Abstract
Background
RNA interference (RNAi) is an evolutionarily conserved process that functions to inhibit gene expression. The use of RNAi in mammals as a tool to study gene function has rapidly developed in the last couple of years since the discovery that the function-inhibiting units of RNAi are short 21–25 nt double-stranded RNAs (siRNAs) derived from their longer template. The use of siRNAs allows for gene-specific knock-down without induction of the non-specific interferon response in mammalian cells. Multiple systems have been developed to introduce siRNAs into mammals. One of the most appealing of these techniques is the use of vectors containing polymerase III promoters to drive expression of hairpin siRNAs. However, there are multiple limitations to using hairpin siRNA vectors including the observation that some are unstable in bacteria and are difficult to sequence.
Results
To circumvent the limitation of hairpin siRNA vectors we have developed a convergent opposing siRNA expression system called pHippy. We have generated pHippy vectors or expression cassettes that knock down the expression of both reporter and endogenous genes. As a proof of principle that pHippy can be used to generate random siRNA libraries, we generated a small siRNA library against PGL3 luciferase and demonstrated that we could recover functional siRNAs that knock down PGL3 luciferase.
Conclusions
siRNA is a powerful tool to study gene function. We have developed a new vector with opposing convergent promoters for the expression of siRNAs, which can be used to knock down endogenous genes in a high throughput manner or to perform functional screening with random or cDNA-derived siRNA libraries.
Background
RNA interference is an evolutionarily conserved process that functions to inhibit gene expression [1,2]. The phenomenon of RNAi was first described by Fire et al. six years ago. In seminal experiments they injected double stranded RNA (dsRNA) into Caenorhabditis elegans, which led to efficient sequence-specific gene silencing of the mRNA that was complimentary to the dsRNA [3]. RNAi has also been described in plants as a phenomenon called post-transcriptional gene silencing (PTGS) which is likely used as a viral defense mechanism [4-6]. Introduction of long dsRNA into a variety of organisms such as Drosophila, Trypanosoma, and pre-implanted mouse oocytes has been shown to specifically inhibit the complimentary mRNA [5-7]. However, in somatic mammalian cells long dsRNA also induces the interferon response, which globally inhibits translation by induction of the kinase, PKR, and 2', 5'-oligoadenylate synthetase. This has limited the use of RNAi as a tool to study gene function in mammalian cells [8,9].
The first indication that the molecules that regulate PTGS were short RNAs processed from longer dsRNA was the identification of short 21 nt to 22 nt dsRNA derived from the longer dsRNA in plants [6]. This observation was recapitulated in Drosophila embryo extracts where long dsRNA was found processed into 21–25 nt short RNA by the RNase III type enzyme, Dicer [9-11]. These observations led Elbashir et al. to test if synthetic 21–25 nt synthetic dsRNAs function to specifically inhibit gene expression in Drosophila embryo lysates and mammalian cell culture [9-11]. They demonstrated that siRNA had the ability to specifically inhibit gene expression in mammalian cell culture without induction of the interferon response. These observations led to the development of many techniques for the specific knockdown of genes in mammalian cell culture.
Of these techniques, plasmid-based systems that generate hairpin siRNAs are very appealing [12-15]. These vectors are fairly inexpensive and have been shown to inhibit multiple genes both transiently and in long-term experiments. However, hairpin vectors suffer from multiple limitations. Hairpins can be hard to synthesize in bacteria, difficult to sequence, and the oligos needed to generate them can be costly and error-prone [14,16]. In addition, the hairpin length and sequence can affect the ability of the siRNA to inhibit gene expression [12,17]. Although cDNA-based siRNA libraries have been generated in hairpin vectors, generation of these libraries is technically difficult and heterologous hairpin loops have to be added to the siRNA [18,19]. The latter may increase off-target effects of the siRNAs generated from these strategies.
To circumvent the limitations of hairpin siRNA expression vectors, we have generated a siRNA expression vector, which expresses siRNAs from convergent opposing promoters. We have named this vector "HI inverted U6 promoter plasmid," or pHippy. Short double-stranded oligonucleotides can be easily and efficiently introduced into this vector to knock down any mRNA, or siRNA expression cassettes can be generated in a high throughput manner by PCR. We demonstrate that pHippy can generate functional siRNAs in mammalian cells that knock down the expression of both ectopic reporters and endogenous genes. Because both strands of the siRNA are transcribed from the same template DNA, random or cDNA-derived siRNA libraries can easily be generated in pHippy. As a proof of principle we generated a random pool of siRNAs against PGL3 luciferase and screened this library for silencing activity. This library was deconvoluted to identify functional vectors that have the original siRNA sequence against PGL3 luciferase. Although this is only a partial random library to a previous efficacious siRNA, this demonstrates that random siRNA libraries can be used to functionally identify genes.
