Abstract
Telomeric RNAs (TERRAs) are UUAGGG repeat-containing RNAs that are transcribed from the subtelomere towards the telomere. The precise genomic origin of TERRA has remained elusive. Using a whole-genome RNA-sequencing approach, we identify novel mouse transcripts arising mainly from the subtelomere of chromosome 18, and to a lesser extend chromosome 9, that resemble TERRA in several key aspects. Those transcripts contain UUAGGG-repeats and are heterogeneous in size, fluctuate in abundance in a TERRA-like manner during the cell cycle, are bound by TERRA RNA-binding proteins and are regulated in a manner similar to TERRA in response to stress and the induction of pluripotency. These transcripts are also found to associate with nearly all chromosome ends and downregulation of the transcripts that originate from chromosome 18 causes a reduction in TERRA abundance. Interestingly, downregulation of either chromosome 18 transcripts or TERRA results in increased number of telomere dysfunction-induced foci, suggesting a protective role at telomeres.
 Telomeric RNAs (TERRAs) are known to be transcribed towards the telomere from subtelomeric regions, however, their precise genomic origins are unclear. Here López de Silanes et al. identify novel transcripts that originate from the subtelomeric region of mouse chromosome 18 and behave as bona fide TERRAs.
Telomeric RNAs (TERRAs) are known to be transcribed towards the telomere from subtelomeric regions, however, their precise genomic origins are unclear. Here López de Silanes et al. identify novel transcripts that originate from the subtelomeric region of mouse chromosome 18 and behave as bona fide TERRAs.
In spite of the heterochromatic nature of telomeres and of the silencing of reporter genes in their vicinity1, telomeres are transcribed2,3. In particular, the telomeric C-strand is transcribed by RNA polymerase II, giving rise to UUAGGG repeat-containing transcripts known as TERRA. TERRAs are thought to be transcribed from the subtelomere towards the telomere2. They are reported to contain a telomeric track, as well as sequences arising from subtelomeric regions, although the full-sequence remains unknown2,3,4. TERRA are nuclear long noncoding RNAs of a heterogeneous length (from 0.2 kb to 9 kb in human and mouse cells)2,3. Although several causes have been postulated to explain this heterogeneity5,6, its origin is still unknown owing to the lack of a full TERRA sequence. RNA-fluorescence in situ hybridization (FISH) experiments have indicated that some of the TERRA transcripts co-localize or associate with telomeres2,3,7,8,9. In addition, TERRA are bound by a complex array of proteins, including components of the shelterin complex such as TRF1 and TRF2, as well as different members of the hnRNP family7,10.
In human cells, a TERRA promoter was described to consist of a 61-29-37 repeat present in 20 different chromosomes4. This promoter is methylated in a DNMT1- and DNMT3b-dependent manner as cells doubly deficient for DNMT1 and DNMT3a/3b display higher TERRA levels4,11. In yeast, a TERRA transcription start site was identified in chromosome 1, although the promoter region was not characterized12. Both in the case of human and yeast promoters, demonstration that transcripts arising from these subtelomeric regions co-localize with canonical TERRA transcripts by RNA-FISH experiments is still lacking. This very limited information on TERRA sequences and promoters represents a difficulty for understanding TERRA role and regulation. In this regard, biochemical in vitro assays suggested that TERRA might regulate telomere replication by coordinating the cell cycle-dependent exchange of the RPA and Pot1 proteins through the sequestration of hnRNP A1 from telomeres13. Recently, TERRA was proposed to be induced by telomere shortening and nucleate telomerase at short telomeres14. TERRA has been also proposed to inhibit telomerase in vitro and to have a role in telomeric heterochromatin3,8,10,15.
In mice, no TERRA promoters have been described to date, which has hampered understanding the role of TERRA in vivo via the generation of genetically modified mouse models. In contrast to humans, mouse TERRA levels are reduced in cells deficient for DNMT1 and DNMT3a/3b, thus anticipating differences with the human promoter3. Here we set to identify the mouse TERRA sequence and promoter. To this end, we performed a whole-subtelomere analysis and identified novel transcripts that resemble TERRA in many different attributes. These transcripts arise predominantly from the subtelomere of chromosome 18. Chromosome 18 transcripts contain UUAGGG-repeats, are found at telomeres with the same frequency than TERRA transcripts, are regulated throughout the cell cycle in a TERRA-like manner, are bound by the same RNA-binding proteins that bind TERRA7, are regulated by stresses in a similar manner to TERRA and are induced upon induction of pluripotency-like TERRA. Chromosome 18 transcripts can bind to chromosome 18 telomeres but also to the rest of the chromosome ends, although not at once. Importantly, downregulation of chromosome 18 transcripts causes downregulation of TERRA. Interestingly, the downregulation of either chromosome 18 transcripts or TERRA results in similar induction of telomere dysfunction-induced foci (TIF). Finally, we identify the promoter region for the transcription of chromosome 18 transcripts resembling TERRA.
Results
Identification of novel transcripts that resemble TERRA
Here we set out to identify the genomic locus of mouse TERRA. To this end, we carried out whole-genome RNA-deep sequencing of six mouse samples. In particular, we used three poly(A)+ RNA samples derived from pMEFs, C2C12 and NS1 cells (in mouse cells, TERRA is enriched around 2.5 times in polyA(+) fractions relative to the polyA (-) (Supplementary Fig. 1A), and the rest of the samples were further enriched in TERRA levels: two by RNA-immunoprecipitation (IP) of hnRNP A1 and HuR (known to bind TERRA7) followed by isolation of the immunoprecipitated RNA, and the remaining sample by biotin pull-down of DNA-free nuclear fractions from pMEFs with a biotinylated antisense TERRA oligo consisting of eight CCCTAA repeats as bait followed by RNA isolation (see Supplementary Fig. 1B for TERRA enrichment in the pull-down with the telomeric oligo compared with a control RNA oligo). RNA from each individual sample was next used for construction of independent sequencing libraries and sequenced (Methods).
We performed quality check of both RNA-seq reads and the alignments. Reads in all samples were enriched in intragenic regions (intronic+exonic) as compared with intergenic regions (Supplementary Fig. 1C). We found enrichment in reads aligned to the PTMA mRNA (a well-established target of HuR16) in the HuR-IP sample compared with the other samples (Supplementary Fig. 1D). Cyclin D2, the mRNA with the greatest enrichment in reads upon hnRNP A1-IP was also enriched in the HuR-IP sample (Supplementary Fig. 1D). In the biotin pull-down sample, we found a 400 thousand-fold increase in the number of reads containing (TTAGGG)4–6 repeats compared with the other samples, validating its enrichment in UUAGGG-containing transcripts (Supplementary Fig. 1E).
As TERRA transcripts are 0.2–10 kb in length and part of this length corresponds to the telomere, we first focused our analysis on a 30-kb region adjacent to the telomere of each chromosome (centromere-adjacent telomeres were not studied because they are not sequenced). As expected, the number of reads aligned in this 30-kb region (from now on ‘subtelomere region’) was not very abundant compared with regions rich-in-annotated coding genes, indicating lower transcription at subtelomeres in accordance with their heterochromatic nature1.
In the latest mouse assembly mm10, most of the chromosomes are sequenced until the telomere. Chromosome 4, 6, X and Y remain to be fully sequenced (Supplementary Fig. 2A) but we found the presence of contiguous TTAGGG-repeats in the 3′ends of chromosomes 6 and X sequences and decided to include them in the screening. Figure 1 shows the RNA-seq read density at the subtelomere of each chromosome from the biotin pull-down sample (the one with higher UUAGGG-read content), as well as from the sum of all samples (see the other individual samples in Supplementary Fig. 2B). Many subtelomeres display little or no transcription independently of how the TERRA enrichment was achieved (for example, chromosomes 1, 2, 4, 5, 7, 12, 14 and 15; Fig. 1a and Supplementary Fig. 2B). Nevertheless, we decided to include these subtelomeres in further analyses in case they could be low contributors of TERRA. We next confirmed the presence of transcription at these subtelomeres by PCR using two sets of primers per subtelomere. We designed primers in regions showing RNA-seq read alignments, prioritizing those regions with reads from the pull-down sample that had the highest UUAGGG-read content. We could not design primers for chromosome 3 because of the presence of a coding gene in the entire region (Wls gene and Supplementary Fig. 2). We did not detect transcription at the subtelomeres of chromosomes 5, 7, 12, 14 and 15, in agreement with the low abundance and scattered RNA-seq reads (Fig. 1a and Supplementary Fig. 2A; Table 1). PCR was also negative for chromosome 13. Detection of RNA-reads in this region may be explained by the presence of an annotated Ref Seq in the 5′-end subtelomeric region (NM_001037925; Fig. 1a and Supplementary Fig. 2A; Table 1). We detected transcription by PCR at the subtelomeres of chromosomes 1 and 2 in spite of their low RNA-reads, as well as at the subtelomeres of chromosomes 6, 8, 9, 10, 11, 16, 17, 18, 19 and X, and selected them for further analysis.
