellulosic hydrolysate, SGH; switchgrass hydrolysate, CSH; corn stover hydrolysate.
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in 6% glucan loading ACSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least 3 biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in 9% glucan loading ACSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-AHP CSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-AHP SGH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-6% glucan loading ACSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-detoxified AHP CSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-detoxified AHP SGH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-80% dilute acid pretreated hydrolysate #1 relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-80% dilute acid pretreated hydrolysate #2 relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-IL SGH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Hydrolysate-tolerant YB-210/GLBRCY0 engineered with XR/XDH and evolved for aerobic xylose metabolism does not ferment xylose anaerobically. The YB-210/Y0 strain engineered with XYL1, 2 and 3 genes from S. stipitis and aerobically-evolved (GLBRCY73) was cultured in bioreactors and evaluated for consumption of xylose in aerobic YPDX (A), anaerobic YPDX (B) and anaerobic ACSH (C) media as described in Materials and Methods. Concentrations (g/L) of glucose (green), xylose (red), dry cell weight (black) and ethanol (blue) are averages and standard deviations from two independent biological replicates.
(TIF)
GLBRCY133 (Y128) cell extracts display reduced in vitro xylose reductase activity similar to GRE3 deletion strains. The indicated strains were cultured aerobically in YPD, harvested and prepared for in vitro xylose reductase activity assays as described in Materials and Methods. Xylose and NADPH were added to each extract, and then rates of change in absorbance at 340 nm were measured to determine the Units of enzymatic activity normalized to mg of total protein in the cellular extract. The graph displays the average percent of in vitro xylose reductase activities and standard deviations of indicated strains relative to GLBRCY132 (marker rescued GLBRCY127, which contains wild-type GRE3) in biological duplicate.
(TIF)
Wild and domesticated S. cerevisiae strains used in phenotypic growth studies.
(XLSX)
Acknowledgments
We thank Drs. Jeff Lewis, Justin Fay, and Clete Kurtzmann for S. cerevisiae strains, William Bothfeld, Mick McGee, Brendan Thomson and Robert Zinkel for technical support, Novozymes for providing enzymes, Dr. Brian Burger and James Hose for protocols and technical advice, our industrial collaborator for dilute acid hydrolysates, Drs. Tom Jeffries, Chris Hittinger and Jeff Piotrowski for scientific advice, and Dr. Donna Bates for comments on this manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded in part by the Department of Energy (DOE) Great Lakes Bioenergy Research Center (DOE BER Office of Science BER DE-FC02-07ER64494). The portion of work conducted by the Joint BioEnergy Institute was supported by the Office of Science, Office of Biological and Environmental Research, of the United States Department of Energy under Contract No. DE-AC02-05CH11231. The portion of the work conducted by the Advanced Biofuels Process Demonstration Unit was funded by support from Office of Biomass Program within the United States DOE’s Office of Energy Efficiency and Renewable Energy, and also the funding support from the American Recovery and Reinvestment Act. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sheehan JJ (2009) Biofuels and the conundrum of sustainability. Curr Opin Biotechnol 20: 318–324. [DOI] [PubMed] [Google Scholar]
- 2. Wyman CE (2007) What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol 25: 153–157. [DOI] [PubMed] [Google Scholar]
- 3. Pauly M, Keegstra K (2008) Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J 54: 559–568. [DOI] [PubMed] [Google Scholar]
- 4. Jeffries TW, Jin YS (2004) Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biotechnol 63: 495–509. [DOI] [PubMed] [Google Scholar]
- 5. Runquist D, Hahn-Hagerdal B, Radstrom P (2010) Comparison of heterologous xylose transporters in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim SR, Park YC, Jin YS, Seo JH (2013) Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol Adv 31: 851–861. [DOI] [PubMed] [Google Scholar]
- 7. van Maris AJ, Winkler AA, Kuyper M, de Laat WT, van Dijken JP, et al. (2007) Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv Biochem Eng Biotechnol 108: 179–204. [DOI] [PubMed] [Google Scholar]
- 8. Hahn-Hagerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF (2007) Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol 74: 937–953. [DOI] [PubMed] [Google Scholar]
- 9. Laluce C, Schenberg AC, Gallardo JC, Coradello LF, Pombeiro-Sponchiado SR (2012) Advances and developments in strategies to improve strains of Saccharomyces cerevisiae and processes to obtain the lignocellulosic ethanol–a review. Appl Biochem Biotechnol 166: 1908–1926. [DOI] [PubMed] [Google Scholar]