Results and discussion
Design of an opposing convergent siRNA expression vector, pHippy
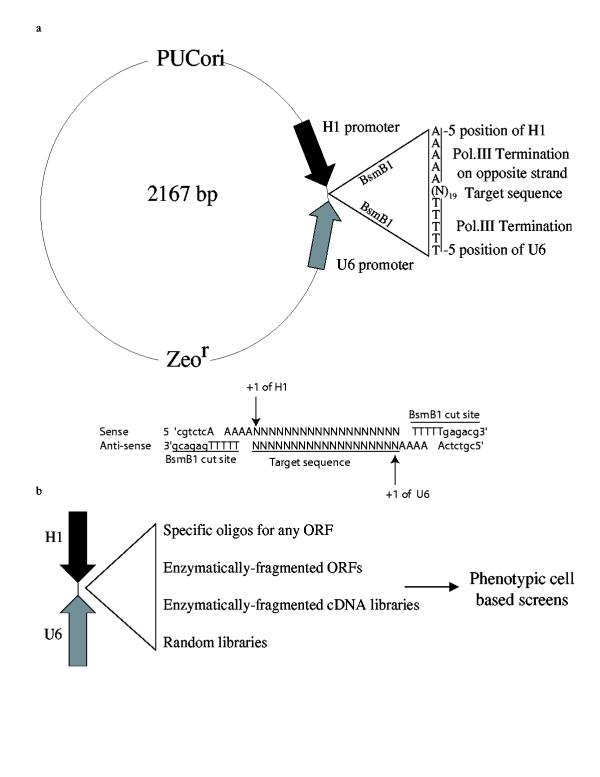
pHippy contains two opposing polymerase III (pol. III) promoters to drive the expression of both strands of a template DNA cloned in between the promoters. To circumvent generating an inverted repeat, which can cause plasmid instability in E. coli, the human H1 and human U6 promoters were used instead of two H1 or U6 promoters. Both the H1 and U6 promoter were modified to contain a five Thymidine pol. III termination sequence at the -5 to -1 position and a BsmBl site at the -12 to -6 position (Fig. 1a). This does not appear to affect the ability of either promoter to drive expression of RNA nor does it appear to affect the transcriptional start site. The BsmBl cuts 5 bases downstream of its recognition site and in the case of pHippy will leave 4 Thymidine 3' overhangs at -1 to -4 of both U6 and H1 promoters. Digestion of pHippy with BsmB1 renders the two ends incompatible for ligation and hence gives a very low level of background when ligated. More than 25 double stranded oligos have been cloned into pHippy with nearly 100% cloning efficiency (data not shown). The target sequence for any mRNA can be cloned into pHippy and the DNA will be transcribed from both strands and form a double stranded RNA with two 5'Uridine overhangs, which closely resembles functional siRNA produced by Dicer [11,12,20,21]. This vector system can be used to generate siRNA to target any transcript or open reading frame (ORF) (Fig. 1b). Pools of siRNAs for specific mRNAs can be generated by restriction enzyme or DNase digestion of a cDNA for any gene and subsequent cloning of the small fragments into pHippy (Fig. 1b). This circumvents the need to find specific targets, which can be time consuming and costly. In addition, libraries of siRNAs can be generated by restriction enzyme or DNase digestion of cDNA libraries or by cloning of random oligos into pHippy (Fig. 1b). These libraries can be used to screen populations of transfected cells for phenotypic changes and the cells with the desired phenotype can be recovered, allowing the siRNA construct to be rescued and characterized. This type of screen allows an unbiased means to identify genes involved in diverse biological processes.
Figure 1.
