Abstract
Resting CD4+ T-cell populations from human immunodeficiency virus type 1 (HIV-1)-infected individuals include cells with integrated HIV-1 DNA. In individuals showing suppression of viremia during highly active antiretroviral therapy (HAART), resting CD4+ T-cell populations do not produce virus without cellular activation. To determine whether the nonproductive nature of the infection in resting CD4+ T cells is due to retroviral integration into chromosomal regions that are repressive for transcription, we used inverse PCR to characterize the HIV-1 integration sites in vivo in resting CD4+ T cells from patients on HAART. Of 74 integration sites from 16 patients, 93% resided within transcription units, usually within introns. Integration was random with respect to transcriptional orientation relative to the host gene and with respect to position within the host gene. Of integration sites within well-characterized genes, 91% (51 of 56) were in genes that were actively expressed in resting CD4+ T cells, as directly demonstrated by reverse transcriptase PCR (RT-PCR). These results predict that HIV-1 sequences may be included in the primary transcripts of host genes as part of rapidly degraded introns. RT-PCR experiments confirmed the presence of HIV-1 sequences within transcripts initiating upstream of the HIV-1 transcription start site. Taken together, these results demonstrate that HIV-1 genomes reside within actively transcribed host genes in resting CD4+ T cells in vivo.
Human immunodeficiency virus type 1 (HIV-1) replicates preferentially in activated CD4+ T cells (44). However, at any given time, most of the CD4+ T cells in the body are in a profoundly quiescent state that is not fully permissive for viral replication. In resting CD4+ T cells, replication is restricted at several different steps in the virus life cycle. For R5 viruses, entry is restricted due to the absence of CCR5 on most resting CD4+ T cells, particularly those belonging to the naïive subset (48, 52). Reverse transcription is kinetically restricted in resting CD4+ T cells (49, 69) and can take as long as 3 days to complete (49). There is also a block at the level of nuclear import of the preintegration complex (PIC) (9). Despite the karyophilic properties of the PIC that allow nuclear import in some nondividing cells, such as macrophages (8, 23, 27, 70), import does not readily occur in resting CD4+ T cells, possibly because of the absence in resting lymphocytes of sufficient levels of ATP to drive the import of large structures, such as the PIC (9, 38). Thus, the infection does not proceed to the point of integration. As a result, in untreated patients, most of the HIV-1 DNA in resting CD4+ T cells is linear, unintegrated viral DNA localized to the cytoplasm (9, 10, 14). This unintegrated HIV-1 DNA represents a labile, inducible reservoir for the virus; if a T cell is activated before the PIC decays, then subsequent steps in the life cycle can occur and virus particles can be produced (10, 38, 49, 61, 69).
Although direct infection of resting CD4+ T cells does not generally proceed to integration, low levels of integrated HIV-1 DNA can be demonstrated in purified resting CD4+ T-cell populations from infected individuals (14, 16). This apparent paradox can be explained by assuming that resting CD4+ T cells with integrated HIV-1 DNA are actually derived from activated CD4+ T cells that became infected and then reverted back to a resting memory state. Reversion to a memory state is a normal physiologic process that generates the long-lived cells required for immunologic memory (reviewed in reference 34). Most of the resting CD4+ T cells that carry integrated HIV-1 DNA in vivo have a memory phenotype (6, 14, 48). The presence of integrated HIV-1 DNA in long-lived memory T cells provides a mechanism for long-term viral persistence even in patients who are on highly active antiretroviral therapy (HAART) and who show complete suppression of detectable viremia (13, 20, 21, 47, 57, 67).
