Abstract
Noise-induced hearing loss (NIHL) is a prevalent health risk. Inbred mouse strains 129S6/SvEvTac (129S6) and MOLF/EiJ (MOLF) show strong NIHL resistance (NR) relative to CBA/CaJ (CBACa). In this study, we developed quantitative trait locus (QTL) maps for NR. We generated F1 animals by intercrossing (129S6 × CBACa) and (MOLF × CBACa). In each intercross, NR was recessive. N2 animals were produced by backcrossing F1s to their respective parental strain. The 232 N2-129S6 and 225 N2-MOLF progenies were evaluated for NR using auditory brainstem response. In 129S6, five QTL were identified on chromosomes (Chr) 17, 18, 14, 11, and 4, referred to as loci nr1, nr2, nr3, nr4, and nr5, respectively. In MOLF, four QTL were found on Chr 4, 17, 6, and 12, referred to as nr7, nr8, nr9, and nr10, respectively. Given that NR QTL were discovered on Chr 4 and 17 in both the N2-129S6 and N2-MOLF cross, we generated two consomic strains by separately transferring 129S6-derived Chr 4 and 17 into an otherwise CBACa background and a double-consomic strain by crossing the two strains. Phenotypic analysis of the consomic strains indicated that whole 129S6 Chr 4 contributes strongly to mid-frequency NR, while whole 129S6 Chr 17 contributes markedly to high-frequency NR. Therefore, we anticipated that the double-consomic strain containing Chr 4 and 17 would demonstrate NR across the mid- and high-frequency range. However, whole 129S6 Chr 17 masks the expression of mid-frequency NR from whole 129S6 Chr 4. To further dissect NR on 129S6 Chr 4 and 17, CBACa.129S6 congenic strains were generated for each chromosome. Phenotypic analysis of the Chr 17 CBACa.129S6 congenic strains further defined the NR region on proximal Chr 17, uncovered another NR locus (nr6) on distal Chr 17, and revealed an epistatic interaction between proximal and distal 129S6 Chr 17.
Keywords: QTL, 129S6, MOLF, noise-induced hearing loss
INTRODUCTION
The availability of inbred mouse strains with diverse auditory phenotypes provides a powerful tool to study genetic and environmental influences on cochlear function. An inbred mouse strain represents a collection of genetically identical mice that have been maintained by brother-to-sister mating for more than 40 generations. During this process, one inbred mouse strain may become particularly resistant to a specific environmental stress, while another inbred strain may be especially susceptible. This source of genetic variability provides an opportunity to identify the genes underlying these phenotypic differences forming the basis of quantitative trait loci (QTL) mapping (Lander and Botstein 1989). Once a chromosome of interest is identified in a QTL mapping cross, selective mating strategies can be employed to transfer this chromosome from the donor strain into a host recipient strain to determine how this individual chromosome contributes to the phenotypic trait. These chromosome substitution strains, or consomic strains, can then be utilized to generate congenic strains containing progressively smaller portions of the donor chromosome, allowing for the fine mapping of a single QTL locus (Flint et al. 2005). These strategies have facilitated genetic dissection of numerous complex traits including age-related hearing loss (ahl) (Noben-Trauth and Johnson 2009).
Like age-related hearing loss, noise-induced hearing loss (NIHL) is a serious and widespread health problem (Nelson et al. 2005). It has been estimated that 22 million adults (ages 20 to 69) in the USA have a permanent hearing loss resulting from occupational (including active military service) and recreational noise exposure (NIDCD 2000). In addition to adults, hearing loss is on the rise in children, raising concerns regarding recreational noise exposure in this age group (Rabinowitz 2012). Within a given population, individuals vary in the degree of NIHL experienced in response to acoustic overstimulation (Lu et al. 2005), suggesting that some individuals have “tender” ears while others have “tough” ears. Susceptibility or resistance to NIHL is amenable to genetic analysis in mice given the availability of inbred mouse strains that show large interstrain variations in response to noise exposure. Anatomical and physiological commonalities between the human and mouse auditory system make mice a convenient model system.
Work on NIHL has identified two mouse strains, 129S6/SvEvTac (Yoshida et al. 2000) and MOLF/EiJ (Candreia et al. 2004) (referred to as 129S6 and MOLF, respectively) that demonstrate remarkable noise resistance (NR) relative to the normal, good hearing reference strain CBA/CaJ (referred to as CBACa) (Zheng et al. 1999). Toward the goal of identifying genes that contribute to NR, we used noninvasive auditory brainstem response (ABR) testing to provide a detailed characterization of NIHL for 129S6, MOLF, CBACa, and their F1 and N2 offspring. We report that NR in 129S6 and MOLF is inherited recessively. Using QTL mapping followed by construction of consomic and congenic strains, we have identified and refined the chromosomal locations of ten NR loci (nr1-10) and discovered epistatic interactions between the NR regions.
MATERIALS AND METHODS
Animals and Study Sites
Studies were conducted at laboratory sites in Seattle and Boston.
Seattle
All animals entering our protocols were born and reared at the Seattle site under specific pathogen free conditions in a facility approved by the American Association for Accreditation of Laboratory Animal Care. Three inbred strains of mice were utilized in this study: (1) CBA/CaJ (strain 000654, Jackson Labs) referred to as CBACa, (2) MOLF/EiJ (strain 000550, Jackson Labs) referred to as MOLF, and (3) 129S6/SvEvTac (Taconic Labs) referred to as 129S6. Breeding pairs were established to provide inbred parental mice, F1 intercross hybrids, and N2 progeny. The following groups of mice were characterized at the Seattle site: (1) CBACa and 129S6 parentals, (2) CBACa129S6F1 hybrids, (3) CBACa.129S6 consomic strains, and (4) CBACa.129S6 congenic strains.
Boston
The following groups of mice were characterized at the Boston site: (1) CBACa and MOLF parentals, (2) CBACaMOLFF1 hybrids, and (3) all N2 mice from the [(129S6 × CBACa) × 129S6] and [(MOLF × CBACa) × MOLF] QTL mapping crosses. All mice analyzed at the Boston site were shipped by air from the Seattle site. Prior to shipping, the N2 mice were tail-clipped to facilitate genetic analysis in Seattle. Threshold sensitivity of animals transported from Seattle was indistinguishable from animals received directly from commercial labs. On arrival in Boston, animals were held 4–7 days without treatment prior to noise exposure. All animal procedures were approved by the Institutional Animal Care and Use Committees of the Massachusetts Eye and Ear Infirmary and the University of Washington.
Acoustic Exposures
The noise exposure chambers were very similar at the Seattle and Boston sites. Mice were noise exposed at 4–6 weeks as described below, returned to the animal care facility, and held without further treatment for an additional 2 weeks before physiologic testing to quantify noise-induced permanent threshold shifts (PTS). Sound exposures were delivered to awake animals using procedures described in detail previously (Kujawa and Liberman 1997). Four to eight animals, held one per wire mesh cell in a slowly rotating subdivided cage suspended directly below the horn of the sound-delivery loudspeaker, were exposed in each session. An age-matched CBACa mouse was included in each exposure session as a continuity control. The acoustic overexposure was an octave band of noise (8–16 kHz) presented continuously for 2 h at 103 dB SPL. This exposure was chosen to maximize the measured difference in PTS between CBACa and 129S6 (Yoshida et al. 2000).
Auditory Testing
Cochlear function was tested in unexposed and exposed mice using ABR. Mice were anesthetized with a combination of xylazine (10 mg/kg) and ketamine (100 mg/kg), placed in a sound-attenuated chamber, and ABRs were recorded with needle electrodes in the scalp. Open and closed-field ABR acoustics were used in Seattle and Boston, respectively, with small differences in ABR stimulus parameters causing systematic differences in thresholds between the two sites. However, all of the relevant data were extracted from comparisons made between animals tested at the same site, i.e., all N2 animals were characterized in Boston, while all consomic and congenic lines were characterized in Seattle.
In Seattle, sound pressure levels were calibrated daily with a condenser microphone at the position of the mouse’s ear. ABRs were evoked with tone pips, with 1 ms cos2 rise/fall, presented at 13.3/s. Each mouse analyzed at the Seattle site was characterized twice by ABR: first, to collect an auditory baseline prior to noise exposure and then 2 weeks after the noise exposure. A PTS value was calculated for each mouse by comparing the difference between the pre- and postexposure ABR thresholds. In Boston, a closed-field acoustic system was used with a miniature electret microphone at the end of a probe tube to measure sound pressure in situ before each mouse was tested. Tone pips with 0.5-ms cos2 rise/fall were delivered at 30/s. At both sites, responses were amplified (×1,000), filtered (0.3–3.0 kHz), and digitized. Level was incremented in 5-dB steps from below threshold up to 80 dB SPL. Thresholds were determined, by visual inspection of stacked waveforms, as the lowest intensity evoking a reproducible wave, typically wave V. When responses were absent at the highest level, a threshold value 5 dB greater was assigned. To establish the QTL maps, a baseline preexposure ABR was used to characterize each N2 mouse. Two weeks after the noise exposure, these same N2 mice were again characterized by ABR to measure the postexposure value, which was used as the phenotype for the QTL analysis. To characterize and compare NR between CBACa, the CBACaMOLFF1 hybrids, and MOLF, the mice were separated into pre- and postexposure groups. For CBACa, 170 mice were utilized; baseline preexposure values were established using 50 mice, while postexposure values were collected on 120 mice. For the CBACaMOLFF1 hybrids, 30 mice were employed, 15 for preexposure and 15 for postexposure measures. For MOLF, 40 mice were examined, 26 for preexposure and 14 for postexposure measures. The PTS values were generated by comparing the pre- and postexposure values between the groups.
Genetic Crosses and QTL Mapping
CBACa was crossed with either 129S6 or MOLF to generate CBACa129S6F1 or CBACaMOLFF1 hybrids, which were then backcrossed to the 129S6 or MOLF parental strain, respectively. F1 and N2 mice were noise exposed and screened for NR as described above. Tail DNA was isolated from 232 and 225 N2-129S6 and N2-MOLF mice, respectively, using standard protocols. The N2 DNAs were analyzed with 84 simple-sequence length polymorphic (SSLP) markers (Applied Biosystems) also referred to as microsatellite markers that were polymorphic between CBACa, 129S6, and MOLF, providing a full genome scan at approximately 20 cM resolution. Following the initial genome scan, additional markers in chromosomal areas of interest were genotyped in the N2 DNAs.
Statistics
Auditory Evaluations
Statistical analysis was performed using PRISM (GraphPad Software, Inc.). When appropriate, an ANOVA was performed followed by Bonferroni post hoc comparisons to detect differences between strains or genotypes.