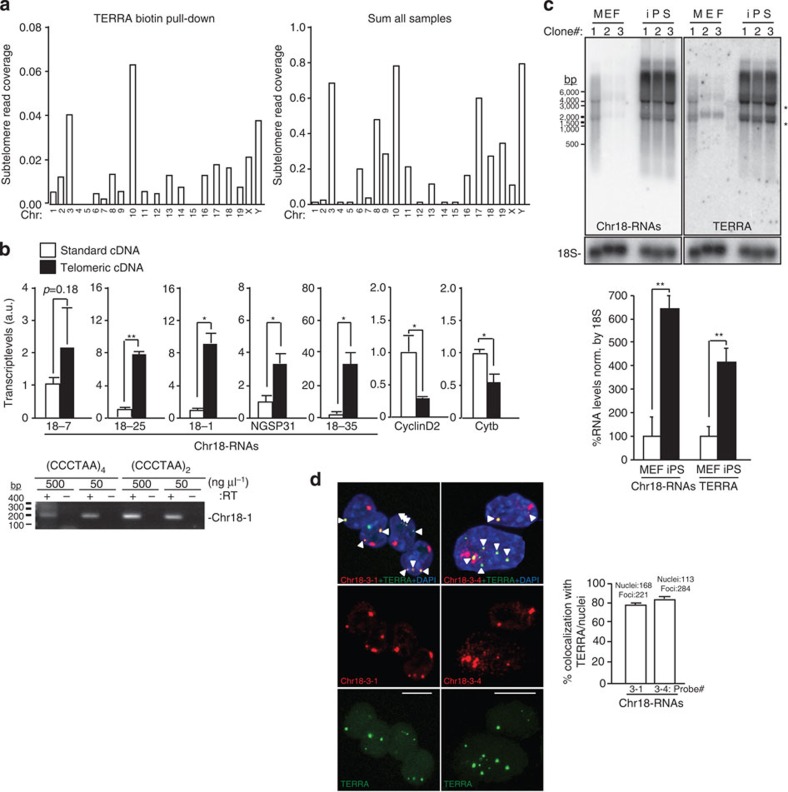
Figure 1. Identification of UUAGGG-containing transcripts that arise from the subtelomere of chromosome 18 that resembles TERRA.
(a) Graphs show the read density corresponding to (left) the TERRA biotin pull-down sample or (right) to the sum of all sample in a 30-kb region adjacent to the telomere of each chromosome. Note that chromosome 4 and Y are not sequenced until the telomere. (b) Reverse-transcribed RNA prepared with either an oligo complementary to the telomeric repeat (4xCCCTAA; ‘telomeric cDNA’) or with random hexamers (‘standard cDNA) was used for quantitative PCR detection of chromosome 18 transcripts and the non-telomeric genes Cyclin D2 and Cytochome b. Data provided are the mean values±s.e.m. from three different iPS clones. (Bottom) Reverse-transcribed RNA prepared with two different oligos complementary to the telomeric repeat (CCCTAA)4 or (CCCTAA)2 were used for PCR detection of chromosome 18 transcripts using primer Chr18-1. Two different concentrations of oligos were used. RT(-) reactions (performed in the absence of reverse transcriptase) are shown to exclude possible amplifications because of genomic DNA contamination (see full gel in Supplementary Note 1). (c) Northern blotting using 32P-dCTP-labelled probes targeting either transcripts arising from the subtelomere of chromosome 18 (Chr18-RNAs) or TERRA’s telomeric track (TERRA) in three independent clones of pMEF and iPS; hybridization of 18S was included as a loading control. Both northern blots were done on the same membrane, first chromosome 18 probe and, after stripping, TERRA probe. *Unspecific band due to cross-hybridization with rRNA 18S and 28S. (Graph) Northern blot quantification. Mean values±s.e.m. from the three different clones are indicated. (d) Confocal microscopy images of double RNA-FISH preparations using probes targeting either chromosome 18-RNAs (probes 18-3-1 and 18-3-4; red) or TERRA’s telomeric track (green). (Graph) The percentages of co-localization per nuclei of chromosome 18 probes with TERRA spots are represented (mean±s.d., n=number of nuclei; three independent experiments). Total number of foci and nuclei used for the analysis are indicated. Scale bar, 10 μm. The Student’s t-test was used for all statistical analysis (*P<0.05 and **P<0.001).
Table 1. Summary of TERRA test carried out on transcripts arising from the subtelomere of all chromosomes.
Next, we examined whether these transcripts were transcribed until the telomere-like TERRA. To this end, we checked for the presence of UUAGGG-repeats within the transcripts by three different means. First, we prepared two different types of cDNAs, one using random primers that we named ‘standard cDNA’ and the other enriched in telomeric repeats, which we named ‘telomeric cDNA’, obtained with an oligo complementary to the telomeric repeat (4 × CCCTAA). By using the telomeric repeat oligo, RNAs containing UUAGGG-repeats will be preferentially converted into cDNA compared with other RNAs that do not contain these repeats. We then performed quantitative PCR on these cDNAs using specific primers for the detection of the candidate transcripts. Only primers detecting transcripts arising from chromosome 18 subtelomere were significantly enriched in the ‘telomeric cDNA’ compared with the ‘standard cDNA’, thus indicating the presence of UUAGGG-repeats within these transcripts (Fig. 1b). Primers detecting other regions of chromosome 18 RNAs that were characterized by RACE (Rapid Amplification of cDNA Ends; see below) were also enriched (primer 18-7, NGSP31 and 18–35; primer position in Supplementary Fig. 3). Cyclin D2 and Cytochrome b, two genes that do not contain telomeric repeats, were more enriched in the ‘standard cDNA’ (Fig. 1b). Amplicons obtained with primers against ‘region 1’ (Chr18-1) using two different types of telomeric cDNAs are also shown (Fig. 2b, bottom). In contrast to chromosome 18 transcripts, RNAs arising from other highly transcribed subtelomeres (chromosomes 8, 10 and 17; Fig. 1a) were not enriched in the ‘telomeric cDNA’ (Supplementary Fig. 4). Similarly, transcripts arising from other PCR positive subtelomeres were not enriched. Nevertheless, owing to the fact that the differences were not significant in the case of chromosomes 6, 9, 19 and X-RNAs, we decided to include them in further analyses (Table 1).