Resting CD4+ T-cell populations from patients on HAART generally do not release virus unless the cells are stimulated in some manner (12, 14, 16, 28). Even when reverse transcriptase (RT) PCR assays sensitive to 50 copies of HIV-1/ml are used, the production of HIV-1 virions cannot be detected (12). Some form of activating stimulus is needed to induce HIV-1 gene expression from infected resting CD4+ T cells. Treatment of resting CD4+ T cells with mitogens (13, 21, 67), cytokines (15, 56, 65), or phorbol esters (37, 40) can induce virus production. HIV-1 nef can induce macrophage production of factors that increase the permissivity of resting CD4+ T cells to productive infection (63). In vivo, activating stimuli may be supplied in the context of lymphoid tissue microenvironments (18, 63, 71), particularly in patients who are viremic. Viremia is associated with increased levels of T-cell activation, and in viremic patients, infected CD4+ T cells with a resting phenotype display abnormal patterns of gene expression that may allow some degree of virus production (12, 71). Nevertheless, in patients who are on HAART and who have shown prolonged suppression of viremia, purified resting CD4+ T cells from the blood produce little if any virus without stimulation.
The presence of integrated viral genomes in cells that do not produce virus raises the question of how virus production is curtailed in these cells. Clearly, some cells may contain viral genomes that are defective. Among resting CD4+ T cells, the frequencies of cells harboring replication-competent HIV-1 that can be rescued by cellular activation are on the order of 10−6 to 10−7, at least 100-fold lower than the frequencies of cells harboring integrated HIV-1 DNA (13, 14). These findings suggest that most of the integrated HIV-1 DNA in resting CD4+ T cells exists in an irreversibly nonproductive state, while a minor fraction exists in a reversibly nonproductive (latent) state. Although posttranscriptional regulatory mechanisms for limiting virus production have been proposed (43, 50), it is difficult to demonstrate functional HIV-1 mRNAs, particularly multiply spliced RNAs, in rigorously purified populations of resting CD4+ T cells from infected individuals on HAART (28). The bulk of the evidence suggests that the absence of virus production by resting CD4+ T cells with integrated HIV-1 DNA is due to factors affecting transcriptional initiation (5, 17, 24, 32, 33, 45, 64) or elongation (1, 25, 29-31, 35). For example, resting cells lack nuclear forms of the activation-dependent host transcription factors NF-κB and NFAT, which are important for gene expression from the HIV-1 long terminal repeat (LTR) (5, 17, 45, 64).
Recently, there has been considerable interest in the idea that the nonproductive nature of infection in resting CD4+ T cells may reflect proviral integration into chromosomal sites that are or that become repressive for transcription (26, 32, 33). Resting CD4+ T cells are profoundly quiescent cells with densely heterochromatic nuclei (19). The silencing of important genes in T cells involves changes in chromatin structure or repositioning to heterochromatic regions (58). Elegant studies by Jordan et al. have suggested that HIV-1 latency may involve integration into regions of heterochromatin (32). Transformed cell lines infected in vitro and then selected for reversibly nonproductive infection showed preferential integration into centromeric regions known to be repressive for transcription. In contrast, in vitro infections of T-cell lines conducted without selection revealed preferential integration into transcribed genes (54, 68).
The nature of HIV-1 integration sites in resting CD4+ T cells from infected individuals has not been analyzed yet. We used inverse PCR to characterize integration sites in resting CD4+ T cells from patients on HAART and to explore whether the nonproductive nature of infection in resting CD4+ T cells is related to the characteristics of the integration sites.
MATERIALS AND METHODS
Purification of resting CD4+ T cells.
Peripheral blood was obtained from patients who achieved and maintained suppression of viral replication to below the limit of detection of ultrasensitive clinical assays (<50 copies of HIV-1 RNA/ml of plasma) while on HAART. Informed consent was obtained from all patients. This study was approved by an institutional review board.
Resting CD4+ T cells were purified by using a previously described two-stage process (14, 16, 21). Peripheral blood mononuclear cells were negatively selected to remove CD8+ T cells, B cells, monocytes, NK cells, and activated CD4+ T cells by using appropriate monoclonal antibodies and magnetic beads conjugated with antibodies to mouse immunoglobulin G (14, 21). The depletion of activated CD4+ T cells was accomplished by using antibodies to both early (CD69 and CD25) and late (HLA-DR) activation markers. Further purification of resting CD4+ T cells was accomplished by sorting for small lymphocytes with high levels of CD4 and low levels of HLA-DR surface expression (14, 21). The resulting populations of resting CD4+ T cells showed <1% contamination with activated cells. In some experiments, the CD45RO+ CD45RA− subset of resting CD4+ T cells was isolated by including anti-CD45RA antibody in the initial depletion and then sorting for CD4+ CD45RO+ HLA-DR− cells. The purity of these populations was ∼95%.