QTL Maps
Although the N2-129S6 mice were phenotyped on a quantitative scale, we performed QTL analysis using binary versions of the phenotypes. The justification for this choice was twofold. First, the observed phenotype distributions appeared bimodal and skewed (presumably due to the relatively high noise exposure level which rendered the CBACa mice nonresponsive at the highest test levels at some test frequencies), so analysis of quantitative phenotypes using the typical normal model was not entirely appropriate. Second, the nature of the underlying phenotypes could be viewed in a qualitative manner with individuals classified either as nonresistant or resistant for each of the frequency levels at which they were observed. Cutoffs were assigned at each frequency such that roughly one third of the individuals were scored as resistant. Each N2 mouse is assigned a binary phenotype score of 0 (nonresistant) or 1 (resistant) at each test frequency. The QTL analysis was performed using these scores to look for association at each individual frequency (8, 12, 16, 24, and 32 kHz). Next, the binary scores are averaged across a set of frequencies to determine if the N2 mouse is nonresistant or resistant. For example, if an N2 mouse received scores of 0, 1, 1, at 12, 16, and 24 kHz, the mouse would be scored as resistant. An additional QTL analysis was performed using these binary averages across various frequency combinations. The genome scan data was analyzed with NR status as a binary trait, using a model for binary diseases (Xu and Atchley 1996). At each marker locus, we assessed fit of the single QTL model adjusted for sex, allowing different disease penetrance for each genotype group, versus the sex-adjusted no QTL model, using a single penetrance value regardless of genotype. LOD scores were obtained as the log10 likelihood ratio comparing the single QTL and no QTL models. Significance thresholds were obtained by permutation in which data from the genome scan was reanalyzed 10,000 times, randomly shuffling phenotypes while keeping genotypes fixed (Churchill and Doerge 1994). LOD scores for genome-wide significance were obtained at the 99th and 95th percentiles of the permutation distributions, while the 75th percentile was taken as a suggestive threshold. Genome scans and permutations were performed using the R/qtl package (Broman et al. 2003).
129S6 Consomic and Congenic Strains
To produce a consomic strain, the donor 129S6 strain carrying the NR QTL on Chr 4 or 17 was backcrossed progressively to the recipient CBACa host strain. At each generation, offspring were selected genetically using the SSLP markers described above to ensure that 129S6 Chr 4 or 17 was transferred completely and to select for mice containing the highest percentage of CBACa genetic material on the remaining chromosomes. The resulting two consomic strains were referred to as CBA/CaJ-Chr 4129S6/SvEvTac/Tem and CBA/CaJ-Chr 17129S6/SvEvTac/Tem, where “Tem” indicates that the strains were created in the Tempel Lab. These two strains were then crossed to generate the CBA/CaJ-Chr 4129S6/SvEvTac Chr 17129S6/SvEvTac/Tem consomic strain. Congenic strains were generated by crossing the CBA/CaJ-Chr 4129S6/SvEvTac/Tem and CBA/CaJ-Chr 17129S6/SvEvTac/Tem consomic mice to CBACa and screening offspring with SSLP markers on Chr 4 and 17.
RESULTS
129S6 and MOLF Demonstrate Strong NR Relative to CBACa
129S6 Versus CBACa
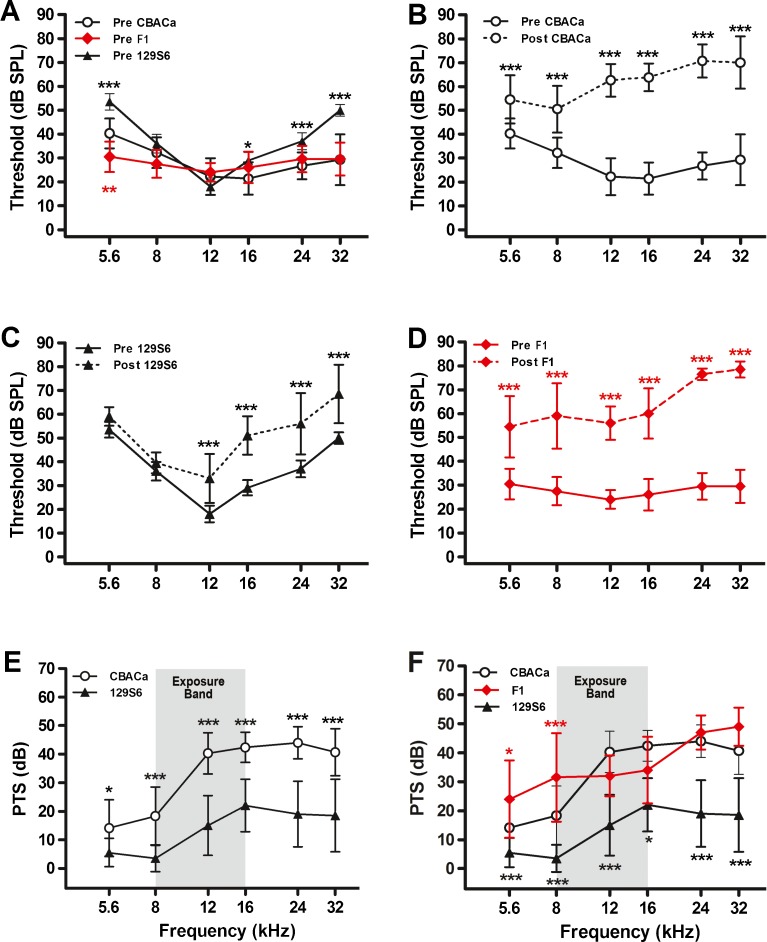
Figure 1A displays preexposure threshold levels for CBACa, 129S6, and CBACa129S6F1 at 4 weeks of age. This baseline measure indicates that CBACa and the F1 hybrids are similar at most frequencies (except 5.6 kHz), while 129S6 shows elevated thresholds relative to CBACa at several frequencies particularly in the low (5.6 kHz) and high (24 and 32 kHz) frequency range. Threshold elevation in 129S6 relative to CBACa is consistent and stable within the 4–8 weeks of experimental time frame of this study (data not shown). Two weeks after exposure to an octave band noise (8–16 kHz) at 103 dB for 2 h, CBACa exhibits dramatic pre- versus postexposure threshold increases at all frequencies (5.6, 8, 12, 16, 24, and 32 kHz) (Fig. 1B). Like CBACa, 129S6 also exhibits pre- versus post-threshold increases following noise exposure but not to the same extent as CBACa and only in the mid and upper frequencies (12, 16, 24, and 32 kHz) (Fig. 1C). The F1 hybrids demonstrate marked pre- versus postexposure threshold increases at all frequencies (Fig. 1D). To calculate PTS (Fig. 1E, F), the preexposure threshold was subtracted from the postexposure threshold at each frequency. 129S6 shows dramatically less PTS than CBACa (Fig. 1E) as reported previously (Yoshida et al. 2000). 129S6 demonstrates PTS ranging from near zero to 22 dB, depending on the frequency, while CBACa shows PTS ranging from 14 to 44 dB. These findings confirm that 129S6 demonstrates exceptional NR relative to CBACa. To examine the inheritance pattern of 129S6 NR, the CBACa and 129S6 mice were mated to generate CBACa129S6F1 hybrids. PTS in the F1 hybrids was much more CBACa-like than 129S6-like (Fig. 1F), suggesting that NR segregates as a recessive trait.
FIG. 1.
Noise resistance in CBACa versus 129S6. Pre- and postexposure ABR thresholds were measured for CBACa (n = 29), 129S6 (n = 10), and their CBACa129S6F1 hybrids (n = 10). The noise exposure band (8–16 kHz) is highlighted in gray. Data are expressed as means (+SD). Significance is indicated by asterisks: *p < 0.05, **p < 0.01, and ***p < 0.001 in a Bonferroni post hoc test following a two-way ANOVA. A Preexposure thresholds for 129S6 are elevated relative to CBACa and the F1 hybrids at several frequencies. The overall strain effect in the ANOVA is F2,276 = 49, p < 0.0001. The black and red asterisks denote p values comparing CBACa to 129S6 and CBACA to the F1 hybrids, respectively. B For CBACa, postexposure thresholds are markedly elevated across all frequencies compared to preexposure values. The overall strain effect in the ANOVA is F1,336 = 1492, p < 0.0001. C For 129S6, postexposure thresholds are elevated in the mid- and high-frequency range compared to preexposure values. The overall strain effect in the ANOVA is F1,108 = 118, p < 0.0001. D For the F1 hybrids, postexposure thresholds are markedly elevated across all frequencies compared to preexposure values. The overall strain effect in the ANOVA is F1,108 = 639, p < 0.0001. E While both strains exhibit PTS, the shift for 129S6 is dramatically less than CBACa. The overall strain effect in the ANOVA is F1,222 = 241, p < 0.0001. F PTS values are similar between CBACa and CBACa129S6F1 hybrids at most frequencies. The overall strain effect in the ANOVA is F2,276 = 145, p < 0.0001. The black and red asterisks denote p values comparing CBACa to 129S6 and CBACA to the F1 hybrids, respectively.
MOLF Versus CBACa
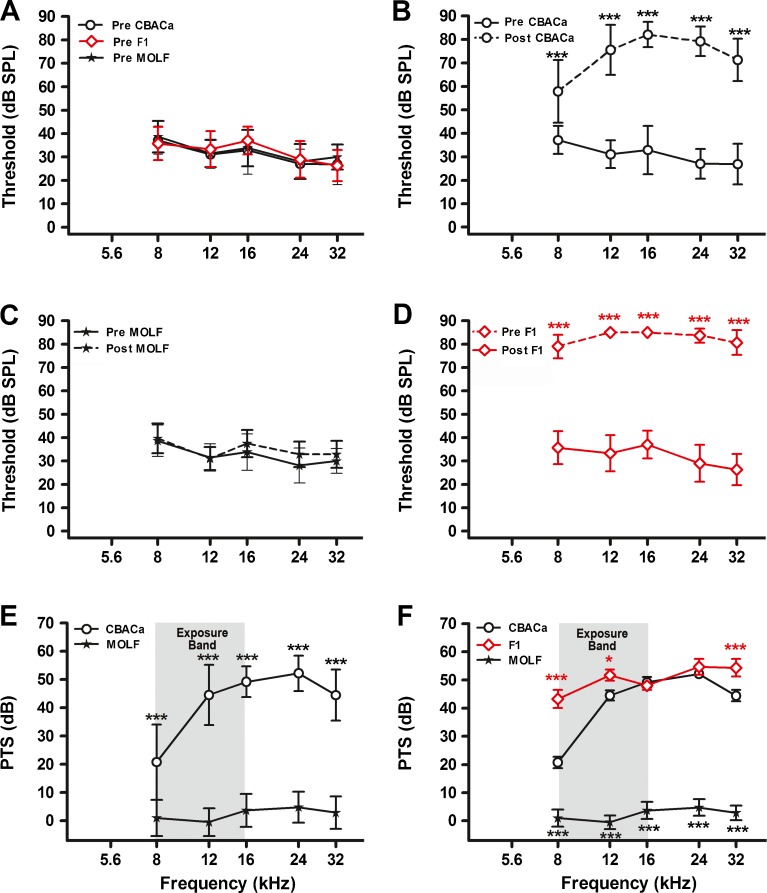
MOLF is another mouse strain reported to have a high level of NR (Candreia et al. 2004). Preexposure threshold levels for CBACa, MOLF, and CBACaMOLFF1 are nearly identical at 4 weeks of age (Fig. 2A). Following noise exposure, CBACa exhibits dramatic pre- versus postexposure threshold increases at all frequencies (8, 12, 16, 24, and 32 kHz) (Fig. 2B). Unlike CBACa, MOLF does not exhibit significant pre- versus post-threshold increases at any experimental frequency (Fig. 2C). Like CBACa, the F1 hybrids demonstrate marked pre- versus postexposure threshold increases at all frequencies (Fig. 1D). The interstrain differences in PTS are dramatic, with MOLF showing less than 5 dB PTS at all frequencies compared to a maximum of 52 dB in CBACa (Fig. 2E). To examine the inheritance pattern of MOLF NR, the CBACa and MOLF mice were mated to generate CBACaMOLFF1 hybrids. Again, PTS in the F1 hybrids was much more CBACa-like than MOLF-like (Fig. 2F), suggesting that NR segregates as a recessive trait.