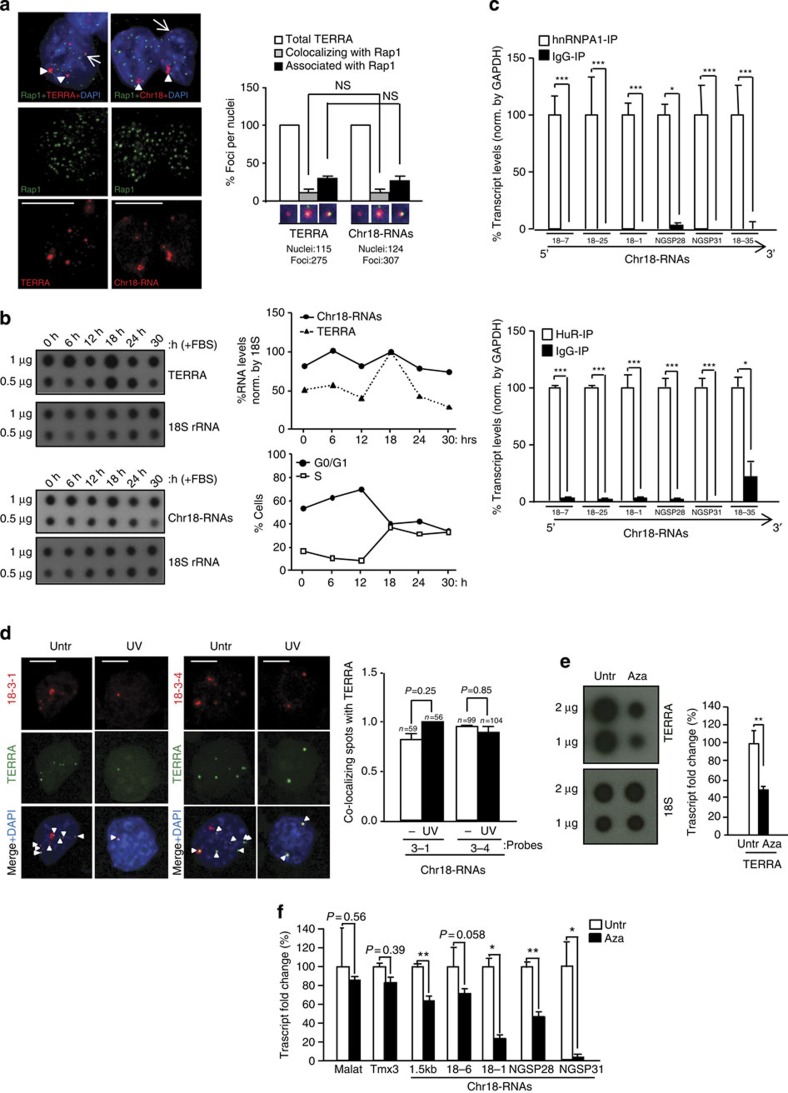
Figure 2. Transcripts arising from the subtelomere of chromosome 18 show a ‘TERRA behaviour’.
(a) Immunofluorescence to detect the telomere marker Rap1 (green) followed by RNA-FISH to detect either TERRA or chromosome 18-transcripts (red). (Graph) Percentages of Rap1 foci co-localizing/associating with TERRA or chromosome 18-RNAs per nuclei (mean±s.e.m., n=number of nuclei; three different antibodies were used for telomere detection (Rap1, TPP1 and TRF1); see the results of the other two in Supplementary Fig. 8A and B). (b) Upon pMEF synchronization, TERRA and chromosome 18-RNA levels were measured by RNA dot-blot at different time points upon serum release; 18S serves as a loading control. (Top graph) Quantification of transcripts levels normalized by 18S. (Bottom graph) Percentage of cells in G0/G1 and S phase upon serum release. (c) Immunoprecipitation (IP) assay with antibodies recognizing hnRNP A1 or HuR followed by qRT–PCR for chromosome 18-RNAs detection using primers against different regions. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was used for normalization. Data were compared with respect an IgG-IP (mean values±s.e.m. from three different iPS clones). (d) Cells were irradiated with ultraviolet C and, upon recovery, fixed for RNA-FISH. Confocal microscopy images of double RNA-FISH using probes targeting either chromosome 18-RNAs (probes 18-3-1 and 18-3-4; red) or TERRA’s telomeric track (green) are shown. (Graph) Percentages of co-localizing foci per nuclei (mean values +s.e.m., n=number of nuclei; two different probes were used to detect chromosome 18-RNAs). (e) Upon 5′Azacytidine treatment, RNA was isolated and use for (left) TERRA detection by RNA dot-blot with a probe against the telomeric track; 18S serves as loading control. (Graph) TERRA quantification normalized by 18S (mean values±s.d., n=three replicates). (f) Quantification chromosome 18 transcripts by qRT–PCR using primers targeting different regions (mean values±s.d., n=three replicates). Tmx3 is the coding gene closest to chromosome 18 telomere and Malat1 a long-noncoding RNA located in a non-subtelomeric region. Student’s t-test was used in all statistical analysis (*P<0.05, **P<0.001 and ***P<0.0001). Total number of foci and nuclei are indicated in the corresponding panels. Arrowheads and arrows indicate co-localization and association events (partial co-localizations), respectively. Untr, untreated. Scale bar, 5 μm. NS, not significant.
Second, we studied the northern blot ‘signature’ of the different transcripts to see whether they showed the characteristic ‘TERRA smear’ obtained when using a probe against the UUAGGG-track. Only probes detecting transcripts arising from chromosome 18 subtelomere showed a smear similar to TERRA (Fig. 1c). Moreover, these transcripts were increased in iPS cells compared with their parental MEFs, as described for TERRAs17 (Fig. 1c). Transcripts arising from chromosome 9 and X subtelomeres showed a partial TERRA-like smear, but they did not increase in iPS cells compared with pMEF (Supplementary Fig. 5A). Probes against chromosome 6-, 13- and 19-RNAs only detected background signal (Table 1). The highly transcribed subtelomeres of chromosomes 8, 10 and 17 did not display the characteristic TERRA signature (Supplementary Fig. 5B).
Third, we tested whether these subtelomere-originated transcripts did co-localize with TERRA transcripts using RNA-FISH. To this end, we prepared RNA-FISH probes against these transcripts and co-hybridized them with a TERRA probe targeting the UUAGGG-repeat (Supplementary Fig. 3). Probes were tested not to give signal upon RNase treatment as well as when using a probe against the negative strand (Supplementary Fig. 6A). In agreement with the known nuclear localization of TERRA3 (Fig. 1d and Supplementary Fig. 7A), probes targeting chromosome 18-RNAs rendered signals restricted to the nuclear compartment (Fig. 1d). We confirmed the nuclear localization of all the chromosome 18 transcripts by quantitative reverse transcription PCR (qRT–PCR; Supplementary Fig. 7B). Malat1 and cytochrome b served as controls for the nuclear and cytoplasmic fractions, respectively (Supplementary Fig. 7B). After confocal analysis of the RNA-FISH signals, we found that 80% of TERRA transcripts co-localized with chromosome 18 transcripts (Fig. 1d, graph). RNA-FISH of Tmx3, the closest annotated coding gene to chromosome 18 telomere, did not co-localized with the TERRA probe (Supplementary Fig. 6B).
Chromosome 9-RNAs also co-localized with TERRA spots despite these RNAs did not display clear TERRA features in the previous tests (Supplementary Fig. 6C). Nevertheless, the signal was very weak and showed a markedly lower co-localization than that obtained with chromosome 18-RNAs, with only 12% of TERRA spots co-localizing with chromosome 9 transcripts (Supplementary Fig. 6C, graph; probe controls can be found in Supplementary Fig. 6A,C and D). This suggests that they maybe low contributors of TERRA. RNA-FISH signals from chromosome X-RNAs rendered a panuclear staining with no detectable spots (Table 1). Finally, transcripts arising from the highest transcribed subtelomeres (chromosome 8, 10 and 17) did not co-localize with TERRA spots. Instead, we detected few spots showing associations (no co-localization) with TERRA, also seen with the sense probe, for chromosome 8-RNAs and cytoplasmic signal for chromosome 17-RNAs (Table 1 and Supplementary Fig. 6E).
Together, the different tests performed strongly support that transcripts arising from the subtelomeres of chromosome 18 contain UUAGGG-repeats and appear to be the main contributors of TERRA.
Chromosome 18 transcripts behave as TERRA transcripts
To further test whether chromosome 18-RNAs are part of TERRA, we measured different cellular parameters. TERRAs are known to associate with telomeres2,3,8 (Fig. 2a). Immunodetection of the telomere-binding protein Rap1, a component of telomeres18, followed by chromosome 18-RNAs detection by RNA-FISH showed that chromosome 18-RNAs co-localize with Rap1 (Fig. 2a). Importantly, we obtained similar frequencies of co-localization and association events of both chromosome18-RNAs and TERRA with Rap1 (Fig. 2a, graph). Similar results were obtained when using TPP1 and TRF1 for telomere detection (Supplementary Fig. 8A, B). In human cells, downregulation of Upf1, which is part of the nonsense-mediated mRNA decay core machinery, results in increased cells displaying telomere localization of TERRA2. In mouse cells, however, the number of cells displaying TERRA or chromosome 18-RNAs localization at telomeres did not change upon Upf1 downregulation (Supplementary Fig. 8C, left graph). Furthermore, the number of co-localization events per nuclei was similar for both TERRA and Chromosome 18 probes (Supplementary Fig. 8C, right graph). Chromosome 9-RNAs associated with telomeres but co-localization was not observed (Supplementary Fig. 8D and graph). This could be due to the strong weakening of RNA-FISH signal upon the TRF1 immunofluorescence. We next performed cell cycle analysis of both chromosome 18-RNAs and TERRA expression and found that they are regulated in a similar manner throughout the cell cycle. In particular, upon synchronization of pMEFs, both chromosome 18-RNAs and TERRA accumulate in early G1, as described in human cells19 but they decline during G1 progression to peak again in early S phase (Fig. 2b).