Analysis of HIV-1 integration sites.
A modification of a previously described inverse PCR strategy was used for analysis of HIV-1 integration sites (14, 16). Genomic DNA from highly purified resting CD4+ T cells was digested with PstI, which cuts frequently in genomic DNA but only once in most HIV-1 isolates, at nucleotide (nt) 1419 in gag (HXB2R coordinates) (39). Digested DNA was ligated under dilute conditions favoring intramolecular ligation. Circularized DNA was amplified with outwardly directed primers in conserved regions of the HIV-1 LTR and gag: outer LTR primer, 5′-TAACCAGAGAGACCCAGTACAGGC-3′, nt 468 to 445 in the LTR; outer gag primer, 5′-GGTCAGCCAAAATTACCCTATAGTG-3′, nt 1170 to 1194 in gag. This amplification captured the junction between the 5′ end of the viral genome and host cell DNA. PCR conditions were as follows. DNA was denatured at 94°C for 3 min. This step was followed by 30 cycles at 94°C for 30 s, 65°C for 1 min, and 68°C for 2 min. A final extension was carried out for 4 min at 68°C. A second PCR was carried out with the same cycling parameters and a nested set of outwardly directed, HIV-1-specific primers (inner LTR primer, 5′-TGGTACTAGCTTGAAGCACCATCCA-3′, nt 152 to 128 in the LTR; inner gag primer, 5′-TGTTAAAAGAGACCATCAATGAGGAAG-3′, nt 1388 to 1414 in gag). The nested reaction produced bands visible on agarose gels. The bands were eluted, cloned, and sequenced. By using the UCSC Bioinformatics Human Genome database (http://www.genome.ucsc.edu; July 2003), the human genomic sequence in each inverse PCR product was identified as a unique best-hit Blat ranking joined directly to the end of the 5′ LTR of HIV-1. A similar inverse PCR strategy that involved an initial digestion with NdeI was also used.
Analysis of gene expression in resting CD4+ T cells by RT-PCR.
RNA was isolated (RNeasy minikit; Qiagen) from highly purified resting CD4+ T cells and phytohemagglutinin (PHA)-activated CD4+ T lymphoblasts from uninfected donors or from selected cell lines or primary cells from other tissues. RNA was treated with DNase and then reversed transcribed into cDNA by using random hexamers and Superscript II RNase H− RT (Invitrogen) at 42°C for 1 h, followed by incubation at 70°C for 15 min and cooling to 4°C. The resulting cDNA was used in PCRs with gene-specific primers designed to span an intron so that amplification of genomic DNA would not occur. The amplification conditions were as follows: 94°C for 2 min; 30 cycles at 94°C for 30 s, 60°C for 1 min, and 68°C for 2 min; and a final extension at 68°C for 4 min. In each experiment, control reactions from which RT was omitted were run to ensure that genomic DNA was not amplified.
Analysis of transcription from upstream cellular genes.
RNA was isolated from highly purified resting CD4+ T cells from HIV-1-positive donors who had shown suppression of viremia to <50 copies/ml while on HAART as described above. For each analysis, 106 sorted resting CD4+ T cells were lysed in 350 μl of lysis buffer (RNeasy minikit), and total RNA was isolated according to the manufacturer's protocol. RNA was treated with DNase (DNase I, amplification grade; Invitrogen) to remove genomic DNA. RNA was then reverse transcribed as described above with a 2 μM concentration of an HIV-specific primer (HIV-RT, 5′-AGTCGCCGCCCCTCGCCTCCTGC-3′, nt 720 to 742 in gag), 130 ng of random hexamers (Invitrogen), 1 μl of RNase inhibitor (RNAguard; Amersham Pharmacia Biotech), and Superscript II according to the manufacturer's protocol. One-fourth of the isolated RNA was used for each RT reaction. Control reactions in which RT was omitted were run to ensure that genomic DNA was not amplified.