Depiction of the pHippy dual siRNA expression vector and some of its possible uses. a, pHippy has convergent opposing Human H1 and U6 polymerase III promoters that drive expression of both strands of any template cloned in between the BsmB 1 cloning sites. pHippy also contains the Puc origin and the Zeocin-resistance gene for propagation and replication in bacteria. As depicted the H1 and U6 promoters were modified to contain a polymerase III termination signal (TTTTT) between the -5 to -1 position of the promoter, and BsmBl sites. BsmBl is a type II restriction enzyme, which cuts outside of its recognition sequence and will in the case of pHippy leaves 3' TTTT overhangs on both strands of the plasmid as depicted. 19 nt target siRNA can be cloned into pHippy as double stranded oligos by addition of AAAA to the 5' ends of the oligos as depicted b. b, Proposed uses for the pHippy siRNA vector system.
pHippy efficiently inhibits the ectopic expression of reporter genes
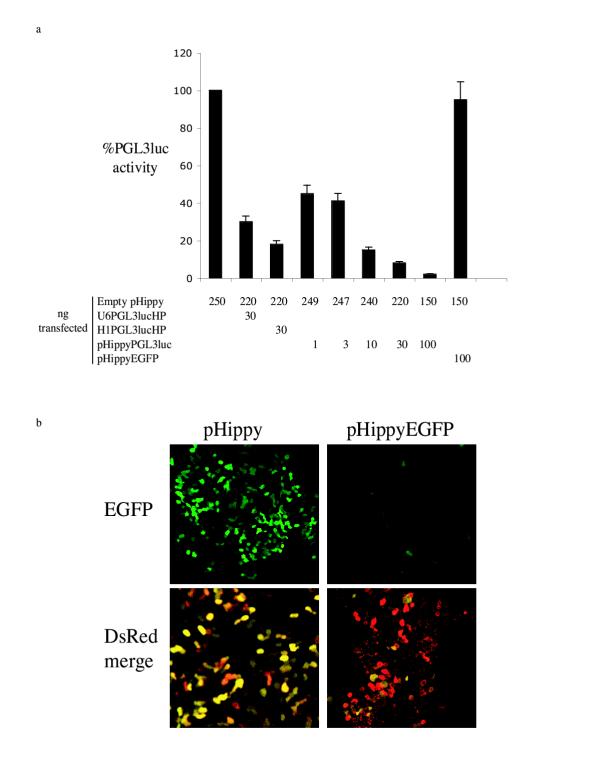
To determine if pHippy produces functional siRNAs, we generated a pHippy vector against PGL3 luciferase (pHippyPGL3luc) from a previously reported synthetic siRNA directed against PGL3 luciferase [9]. Using the same target sequence against PGL3 luciferase, we made two hairpin siRNA vectors, with one driven from the U6 promoter (U6PGL31ucHP) and one driven from the H1 promoter (HIPGL3lucHP). To determine the efficiency of inhibition of the PGL3 luciferase target by the various siRNA vectors, 293T cells were transfected with a cocktail of PGL3 luciferase, Renilla luciferase, and a siRNA vector driving a hairpin or a pHippy siRNA. After normalization for transfection with Renilla luciferase, the empty pHippy vector gave similar luciferase levels as cells transfected with luciferase alone, and this level was set to 100% luciferase activity (Fig 2a). Both of the vectors that express hairpin RNAs against PGL3 luciferase significantly inhibited luciferase expression (Fig. 2a). The pHippy vector with the target sequence against PGL3 luciferase also inhibited expression of PGL3 luciferase. Moreover, at the same concentration of DNA transfected, pHippyPGL31uc is more efficient at inhibiting Luciferase expression than either of the hairpin vectors. Specifically, at 30 ng of DNA transfected PHippyPGL31uc is 2–4 times more efficient than inhibition of PGL3 luciferase by the vectors that generate hairpin RNAs. Although pHippy-generated siRNA appears to be more efficient than hairpin generated siRNA, a more detailed side by side comparison would have to be conducted to establish specific activities.
Figure 2.
a, Luciferase assay to determine the efficiency of target suppression by the pHippy vector. Hairpin siRNA expression vectors were generated against PGL3 luciferase with either the U6 (U6GPL31ucHP) or H1 (HlPGL31ucHP) promoters. For comparison, pHippy siRNA vectors were generated against PGL3 luciferase (pHippyPGL31uc) or against EGFP (pHippyEGFP). 293T cells were transfected with the constructs shown and assayed for luciferase activity 24 hours later. The absolute levels of PGL3 luciferase were ~100,000 relative light units (RLUs) which was set at 100%. All experiments were normalized for transfection efficiency with an expression vector for Renilla luciferase. The average normalized PGL3 luciferase levels and standard deviation are shown for 3 experiments. b, EGFP fluorescent assay to monitor the suppression of protein expression by pHippyEGFP. 293T cells were co-transfected with empty pHippy or pHippyEGFP and expression vectors for DsRED and EGFP. Confocal image of EGFP and EGFP merged with DsRED are shown. One representative experiment is shown.