We also determined whether chromosome 18 transcripts were bound by hnRNP A1 and HuR, two of the RNA-binding proteins that bind TERRA7,10. RNA-IP experiments performed in conditions that preserved the RNA–protein interaction followed by transcript detection using qRT–PCR showed that chromosome 18 transcripts are bound by both hnRNP A1 and HuR, as indicated by the relative enrichment of these transcripts in their corresponding IP when compared with the IgG-IP (Fig. 2c). hnRNP A2B1, also known to bind TERRA7, binds to chromosome 18-RNAs but only in those regions further from the telomere (Supplementary Fig. 9A). Interestingly, chromosome 9-RNAs were bound by HuR and hnRNP A2B1 but not by hnRNP A1, indicating different binding preferences to chromosome 18 RNAs and TERRA (Supplementary Fig. 9B). Chromosome 8, 10 and 17 transcripts were not bound by these proteins with the exception of chromosome 17 transcripts, which showed hnRNP A2B1 binding (Supplementary Fig. 9C).
To further characterize chromosome 18-transcripts, we studied their response to exogenous stresses. We first tested TERRA nuclear localization upon ultraviolet C irradiation as some TERRA-binding proteins translocate to the cytoplasm upon ultraviolet C20. The nuclear localization of TERRA did not change in response to ultraviolet C but the number of TERRA spots decreased, in agreement with the shutdown of transcription in response to ultraviolet C (Fig. 2d)21. Chromosome 18-transcript behaved like TERRA (Fig. 2d). Importantly, co-localization between TERRA and chromosome 18-RNAs remained unaffected upon ultraviolet C treatment, further supporting that chromosome 18-transcripts are part of TERRA (Fig. 2d, graph). Mouse cells deficient for DNA methyltransferase 1 (DNMT1) or DNMT3a/3b show lower TERRA levels than wild-type controls3. In agreement with this, 5′azacytidine (DNMT inhibitor) decreased both TERRA (Fig. 2e) and chromosome 18-RNAs levels (Fig. 2f).
The fact that chromosome 18-RNAs contain UUAGGG-repeats and behave as TERRA in many cellular assays strongly supports that these transcripts are genuine TERRA transcripts. From now on, we will refer to these transcripts ‘chromosome 18-TERRAs’.
Chromosome 18-TERRAs’ promoter and transcription start sites
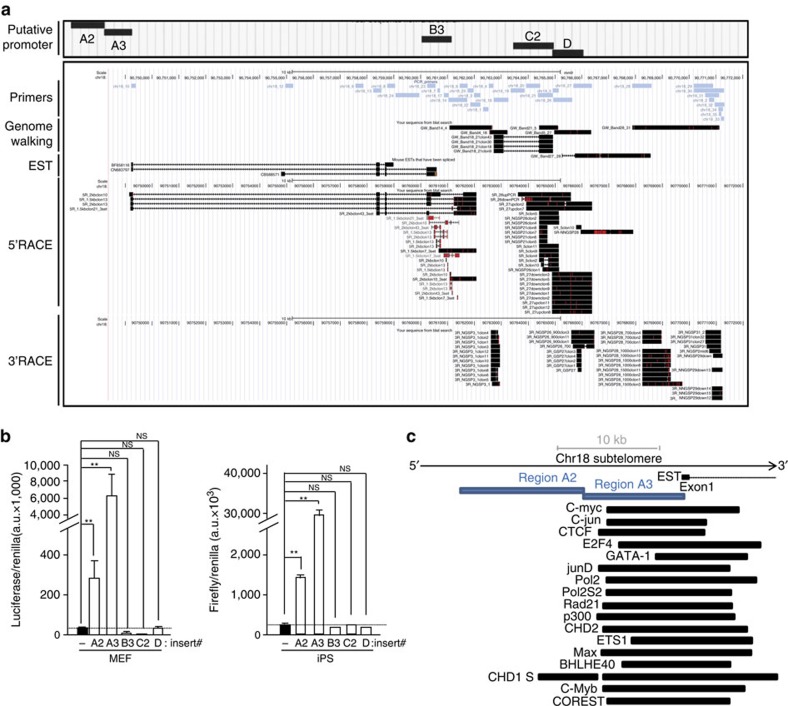
We next set out to determine the transcription start and termination sites of these transcripts by carrying out 5′ and 3′ RACE experiments, respectively. Besides employing ‘region 1’ of chromosome 18 (region amplified by primer chr18-1) as starting point of RACE experiments, we used other regions within this subtelomere that were positive for transcription (Supplementary Fig. 10A). Forward and reverse primers from different sets of primers were used together to generate longer amplicons, then sequenced and named ‘Genome walking’ sequences (Fig. 3a). We cloned approximately ten of these sequences and seventy-five sequences upon 5′RACE and 3′RACE assays that preferentially aligned at chromosome 18 subtelomere (Fig. 3a). Regarding the 3′RACE clones, we were able to undoubtedly map most of them, being the ones closer to the telomere only 250 bases away. Three of the 3′RACE clones contained multiple non-contiguous TTAGGG repeats within their sequence and were mapped right at the telomere, although the alignment was not perfect (Supplementary Fig. 10B and C). Possible explanations to this imperfect alignment could be the generation of PCR/sequencing artefacts because of the repetitive nature of these sequences or to miss-sequencing of the reference genome (for example, see identical mismatch in 5′R-27up clones from 5′RACE sequences). RACE experiments unveiled important features of chromosome 18-TERRAs. In particular, these transcripts have exons and undergo alternative splicing (see for instance the skipping of exon 4 in one of the top 5′RACE sequences; Fig. 3a). Moreover, they have multiple transcription start and termination sites (Fig. 3a). Both the multiple transcription start sites and the alternative splicing may explain the TERRA length heterogeneity (‘TERRA smear’ on northern blots).
Figure 3. Identification of transcription initiation and termination sites of chromosome 18-RNAs as well as their promoter.
(a) UCSC snapshot depicting, from top to bottom, putative promoter regions (A2, A3, B3, C2 and D), genomic scale, genomic position, primer position, ‘genome walking’ transcripts, annotated mouse EST and the sequences cloned upon 5′RACE and 3′RACE experiments (RACE: Rapid Amplification of CDNA Ends; red lines indicate mismatches with respect to the reference genome). (b) The promoter-free vector pGL3 (firefly luciferase reporter system) containing the different putative promoter regions was transiently cotransfected into iPS or their parental pMEFs cells along with pGL4-Renilla (used to normalize for transfection efficiency); 48 h later, protein was extracted and used for the detection of firefly and renilla luciferase activities. Graph shows the relative fold increase in firefly luciferase activity seen in the pGL3-containing promoter regions relative to the empty vector after normalization to renilla activity. Mean values±s.d., n=3 technical replicates from one representative experiment (two independent transfections were performed in pMEFs and three in iPS; the activity of regions A2 and A3 was significant different compared with the other regions in all transfections). Student’s t-test was used for statistical analysis (*P<0.05 and **P<0.001). (c) Diagram showing, from top bottom, genomic scale, annotated EST, position of promoter regions A2 and A3 and examples of transcription factor-binding sites from ChIP data from the Stanford/Yale/ENCODE Project. NS, not significant.
RACE sequences mapped within a genomic area of 20 kb adjacent to the telomere (Fig. 3a). This region is conserved in mammals and contains different transcriptional signals such as CAGE tags and histone marks of active transcription (Supplementary Fig. 11 or blat RACE sequences (Supplementary Information) in the UCSC (University of California, Santa Cruz) link (Methods)). There is an annotated mouse expressed-sequence-tag (EST) within this region and some of the RACE sequences overlapped with it (Supplementary Fig. 11). External RNA-seq data from the ENCODE Project22,23 show cluster of reads overlapping with the main chromosome 18-TERRA exons and that these transcripts are tissue specific and developmentally regulated (Supplementary Fig. 11).