FIG. 2.
Noise resistance in CBACa versus MOLF. Pre- and postexposure ABR thresholds were measured for CBACa (preexposure group n = 50, postexposure group n = 120), MOLF (preexposure group n = 26, postexposure group n = 14), and their CBACaMOLFF1 hybrids (preexposure group n = 15, postexposure group n = 15). The noise exposure band (8–16 kHz) is highlighted in gray. Data are expressed as means (+SD). Significance is indicated by asterisks: *p < 0.05, **p < 0.01, and ***p < 0.001 in a Bonferroni post hoc test following a two-way ANOVA. A Preexposure thresholds are nearly identical between CBACa, MOLF, and the F1 hybrids. B For CBACa, postexposure thresholds are markedly elevated across all frequencies compared to preexposure values. The overall strain effect in the ANOVA is F1,840 = 3958, p < 0.0001. C For MOLF, postexposure thresholds are not significantly different from preexposure values. D For the F1 hybrids, postexposure thresholds are markedly elevated across all frequencies compared to preexposure values. The overall strain effect in the ANOVA is F1,139 = 3038, p < 0.0001. E Unlike CBACa, MOLF does not exhibit significant PTS at any test frequency. The overall strain effect in the ANOVA is F1,660 = 1205, p < 0.0001. F PTS values are similar between CBACa and CBACaMOLFF1 hybrid mice from 12–24 kHz. The overall strain effect in the ANOVA is F2,730 = 892, p < 0.0001. The black and red asterisks denote p values comparing CBACa to MOLF and CBACA to the F1 hybrids, respectively.
QTL Maps Define NR Chromosomes in 129S6 and MOLF
129S6 Cross
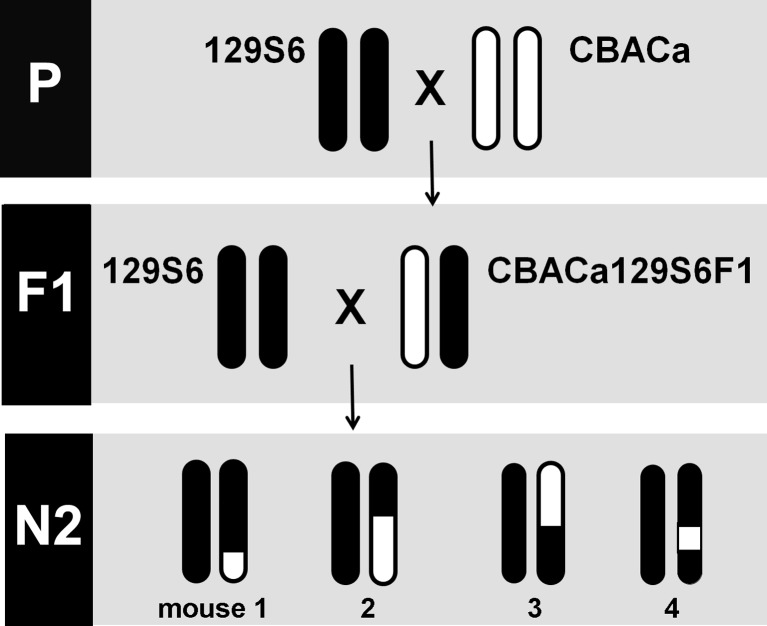
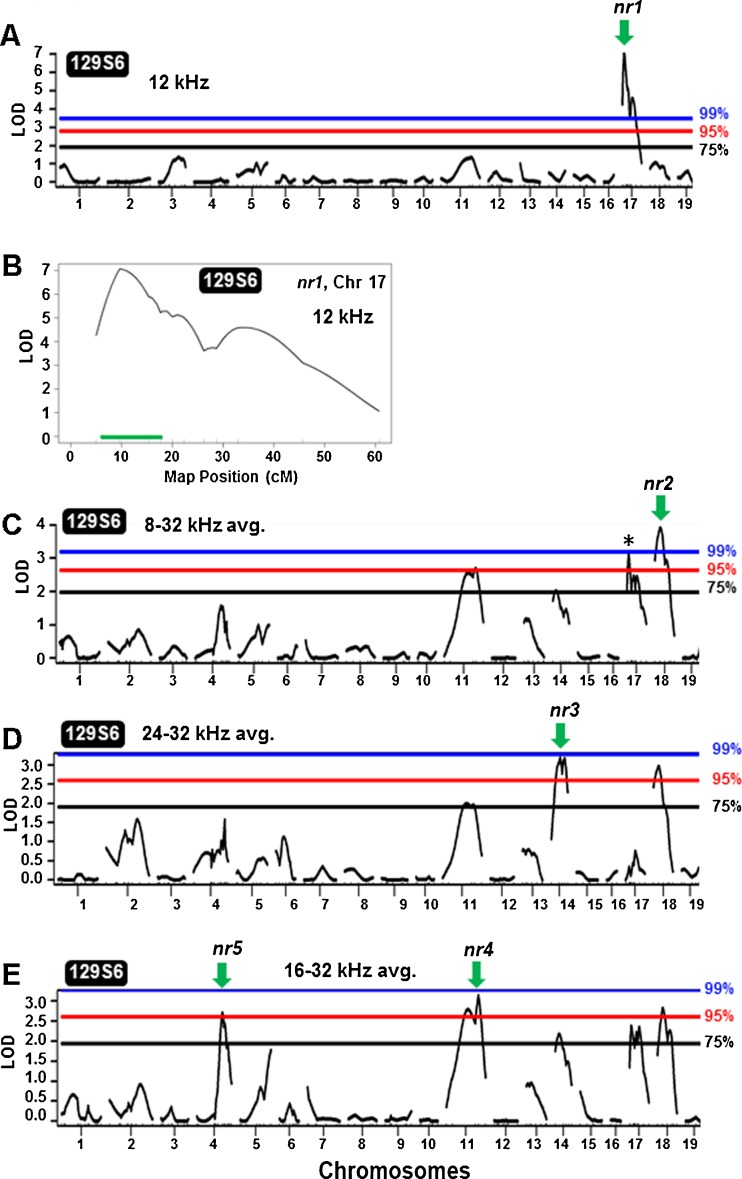
To generate a QTL map of the 129S6 NR trait, the CBACa129S6F1 hybrids were backcrossed to 129S6 parental mice to generate 232 N2 mice (Fig. 3) that were analyzed for NR and genetic markers. Based on the post-threshold value at each frequency, mice were divided into nonresistant or resistant groups and the QTL analysis was conducted using a binary approach with either single frequencies or with multiple frequency averages (see “Materials and Methods”). As summarized in Table 1, QTL for NR were detected by interval mapping analysis on five chromosomes: Chr 4 (nr5), Chr11 (nr4), Chr 14 (nr3), Chr17 (nr1), and Chr18 (nr2). These QTL are designated nr1-5 for noise resistance locus 1–5, in order of decreasing LOD score. As anticipated, the QTL represented positive associations between NR and the 129S6/129S6 homozygous genotype. Based on crossover events in the N2 population, a centimorgan (cM) map was derived for the [(129S6 × CBACa) × 129S6] cross on these five chromosomes. On Chr 4, a LOD score of 2.71 (p = 0.0471) was detected at 76.2 cM near marker D4Mit74 using 16, 24, and 32 kHz. On Chr 11, a LOD score of 3.15 (p = 0.0193) was detected at 92.9 cM near marker D11Mit126 using 16, 24, and 32 kHz. Also located near D11Mit126 is the ahl8 QTL identified as Fscn2 (fascin homologue 2, actin-bundling protein, retinal) (Shin et al. 2010). Given that ahl8 is associated with a G-to-A transition (G326A, rs26996001) in Fscn2, we sequenced this nucleotide in 129S6 versus CBACa as previously described (Shin et al. 2010) and determined that both strains carry the wildtype G326A allele (data not shown). However, this analysis does not exclude the association of nr4 with other polymorphisms in Fscn2. On Chr 14, a LOD score of 3.18 (p = 0.0152) was detected at 35.5 cM in the vicinity of marker D14Mit39 using 24 and 32 kHz. On Chr 17, a highly significant LOD score of 7.07 (p < 0.001) was detected at 9.62 cM near marker D17Mit113 using 12 kHz. The same nr1 Chr 17 QTL was observed at 9.62 cM (LOD 3.13, p = 0.0169) using 8, 12, 16, 24, and 32 kHz. On Chr 18, a LOD score of 3.91 (p = 0.0027) was detected at 21.0 cM near marker D18Mit194 using 8, 12, 16, 24, and 32 kHz. Figure 4A provides a graph of the one-dimensional genome-wide QTL scan displaying the highly significant nr1 QTL peak on Chr 17 (LOD 7.07) using 12 kHz. A confidence interval plot from the nr1 Chr 17 QTL is shown in Figure 4B highlighting a tight (9 cM) candidate gene confidence interval. Graphs of the one-dimensional genome-wide QTL scans for nr2, nr3, nr4, and nr5 on Chr 18, 14, 11, and 4, respectively, are shown in Figure 4C–E.
FIG. 3.
QTL genetic mapping cross. The cartoon depicts the QTL genetic mapping cross using one representative autosome. The 129S6 (filled chromosomes) and CBACa (open chromosomes) parental strains were mated to produce CBACa129S6F1 hybrids. The F1 mice were then backcrossed to 129S6 to generate 232 N2-129S6 mice. Each N2 mouse inherits two chromosomes: (1) an 129S6 chromosome from the 129S6 homozygous parent and (2) a potentially recombinant chromosome from the CBACa129S6F1 hybrid parent. Each N2 mouse carries a unique genetic background that depends on the crossover points in the CBACa129S6F1 hybrid parent.
TABLE 1.