- 10. Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, et al. (2013) Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One 8: e57048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Madhavan A, Srivastava A, Kondo A, Bisaria VS (2012) Bioconversion of lignocellulose-derived sugars to ethanol by engineered Saccharomyces cerevisiae. Crit Rev Biotechnol 32: 22–48. [DOI] [PubMed] [Google Scholar]
- 12. Demeke MM, Dietz H, Li Y, Foulquie-Moreno MR, Mutturi S, et al. (2013) Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 6: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bera AK, Ho NW, Khan A, Sedlak M (2011) A genetic overhaul of Saccharomyces cerevisiae 424A(LNH-ST) to improve xylose fermentation. J Ind Microbiol Biotechnol 38: 617–626. [DOI] [PubMed] [Google Scholar]
- 14. Van Vleet JH, Jeffries TW, Olsson L (2008) Deleting the para-nitrophenyl phosphatase (pNPPase), PHO13, in recombinant Saccharomyces cerevisiae improves growth and ethanol production on D-xylose. Metab Eng 10: 360–369. [DOI] [PubMed] [Google Scholar]
- 15. Young EM, Tong A, Bui H, Spofford C, Alper HS (2014) Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc Natl Acad Sci U S A 111: 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barrick JE, Lenski RE (2013) Genome dynamics during experimental evolution. Nat Rev Genet 14: 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diao L, Liu Y, Qian F, Yang J, Jiang Y, et al. (2013) Construction of fast xylose-fermenting yeast based on industrial ethanol-producing diploid Saccharomyces cerevisiae by rational design and adaptive evolution. BMC Biotechnol 13: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scalcinati G, Otero JM, Van VleetJR, Jeffries TW, Olsson L, et al. (2012) Evolutionary engineering of Saccharomyces cerevisiae for efficient aerobic xylose consumption. FEMS Yeast Res 12: 582–597. [DOI] [PubMed] [Google Scholar]
- 19. Klimacek M, Kirl E, Krahulec S, Longus K, Novy V, et al. (2014) Stepwise metabolic adaption from pure metabolization to balanced anaerobic growth on xylose explored for recombinant Saccharomyces cerevisiae. Microb Cell Fact 13: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blanch HW, Simmons BA, Klein-Marcuschamer D (2011) Biomass deconstruction to sugars. Biotechnol J 6: 1086–1102. [DOI] [PubMed] [Google Scholar]
- 21. Piotrowski JS, Zhang Y, Bates DM, Keating DH, Sato TK, et al. (2014) Death by a thousand cuts: the challenges and diverse landscape of lignocellulosic hydrolysate inhibitors. Front Microbiol 5: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Almeida JR, Runquist D, Sanchez i Nogue V, Liden G, Gorwa-Grauslund MF (2011) Stress-related challenges in pentose fermentation to ethanol by the yeast Saccharomyces cerevisiae. Biotechnol J 6: 286–299. [DOI] [PubMed] [Google Scholar]
- 23. Liu ZL (2006) Genomic adaptation of ethanologenic yeast to biomass conversion inhibitors. Appl Microbiol Biotechnol 73: 27–36. [DOI] [PubMed] [Google Scholar]
- 24. Rasmussen H, Sorensen HR, Meyer AS (2014) Formation of degradation compounds from lignocellulosic biomass in the biorefinery: sugar reaction mechanisms. Carbohydr Res 385: 45–57. [DOI] [PubMed] [Google Scholar]
- 25. Sato TK, Liu T, Parreiras LS, Williams DL, Wohlbach DJ, et al. (2014) Harnessing Genetic Diversity in Saccharomyces cerevisiae for Fermentation of Xylose in Hydrolysates of Alkaline Hydrogen Peroxide-Pretreated Biomass. Appl Environ Microbiol 80: 540–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ouellet M, Datta S, Dibble DC, Tamrakar PR, Benke PI, et al. (2011) Impact of ionic liquid pretreated plant biomass on Saccharomyces cerevisiae growth and biofuel production. Green Chemistry 13: 2743–2749. [Google Scholar]
- 27. Balan V, Bals B, Chundawat SP, Marshall D, Dale BE (2009) Lignocellulosic biomass pretreatment using AFEX. Methods Mol Biol 581: 61–77. [DOI] [PubMed] [Google Scholar]
- 28. Lau MW, Gunawan C, Balan V, Dale BE (2010) Comparing the fermentation performance of Escherichia coli KO11, Saccharomyces cerevisiae 424A(LNH-ST) and Zymomonas mobilis AX101 for cellulosic ethanol production. Biotechnol Biofuels 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chundawat SP, Vismeh R, Sharma LN, Humpula JF, da Costa Sousa L, et al. (2010) Multifaceted characterization of cell wall decomposition products formed during ammonia fiber expansion (AFEX) and dilute acid based pretreatments. Bioresour Technol 101: 8429–8438. [DOI] [PubMed] [Google Scholar]
- 30. Koppram R, Albers E, Olsson L (2012) Evolutionary engineering strategies to enhance tolerance of xylose utilizing recombinant yeast to inhibitors derived from spruce biomass. Biotechnol Biofuels 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ismail KS, Sakamoto T, Hatanaka H, Hasunuma T, Kondo A (2013) Gene expression cross-profiling in genetically modified industrial Saccharomyces cerevisiae strains during high-temperature ethanol production from xylose. J Biotechnol 163: 50–60. [DOI] [PubMed] [Google Scholar]