To determine if pHippy generates sequence-specific inhibitory siRNAs, a vector was designed against EGFP. pHippyEGFP did not inhibit the expression of PGL3 Luciferase (Fig. 2a). However, it inhibited the expression of EGFP (Fig. 2b). This not only establishes that the inhibition of expression of EGFP and Luciferase is gene-specific, but also demonstrates that pHippy can theoretically be used to knock down the expression of any gene. Supportingly, to date we have knocked down expression of more than 10 unique genes with pHippy (data not shown). We have not observed off-target effect of these siRNAs, though we have not systematically screened for such potential effects. We speculate that siRNAs generated by pHippy are less likely to generate off-target effects because there are no heterologous sequences included in the siRNA, such as a hairpin loop. On average we have found that 3–4 constructs with unique target sequences have to be tested to obtain functional siRNA, which is very similar to other siRNA systems.
pHippy efficiently inhibits the expression of endogenous genes
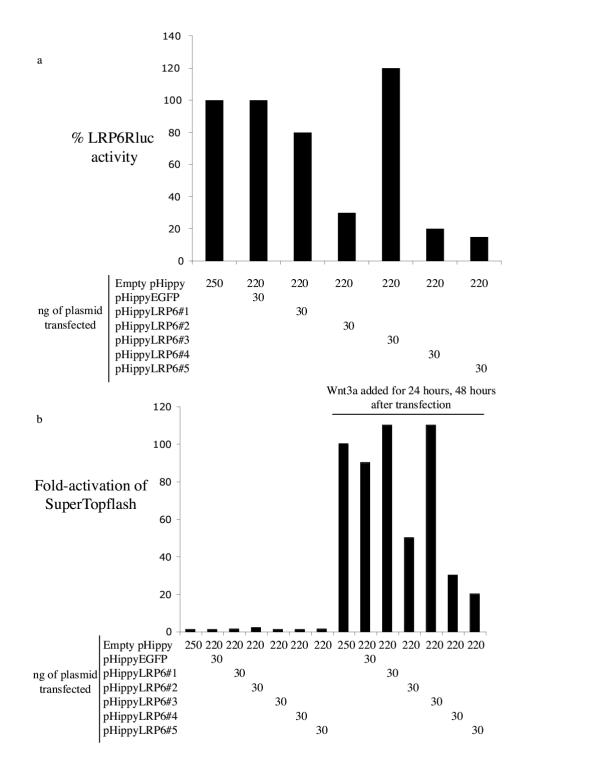
To determine if pHippy can be used to inhibit the expression of endogenous genes, five pHippy constructs were generated against the human Low-density Lipoprotein Receptor-related Protein 6 (LRP6). To first demonstrate that the constructs could inhibit expression of ectopic LRP6, they were screened for their ability to inhibit expression of a fusion protein consisting of LPR6 and Renilla luciferase (LRP6Rluc). This fusion of a target gene to Renilla luciferase allows for rapid quantitative assessment of the efficiency of any given siRNA construct. Three of the five pHippy constructs generated against LRP6 inhibit LRP6Rluc expression by more than 50% (Fig. 3a). To test if expression of endogenous LRP6 was also inhibited, we determined if any of the pHippy constructs altered an indirect readout of the biological function of LRP6. Briefly, LRP6 is part of the Wnt receptor signaling complex, and it is required to receive and transduce Wnt signaling to downstream components in the Wnt signaling cascade [22]. Thus, the biological function of LRP6 culminates in the activation of β-catenin-mediated transcription, which can be efficiently measured by activation of the Super(8X)Topflash reporter [23]. To test if LRP6 is required for Wnt signal transduction in human cells, 293T cells were co-transfected with a cocktail of Super(8X)Topflash, Renilla luciferase, and the five pHippy constructs directed against LRP6 or control pHippy constructs. 24 hours after transfection the cells were treated with Wnt3a-conditioned media to activate Wnt signaling and thus the expression of Super(8X)Topflash, then cultured for an additional 24 hours. After normalization for transfection with Renilla luciferase, the cells transfected with empty pHippy or pHippyEGFP were set to 1 fold activation (Fig. 3b). Treatment with Wnt3a increases the reporter activation about 100-fold (Fig. 3b). Co-transfection of some of the pHippyLPR6 constructs inhibited Wnt3a activation of Super(8X)Topflash more than 50% (Fig 3b). Strikingly, there was a strict correlation between the ability of the pHippyLRP6 constructs to inhibit the expression of LRP6Rluc and the ability to inhibit Wnt3a activation of Super(8X)Topflash (Fig. 3a,3b). These experiments demonstrate that pHippy constructs have the ability to inhibit the expression and thus the biological function of endogenous genes.