NR QTL in 129S6
| Frequencies (kHz) associated with NR | QTL | Chr | Position (cM) | LOD | p value | 95 % CI (cM) | Marker nearest to peak LODa | Location of marker on GRCm38 (Mb) |
|---|---|---|---|---|---|---|---|---|
| 16, 24, 32 | nr5 | 4 | 76.2 | 2.71 | 0.0471 | 67–97 | D4Mit74 b | 114.35 |
| 16, 24, 32 | nr4 | 11 | 92.9 | 3.15 | 0.0193 | 39–105 | D11Mit126 c | Not available |
| 24, 32 | nr3 | 14 | 35.5 | 3.18 | 0.0152 | 23–57 | D14Mit39 d | 68.55 |
| 12 | nr1 | 17 | 9.62 | 7.07 | <0.001 | 7–16 | D17Mit113 e | 11.98 |
| 8,12,16,24,32 | nr1 | 17 | 9.62 | 3.13 | 0.0169 | 5–44 | D17Mit113 e | 11.98 |
| 8,12,16,24,32 | nr2 | 18 | 21.0 | 3.91 | 0.0027 | 6–41 | D18Mit194 f | 43.66 |
| 12, Congenicg | nr6 | 17 | 32.9h | NA | NA | NA | NA | NA |
Consortium Mouse Build 38, Dec. 2011. The marker cM position is derived from recombination events in the N2 [(129S6 × CBACa) × 129S6] cross
CI confidence interval, cM centiMorgan, GRCm38 genome reference
aMarker genotyped in the cross that is in the closest proximity to the peak LOD
b D4Mit74 maps at 75.5 cM
c D11Mit126 maps at 92.9 cM
d D14Mit39 maps at 32.9 cM
e D17Mit113 maps at 9.62 cM
f D18Mit194 maps at 20.4 cM
gThe NR nr6 locus was identified in the CBACa.129S6-(D17Mit119-D17Mit1)/Tem congenic strain, which showed NR at 5.6 and 12 kHz
hThe nr6 region defined by the CBACa.129S6-(D17Mit119-D17Mit1)/Tem congenic strain would include marker D17Mit119D at 32.9 cM and all chromosomal regions distal to this marker
FIG. 4.
129S6 noise resistance QTL genome scans. A, C, D, and E show one-dimensional genome-wide QTL scans. The blue, red, and black bars denote the 99, 95, and 75 % permutation thresholds, respectively. The arrows indicate peak LOD curves with green-colored arrows indicating NR is associated with 129S6/129S6. A 129S6 Chr 17 contains a highly significant NR QTL peak (LOD, 7.07) using 12 kHz, designated as nr1. B The confidence interval plot for 129S6 nr1 QTL on Chr 17. The green bar represents the confidence interval for nr1. C 129S6 Chr 18 shows a NR QTL peak (LOD 3.91) using the 8, 12, 16, 24, and 32 kHz average, designated as nr2. This plot also displays the nr1 Chr 17 QTL peak with a LOD 3.13 (asterisk) reported in Table 1. D 129S6 Chr 14 shows a QTL peak (LOD 3.18, nr3) using 24 and 32 kHz. E 129S6 Chr 4 (LOD 2.71, nr5) and 129S6 Chr 11 (LOD 3.15, nr4) peak QTL were both detected using 16, 24, and 32 kHz.
MOLF Cross
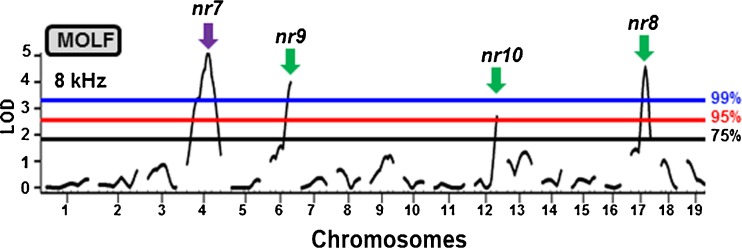
To generate a QTL map of the MOLF NR trait, the CBACaMOLFF1 hybrids were backcrossed to MOLF parental mice to generate 225 N2 mice that were analyzed for NR and genetic markers. A binary QTL approach was employed as described above for 129S6. As summarized in Table 2, QTL for NR were detected by interval mapping analysis on four chromosomes: Chr 4 (nr7), Chr 6 (nr9), Chr 12 (nr10), and Chr 17 (nr8). These QTL are designated nr7-10 for noise resistance locus 7–10, in order of decreasing LOD score. As expected, the QTL on Chr 6, 12, and 17 represented a positive association between NR and the MOLF/MOLF homozygous genotype. Surprisingly, the QTL on Chr 4 represented a positive association between NR and the CBACa/MOLF heterozygous genotype. Based on crossover events in the N2 population, a centimorgan (cM) map was derived for the [(MOLF × CBACa) × MOLF] cross. For all four chromosomes, the same QTL were observed when using 8 kHz or 8, 12, 16, 24, and 32 kHz (Table 2). In each case, the peak LOD score was noted using 8 kHz as described below. On Chr 4, a LOD score of 5.12 (p = 0.0003) was detected at 51.2 cM near marker D4Mit9. On Chr 6, a LOD score of 3.99 (p = 0.0028) was detected at 67.0 cM near marker D6Mit198. On Chr 12, a LOD score of 2.68 (p = 0.0412) was detected at 53.0 cM near marker D12Mit16. On Chr 17, a LOD score of 4.57 (p = 0.0009) was detected at 44.5 cM near marker D17Mit93. Figure 5 is a graph of the one-dimensional genome-wide QTL scan displaying the nr7-10 QTL on Chr 4, 6, 12, and 17, respectively, using 8 kHz.
TABLE 2.
NR QTL in MOLF
| Frequencies (kHz) associated with NR | QTL | Chr | Position (cM) | LOD | p value | 95 % CI (cM) | Marker nearest to peak LODa | Location of marker on GRCm38 (Mb) |
|---|---|---|---|---|---|---|---|---|
| 8 | nr7 | 4 | 51.2 | 5.12 | 0.0003 | 35–65 | D4Mit9 b | 95.07 |
| 8,12,16,24,32 | nr7 | 4 | 49.2 | 4.34 | 0.0011 | 30–62 | D4Mit9 b | 95.07 |
| 8 | nr9 | 6 | 67.0 | 3.99 | 0.0028 | 56–67 | D6Mit198 c | Not available |
| 8,12,16,24,32 | nr9 | 6 | 67.0 | 3.41 | 0.0092 | 56–67 | D6Mit198 c | Not available |
| 8 | nr10 | 12 | 53.0 | 2.68 | 0.0412 | 13–53 | D12Mit16 d | 109.78 |
| 8,12,16,24,32 | nr10 | 12 | 53.0 | 2.31 | 0.0968 | 2–53 | D12Mit16 d | 109.78 |
| 8 | nr8 | 17 | 44.5 | 4.57 | 0.0009 | 38–52 | D17Mit93 e | 73.80 |
| 8,12,16,24,32 | nr8 | 17 | 44.5 | 4.36 | 0.0011 | 38–51 | D17Mit93 e | 73.80 |
Consortium Mouse Build 38, Dec. 2011. The marker cM position is derived from recombination events in the N2 [(MOLF x CBACa) x MOLF] cross
CI confidence interval, cM centiMorgan, GRCm38 genome reference
aMarker genotyped in the cross that is in the closest proximity to the peak LOD
b D4Mit9 maps at 44.5 cM
c D6Mit198 maps at 67.0 cM
d D12Mit16 maps at 53.0 cM
e D17Mit93 maps at 44.5 cM
FIG. 5.
MOLF noise resistance QTL genome scans. The blue, red, and black bars denote the 99, 95, and 75 % permutation thresholds, respectively. The arrows indicate peak LOD curves with green-colored arrows indicating NR is associated with MOLF/MOLF, while the purple-colored arrow represents NR association with CBACa/MOLF. The four peak MOLF NR QTL [Chr 4 (nr7), Chr 6 (nr9), Chr 12 (nr10), and Chr 17 (nr8)] were all detected using 8 kHz and can therefore all be represented on the same plot.
129S6 to MOLF Comparison
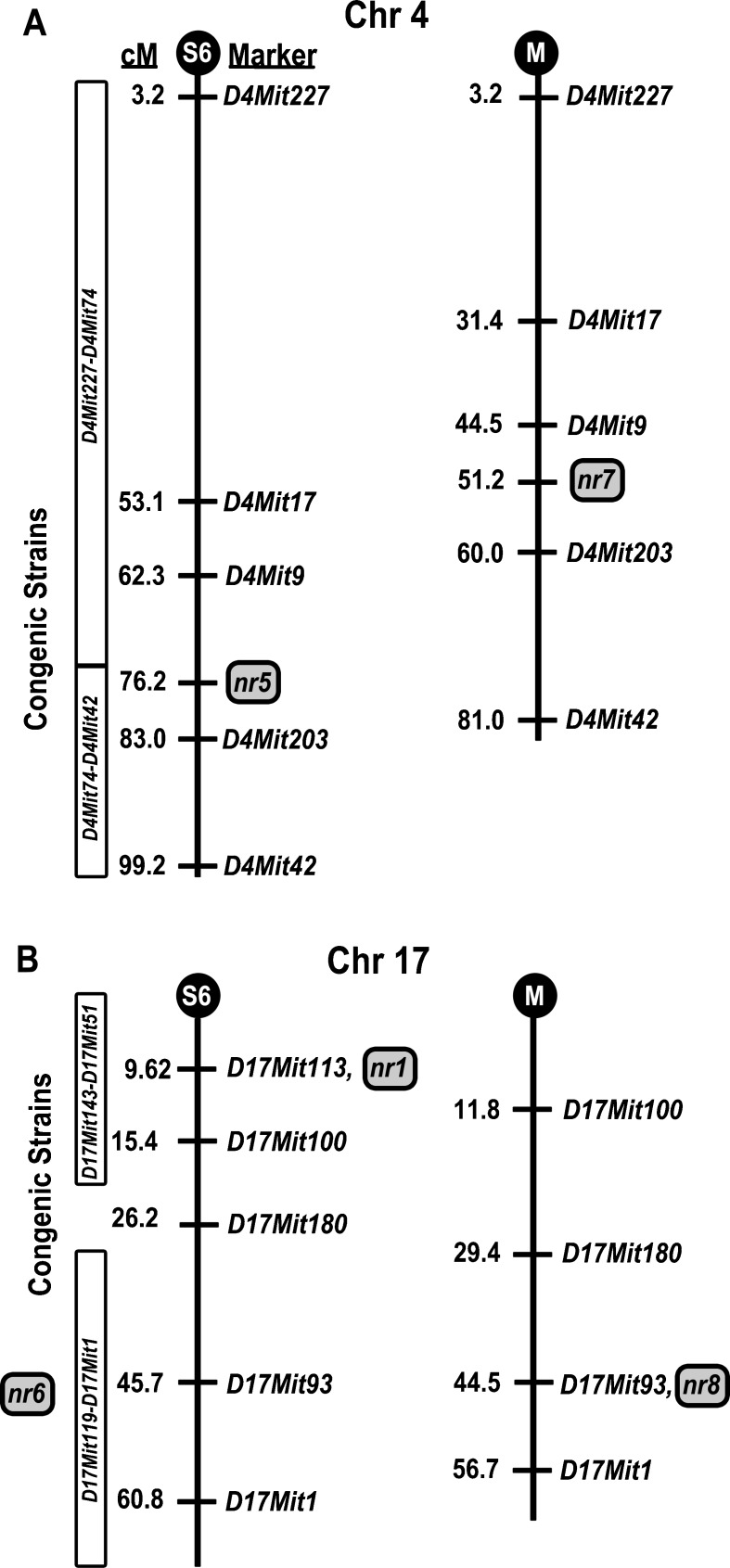
Chr 4 and 17 harbor NR QTL in both the 129S6 and MOLF cross. Do the Chr 4 and 17 QTL represent the same loci in both crosses? For Chr 4, the recombination frequency varied between [(129S6 × CBACa) × 129S6] and [(MOLF × CBACa) × MOLF] as indicated by the cross-derived cM maps containing markers (D4Mit227, D4Mit17, D4Mit9, D4Mit203, D4Mit42) genotyped in both crosses (Fig. 6A). While the 129S6 cM map is more expanded than the MOLF map, the peak LOD in both crosses is located between markers D4Mit9 and D4Mit203, suggesting that the QTL on Chr 4 in 129S6 (nr5) and MOLF (nr7) may represent the same or closely linked loci. As noted in the 129S6 cross, the NR phenotype on Chr 4 is associated with the homozygous 129S6/129S6 genotype. However, in the MOLF cross, the NR phenotype on Chr 4 is associated with the heterozygous CBACa/MOLF genotype, suggesting that the CBACa allele offers more NR than the MOLF allele. If the same gene is underlying the Chr 4 NR in both crosses, we might have an allelic series of gene function for NR: 129S6 > CBACa > MOLF. For Chr 17, the recombination frequency was similar between [(129S6 × CBACa) × 129S6] and [(MOLF × CBACa) × MOLF] as suggested by the cross-derived cM maps containing markers (D17Mit100, D17Mit180, D17Mit93, D17Mit1) genotyped in both crosses (Fig. 6B). As mentioned above, the 129S6 Chr 17 LOD of 7.07 was detected at 9.62 cM while the peak MOLF Chr LOD was positioned at 44.5 cM, indicating that the two QTL (nr1 and nr8) are distinct loci.