- 32. Garcia Sanchez R, Karhumaa K, Fonseca C, Sanchez Nogue V, Almeida JR, et al. (2010) Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnol Biofuels 3: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mortimer RK, Johnston JR (1986) Genealogy of principal strains of the yeast genetic stock center. Genetics 113: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin M, Sarks C, Gunawan C, Bice BD, Simonett SP, et al. (2013) Phenotypic selection of a wild Saccharomyces cerevisiae strain for simultaneous saccharification and co-fermentation of AFEXTM pretreated corn stover. Biotechnol Biofuels 6: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwalbach MS, Keating DH, Tremaine M, Marner WD, Zhang Y, et al. (2012) Complex physiology and compound stress responses during fermentation of alkali-pretreated corn stover hydrolysate by an Escherichia coli ethanologen. Appl Environ Microbiol 78: 3442–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li M, Foster C, Kelkar S, Pu Y, Holmes D, et al. (2012) Structural characterization of alkaline hydrogen peroxide pretreated grasses exhibiting diverse lignin phenotypes. Biotechnol Biofuels 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Banerjee G, Car S, Liu T, Williams DL, Meza SL, et al. (2012) Scale-up and integration of alkaline hydrogen peroxide pretreatment, enzymatic hydrolysis, and ethanolic fermentation. Biotechnol Bioeng 109: 922–931. [DOI] [PubMed] [Google Scholar]
- 38. Li C, Tanjore D, He W, Wong J, Gardner JL, et al. (2013) Scale-up and evaluation of high solid ionic liquid pretreatment and enzymatic hydrolysis of switchgrass. Biotechnol Biofuels 6: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sherman F (2002) Getting started with yeast. Methods Enzymol 350: 3–41. [DOI] [PubMed] [Google Scholar]
- 40. Wohlbach DJ, Kuo A, Sato TK, Potts KM, Salamov AA, et al. (2011) Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. Proc Natl Acad Sci U S A 108: 13212–13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Voth WP, Richards JD, Shaw JM, Stillman DJ (2001) Yeast vectors for integration at the HO locus. Nucleic Acids Res 29: E59–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Amador-Noguez D, Brasg IA, Feng XJ, Roquet N, Rabinowitz JD (2011) Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum. Appl Environ Microbiol 77: 7984–7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buescher JM, Moco S, Sauer U, Zamboni N (2010) Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal Chem 82: 4403–4412. [DOI] [PubMed] [Google Scholar]
- 45. Witteveen C, Busink R, Van de Vondervoort P, Dijkema C, Swart K, et al. (1989) L-Arabinose and D-xylose catabolism in Aspergillus niger. J Gen Microbiol 135: 2163–2171. [Google Scholar]
- 46. Kuhn A, van Zyl C, van Tonder A, Prior BA (1995) Purification and partial characterization of an aldo-keto reductase from Saccharomyces cerevisiae. Appl Environ Microbiol 61: 1580–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jin YS, Jeffries TW (2003) Changing flux of xylose metabolites by altering expression of xylose reductase and xylitol dehydrogenase in recombinant Saccharomyces cerevisiae. Appl Biochem Biotechnol 105–108: 277–286. [DOI] [PubMed] [Google Scholar]
- 48. Zor T, Selinger Z (1996) Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem 236: 302–308. [DOI] [PubMed] [Google Scholar]
- 49. Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, et al. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- 50. van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, et al. (2000) An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol 26: 706–714. [DOI] [PubMed] [Google Scholar]
- 51. Fay JC, Benavides JA (2005) Hypervariable noncoding sequences in Saccharomyces cerevisiae. Genetics 170: 1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lewis JA, Elkon IM, McGee MA, Higbee AJ, Gasch AP (2010) Exploiting natural variation in Saccharomyces cerevisiae to identify genes for increased ethanol resistance. Genetics 186: 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liti G, Carter DM, Moses AM, Warringer J, Parts L, et al. (2009) Population genomics of domestic and wild yeasts. Nature 458: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brat D, Boles E, Wiedemann B (2009) Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microbiol 75: 2304–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Karhumaa K, Hahn-Hagerdal B, Gorwa-Grauslund MF (2005) Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22: 359–368. [DOI] [PubMed] [Google Scholar]
- 56. Kuyper M, Winkler AA, van Dijken JP, Pronk JT (2004) Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res 4: 655–664. [DOI] [PubMed] [Google Scholar]