Figure 3.
Assays to determine if pHippy can be used to inhibit endogenous genes. a, Renilla luciferase assays to determine if any of 5 unique pHippy constructs against Human LRP6 inhibit activity of a fusion of LRP6 to Renilla luciferase. 293T were co-transfected with the vectors shown and assayed for luciferase activity 48 hours later. Cells co-transfected with LRP6Rluc and empty pHippy were set to 100%. All experiments were normalized for transfection with PGL3 luciferase. The average normalized Renilla luciferase levels and standard deviation are shown for 3 experiments. b, Determination of whether inhibition of endogenous LRP6 inhibits Wnt3a activation of the Wnt/β-catenin signaling pathway. 293T cells were co-transfected with the vectors shown, Super(8X)Topflash (to measure Wnt/β-catenin-signaling), and a constitutive expression vector for Renilla luciferase. 24 hours after transfection the cells were treated with Wnt3a conditioned media for 24 hours and then assayed for luciferase activity. Untreated cells transfected with empty vector were set to 1 fold activation of Super(8X)Topflash, which corresponds to ~10,000 RLUs. Cells treated with Wnt3a induce about 100 fold-activation of luciferase as shown. All experiments were normalized for transfection efficiency with the Renilla luciferase expression vector. One representative experiment is shown.
pHippy expression cassettes can easily be generated in a high throughput manner by PCR
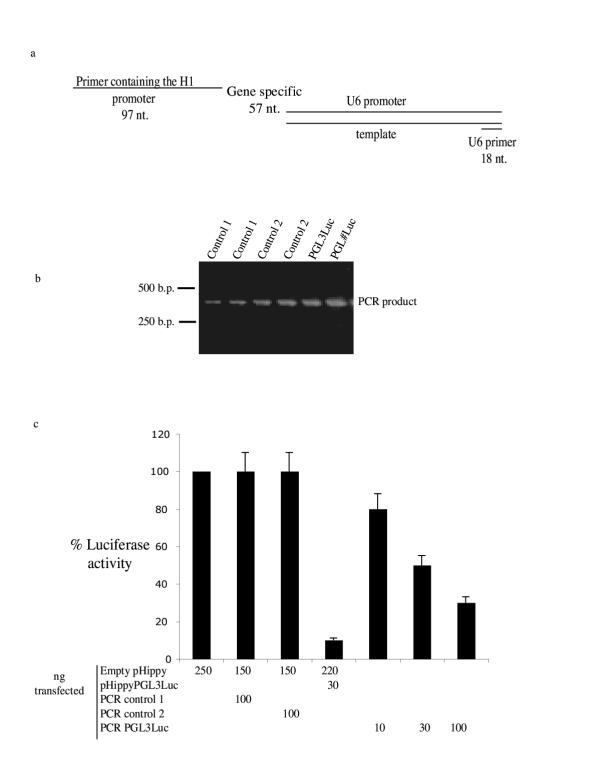
To generate pHippy siRNA constructs in a rapid manner, a PCR method was devised that incorporates the U6 and H1 promoters from pHippy on either end of a PCR product. A gene-specific primer for a target gene can be sandwiched between the two convergent promoters. To develop this system three oligonucleotides were synthesized: 1) a 97 nt primer consisting of the entire H1 promoter from pHippy, 2) a gene-specific primer containing 18 nt of complimentary sequence to both the H1 and U6 promoters and 21 nt of gene-specific (PGL3 luciferase) or random control sequences, and 3) an 18 nt anti-sense primer to the human U6 promoter (Fig. 4a). After PCR with H1, U6, and limiting amounts of the gene-specific primer using a U6 template, a single robust band was generated for all of the different gene-specific primers of the appropriate size (Fig. 4b). These PCR products were cleaned with a nucleospin column and co-transfected into 293T cells with the PGL3 luciferase reporter and Renilla luciferase as a normalization control. Neither of the control PCR products significantly inhibited PGL3 luciferase activity (Fig. 4c). However, the PCR product generated against PGL3 luciferase inhibited luciferase in a dose-dependent manner (Fig. 4c). Although the pHippy PCR cassette against PGL3 luciferase inhibited PGL3 luciferase expression, it was not as efficient as the pHippy vector directed against PGL3 luciferase (Fig. 4c). This may be due to faster degradation of the PCR product and might be circumvented by additional sequences on the 5' and 3' ends of the PCR product.