FIG. 6.
129S6 and MOLF NR loci comparison for Chr 4 and 17. Genetic maps were derived from the [(129S6 × CBACa) × 129S6] and [(MOLF × CBACa) × MOLF] crosses. The vertical line represents the chromosome with markers genotyped in both crosses to the right and the cM distance separating the markers to the left. The black circle at the top of the chromosome represents the centromere: “S6” indicates the 129S6 cross and “M” denotes the MOLF cross. The best estimates for the locations of the nr1, nr5, nr6, nr7, and nr8 QTL are shown by a gray-colored box. CBACa.129S6 congenic strains used in defining candidate NR regions are shown to the left of the 129S6 chromosome map. A Map comparison for Chr 4. B Map comparison for Chr 17.
129S6 Chr 4 and 17 Contribute to NR in Consomic Strains
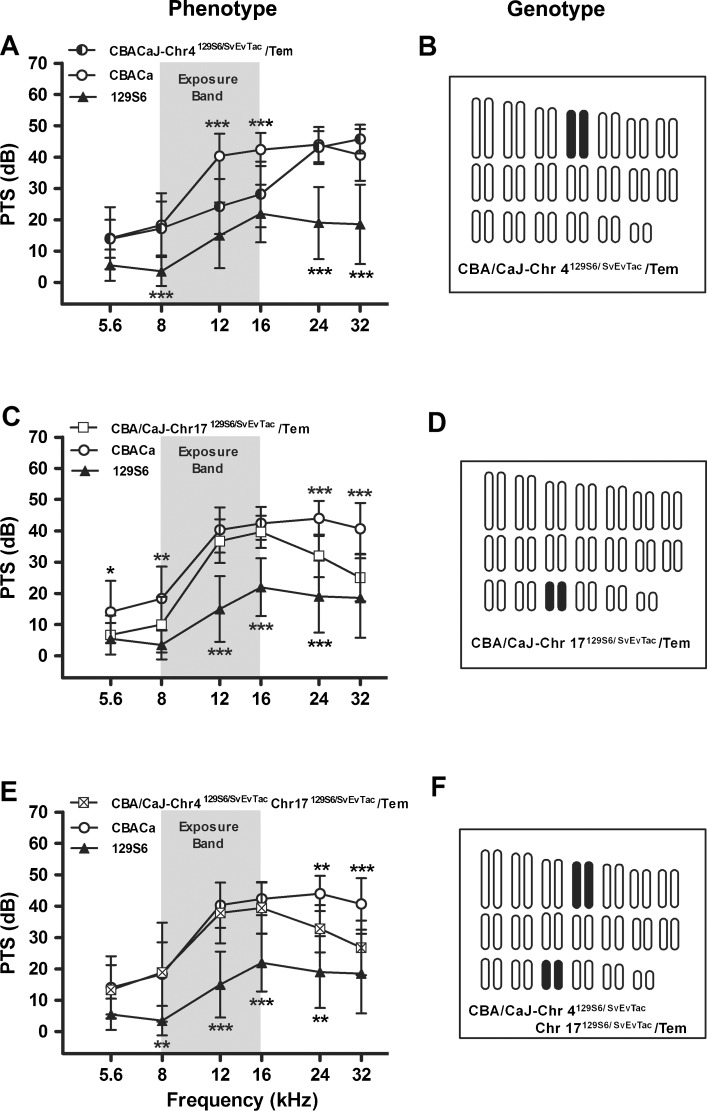
Based on the QTL findings for Chr 4 and 17, we created three consomic 129S6 strains containing either Chr 4, Chr 17, or Chr 4 and 17 in a CBACa background (CBA/CaJ-Chr 4129S6/SvEvTac/Tem, CBA/CaJ-Chr 17129S6/SvEvTac/Tem, and CBA/CaJ-Chr 4129S6/SvEvTac Chr 17129S6/SvEvTac/Tem, respectively) (Fig. 7). Each consomic strain was evaluated to determine the NR contribution carried by 129S6 Chr 4 and 17 in isolation or combination. A PTS comparison among CBACa, 129S6, and each consomic strain is summarized in Figure 7.
FIG. 7.
Chromosome 4 and 17 consomic strains. PTS for three consomic strains are compared to the parental CBACa (n = 29) and 129S6 (n = 10) strains (A, C, E), displayed with the chromosome content of each consomic strain (B, D, F). 129S6 versus CBACa chromosomal segments are depicted by filled versus open chromosomes, respectively. The noise exposure band (8–16 kHz) is highlighted in gray. Data are expressed as means (±SD). A p value asterisk located above a CBACa data point represents the statistical comparison between CBACa and the consomic strain of interest, while an asterisk below a 129S6 data point represents the comparison between 129S6 and the consomic strain. *p < 0.05, **p < 0.01, and ***p < 0.001 in a Bonferroni post hoc test following a two-way ANOVA. A 129S6 Chr 4 appears to contribute to NR at 12 and 16 kHz in the CBA/CaJ-Chr 4129S6/SvEvTac/Tem strain (n = 18). The overall strain effect in the ANOVA is F2,324 = 147, p < 0.0001. B This consomic strain contains 129S6 Chr 4 in a CBACa background. C 129S6 Chr 17 appears to contribute to NR at 5.6, 8, 24, and 32 kHz in the CBA/CaJ-Chr 17129S6/SvEvTac/Tem strain (n = 15). The overall strain effect in the ANOVA is F2,306 = 132, p < 0.0001. D This consomic strain contains 129S6 Chr 17 in a CBACa background. E The consomic strain (n = 8) containing 129S6 Chr 4 and 17, demonstrates NR at 24 and 32 kHz. The overall strain effect in the ANOVA is F2,264 = 94, p < 0.0001. F This consomic strain contains 129S6 Chr 4 and 17 in a CBACa background.
Chr 4 Consomic Strain
Whole 129S6 Chr 4 contributes to NR in the mid-frequency range. At 5.6, 8, 24, and 32 kHz (Fig. 7A), the CBA/CaJ-Chr 4129S6/SvEvTac/Tem mice (Fig. 7B) show PTS values nearly identical to CBACa. However, at 12 and 16 kHz, the CBA/CaJ-Chr 4129S6/SvEvTac/Tem PTS values are significantly different from CBACa but similar to 129S6. These findings indicate that the whole 129S6 Chr 4 contributes strongly to NR at 12 and 16 kHz.
Chr 17 Consomic Strain
Whole 129S6 Chr 17 contributes strongly to NR in the high-frequency range and significantly at low frequencies. At 12 and 16 kHz (Fig. 7C), the CBA/CaJ-Chr 17129S6/SvEvTac/Tem mice (Fig. 7D) show PTS values similar to CBACa. However, at 24 kHz, the CBA/CaJ-Chr 17129S6/SvEvTac/Tem PTS values deviate from CBACa, and at 5.6, 8, and 32 kHz, the CBA/CaJ-Chr 17129S6/SvEvTac/Tem PTS values are similar to 129S6. These findings indicate that the whole 129S6 Chr 17 contributes strongly to NR at 24 and 32 kHz as well as contributing protection at 5.6 and 8 kHz.
Chr 4 and 17 Double-Consomic Strain
Data from the double consomic strain indicates that whole 129S6 Chr 4 and Chr 17 interact. Given that the CBA/CaJ-Chr 4129S6/SvEvTac/Tem consomic strain demonstrates robust NR in the mid frequencies and the CBA/CaJ-Chr 17129S6/SvEvTac/Tem consomic strain demonstrates marked NR in the high frequencies, we expected that the CBA/CaJ-Chr 4129S6/SvEvTac Chr 17129S6/SvEvTac/Tem consomic strain would show strong NR across the mid and high frequencies. However, as plotted in Figure 7E, the CBA/CaJ-Chr 4129S6/SvEvTac Chr 17129S6/SvEvTac/Tem consomic strain (Fig. 7F) displays NR only at 24 and 32 kHz presumably contributed by whole Chr 17. The presence of whole 129S6 Chr 17 in the CBA/CaJ-Chr 4129S6/SvEvTac Chr 17129S6/SvEvTac/Tem strain appears to mask the mid-frequency NR contribution of whole 129S6 Chr 4.
Chr 4 and 17 Congenic Strains Further Delineate NR Regions
To narrow the NR regions of 129S6 Chr 4 and 17, the CBA/CaJ-Chr 4129S6/SvEvTac/Tem and CBA/CaJ-Chr 17129S6/SvEvTac/Tem consomic strains were used in marker-assisted mating strategies to generate two Chr 4 congenic strains [CBACa.129S6-(D4Mit227-D4Mit74)/Tem and CBACa.129S6-(D4Mit74-D4Mit42)/Tem] and three Chr 17 congenic strains [CBACa.129S6-(D17Mit143-D17Mit51)/Tem), CBACa.129S6-(D17Mit143-D17Mit100)/Tem, and CBACa.129S6-(D17Mit119-D17Mit1)/Tem].