PCR amplification of the resulting cDNA was carried out with Expand high-fidelity PCR enzyme, a 1 μM concentration of each gene-specific primer, a 0.2 μM concentration of each deoxynucleoside triphosphate, and 5 μl of cDNA in a 50-μl reaction volume. Primers used for amplification were as follows: HIV-START5′, 5′-GGGTCTCTCTGGTTAGACCAGATCTGAGCC-3′, nt 454 to 483, repeat region; HIV-UP5′, 5′-GGCGAGCCCTCAGATCCTGC-3′, nt 406 to 425, U3 region; and HIV-3′, 5′-CAGCAAGCCGAGTCCTGCGTCG-3′, nt 687 to 708 in gag.
Single-round PCRs of 35 cycles were carried out with either primers HIV-UP5′ and HIV-3′ or primers HIV-START5′ and HIV-3′. Heminested PCRs were also performed. Outer PCRs were carried out with either primers HIV-UP5′ and HIV-RT or primers HIV-START5′ and HIV-RT (35 cycles). Inner PCRs were carried out with either primers HIV-UP5′ and HIV-3′ or primers HIV-START5′ and HIV-3′ (30 cycles). Cycling parameters were as follows: for single-round and outer PCRs: (i) denaturation for 2 min at 94°C; (ii) 35 cycles at 94°C for 45 s, 60°C for 50 s, and 72°C for 1 min; and (iii) a final extension at 72°C for 5 min. Nested PCRs were carried out with the same parameters for 30 cycles.
PCR products were cloned by TOPO TA cloning (Invitrogen) and sequenced. The specificity of the PCR was confirmed by Southern blot hybridization with an HIV-1-specific probe (5′-CTGCTAGAGATTTTCCACACTGAC-3′, nt 612 to 635, U5 region) and by sequence analysis.
Statistical analysis of integration sites.
The P values for the integration environment and the correlation between host gene transcriptional direction and HIV-1 orientation were calculated based on the chi-square distribution. The significance of integration site clustering was inferred from the Poisson distribution. The correlation between integration sites and host transcription start sites was calculated by using nonparametric runs test of randomness (MINITAB Inc.).
RESULTS
Cloning of HIV-1 integration sites from resting CD4+ T cells of patients on HAART.
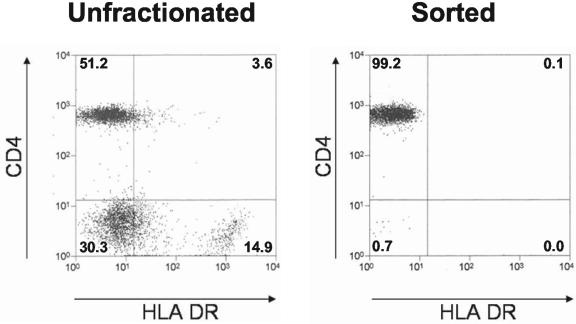
To determine whether the nonproductive nature of HIV-1 infection of resting CD4+ T cells is due to transcriptional repression resulting from integration into regions of heterochromatin, we cloned integration sites from highly purified resting CD4+ T cells from the peripheral blood of infected patients who showed long-term suppression of viremia while on HAART. In such patients, other more labile reservoirs decay, leaving the stable integrated form of HIV-1 in resting memory CD4+ T cells as the principal reservoir (3, 14, 21). A two-stage purification procedure gave preparations of resting CD4+ T cells that contained <0.1% contamination with activated (HLA-DR+) cells (Fig. 1). Resting CD4+ T cells purified in this way do not produce virus without cellular activation (12, 14, 16, 21, 28).
FIG. 1.