Figure 4.
A high throughput system to generate pHippy expression cassettes. a, A multiple primer PCR system was used to generate PCR products that express siRNA after transfection into cells. A 5' primer (U6 primer) was designed to human U6. A second primer (gene specific) was designed with sequences that are complementary to the 3' end of the human 6 promoter, the target siRNA sequence, and a sequence that is complementary to the 3' of the human H1 promoter. A third primer was designed that contains the human H1 promoter and is complementary to the gene specific primer. b, An agarose gel stained with ethidium bromide demonstrates the robustness of the PCR method for several different gene specific primers. c, A luciferase assay as described in the legend to Figure 3b to determine if the PCR products inhibit PGL3 luciferase activity.
Generation of random siRNA-libraries in pHippy
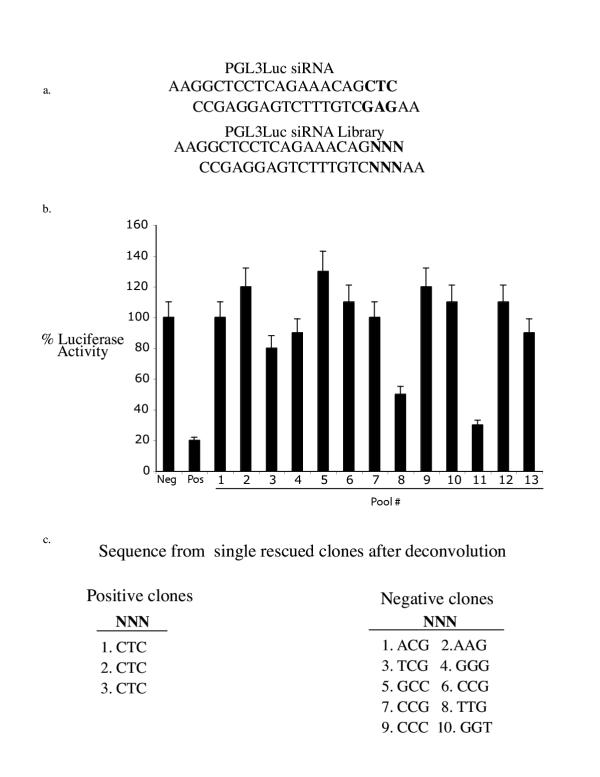
The pHippy system is well-suited for generation of cDNA or random siRNA libraries because both strands of DNA template are transcribed to generate siRNA. To determine if pHippy could in principle be used for a random screen, a random library of siRNAs based on PGL3 luciferase was generated (Fig. 5a). This library was generated by randomizing 3 nt in the functional PGL3 siRNA described in Figure 2 and corresponds to a library of 64 possible siRNAs. To determine if this library could be screened to recover siRNA activity, 130 randomly chosen clones from E. coli containing the library were picked and pooled in groups of 10. These 13 pools were screened for their ability to inhibit PGL3 luciferase activity. The library consists of a maximum of 64 possible siRNAs and only 2 of the pools from the 130 clones would be predicted to inhibit PGL3 luciferase activity. In agreement with this calculation, pools 8 and 11 had significant inhibitory activity (Fig. 5b). These pools of 10 were further reduced to single clones and each individual clone was re-screened for inhibitory activity. Pool 8 contained one clone that had inhibitory activity and pool 11 contained two clones with inhibitory activity. Sequencing of these three clones revealed that these clones contained the original sequence against PGL3 (Fig 5c). In contrast, 10 clones that did not inhibit PGL3 luciferase activity had random sequences at the mutant positions (Fig. 5c).
Figure 5.