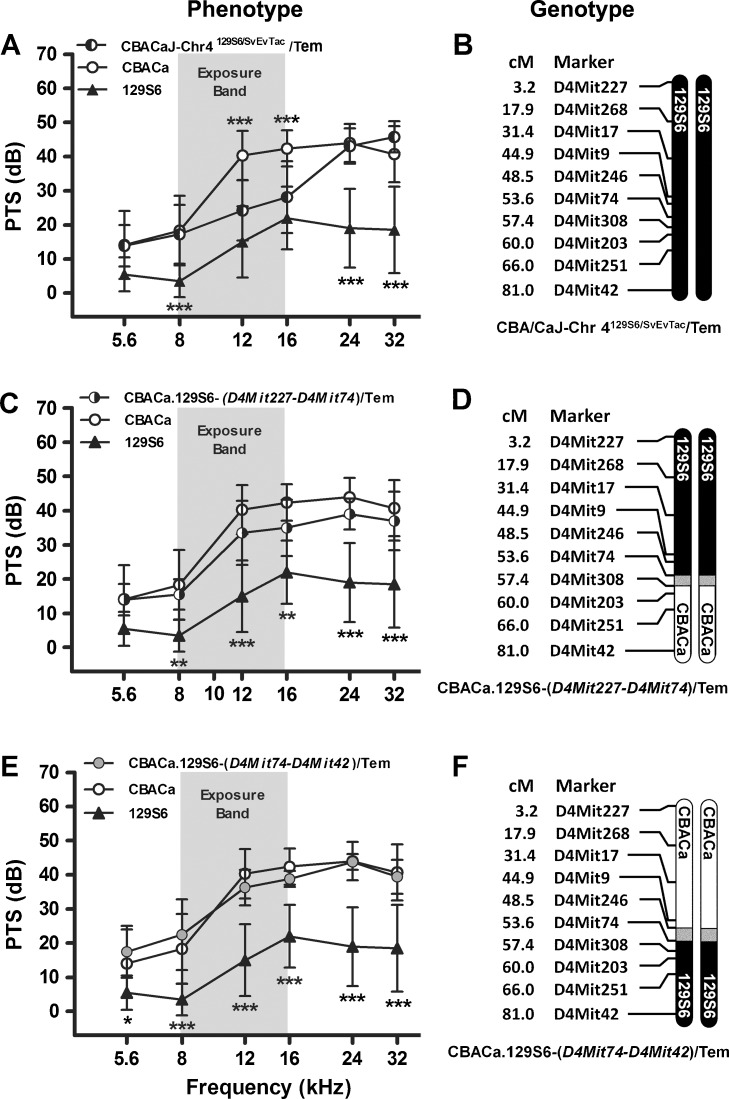
Chr 4 Congenic Strains
To facilitate comparison, the PTS phenotype and genotype of the CBA/CaJ-Chr 4129S6/SvEvTac/Tem consomic strain are shown in Figure 8A, B, respectively. As mentioned previously, the CBA/CaJ-Chr 4129S6/SvEvTac/Tem consomic strain contributes to NR at 12 and 16 kHz (Figs. 7A and 8A). The CBACa.129S6-(D4Mit227-D4Mit74)/Tem strain demonstrates PTS values not significantly different from CBACa (Fig. 8C), suggesting that the 129S6 homozygous region proximal to D4Mit74 (Fig. 8D) is not sufficient to support NR at 12 and 16 kHz. Similarly, the CBACa.129S6-(D4Mit74-D4Mit42)/Tem strain shows PTS values not significantly different from CBACa (Fig. 8E), suggesting that the 129S6 homozygous region distal to D4Mit74 (Fig. 8F) is not sufficient to support NR at 12 and 16 kHz. One interpretation of these results is that genetic elements present on both the proximal and distal segments of 129S6 Chr 4 cooperate to generate NR in the mid-frequency range in the CBA/CaJ-Chr 4129S6/SvEvTac/Tem consomic strain.
FIG. 8.
Chromosome 4 congenic strains. PTS (A, C, E) for two Chr 4 congenic strains are compared to the parental CBACa (n = 29, open circle) and 129S6 (n = 10, triangle) strains, displayed with the recombinant crossover event defining each strain (B, D, F). 129S6 and CBACa chromosomal segments are depicted by filled and open chromosomes, respectively. The gray regions represent uncharacterized DNA near the crossover point. SSLP markers used to characterize the strain and the cM (centiMorgan) position for each marker are indicated. The noise exposure band (8–16 kHz) is highlighted in gray. Data are expressed as means (±SD). A p value asterisk located above a CBACa data point represents the statistical comparison between CBACa and the consomic or congenic strain of interest, while an asterisk below a 129S6 data point represents the comparison between 129S6 and the strain. *p < 0.05, **p < 0.01, and ***p < 0.001 in a Bonferroni post hoc test following a two-way ANOVA. A 129S6 Chr 4 appears to contribute to NR at 12 and 16 kHz in the CBA/CaJ-Chr 4129S6/SvEvTac/Tem strain (n = 18). PTS data are the same as those shown in Fig. 6A, duplicated here to allow comparison between the two congenic strains generated from the CBA/CaJ-Chr 4129S6/SvEvTac/Tem consomic strain. B This consomic strain contains a nonrecombinant 129S6 Chr 4. C The NR phenotype in strain CBACa.129S6-(D4Mit227-D4Mit74)/Tem (n = 10) is not significantly different from CBACa. D This congenic strain contains a crossover between D4Mit74 and D4Mit308. E The NR phenotype in strain CBACa.129S6-(D4Mit74-D4Mit42)/Tem (n = 8) is not significantly different from CBACa. F This congenic strain is defined by a crossover between D4Mit246 and D4Mit74.
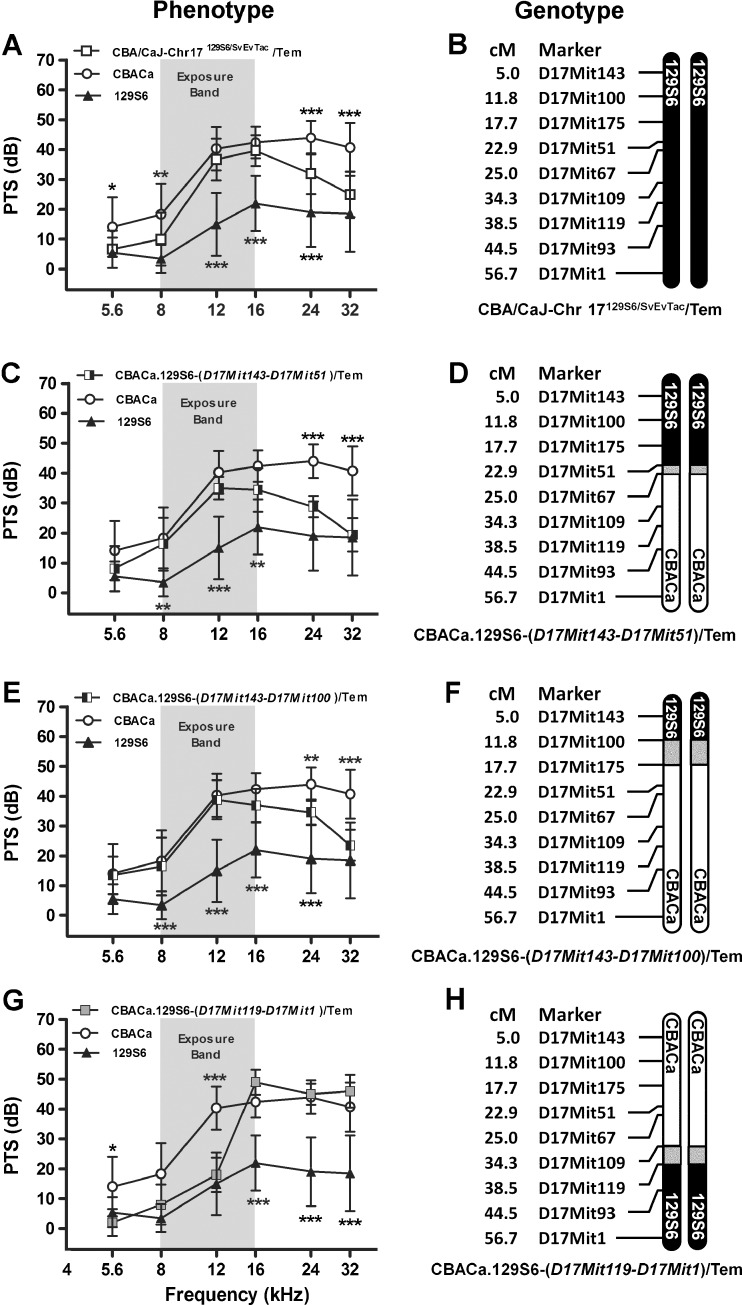
Chr 17 Congenic Strains
To facilitate comparison, the PTS phenotype and genotype of the CBA/CaJ-Chr 17129S6/SvEvTac/Tem consomic strains are shown in Figure 9A, B, respectively. As mentioned previously, the CBA/CaJ-Chr 17129S6/SvEvTac/Tem consomic strain contributes to NR at 5.6, 8, 24, and 32 kHz (Figs. 7C and 9A). The CBACa.129S6-(D17Mit143-D17Mit51)/Tem congenic strain shows significant NR at 24 and 32 kHz but not at 5.6 and 8 kHz (Fig. 9C), suggesting that the 129S6 homozygous region proximal to D17Mit51 (Fig. 9D) is sufficient to contribute to NR at high but not low frequencies. Further fragmentation of proximal Chr 17 resulted in the CBACa.129S6-(D17Mit143-D17Mit100)/Tem strain. At 24 and 32 kHz, the PTS values for strain CBACa.129S6-(D17Mit143-D17Mit100)/Tem remain significantly more NR than CBACa (Fig. 9E), indicating that the 129S6 homozygous region proximal to D17Mit100 (Fig. 9F) contains one or more loci contributing to high-frequency NR. We also generated a third Chr 17 congenic strain CBACa.129S6-(D17Mit119-D17Mit1)/Tem containing a homozygous 129S6 region distal to D17Mit119. Surprisingly, this congenic strain demonstrated marked NR at 12 kHz (Fig. 9G), which was not detected in the CBA/CaJ-Chr 17129S6/SvEvTac/Tem consomic strain suggesting that 129S6 proximal Chr 17 may mask the mid-frequency contribution of 129S6 distal Chr 17. These findings denote the presence of a second NR QTL on distal Chr 17 (Fig. 9H) contributing to mid-frequency NR designated nr6 (Table 1). Alignment of the 129S6/129S6 homozygous area in the CBACa.129S6-(D17Mit119-D17Mit1)/Tem strain with the 129S6 and MOLF cM maps (Fig. 6B) demonstrates that the 129S6/129S6 region in the congenic strain overlaps with the MOLF/MOLF nr8 QTL region highlighting the possibility that 129S6 and MOLF may share the same NR QTL (nr6 and nr8, respectively) on distal Chr 17.
FIG. 9.