Purification of resting CD4+ T cells from patients on HAART. Flow cytometric analysis of unfractionated peripheral blood mononuclear cells (left panel) and highly purified resting CD4+ T lymphocytes (right panel) stained with phycoerythrin-conjugated anti-CD4 and fluorescein isothiocyanate-conjugated anti-HLA-DR antibodies. Numbers indicate the percentages of cells in each quadrant.
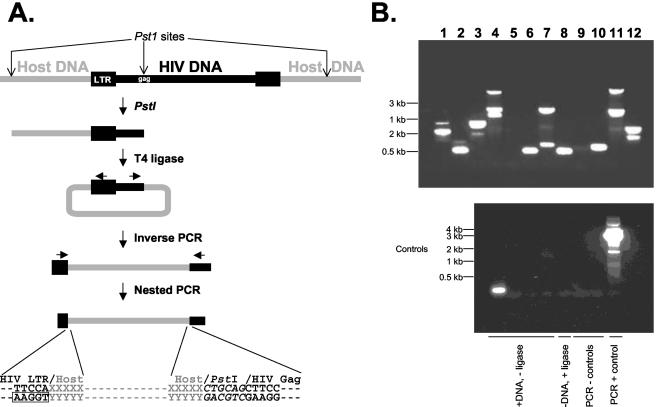
HIV-1 integration sites and the host cell DNA sequences immediately upstream were amplified by inverse PCR (Fig. 2A). Genomic DNA from purified resting CD4+ T cells was digested with PstI or NdeI. The restricted DNA was diluted and then ligated under conditions favoring intramolecular ligation. The resulting circles were used as templates in PCRs with outwardly directed primers located in conserved regions within the terminal fragment of the viral genome. This amplification captured the junction between the viral genome and upstream host cell DNA. A second PCR with a nested set of outwardly directed primers provided sufficient sensitivity to detect rare single-copy integration events. These were visible as bands on an agarose gel suitable for cloning (Fig. 2B). Because integration is random with respect to PstI or NdeI sites, each integration event gives rise to a band of a distinct size, reflecting the distance to the nearest upstream restriction site in the host DNA (Fig. 2A). Distinct sets of bands were obtained in separate PCRs from the same patient (Fig. 2B). Controls for the inverse PCR included reactions from which ligase was omitted (Fig. 2B). These were generally negative except for occasional spurious reaction products which could be readily identified and excluded by sequence analysis. The positive control for the inverse PCR was a previously described LTR-gag-containing plasmid, p1418-PstI (14).
FIG. 2.
Detection of HIV-1 integration sites in resting CD4+ T lymphocytes from patients on HAART by inverse PCR. (A) Inverse PCR strategy for cloning HIV-1 integration sites. Genomic DNA from highly purified resting CD4+ T cells was digested with PstI, which cuts frequently in genomic DNA but only once in most HIV-1 isolates, at nt 1419 in gag. Digested DNA was subjected to intramolecular ligation and nested PCR amplification as described in the text. Boxed nucleotides represent the 5′ end of the LTR. (B) Inverse PCR products from a representative patient. (Top panel) Results of 12 independent inverse PCRs for the same patient. Sequence analysis demonstrated that each band represented a unique integration event from a single infected resting CD4+ T cell. (Bottom panel) Control reactions carried out either without ligase or without DNA.
After cloning and sequencing, integration sites were validated by sequence analysis. Integration sites were considered valid only when they contained a direct junction between the 5′ end of the HIV-1 LTR, beginning with the sequence 5′-TGGAA, and a sequence that was uniquely homologous to a continuous segment of the human genome (Fig. 2A). The AC dinucleotide found at the 5′ end of linear, unintegrated HIV-1 DNA (41, 46, 60) is removed during the integration reaction and is absent except in about ∼1/16 of the integration sites where host nucleotides of this sequence occur fortuitously. Based on genome region-specific sequence data (11, 53, 55), we estimate that short (<4-kb), clonable PstI fragments represent 30 to 40% of the total sequences in euchromatic, centromeric, and telomeric regions of the human genome. Therefore, this method does not introduce an obvious bias for or against integration sites in particular regions of the genome. Integration sites identified with PstI or NdeI had similar characteristics.