A siRNA screen was conducted with a small siRNA library against PGL3 luciferase. a, Depiction of the small siRNA library against PGL3 luciferase. The target sequence for the siRNA against PGL3 siRNA that was tested in Figure 3b is shown. The bases in bold were randomized by synthesizing an oligo with random bases in place of CTC and cloned into the pHippy vector. b, Screening of the siRNA library. 12 pools of 10 bacterial colonies from the random siRNA library against PGL3 luciferase were screened for their ability to inhibit PGL3 luciferase activity as described in the legend to Figure 3b. c, The sequences of the siRNA single clones, which did or did not inhibit PGL3 luciferase. Pools 8 and 11 were broken down into individual clones and re-screened for inhibitory activity. Pool 8 contained 1 positive clone and Pool 11 contained 2 positive clones. The sequences of all three positive clones is shown on the left and corresponds to the original PGL3 siRNA. The sequences of 10 negative clones are shown on the right.
These experiments demonstrate that the pHippy system can be used for random siRNA screens. Specifically, libraries can be generated where all of the 21 nt of the siRNA are random. This library would encompass multiple targets in every gene in the human genome and could be used for phenotypic single cell assays to identify genes required for the screened phenotype, without first knowing the siRNA sequence. For instance, a random siRNA library could be used to identify genes required for Wnt signaling by screening for siRNAs that inhibit the ability of Wnt to activate Super(8X)Topflash.
Conclusions
pHippy is an siRNA expression vector, which utilizes convergent promoters to generate functional siRNAs by driving expression from both strands of the same template DNA. In tests to date, pHippy vectors directed against PGL3 luciferase knock down the expression of luciferase better than hairpin RNAs. This is likely because pHippy produces functional siRNA that do not need to be cleaved by Dicer. In addition, pHippy vectors against any target gene are easier to generate than with hairpin vectors. Specifically, the oligos needed to generate pHippy siRNA vectors are shorter and more cost effective, and the final clones are easier to identify and sequence. pHippy expression cassettes can be generated in a high throughput manner by PCR without the need to propagate through bacteria.
We have used pHippy to knock out the expression of an endogenous Wnt pathway co-receptor, as scored by measuring effects on a Wnt pathway reporter, thereby validating this approach for endogenous targets. siRNAs synthesized from pHippy are predicted to have fewer off-target effects than some siRNA approaches since pHippy-derived siRNAs have no extraneous nucleotides, a feature one might design into vectors developed with gene therapy as a goal.
One of the biggest advantages is that pHippy can be used to generate random and cDNA siRNA expression libraries, which can be used to identify genes involved in any biological phenomena that can be screened by phenotype. As a proof of principle we screened a small random siRNA library for siRNAs that inhibit PGL3 luciferase activity, and were able to recover functional siRNAs that knocked down activity of this target. This approach can now be used to identify siRNAs that modulate diverse biological phenomena.
Methods
Cell culture and transfection
293T cells were grown in DMEM supplemented with 10% FBS and 1% Penicillin/Streptomycin under standard conditions. All transfections were performed in 24 well plates with Lipofectamine Plus or 2000 (Invitrogen) according to the manufacturer's specifications.
Confocal microscopy
2.5. × l05 293T cells were seeded in 12 well plates and transfected with 10 ng of pEGFPNl (Clontech), pDSREDNl (Clontech), and 30 ng of the indicated siRNA construct. 24 hours after transfection the cells were removed from the plates with PBS and live cells were sandwiched between cover-slips and glass slides and visualized for fluorescence using the appropriate lasers and filters to visualize EGFP and DSRED.
Reporter assay
Luciferase assays were preformed according to the Dual luciferase assay specifications (Promega). In all cases 293T cells were transfected with 10 ng of CMV-PGL31uciferase and 100 pg of pRLCMV (Promega), and the cells were harvested 24 hours later and assayed for luciferase activity in 96 well plate in a Berthold 96 V luminometer. Super(8X)Topflash reporter assays were performed as described [23]. 293T cells seeded in 24 well plates were transfected with 10 ng of Super(8X)Topflash, 100 pg of pRLCMV (Promega), and the indicated amount of effecter plasmids. The concentrations of all transfections were brought up to a total of 250 ng with the vector CS2+. Assays were performed as described [23] and according to the Dual luciferase assay specifications (Promega).
Multiple primer PCR
pHippy siRNA expression cassettes were generated by a single step multiple primer PCR. In short, a 10 ng of plasmid containing the human U6 promoter was used as template for PCR in a 50 μl reaction containing 2 μl of 10 pm/μl U6 primer, 2 μl of a primer encompassing the entire H1 promoter, 2 μl of 0.01 pm/μl of the gene-specific linker primer, 10 μl of 2 mM DNTPs, 10 μl of advantage buffer, and 0.5 μl tag-advantage (Clontech). The PCR products were generate by 30 cycles of touchdown PCR program that ramped down from 60°C to 50°C.