- 57. Lee SM, Jellison T, Alper HS (2012) Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 78: 5708–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ni H, Laplaza JM, Jeffries TW (2007) Transposon mutagenesis to improve the growth of recombinant Saccharomyces cerevisiae on D-xylose. Appl Environ Microbiol 73: 2061–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Walfridsson M, Hallborn J, Penttila M, Keranen S, Hahn-Hagerdal B (1995) Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl Environ Microbiol 61: 4184–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Traff KL, Jonsson LJ, Hahn-Hagerdal B (2002) Putative xylose and arabinose reductases in Saccharomyces cerevisiae. Yeast 19: 1233–1241. [DOI] [PubMed] [Google Scholar]
- 61. Traff KL, Otero Cordero RR, van Zyl WH, Hahn-Hagerdal B (2001) Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl Environ Microbiol 67: 5668–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Toivari MH, Salusjarvi L, Ruohonen L, Penttila M (2004) Endogenous xylose pathway in Saccharomyces cerevisiae. Appl Environ Microbiol 70: 3681–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wenger JW, Schwartz K, Sherlock G (2010) Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet 6: e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yamanaka K (1969) Inhibition of D-xylose isomerase by pentitols and D-lyxose. Arch Biochem Biophys 131: 502–506. [DOI] [PubMed] [Google Scholar]
- 65. Kuyper M, Hartog MM, Toirkens MJ, Almering MJ, Winkler AA, et al. (2005) Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res 5: 399–409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Domesticated strains of S. cerevisiae grow poorly in lignocellulosic hydrolysates. Representative aerobic growth profiles of lab strains BY4741 (A) and CEN.PK2 (B) cultured in 96-well plates with hydrolysates made from various pretreated lignocellulose hydrolysates (see Materials and Methods) and supplemented with yeast extract and peptone (YP) are shown by plotting cell density (optical density at 595 nm) every 20 min for 24 h. ACSH; 6% glucan loading AFEX pretreated corn stover hydrolysate, AHP; Alkaline Hydrogen Peroxide pretreatment, IL; Ionic Liquid ([C2mim][OAc]) pretreated, Dil. Acid; Dilute Acid pretreated lignocellulosic hydrolysate, SGH; switchgrass hydrolysate, CSH; corn stover hydrolysate.
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in 6% glucan loading ACSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least 3 biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in 9% glucan loading ACSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-AHP CSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-AHP SGH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-6% glucan loading ACSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-detoxified AHP CSH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-detoxified AHP SGH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-80% dilute acid pretreated hydrolysate #1 relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-80% dilute acid pretreated hydrolysate #2 relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Bar graph displaying average growth rates (grey bars) of wild and domesticated S. cerevisiae strains in YP-IL SGH relative to YPD. Averages and standard deviations (black bars) are calculated from at least biological replicates. The row location for NRRL YB-210/GLBRCY0 strain used in this study is identified by opposite coloration (average growth rate in black, standard deviation in grey).
(TIF)
Hydrolysate-tolerant YB-210/GLBRCY0 engineered with XR/XDH and evolved for aerobic xylose metabolism does not ferment xylose anaerobically. The YB-210/Y0 strain engineered with XYL1, 2 and 3 genes from S. stipitis and aerobically-evolved (GLBRCY73) was cultured in bioreactors and evaluated for consumption of xylose in aerobic YPDX (A), anaerobic YPDX (B) and anaerobic ACSH (C) media as described in Materials and Methods. Concentrations (g/L) of glucose (green), xylose (red), dry cell weight (black) and ethanol (blue) are averages and standard deviations from two independent biological replicates.
(TIF)
GLBRCY133 (Y128) cell extracts display reduced in vitro xylose reductase activity similar to GRE3 deletion strains. The indicated strains were cultured aerobically in YPD, harvested and prepared for in vitro xylose reductase activity assays as described in Materials and Methods. Xylose and NADPH were added to each extract, and then rates of change in absorbance at 340 nm were measured to determine the Units of enzymatic activity normalized to mg of total protein in the cellular extract. The graph displays the average percent of in vitro xylose reductase activities and standard deviations of indicated strains relative to GLBRCY132 (marker rescued GLBRCY127, which contains wild-type GRE3) in biological duplicate.
(TIF)
Wild and domesticated S. cerevisiae strains used in phenotypic growth studies.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.