Chromosome 17 congenic strains. PTS (A, C, E, G) for three Chr 17 congenic strains are compared to the parental CBACa (n = 29) and 129S6 (n = 10) strains, displayed with the recombinant crossover event defining each strain (B, D, F, H). Other display conventions are as described for Figure 8. A 129S6 Chr 17 appears to contribute to NR at 5.6, 8, 24, and 32 kHz in the CBA/CaJ-Chr 17129S6/SvEvTac/Tem strain (n = 15). PTS data are the same as those shown in Figure 7C, duplicated here to allow comparison among all three congenic strains generated from the CBA/CaJ-Chr 17129S6/SvEvTac/Tem consomic strain. B This consomic strain contains a nonrecombinant 129S6 Chr 17. C The CBACa.129S6-(D17Mit143-D17Mit51)/Tem (n = 8) strain demonstrates NR at 24 and 32 kHz. The overall strain effect in the ANOVA is F2,264 = 100, p < 0.0001. D This congenic strain contains a crossover between D17Mit51 and D17Mit67. E The CBACa.129S6-(D17Mit143-D17Mit100)/Tem strain (n = 13) demonstrates NR at 24 and 32 kHz. The overall strain effect in the ANOVA is F2,294 = 133, p < 0.0001. F This congenic strain contains a crossover between D17Mit100 and D17Mit175. G The CBACa.129S6-(D17Mit119-D17Mit1)/Tem strain (n = 5) demonstrates significant NR at 5.6 and 12 kHz. H This congenic strain is defined by a crossover between D17Mit109 and D17Mit119. The overall strain effect in the ANOVA is F2,246 = 82, p < 0.0001.
Chr 4 and 17 129S6 NR QTL Interact
As described above, two strong interactions were observed when analyzing the consomic and congenic 129S6 lines (Table 3). First, the mid-frequency NR contribution of 129S6 whole Chr 4 (Fig. 7A) is not apparent (Fig. 7E) if 129S6 whole Chr 17 is present in the same animal. Second, in the congenic strains, the 12 kHz NR contribution of distal 129S6 Chr 17 (Fig. 9G) is not apparent until 129S6 homozygosity on proximal 17 is replaced by CBACa (Fig. 9H).
TABLE 3.
129S6 epistatic interaction summary
| Genotype | NR phenotype (kHz) | Interaction |
|---|---|---|
| Whole Chr 4 | 12, 16 | Masked by whole Chr 17 |
| Distal Chr 17 | 12 | Masked by proximal Chr 17 |
DISCUSSION
In this paper, we characterized the NR phenotype of the CBACa, 129S6, and MOLF inbred mouse strains and established that NR in 129S6 and MOLF is inherited in an autosomal recessive manner. Genotypic and phenotypic analysis of N2 progeny identified nine NR QTL: five QTL in 129S6, nr1 (Chr 17), nr2 (Chr 18), nr3 (Chr 14), nr4 (Chr 11), and nr5 (Chr 4) and four QTL in MOLF, nr7 (Chr 4), nr8 (Chr 17), nr9 (Chr 6), and nr10 (Chr 12). Congenic strain construction in 129S6, identified a tenth NR locus (nr6) on distal Chr 17. The 129S6 and MOLF NR QTL on Chr 4 (nr5 and nr7) and Chr 17 (nr6 and nr8), respectively, map in a similar chromosomal vicinity and may represent the same or closely linked genes. Phenotypic analysis of the consomic and congenic strains revealed two epistatic interactions, one between Chr 4 and 17 and another between proximal and distal 17.
Given that both 129S6 and MOLF demonstrate robust NR, it is not surprising that the strains share NR regions in common on Chr 4 and 17. However, in addition to shared regions, 129S6 and MOLF contain unique NR loci—an outcome that is consistent with differences between the two strains in baseline cochlear thresholds and NR behavior. First, 129S6 shows elevated preexposure thresholds compared to CBACa, while baseline measures of MOLF are nearly identical with CBACa. Second, while 129S6 and MOLF both show NR, the nature of the resistance is different between the strains. 129S6 demonstrates intensity-independent NR, while MOLF shows intensity-dependent NR. For MOLF, the critical noise exposure intensity level, i.e., the exposure level above which PTS begins to grow very rapidly, is elevated by roughly 3–6 dB with respect to that seen in CBACa (Kujawa et al. 2007). In contrast, for 129S6, there appears to be no critical level, as the slope of the PTS growth versus exposure level function is extremely low (<2 dB PTS growth/dB increase in exposure SPL) compared to that seen in CBACa (~8 dB PTS/dB SPL) (Yoshida et al. 2000; Kujawa et al. 2007). Another genome-wide analysis of NR has been conducted using a four-way cross with 20-month old mice exposed to white noise (2–22 kHz) for 2 h at 110 dB SPL (Schacht et al. 2012). Loci associated with NR detected in the four-way cross do not map to the same chromosomal regions as the QTL described in our study. This may be expected given the different noise exposure bands and the different noise-exposure ages of the mice, which could impact noise vulnerability (Ohlemiller et al. 2011).
In thinking about the genetic basis of NR, it is also important to consider the nature of the underlying damage, which will differ as a function of the severity of the PTS. In the present study, we exposed mice at 103 dB SPL for 2 h. In CBACa, a prior study shows that this exposure will produce minimal hair cell loss, outside of the extreme basal regions that are tuned to cochlear frequencies well above those tested here (Wang et al. 2002). It is likely that the functionally important structural change underlying the PTS we observed was damage to the stereocilia bundles on both inner and outer hair cells (Wang and Liberman 2002). Although the precise mechanisms underlying this stereocilia damage are unclear, there is evidence that reactive oxygen species are important in the generation of noise-induced PTS (Henderson et al. 2006) and that manipulations to the cochlea’s redox pathways can be protective against noise exposure (Yamasoba et al. 1999; Ohinata et al. 2000; Abi-Hachem et al. 2010; Fetoni et al. 2013). There is also likely damage to the fibrocytes in the spiral ligament, which could potentially impact the generation of endolymphatic potential (EP). However, prior study of exposures to even higher levels (112 dB for 2 h) of the same noise band suggests that any noise-induced EP changes in CBACa are transient (Hirose and Liberman 2003). Acoustic overexposure in mice has been shown to induce an inflammatory response involving macrophages that multiply and move into the lateral wall and spiral ligament (Hirose and Liberman 2003; Hirose et al. 2005), and there is evidence that modulation of the cochlea’s inflammatory response can be protective against the type of noise exposure used in this study (Peppi et al. 2011). In this regard, it is interesting that the cochlear macrophage antigen CD68 (Cd68) (Hirose and Liberman 2003; Hirose et al. 2005) is located on Chr11 (69 Mb) within the confidence intervals (CI) of nr4, and that allograft inflammatory factor 1 (Aif1) (Hirose et al. 2005) is positioned on Chr 17 (35 Mb) in the region of nr1. Also, a monocyte chemoattractant protein known as Ccl2 (chemokine C-C motif ligand 2), which is markedly upregulated following acoustic injury (Sautter et al. 2006), is located on Chr 11 (82 Mb) within the CI of nr4. These genes may contribute to NR in 129S6 by modulating inflammatory responses within the cochlea following acoustic trauma. Finally, although there is also significant loss of cochlear nerve synapses following this noise exposure on the inner hair cells throughout the mid- and high-frequency regions (Kujawa and Liberman 2009), this partial neuropathy will not affect ABR thresholds, and thus was not an important factor driving our QTL analysis.
Candidate genes known to result in auditory impairment in mice or humans are present within the QTL CI falling into a variety of gene families (Johnson 2014) as summarized in Table 4. Candidate genes and regions on Chr 17 are considered in greater detail below. On proximal Chr 17, the 129S6 peak QTL is located at D17Mit113 (12.0 Mb) with a 9 cM 95 % CI. Another auditory QTL designated Phl1 for progressive hearing loss mapped in 101/H mice also shows a peak LOD at D17Mit113 (Mashimo et al. 2006). These candidate QTL areas on proximal Chr 17 are supported further by NR congenic strain CBACa.129S6-(D17Mit143-D17Mit100)/Tem with a crossover point at D17Mit100 (26.2 Mb). Candidate loci positioned above D17Mit100 include the following genes known to alter auditory function: Nox3 (NADPH oxidase 3) (Flaherty et al. 2011) at 3.64 Mb, Synj2 (synaptojanin 2) (Manji et al. 2011) at 5.94 Mb, Cldn9 (claudin 9) (Nakano et al. 2009) at 23.7 Mb, Noxo1 (NADPH oxidase organizer 1) (Kiss et al. 2006) at 24.7 Mb, and Axin1 (axin 1) (Reed 1937) at 26.1 Mb. On distal Chr 17, the MOLF peak QTL is located at D17Mit93 (73.8 Mb) with a 14 cM 95 % CI. The putative 129S6 NR (nr6) on distal Chr 17 in the CBACa.129S6-(D17Mit119-D17Mit1)/Tem congenic strain is defined by a crossover point at D17Mit119 (66.9 Mb). Because these QTL are present in two distantly related mouse strains, our task of identifying the causal gene(s) may be simplified if in fact the causal gene is the same in both strains. If so, we might expect the underlying mutations within the gene to be different between distantly related 129S6 and MOLF. Another auditory QTL mapped to distal Chr 17 in a B6 × B6.MSMChr17 consomic cross is ahl3 (age-related hearing loss, locus 3) (Nemoto et al. 2004). Fine mapping of the ahl3 region in congenic strains localized ahl3 to a 14-Mb region between D17Mit274 (53.5 Mb) and D17Mit183 (62.7 Mb), while phenotypic analysis of the ahl3 congenic strains suggested this region may also play a role in noise-induced hearing loss (Morita et al. 2007). The close proximity of these three regions (NR QTL in MOLF, NR congenic strain in 129S6, and ahl3) indicates that this area harbors one or more auditory genes.
TABLE 4.
NR QTL and candidate gene overview
| nr | Chr | Candidate genes | Other auditory QTL |
|---|---|---|---|
| nr1 | Proximal 17 | Nox3, Synj2, Cldn9, Noxo1, Axin1, Aif1 | Phl1 |
| nr2 | 18 | Aqp4, Diap1, Pou4f3, Slc12a2, Tcof1, Atp8b1 | ahl6, Nirep |
| nr3 | 14 | Gjb2, Gjb6, Diap3 | NA |
| nr4 | 11 | Col1a1, Ush1g, Actg1, Cd68, Ccl2 | ahl8, Fscn2 |
| nr5, nr7 | 4 | Bsnd, Lepre1, Kcnq4, Gjb3 | NA |
| nr6, nr8 | Distal 17 | Slc5a7,Slc25a41, Slc25a23 | ahl3 |
| nr9 | 6 | Atp2b2 | NA |
| nr10 | 12 | Coch | NA |
In addition to identifying ten nr loci (nr1-10), our studies revealed genetic interactions between 129S6 Chr 4 and 17. In the congenic strains, the 12 kHz NR contribution of distal 129S6 Chr 17 is not detected in the presence of 129S6/129S6 on proximal 17. In the consomic strain, the mid-frequency NR contribution of 129S6 Chr 4 is not apparent if whole 129S6 Chr 17 is present in the same animal. While we cannot be certain which portion of 129S6 Chr 17 is responsible for masking Chr 4, it is possible that proximal Chr 17 accounts for the suppression. If this is the case, the NR region on proximal Chr 17, which in the consomic and congenic strains contributes to NR at high-frequency regions, may be capable of masking the mid-frequency contribution of both Chr 4 and distal Chr 17. This question could be explored experimentally by establishing strains carrying whole 129S6 Chr 4 in the presence of either distal or proximal 129S6 Chr 17. Identification of the QTL underlying NR on Chr 4 and 17 will allow for these interactions to be elucidated at the molecular level. In summary, we have discovered ten chromosomal regions contributing to NR providing a framework upon which candidate genes identified using distinct yet complementary approaches, such as gene microarray expression studies, can be overlaid. These analyses may reveal individual genes critical to the structural integrity of the cochlea and/or gene pathways capable of buffering the cochlea from acoustic trauma.