To identify the promoter region of chromosome 18-TERRA transcripts, we cloned different regions upstream of the main 5′RACE products into promoter-free luciferase reporter vectors (Fig. 3a). Upon transient transfection with the different plasmids, luciferase activity was increased both in MEFs and iPS cells transfected with region A2 and, especially, region A3 compared with the empty vector (Fig. 3b). No activity was found in downstream regions B3, C2 and D. Interestingly, A2 and A3 regions are adjacent and located immediately upstream of the annotated EST (Fig. 3a), contain multiple CAGE tags and are located at the start of transcriptionally active histone marks according to the Caltech/ENCODE chromatin immunoprecipitation assay (ChIP) data22,23 (blat region A2 and A3 (Supplementary Information) in the UCSC session link (Methods)). Interestingly, region A3 overlaps with multiple transcription-binding sites according to ChIP data available from the ENCODE/Stanford/Yale project22,23. Both general transcription factors (TBP, p300, RNA-PolII and CSTF) and early-response transcription factors, such as c-myc and c-jun, bind to the A2-A3 region (Fig. 3c and Table 2 for a complete list). Upon scanning regions A2 and A3, we did not find CpG islands nor clear matches with the 61-29-37-repeat present at the human promoter4.
Table 2. Transcription factors binding to region A2-A3.
| Transcription factors binding to region A2-A3 |
|---|
| BHLHE40 |
| CHD1 |
| CHD2 |
| C-jun |
| c-Myb |
| c-Myc |
| COREST |
| CTCF |
| E2F4 |
| ETS1 |
| GATA-1 |
| GCN5 |
| HCFC1 |
| JunD |
| MaFk |
| Max |
| Maz |
| Mxi1 |
| NELFe |
| Nrf2 |
| p300 |
| Pol2 |
| Pol2phosphoS2 |
| Rad21 |
| SIN3A |
| SMC3 |
| TBP |
| UBF |
| USF2 |
| ZC3H11A |
| ZKSCAN1 |
| ZNF384 |
| ZNF-MIZD-CP1 |
The data correspond to ChIP data on MEL (Mouse erythroleukemia) and CHC12 (liver cancer) cell lines from the Stanford/Yale/ENCODE Project. Except for the GATA-1, all of these transcriptions factor were ChIP positive for both cell lines.
Chromosomes to which chromosome 18-TERRAs bind
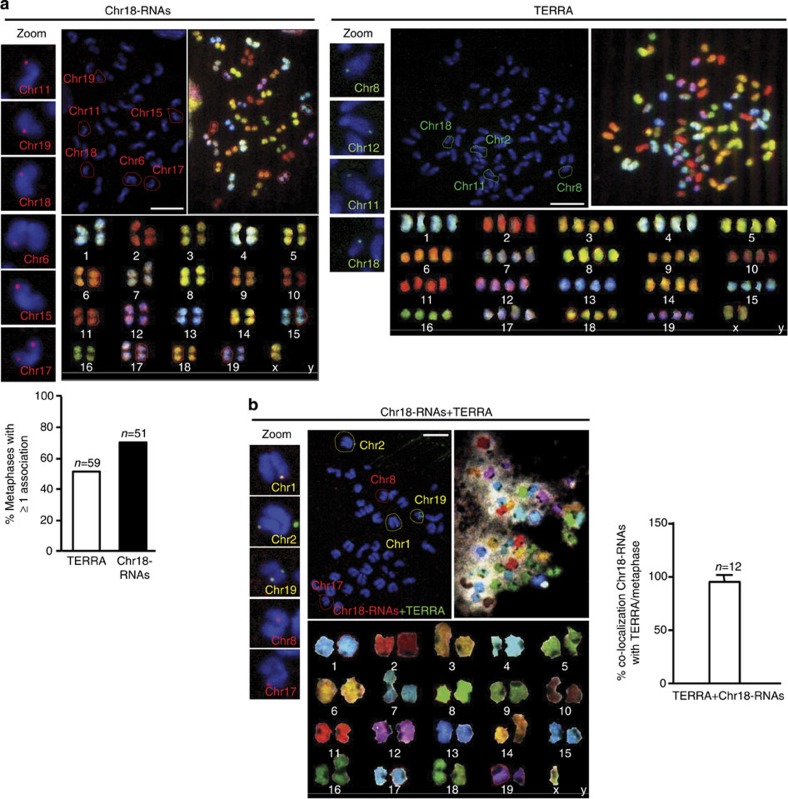
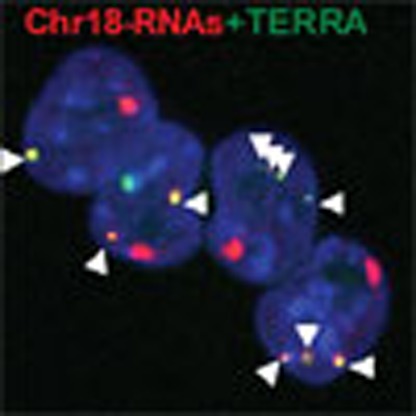
On the basis of the observed associations of chromosome 18-TERRAs to telomeres (Fig. 2a), we next asked whether chromosome 18-TERRAs associate only with the chromosome of origin (chromosome 18) or also with additional chromosomes. To this end, we performed RNA-FISH on metaphases followed by Spectral Karyotyping (SKY). We detected associations of both chromosome 18 RNAs and TERRA to the tip of a subset of chromosomes (Fig. 4a), which were RNase sensitive confirming the RNA nature of these associations. We observed at least one association in 67% of the metaphases when using the chromosome 18 probe and 51% when using the TERRA probe (Fig. 4a, graph). The average number of associations per metaphase varies from one to three for both probes, detecting up to ten associations per metaphase in few cases. Note that the lower number of TERRA spots observed in metaphases compared with interphase is likely due to the detection of only the spots associated with chromosome ends, whereas in interphase both associated and non-associated spots are detected. From the total number of associations, around 60% occur in the tip of the long arm (half of them at the subtelomere rather the telomere) and around 25% in the short arm (half of them adjacent to the centromeric telomere) of chromosomes. The remaining 15% could not be mapped because of artefacts on the metaphase’s shape. Importantly, both probes detected association to the tip of chromosome 18 but also to other chromosomes (Fig. 4a).
Figure 4. Identification of the chromosomes bound by chromosome 18-TERRAs.
(a) RNA-FISH with probes targeting chromosome either 18-RNAs (red) or TERRA’s telomeric track (green) or (b) with both probes (yellow) is shown: (top-middle) RNA-FISH staining in metaphases, (top-right) SKY hybridization and (bottom) chromosome identification by SKY. Those chromosomes showing probe association have been circled and identified in the RNA-FISH preparations according to the SKY information. Zoom of these associations is shown on the left. (Graph a) Percentage of metaphases showing one o more association with either TERRA or with chromosome 18-RNAs (mean±s.e.m.). n=number of metaphases analysed; three independent experiments. Scale bar, 10 μm. (Graph b) Percentage of co-localization of TERRA and chromosome 18-RNAs per metaphases. n=number of metaphases analysed; two independent experiments.
To further confirm the TERRA nature of chromosome 18-RNAs, double RNA-FISH with chromosome 18 and TERRA probes followed by SKY was performed. As expected, we found co-localization events in all metaphases analysed (Fig. 4b). The co-localization of chromosome 18-RNAs with TERRA per metaphase reached almost 100% (Fig. 4b, graph). The detection of associations with the telomere when using both probes dropped compared with when using only one probe most likely due to competition of both probes for nearby positions. Table 3 summarizes the identification of chromosomes associated with chromosome 18-RNAs (red), TERRA transcripts (green) or both (yellow). All chromosomes, except chromosome X, displayed at least once either red and green signals in different metaphase (that is, Chromosomes 3, 5, and so on) or co-localizing signals (chromosomes 1, 2 and so on), suggesting that these transcripts associate virtually with all chromosomes. For chromosome X, we could only detect TERRA association (green signal). Chromosome 9-RNAs were also detected at chromosome ends in metaphases (Supplementary Fig. 12), whereas chromosome 8, 10 and 17-RNAs were not.
Table 3. Summary of the identification of chromosomes associated with chromosome 18 transcripts and TERRA upon RNA-FISH followed by SKY.