RNAi design and recombinant DNA constructs
All recombinant constructs were generated by standard recombinant DNA techniques. The sequences of pHippy will be provided upon request. siRNAs were designed using the web based siRNA design program from the Whitehead Institute web page http://jura.wi.mit.edu/pubint/http://iona.wi.mit.edu/siRNAext/home.php. In general, siRNAs were used from previously published siRNAs or were designed by introducing the corresponding sequence for a given gene into the Whitehead siRNA design program using the following pattern AAN19TT or AAGN17CN2. The output sequences for a given gene that were not complimentary to other genes after blasting were chosen, and the corresponding oligos were designed and ordered. Oligos were based on the Whitehead output sequences and modified for cloning into pHippy by addition of 2 or 4 adenines to the 5' end of the sense and antisense versions of the Whitehead output sequences. Oligos used; PGL3luciferase, sense (5'AAaaggctcctcagaaacagctc3'), antisepses (5'AAaagagctgtttctgaggagcc3'): EGFP, sense (5' AAaagcaagctgaccctgaagttcat3'), antisense (5' AAaaatgaacttcagggtcagcttgc3'): LRP6#1 sense (5'AAaaaggttcccttccacatcct3'), antisense (5'AAaaaggatgtggaagggaacct3'): LRP6#2 sense (5'AAaaaaggttcccttccacatccttt3'), antisense (5'AAaaaaggatgtggaagggaaccttt3'): LRP6#3 sense (5'AAaagaagatggcagccagggct3'), antisense (5'AAaaagccctggctgccatcttc3'): LRP6#4 sense (5'AAaaggcacttacttccctgcaa3'), antisense (5'AAaattgcagggaagtaagtgcc3'): LRP6#5 sense (5'AAaaaaggcacttacttccctgcaatt3'), antisense (5'AAaaaattgcagggaagtaagtgcctt3').
Authors' contributions
AK carried out all of the experiments and drafted the manuscript, and RTM participated in the design and coordination of the research and the preparation and revision of the manuscript. Both authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Bryan White and Stephane Angers for comments on the manuscript. AK is an Associate, and RTM an Investigator, of the HHMI, which supported this work.
Contributor Information
Ajamete Kaykas, Email: akaykas@u.washington.edu.
Randall T Moon, Email: rtmoon@u.washington.edu.
References
- Bernstein E, Denli AM, Hannon GJ. The rest is silence. RNA. 2001;7:1509–21. [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–67. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990; 8:340–4. doi: 10.1016/0167-7799(90)90220-R. [DOI] [PubMed] [Google Scholar]
- Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. Embo J. 1998;17:6739–46. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–2. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell. 1998;95:461–70. doi: 10.1016/S0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. Embo J. 2001;20:6877–88. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Gaudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–58. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99:1443–8. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20:505–8. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Esposito D, Gillette WK, Hartley JL. Blocking oligonucleotides improve sequencing through inverted repeats. Biotechniques. 2003;35:914–920. doi: 10.2144/03355bm02. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–7. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane D, Sugao K, Namiki S, Tanabe M, lino M, Hirose K. Enzymatic production of RNAi libraries from cDNAs. Nat Genet. 2004;36:190–6. doi: 10.1038/ng1290. [DOI] [PubMed] [Google Scholar]
- Sen G, Wehrman TS, Myers JW, Blau HM. Restriction enzyme-generated siRNA (REGS) vectors and libraries. Nat Genet. 2004;36:183–9. doi: 10.1038/ng1288. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Tran N, Cairns MJ, Dawes IW, Arndt GM. Expressing functional siRNAs in mammalian cells using convergent transcription. BMC Biotechnol. 2003;3:21. doi: 10.1186/1472-6750-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer L, Varmus H. Wnt/Wingless signaling through beta-catenin requires the function of both LRP/Arrow and frizzled classes of receptors. BMC Cell Biol. 2003;4:4. doi: 10.1186/1471-2121-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaykas A, Yang-Snyder J, Heroux M, Shah KV, Bouvier M, Moon RT. Mutant Frizzled 4 associated with vitreoretinopathy traps wild-type Frizzled in the endoplasmic reticulum by oligomerization. Nat Cell Biol. 2004;6:52–8. doi: 10.1038/ncb1081. [DOI] [PubMed] [Google Scholar]