Most integrated HIV-1 genomes in resting CD4+ T cells reside in transcription units.
Using the approach described above, we characterized 74 integration sites from 16 patients (Table 1). Each site was unique. Gene-rich chromosomes and gene-dense regions were highly favored as integration sites. Importantly, almost all (93%) of the viral genomes resided within transcription units (Table 1). A total of 76% of the integration sites were in well-characterized genes listed in the curated RefSeq database of molecularly characterized genes (51). Another 18% were within transcription units predicted from sequenced human mRNAs or predicted by the Genscan algorithm. Transcription units occupy about 33% of the human genome (42). Therefore, the fraction of integration sites that lie within genes in vivo in resting CD4+ T cells deviates significantly from that which would occur by random insertion (P < 0.001). Among the 69 integration sites residing within genes, 94% were in introns, probably due to the high percentage of sequence length represented by introns (42).
TABLE 1.
Characteristics of HIV-1 integration sites in infected resting CD4+ T cells
| Chromosome | Patient identification no. | Junctional sequencea | Chromosome locus | Host nt at the junctionb | Integration sitec | Host gene | Description | Orientationd | nt from host transcription start sitee | Host gene expressionf
|
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Resting | With PHA | ||||||||||
| 1 | 21 | AAGGTGTTCC | 1p36.23 | 8158129 | I | RERE | Arg-Glu dipeptide (RE) repeats | − | 428941 (0.92) | + | + |
| 21 | AAGGTTGCCA | 1p36.22 | 9744195 | I | NMNAT1 | Nicotinamide adenylyltransferase | + | 31162 (0.78) | + | + | |
| 149 | AAGGTATGAG | 1p36.11 | 23360670 | I | FUSIP1 | FUS-interacting protein 1 | − | 6447 (0.46) | + | + | |
| 22 | AAGGTCCAAC | 1p34.3 | 35378363 | I | PKD1-like | Polycystic kidney disease 1 related | + | 72287 (0.59) | + | + | |
| 21 | AAGGTGAGGG | 1p34.3 | 35405894 | I | PKD1-like | Polycystic kidney disease 1 related | − | 44772 (0.36) | + | + | |
| 150 | AAGGTCTATA | 1p31.3 | 64357286 | I | KIAA1573 | Unknown function | − | 15771 (0.08) | + | + | |
| 144 | AAGGTCTAAT | 1q21.3 | 151082976 | I | p66beta | Transcription repressor p66 beta | − | 29040 (0.25) | + | + | |
| 153 | AAGGTTCTCC | 1q22 | 151774696 | I | ADAR | Adenosine deaminase, RNA isoform | + | 22765 (0.87) | + | + | |
| 143 | AAGGTCAGAA | 1q32.1 | 196246333 | Int | |||||||
| 2 | 153 | AAGGTGGTTC | 2p23.2 | 28443777 | I | BRE | Brain and reproductive organ expressed | + | 355646 (0.79) | + | + |
| 153 | AAGGTGAGTC | 2q37.3 | 241972574 | I | Predictedg | − | 22723 (0.89) | − | − | ||
| 3 | 21 | AAGGTTAATC | 3p24.1 | 30695399 | I | TGFBR2 | Transforming growth factor β receptor II | − | 72396 (0.85) | + | + |