Loss of chromosome 18-TERRAs induces telomere damage
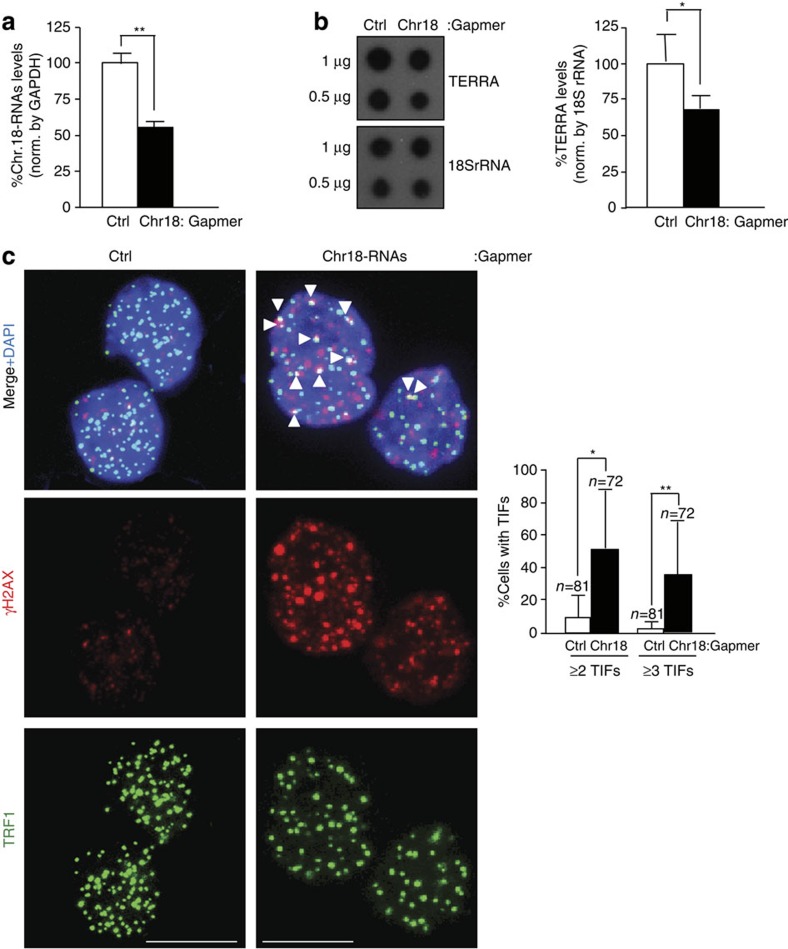
To study the role of chromosome 18-TERRAs at telomeres, we downregulated them using Gapmers-LNA. Significant downregulation was achieved 2 days post transfection as measured by RT–qPCR with a 50% knockdown (Fig. 5a). Importantly, chromosome-18 TERRA downregulation simultaneously decreased TERRA levels as determined by RNA dot-blot with a probe against the telomeric repeat (Fig. 5b), demonstrating that they are contributors of TERRA. Next, we studied the presence of telomere γH2AX DNA damage foci (TIFs). Chromosome 18-TERRA caused a significant increase in the number of cells exhibiting two or three or more TIFs compared with control cells (Fig. 5c). Knockdown of the telomeric track with a Gapmer-LNA (‘TERRA-Gapmer’) also caused a decrease in TERRA levels and a similar increase in the number of cells displaying TIFs (Supplementary Fig. 13A and B). However, the ‘TERRA- Gapmer’ is likely not as specific as the ‘Chr18-Gapmer’ to downregulate TERRA expression as there are other genes that contain telomeric repeats, which could be downregulated by the ‘TERRA-Gapmer’ (for example, gene Xkrx contains multiple TTAGGG repeats, nine of them contiguous). Together, these data suggest that chromosome 18-TERRA transcripts may contribute to protect telomeres from eliciting a DNA damage response.
Figure 5. Downregulation of chromosome 18-TERRAs induces telomere damage.
(a) Cells were transfected with either control Gapmer-LNA (Ctrl) or Gapmer-LNA targeting chromosome 18-RNAs and RNA collected 2 days post transfection. The graph shows the percentage of chromosome 18-RNA levels normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels upon Gapmer-LNA transfection (mean±s.e.m.; three independent transfections). (b) (Left) RNA dot-blot to detect TERRA using a 32P-dCTP-labelled probe; hybridization of 18S rRNA was included as a loading control. (Right) Quantification of the RNA dot-blot signals normalized by 18S rRNA (mean±s.e.m.; three independent transfections). (c) Representative images of TRF1 (green) and γH2AX (red) fluorescence and of the merged images. Co-localization events (arrowheads) indicate telomere dysfunction-induced foci (TIF). Scale bar, 10 μm. (Graph) Percentage of cells with ≥2 or ≥3 TIFs/nuclei (mean±s.d., n=number of nuclei; three independent transfections). Student’s t-test was used for statistical analysis (*P<0.05 and **P<0.001).
Searching for special features in chromosome 18 subtelomere
We next searched for a special signature in the chromosome 18 subtelomere compared with the other subtelomeres. After aligning the full mouse chromosome 18 with the rest of the chromosomes using Lastz and extracting the synteny blocks for the subtelomeric regions, we found conservation of synteny between the subtelomere of chromosome 18 and chromosome 6, 8 and 17 subtelomeres (Supplementary Fig. 14A and Supplementary Methods) but not with chromosome 9 subtelomere, which also produces RNAs with some TERRA features. We also searched for homology of the chromosome 18 promoter region A2 and A3 with the rest of the subtelomeres. Only region A3 displayed a significant homology, which was higher at chromosome 6, 8 and 17 subtelomeres (47–48%; Supplementary Fig. 14B and Supplementary Methods).
We also studied the presence of histone marks for active promoters and enhancers (H3K4me3, H3K4me1 and H3K27ac) at the promoter region A2-A3 of chromosome 18 uploading ChIP data from the ENCODE/LICR project Consortium (Supplementary Fig. 14C). However, these histone marks could also be found at the A3-like regions of chromosome 10 and 17 (Supplementary Fig. 14C). Collectively, these data indicate that chromosome 18 subtelomere did not significantly differ from other transcribed subtelomeres either at the level of conservation of synteny or in the presence of histone marks for active promoters and enhancers.
Discussion
Here we have carried out a whole-subtelomere screening of mouse chromosomes to identify the TERRA locus and discovered a subset of novel transcripts arising from chromosome 18 subtelomere that resemble TERRA in several key features. These transcripts contain telomeric repeats: (i) they are enriched in a telomeric cDNA compared with a standard cDNA, (ii) they render the same sizes as TERRA transcripts on northern blot, (iii) they co-localize with TERRA in interphase and metaphase and (iv) downregulation of these transcripts by Gapmers causes a concomitant decrease in TERRA levels. It is unlikely that the transcription of these transcripts stop right at the start of the telomere (what it is known as ‘αARRET transcripts’) as they render the same northern blot pattern than TERRA (this is not the case for αARRET and TERRA in Schizosaccharomyces pombe24). Of note, transcription of chromosome 18 subtelomere was previously described but whether these RNAs were TERRA was not addressed25.
Transcripts from other mouse subtelomeres failed to show TERRA features except for some features in transcripts arising from the chromosome 9 subtelomere. In particular, although chromosome 9-RNAs were not enriched in the telomeric cDNA fraction and displayed a shorter-size smear on northern blots, they showed co-localization with TERRA spots, although to a lesser extent than chromosome 18-RNAs. This fact, together with the low RNA-FISH signal displayed by chromosome 9-RNAs and the reduced number of spots detected per cell (one versus five o more for chromosome 18-RNAs), suggests that the majority of TERRA transcripts arise from chromosome 18 subtelomere and that only a small fraction may arise from chromosome 9. This is not an unexpected finding as yeast studies showed that although TERRA molecules arose from chromosome 1L, 6R and Y’-element, cells expressed TERRA only from one specific telomere at a given time14. Similarly, mouse chromosome 18 maybe the preferential region for TERRA transcription, although chromosome 9 subtelomeres may take over in some conditions. If chromosome 18 subtelomere is the main source of TERRA transcripts, an interesting question is why TERRA is produced mainly at this chromosome. In humans, the situation might be different as a TERRA promoter was proposed to be present in 20 different chromosomes4. Nevertheless, it remains to be tested whether these transcripts arising from the subtelomere of each of these 20 chromosomes are transcribed until the telomere (as shown here, this is not the case for many of the murine subtelomere-originated transcripts). Interestingly, the differential DNMT dependency of mouse and human TERRA agrees with important differences in their promoters3,4,11. Differences in the factors regulating TERRA at telomeres are also apparent. In particular, Upf1, a nonsense-mediated mRNA decay factor, regulates telomere localization of human TERRA2 but not of mouse TERRA (this work).