Acknowledgments
This work was supported by grants from the NIH including R21DC04983 (SGK), R01DC06305 (BLT), R01DC08577 (SGK), R01DC02739 (BLT), R01DC00188 (MCL), P30DC04661 (BLT), P30DC05209 (MCL), R01GM074244 (KWB), and DoD DM102092 (BLT). Additional support provided by an NSF Graduate Research Fellowship (AM) and a Howard Hughes Medical Institute Medical Research Fellow (AJI).
Conflict of Interest
The authors have declared that no competing interests exist.
Footnotes
Valerie A. Street and Sharon G. Kujawa contributed equally to this work.
Contributor Information
Valerie A. Street, Email: vastreet@uw.edu
Sharon G. Kujawa, Email: Sharon_Kujawa@meei.harvard.edu
Ani Manichaikul, Email: am3xa@eservices.virginia.edu.
Karl W. Broman, Email: kbroman@biostat.wisc.edu
Jeremy C. Kallman, Email: jeremyk44@gmail.com
Dustin J. Shilling, Email: dustinje@mail.med.upenn.edu
Ayaka J. Iwata, Email: iwataa01@uw.edu
Linda C. Robinson, Email: lindarob@u.washington.edu
Carol A. Robbins, Email: crobbins@u.washington.edu
Jin Li, Email: jliyan@u.washington.edu.
M. Charles Liberman, Email: charles_liberman@meei.harvard.edu.
Bruce L Tempel, Phone: (206) 616-4693, Email: bltempel@uw.edu.
References
- Abi-Hachem RN, Zine A, Van De Water TR. The injured cochlea as a target for inflammatory processes, initiation of cell death pathways and application of related otoprotectives strategies. Recent Pat CNS Drug Discov. 2010;5:147–163. doi: 10.2174/157488910791213121. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Candreia C, Martin GK, Stagner BB, Lonsbury-Martin BL. Distortion product otoacoustic emissions show exceptional resistance to noise exposure in MOLF/Ei mice. Hear Res. 2004;194:109–117. doi: 10.1016/j.heares.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetoni AR, De Bartolo P, Eramo SL, Rolesi R, Paciello F, Bergamini C, Fato R, Paludetti G, Petrosini L, Troiani D. Noise-induced hearing loss (NIHL) as a target of oxidative stress-mediated damage: cochlear and cortical responses after an increase in antioxidant defense. J Neurosci: Off J Soc Neurosci. 2013;33:4011–4023. doi: 10.1523/JNEUROSCI.2282-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty JP, Fairfield HE, Spruce CA, McCarty CM, Bergstrom DE. Molecular characterization of an allelic series of mutations in the mouse Nox3 gene. Mamm Genome: Off J Int Mamm Genome Soc. 2011;22:156–169. doi: 10.1007/s00335-010-9309-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- Henderson D, Bielefeld EC, Harris KC, Hu BH. The role of oxidative stress in noise-induced hearing loss. Ear Hear. 2006;27:1–19. doi: 10.1097/01.aud.0000191942.36672.f3. [DOI] [PubMed] [Google Scholar]
- Hirose K, Liberman MC. Lateral wall histopathology and endocochlear potential in the noise-damaged mouse cochlea. J Assoc Res Otolaryngol: JARO. 2003;4:339–352. doi: 10.1007/s10162-002-3036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K, Discolo CM, Keasler JR, Ransohoff R. Mononuclear phagocytes migrate into the murine cochlea after acoustic trauma. J Comp Neurol. 2005;489:180–194. doi: 10.1002/cne.20619. [DOI] [PubMed] [Google Scholar]
- Johnson K (2014) Hereditary hearing impairment in mice. In. http://hearingimpairment.jax.org/index.html: The Jackson Laboratory. Accessed 20 January, 2014
- Kiss PJ, Knisz J, Zhang Y, Baltrusaitis J, Sigmund CD, Thalmann R, Smith RJ, Verpy E, Banfi B. Inactivation of NADPH oxidase organizer 1 results in severe imbalance. Curr Biol: CB. 2006;16:208–213. doi: 10.1016/j.cub.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Kujawa SG, Liberman MC. Conditioning-related protection from acoustic injury: effects of chronic deefferentation and sham surgery. J Neurophysiol. 1997;78:3095–3106. doi: 10.1152/jn.1997.78.6.3095. [DOI] [PubMed] [Google Scholar]
- Kujawa SG, Liberman MC. Adding insult to injury: cochlear nerve degeneration after "temporary" noise-induced hearing loss. J Neurosci: Off J Soc Neurosci. 2009;29:14077–14085. doi: 10.1523/JNEUROSCI.2845-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujawa SG, Liberman MC, Wood ML, Tempel BL (2007) Noise-induced hearing loss variation within and between inbred strains of mice. In: Association for Research in Otolaryngology, Abstract 637
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Cheng X, Li Y, Zeng L, Zhao Y. Evaluation of individual susceptibility to noise-induced hearing loss in textile workers in China. Arch Environ Occup Health. 2005;60:287–294. doi: 10.3200/AEOH.60.6.287-294. [DOI] [PubMed] [Google Scholar]
- Manji SS, Williams LH, Miller KA, Ooms LM, Bahlo M, Mitchell CA, Dahl HH. A mutation in synaptojanin 2 causes progressive hearing loss in the ENU-mutagenised mouse strain Mozart. PLoS One. 2011;6:e17607. doi: 10.1371/journal.pone.0017607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T, Erven AE, Spiden SL, Guenet JL, Steel KP. Two quantitative trait loci affecting progressive hearing loss in 101/H mice. Mamm Genome: Off J Int Mamm Genome Soc. 2006;17:841–850. doi: 10.1007/s00335-004-2438-5. [DOI] [PubMed] [Google Scholar]
- Morita Y, Hirokawa S, Kikkawa Y, Nomura T, Yonekawa H, Shiroishi T, Takahashi S, Kominami R. Fine mapping of Ahl3 affecting both age-related and noise-induced hearing loss. Biochem Biophys Res Commun. 2007;355:117–121. doi: 10.1016/j.bbrc.2007.01.115. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Kim SH, Kim HM, Sanneman JD, Zhang Y, Smith RJ, Marcus DC, Wangemann P, Nessler RA, Banfi B. A claudin-9-based ion permeability barrier is essential for hearing. PLoS Genet. 2009;5:e1000610. doi: 10.1371/journal.pgen.1000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DI, Nelson RY, Concha-Barrientos M, Fingerhut M. The global burden of occupational noise-induced hearing loss. Am J Ind Med. 2005;48:446–458. doi: 10.1002/ajim.20223. [DOI] [PubMed] [Google Scholar]
- Nemoto M, Morita Y, Mishima Y, Takahashi S, Nomura T, Ushiki T, Shiroishi T, Kikkawa Y, Yonekawa H, Kominami R. Ahl3, a third locus on mouse chromosome 17 affecting age-related hearing loss. Biochem Biophys Res Commun. 2004;324:1283–1288. doi: 10.1016/j.bbrc.2004.09.186. [DOI] [PubMed] [Google Scholar]
- NIDCD (2000) Have wise ears for life. In: NIH Publication No. 00–4848. http://www.nidcd.nih.gov/health/hearing/pages/wiseears.aspx: National Institute on Deafness and other Communication Disorders
- Noben-Trauth K, Johnson KR. Inheritance patterns of progressive hearing loss in laboratory strains of mice. Brain Res. 2009;1277:42–51. doi: 10.1016/j.brainres.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Yamasoba T, Schacht J, Miller JM. Glutathione limits noise-induced hearing loss. Hear Res. 2000;146:28–34. doi: 10.1016/S0378-5955(00)00096-4. [DOI] [PubMed] [Google Scholar]
- Ohlemiller KK, Rybak Rice ME, Rellinger EA, Ortmann AJ. Divergence of noise vulnerability in cochleae of young CBA/J and CBA/CaJ mice. Hear Res. 2011;272:13–20. doi: 10.1016/j.heares.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppi M, Kujawa SG, Sewell WF. A corticosteroid-responsive transcription factor, promyelocytic leukemia zinc finger protein, mediates protection of the cochlea from acoustic trauma. J Neurosci: Off J Soc Neurosci. 2011;31:735–741. doi: 10.1523/JNEUROSCI.3955-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz PM (2012) The public health signficance of noise-induced hearing loss. In: Noise-Induced Hearing Loss: Scientific Advances, Springer Handbook of Auditory Research (Le Prell CG, Henderson D, Fay RR, Popper AN (ed), Springer Science and Business Media, LLC, pp 13–25
- Reed SC. The inheritance and expression of fused, a new mutation in the house mouse. Genetics. 1937;22:1–13. doi: 10.1093/genetics/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautter NB, Shick EH, Ransohoff RM, Charo IF, Hirose K. CC chemokine receptor 2 is protective against noise-induced hair cell death: studies in CX3CR1(+/GFP) mice. J Assoc Res Otolaryngol: JARO. 2006;7:361–372. doi: 10.1007/s10162-006-0051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacht J, Altschuler R, Burke DT, Chen S, Dolan D, Galecki AT, Kohrman D, Miller RA. Alleles that modulate late life hearing in genetically heterogeneous mice. Neurobiol Aging. 2012;33(1842):e1815–e1829. doi: 10.1016/j.neurobiolaging.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JB, Longo-Guess CM, Gagnon LH, Saylor KW, Dumont RA, Spinelli KJ, Pagana JM, Wilmarth PA, David LL, Gillespie PG, Johnson KR. The R109H variant of fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of DBA/2 J mice. J Neurosci: Off J Soc Neurosci. 2010;30:9683–9694. doi: 10.1523/JNEUROSCI.1541-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liberman MC. Restraint stress and protection from acoustic injury in mice. Hear Res. 2002;165:96–102. doi: 10.1016/S0378-5955(02)00289-7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hirose K, Liberman MC. Dynamics of noise-induced cellular injury and repair in the mouse cochlea. J Assoc Res Otolaryngol: JARO. 2002;3:248–268. doi: 10.1007/s101620020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Atchley WR. Mapping quantitative trait loci for complex binary diseases using line crosses. Genetics. 1996;143:1417–1424. doi: 10.1093/genetics/143.3.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba T, Schacht J, Shoji F, Miller JM. Attenuation of cochlear damage from noise trauma by an iron chelator, a free radical scavenger and glial cell line-derived neurotrophic factor in vivo. Brain Res. 1999;815:317–325. doi: 10.1016/S0006-8993(98)01100-7. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Hequembourg SJ, Atencio CA, Rosowski JJ, Liberman MC. Acoustic injury in mice: 129/SvEv is exceptionally resistant to noise-induced hearing loss. Hear Res. 2000;141:97–106. doi: 10.1016/S0378-5955(99)00210-5. [DOI] [PubMed] [Google Scholar]
- Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res. 1999;130:94–107. doi: 10.1016/S0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]