| 21 | AAGGTTACAC | 3p21.31 | 49514417 | I | DAG1 | Dystroglycan 1 precursor | + | 47814 (0.73) | + | + | |
| 21 | AAGGTAGAGG | 3q29 | 198226821 | I | DLG1 | Synapse-associated protein 97 | − | 125147 (0.49) | + | + | |
| 4 | 21 | AAGGTATATG | 4p14 | 38495952 | I | Predictedg | − | 37220 (0.69) | − | − | |
| 21 | AAGGTCAAAA | 4q25 | 111007127 | I | FLJ20647 | Hypothetical protein | − | 66890 (0.52) | + | + | |
| 5 | 103 | AAGGTTGACG | 5q11.2 | 54977619 | I | FLJ90709 | Hypothetical protein | − | 46361 (0.54) | + | + |
| 21 | AAGGTTATTC | 5q15 | 96212730 | E | ARTS-1 | Aminopeptidase regulator of shedding | + | 4985 (0.11) | + | + | |
| 142 | AAGGTGCATA | 5q23.1 | 121481991 | E | LOX | Lysyl oxidase preproprotein | − | 8154 (0.68) | − | − | |
| 21 | AAGGTTATGT | 5q23.2 | 127559778 | I | SLC12A2 | Solute carrier family 12 | − | 64079 (0.62) | + | + | |
| 22 | AAGGTCGTAC | 5q31.3 | 139878769 | I | FLJ20288 | Hypothetical protein | − | 68775 (0.47) | + | + | |
| 6 | 21 | AAGGTAATTG | 6p25.1 | 5551277 | I | FARS1 | Phenylalanine-tRNA synthetase | + | 237482 (0.59) | + | + |
| 144 | AAGGTCAGAA | 6p21.33 | 30612175 | I | Predictedg | + | 48696 (0.89) | + | + | ||
| 144 | AAGGTGATGG | 6p21.32 | 32940155 | I | TAP2 | TAP2 | + | 203049 (0.68) | + | + | |
| 79 | AAGGTGACAC | 6p21.31 | 35693844 | I | FKBP51 | FK506-binding protein 51 | + | 9725 (0.09) | + | + | |
| 21 | AAGGTTGTAA | 6p21.2 | 38617237 | I | BTBD9 | KIAA1880 protein | − | 37208 (0.08) | + | + | |
| 21 | AAGGTTTTTG | 6q23.3 | 135304182 | I | HBS1L | HBS1-like | − | 52410 (0.55) | + | + | |
| 21 | AAGGTGATCC | 6q23.3 | 135501140 | I | MYB | v-myb myeloblastosis homolog | + | 18030 (0.48) | + | + | |
| 22 | AAGGTTGATA | 6q23.3 | 135792304 | I | FLJ20069 | Hypothetical protein | + | 7149 (0.03) | − | − | |
| 7 | 21 | AAGGTAATAT | 7q22.3 | 106071860 | I | PIK3CG | Phosphatidylinositol 3-kinase, catalytic, gamma | + | 5405 (0.13) | + | + |
| 8 | 21 | AAGGTGGGTA | 8q24.3 | 144830024 | E | hmRNAh | + | 935 (0.28) | + | + | |
| 9 | 22 | AAGCTGCAGC | 9q21.2 | 75991003 | I | GNAQ | GTP-binding protein, q protein | + | 112487 (0.36) | + | + |
| 22 | AAGGTTACCG | 9q32 | 112854741 | I | Predictedg | + | 46833 (0.43) | − | − | ||
| 10 | 21 | AAGGTAAGGA | 10q25.1 | 111475750 | I | ADD3 | Adducin 3 isoform a | − | 45357 (0.35) | − | − |
| 11 | 21 | AAGGTAAAAA | 11p15.4 | 9744195 | I | NAP1L4 | Nucleosome assembly protein | − | 7800 (0.16) | + | + |
| 21 | AAGGTCACAG | 11q13.1 | 64319021 | I | SF1 | Splicing factor 1 | − | 2574 (0.18) | + | + | |
| 21 | AAGGTATTAT | 11q13.4 | 71857871 | I | SKD3 | Suppressor of K transport defect 3 | + | 14005 (0.10) | + | + | |
| 12 | 143 | AAGGTCGATT | 12p13.32 | 3825992 | I | c12orf6 | Chromosome 12 open reading frame 6 | + | 26809 (0.44) | + | + |