The identification of chromosome 18-TERRA transcripts is important to determine the origin of TERRA’s heterogeneity. It was proposed that TERRA heterogeneity could be due to the existence of different lengths of the transcribed subtelomeric region, to products of TERRA 3′-end processing, or to differential entry of RNA polII into the telomere (reviewed in refs 5, 6). Our findings indicate that TERRA length heterogeneity stems, at least in part, from the use of multiple transcription starts and the existence of alternative splicing. The multiple transcription start sites could originate from the heterochromatic nature of telomeres and subtelomeres (reviewed in refs 1, 26), which in turn may difficult RNA polymerase II accessibility. In support of this notion, relaxation of heterochromatin during nuclear reprogramming increases TERRA transcription17. Further support for the notion that the incapability of RNA polymerase II to perform a full transcription may generate truncated TERRA transcripts comes from: (i) the observation of multiple termination sites and (ii) detection of more RNA-FISH spots when using the chromosome 18 probes than when using the probe against the telomeric track. Additional explanations for this heterogeneity may include transcripts of different sizes arising from other chromosomes (in mouse cells, from chromosome 9), differential 3′-end processing and differential entry of RNA pol II into the telomere.
Downregulation of chromosome 18-TERRA caused a significant increase in the number of TIFs, suggesting a protective role of TERRA at telomeres. This is in agreement with that observed in human cells10. TERRA downregulation may impair sequestering of hnRNP A1 from telomeres and RPA-to-Pot1 switching upon DNA replication, or may diminish the nucleation of telomerase at short telomeres, thus resulting in increased telomeric DNA damage13,14. This protective role of TERRA may be also extended to metaphasic chromosomes as chromosome 18-TERRA is associated to the tip of virtually all metaphasic chromosomes. Intriguingly, we observed an average of two to three associations per metaphase and not to all chromosomes at once. We are not certain whether this is due to technical reasons or to the association of chromosome 18-TERRA with telomeres following a sequential pattern, first to a subset of chromosomes and then exchanging to another subset. The fact that we detect chromosome 18-TERRA transcripts association to also a small subset of telomeres per cell in interphase supports the latter idea.
In summary, here we have identified a subset of novel transcripts arising from the subtelomere of chromosome 18 that are bona fide TERRA transcripts. This finding is of relevance because now we have determined specific regions to modulate and to detect TERRA transcripts levels by different means overcoming the lack of specificity of molecular tools that use the telomeric track as a target. Taking advantage of this, we have been able to identify the promoter that it is driving the transcription of these transcripts, determine one of the causes of TERRA heterogeneity and study the functional role of TERRA at telomeres. Finally, characterization of mouse TERRA locus will allow addressing the role of TERRA in normal development and disease.
Methods
Cells, treatments, transfections and cellular fractionation
MEFs were obtained from wild-type embryos at embryonic day E13.5 (C57BL/6, <6-month old). Mouse C2C12 and NS1 cell lines (ATCC) were cultured according to the ATCC’s recommendations. iPS cells were obtained as reported in ref. 27. Namely, reprogramming of primary MEFs (passage 2) was performed as previously described17, following modifications of a previous protocol28. All procedures performed on mice were revised and approved by the institutional ethical committee. Ultraviolet C treatment was carried out using 60 J m−2 irradiation following by 4 h of recovery. 5-Aza-2′deoxycytidine was used at 1 μM on pMEFs for 3 days. Gapmers-LNAs (Exiqon) were transfected with Dharmafect (Thermo Scientific) at a final concentration of 100 nM and cells collected 2 days post transfection. Gapmer-LNA sequences can be found in Supplementary Information. Nuclear and cytoplasmic fractions were prepared as previously described7.
Cell cycle
pMEFs were synchronized with serum starvation (0.1% fetal bovine serum) for 48 h. The release was done with the addition of 10% fetal bovine serum. Cell cycle position was determined by flow cytometric measurement of nuclear DNA content (using propidium iodide staining).
IP of ribonucleoprotein complexes
The IP assay was performed basically as described in ref. 7 using nuclear extracts from pMEFs, which were previously precleared for 30 min at 4 °C using 15 μg of IgG (Santa Cruz Biotechnology) and 50 μl of protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology). Beads (100 μl) were then incubated (18 h, 4 °C) with 30 μg of antibody and then for 1 h at 4 °C with 1 mg of cell lysate. After extensive washes with NT2 buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM MgCl2 and 0.05% Nonidet P-40) and digestion of proteins in the IP material, the RNA was extracted and used to perform qRT–PCR. Antibodies used were as follows: anti-hnRNP A1 (Sigma, clon 9H10) and HuR (Santa Cruz Biotechnology, 3A2).
Biotin pull-down analysis
Biotin pull-down assays were carried out as described7 using nuclear extracts from pMEFs and antisense TERRA biotinylated transcript consists of eight CCCTAA repeats as a bait; the control biotinlylated transcripts consist of random RNA with no sequence matches with the mouse genome and of the same length than the antisense TERRA transcript.
RNA-deep sequencing and Illumina read alignment
Measure of 150 ng of RNA was randomly fragmented, converted to double-stranded cDNA and subsequently processed through enzymatic treatments of end-repair, dA-tailing, and ligation to adapters as in Illumina's ‘Preparing Samples for Sequencing of mRNA’ manual (Part# 1004898 Rev. A). A fraction of 175–225 bp was extracted from an agarose gel and adapter-ligated library was completed by limited-cycle PCR with Illumina PE primers (15 cycles; Illumina). The resulting purified cDNA library was applied to an Illumina flow cell (Illumina) for cluster generation and sequenced on the Genome Analyzer II. The obtained reads were all single-end 36 bp. Primary data analysis was performed with GAPipeline-1.4 (Illumina) generating FastQ files. The single-end reads were aligned to the reference genome Mus genome assembly mm10 (GRCm38) with TopHat v2.0.4 (ref. 29), using Bowtie 0.12.7 (ref. 30) and Samtools 0.1.16 (ref. 31) allowing up to two initial read mismatches, and up to 20 mappings when multiple alignments are possible (based on alignment scores). Transcripts assembly, estimation of their abundances and differential expression were calculated with Cufflinks 1.3.0 (ref. 29), using the mouse genome annotation data set GRCm38/mm10 from the UCSC Genome Browser32. The expression value of each gene between the conditions is shown as the log10 of the FPKM fold-change (FPKM: fragments per kilobase of transcript per million fragments mapped). Raw data can be access at http://www.ncbi.nlm.nih.gov/sra/ with the identifier SRA059274. Custom CAGE tag track was prepared using the Fantom3 collection of CAGEtag starting sites. The link to UCSC session with custom ‘CAGE tag’ track is: http://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=ilsilanes&hgS_otherUserSessionName=CAGE%20tag.
RACE, telomeric cDNA, reverse transcription–PCR and qRT–PCR
5′and 3′ RACE were performed with the SMART RACE cDNA amplification kit (Clontech). Telomeric cDNA was prepared as a standard cDNA using the first strand cDNA synthesis kit (Invitrogen) but substituting the random hexamers for a C-strand telomeric oligo consisting on four CCCTAA repeats. Reverse transcription–PCR and qRT–PCR assays were performed using standard protocols. Primers used can be found in Supplementary information.
Northern blot and dot-blot
Northern blot and dot-blot analyses were performed using standard protocols. TERRA probe was obtained from a 1.6-kb (TTAGGG)n cDNA insert excised from pNYH3 (kind gift from T. de Lange, Rockefeller University, NY, USA). Probes against chromosome 18 transcripts were prepared from PCR products (see primers used in Supplementary information).
Immunoflorescence, RNA-FISH and SKY
Immunoflorescence alone or followed by RNA-FISH was performed basically as described in ref. 7. Namely, cells grown in superforst coverslips (Thermo Scientific) were placed in cytobuffer (100 mM NaCl/300 mM sucrose/3 mM MgCl2/10 mM Pipes, pH 6.8) for 30 s, washed in cytobuffer with 0.5% Triton X-100 for 30 s, washed in cytobuffer for 30 s, and then fixed for 10 min in 4% paraformaldehyde in PBS. After washing with PBS, cells were blocked overnight at 4 °C in PBS containing 3% (w/v) BSA, 0.1% Tween-20 and 0.8 μg ml−1 RNasin (Promega). Antibodies were incubated in blocking solution for 60 min at 37 °C. The slides were washed twice in PBST, overlaid with secondary antibodies (Molecular Probes) diluted 1:500 for 1 h in blocking solution at 37 °C, washed three times in PBST and postfixed in 4% paraformaldehyde in PBS. The cells were dehydrated in 70, 80, 95 and 100% ethanol, air-dried and hybridized overnight at 50 °C with RNA probe in hybridization buffer (2 × sodium saline citrate (SSC)/50% formamide). Slides were washed two times for 15 min in hybridization buffer at 55 °C, two times for 10 min in 2 × SSC at 55 °C, 10 min in 1 × SSC at 55 °C, 5 min in 4 × SSC at room temperature, 5 min in 4 × SSC containing 0.1% Tween-20 and DAPI (Molecular Probes) at room temperature and 5 min in 4 × SSC at room temperature. Signals were visualized in a confocal ultra-espectral microscope TCS-SP5 (Leica). When RNA-FISH only, dehydration steps were performed right after the first formaldehyde fixation. Mouse SKY was performed after RNA-FISH as previously described33. The coordinates of the metaphases of interest were annotated before SKY. The following antibodies were used: rabbit polyclonal Rap1 (Bethyl, 1:300), rabbit polyclonal TRF1 (in house, 1:500), mouse monoclonal γH2AX (Millipore, 1:800) and rabbit polyclonal TPP1 (kind gift of T. Else, 1:200). RNA-FISH probes were generated from PCR products by in vitro transcription (Ambion) using Cy3-labelled CTP (Amersham; primers available in Supplementary Information). The co-localization events were quantified in five individual confocal layers per nuclei.
Plasmids and reporter assays
For construction of the reporter plasmid, PCR products were prepared with primers spanning the promoter regions of interest (Supplementary Information) and cloned into the plasmid pGL3-basic (Promega). Firefly and Renilla Luciferase activities were measured in cells harvested 2 days after electroporation with the Dual Luciferase Reporter Assay System (Promega), following the manufacturer’s instructions.
Author contributions
I.L.de.S., O.D. and M.A.B. designed the experiments; I.L.de.S., M.L.de.B. and O.D. carried out the experiments and O.G and D.G.P did the bioinformatic analysis.
Additional information
Accession codes: Raw data can be access at http://www.ncbi.nlm.nih.gov/sra/ with the identifier SRA059274.
How to cite this article: López de Silanes, I. et al. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 5:4723 doi: 10.1038/ncomms5723 (2014).
Supplementary Material
Supplementary Figures 1-14, Supplementary Note 1 and Supplementary Methods
Acknowledgments
We are indebted to Stefan Schöeftner and Susana Llanos for reagents and to Manuel Serrano, Maria Elisa Varela and Antonio Maraver for very helpful suggestions and discussion on the manuscript. We thank Diego Megías for confocal image acquisition and to Miguel Ángel Grillo, María del Carmen Carralero and Juan Cruz Cigudosa for the Spectral Karyotyping (SKY). We thank Luis E. Donate for manuscript preparation. External RNA-seq data were generated and analysed by the UW ENCODE group and by the transcriptome group at Cold Spring Harbor Laboratories and the Center for Genomic Regulation (CRG in Barcelona), who are participants in the ENCODE Transcriptome Group. ChIP data of transcription factors binding site were generated and analysed by the laboratories of Michael Snyder at Stanford University and Sherman Weissman at Yale University within the ENCODE Project. Histone marks data belong to the Caltech/ENCODE project in which cell growth, ChIP and Illumina library construction were done in the laboratory of Barbara Wold (California Institute of Technology). Sequencing was done at the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology, initial HiSeq data were generated at Illumina Inc., Hawyard, CA. Cell growth and ChIP of histone marks were carried out by Georgi Marinov, Katherine Fisher, Gordon Kwan, Antony Kirilusha, Ali Mortazavi, Gilberto DeSalvo and Brian Williams. Library Construction, Sequencing and Primary Data Handling by Lorianne Schaeffer, Diane Trout, Igor Antoschechkin (California Institute of Technology), Lu Zhang and Gary Schroth (Illumina Inc.). Data processing and submission by Georgi Marinov and Diane Trout. Research in the Blasco laboratory is funded by the Spanish Ministry of Economy and Competitiveness Projects SAF2008-05384 and CSD2007-00017, the Madrid Regional Government Project S2010/BMD-2303 (ReCaRe), the European Union FP7 Project FHEALTH-2010-259749 (EuroBATS), the European Research Council (ERC) Project GA#232854 (TEL STEM CELL), the Preclinical Research Award from Fundación Lilly (Spain), Fundación Botín (Spain) and AXA Research Fund.
References
- Blasco M. A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8, 299–309 (2007). [DOI] [PubMed] [Google Scholar]
- Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E. & Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 (2007). [DOI] [PubMed] [Google Scholar]
- Schoeftner S. & Blasco M. A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236 (2008). [DOI] [PubMed] [Google Scholar]
- Nergadze S. G. et al. CpG-island promoters drive transcription of human telomeres. RNA 15, 2186–2194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B. & Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 28, 2503–2510 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerhahn S., Iglesias N., Panza A., Porro A. & Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 584, 3812–3818 (2010). [DOI] [PubMed] [Google Scholar]
- Lopez de Silanes I., Stagno d’Alcontres M. & Blasco M. A. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 1, 33 (2010). [DOI] [PubMed] [Google Scholar]
- Farnung B. O., Brun C. M., Arora R., Lorenzi L. E. & Azzalin C. M. Telomerase efficiently elongates highly transcribing telomeres in human cancer cells. PLoS One 7, e35714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R., Brun C. M. & Azzalin C. M. Transcription regulates telomere dynamics in human cancer cells. RNA 18, 684–693 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Norseen J., Wiedmer A., Riethman H. & Lieberman P. M. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35, 403–413 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehezkel S., Segev Y., Viegas-Pequignot E., Skorecki K. & Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 17, 2776–2789 (2008). [DOI] [PubMed] [Google Scholar]
- Pfeiffer V. & Lingner J. TERRA promotes telomere shortening through exonuclease 1-mediated resection of chromosome ends. PLoS Genet. 8, e1002747 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R. L. et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471, 532–536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanelli E., Romero C. A. & Chartrand P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol. Cell 51, 780–791 (2013). [DOI] [PubMed] [Google Scholar]
- Redon S., Reichenbach P. & Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 38, 5797–5806 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A., Kawai T., Yang X., Mazan-Mamczarz K. & Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 24, 1852–1862 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion R. M. et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4, 141–154 (2009). [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 (2005). [DOI] [PubMed] [Google Scholar]
- Porro A., Feuerhahn S., Reichenbach P. & Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell. Biol. 30, 4808–4817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20, 760–769 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I., Spivak G. & Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249 (1987). [DOI] [PubMed] [Google Scholar]
- ENCODE-Project-Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9, e1001046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos J. A. et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 13, 418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A., Wischnewski H., Shchepachev V. & Azzalin C. M. The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 40, 2995–3005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z. et al. Formation of telomeric repeat-containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. J. Cell Sci. 125, 4383–4394 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S. & Blasco M. A. Chromatin regulation and non-coding RNAs at mammalian telomeres. Semin. Cell Dev. Biol. 21, 186–193 (2010). [DOI] [PubMed] [Google Scholar]
- De Bonis M. L., Ortega S. & Blasco M. A. SIRT1 Is Necessary for Proficient Telomere Elongation and Genomic Stability of Induced Pluripotent Stem Cells. Stem Cell Rep. 2, 690–706 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R., Venere M., Yen J. & Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell 1, 245–247 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. & Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez B. et al. Identification of recurrent chromosomal breakpoints in multiple myeloma with complex karyotypes by combined G-banding, spectral karyotyping, and fluorescence in situ hybridization analyses. Cancer Genet. Cytogenet. 169, 143–149 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-14, Supplementary Note 1 and Supplementary Methods