| 152 | AAGGTCAAAG | 12p11.21 | 31748771 | I | hmRNAh | + | 24508 (0.42) | − | + | ||
| 150 | AAGGTGAAAC | 12q13.12 | 49358494 | I | FLJ34278 | Hypothetical protein | − | 173459 (0.99) | + | + | |
| 151 | AAGGTGTCGA | 12q13.13 | 52922906 | I | CBX5 | Heterochromatin protein 1 | + | 16679 (0.93) | + | + | |
| 145 | AAGGTAATGG | 12q22 | 91298430 | I | Predictedg | + | 47946 (0.32) | − | − | ||
| 15 | 146 | AAGGTCACTC | 15q22.2 | 57090553 | I | RNF111 | Ring finger protein 111 | − | 94632 (0.87) | + | + |
| 21 | AAGGTGAATC | 15q23 | 66135863 | I | PIAS1 | Inhibitor of activated STAT | + | 73473 (0.55) | + | + | |
| 153 | AAGGTTATAG | 15q25.3 | 83919694 | I | AKAP13 | A-kinase (PRKA) anchor protein 13 | − | 266055 (0.72) | + | + | |
| 16 | 153 | AAGGTTTTTC | 16p11.2 | 29719156 | I | hmRNAh | + | 522325 (0.81) | − | − | |
| 21 | AAGGTTCCGA | 16p11.2 | 29762139 | I | hmRNAh | − | 479342 (0.74) | − | − | ||
| 21 | AAGGTGAGAA | 16p11.2 | 29847207 | I | KIF22 | Kinesin family member 22 | − | 7512 (0.51) | + | + | |
| 21 | AAGGTCATTG | 16q13 | 57040912 | I | hmRNAh | + | |||||
| 79 | GAGGTTTTAG | 16q22.1 | 67956187 | I | NFATc3 | Nuclear factor of activated T cells | + | 60576 (0.43) | + | + | |
| 17 | 21 | AAGGTGTCAG | 17p13.3 | 2435251 | E | FLJ10543 | Hypothetical protein | − | 10740 (0.80) | + | + |
| 21 | AAGGTGAAAG | 17p11.2 | 16262774 | I | NCoR1 | Nuclear receptor corepressor 1 | − | 56637 (0.31) | + | + | |
| 21 | AAGGTGGAGG | 17p11.2 | 20012102 | I | AKAP10 | A-kinase anchor protein 10 | + | 31056 (0.43) | + | + | |
| 150 | AAGGTGGACC | 17p21.2 | 38936487 | I | TOP2A | DNA topoisomerase II alpha | + | 10739 (0.38) | − | + | |
| 103 | AAGGTGGTTT | 17q21.31 | 42202698 | I | MEOX1 | Mesenchyme homeobox 1 | + | 11398 (0.54) | − | + | |
| 21 | AAGGTGAAAG | 17q25.1 | 73929654 | I | GRB2 | Growth factor receptor-bound 2 | − | 57225 (0.78) | + | + | |
| 21 | AAGGTCACTT | 17q25.1 | 74812346 | I | hmRNAh,i | − | + | + | |||
| 153 | AAGGTGAAGG | 17q25.1 | 74938954 | I | PRPSAP1 | PRPP synthetase associated | − | 8100 (0.19) | + | + | |
| 153 | AAGGTGACTG | 17q25.3 | 76515866 | I | EVER1 | Epidermodysplasia verruciformis 1 | + | 206035 (0.55) | + | + | |
| 21 | AAGGTCAGGC | 17q25.3 | 80511487 | Int | LINE (L1) | ||||||
| 20 | AAGGTGGATC | 17q25.3 | 81407969 | I | TBCD | β-Tubulin cofactor D | − | 19195 (0.10) | + | + | |
| 19 | 99 | AAGGTCTCTA | 19p13.2 | 10153709 | I | DNMT1 | DNA methyltransferase 1 | + | 13102 (0.21) | + | + |
| 21 | AAAGTGATCC | 19p13.2 | 10309206 | I | ICAM3 | Intercellular adhesion molecule 3 | + | 2094 (0.36) | + | + | |
| 21 | AAGGTTATTC | 19p13.11 | 17800839 | I | JAK3 | Janus kinase 3 | − | 15443 (0.84) | + | + | |
| 22 | AAGCTGCAGT | 19q13.11 | 39535086 | I | KIAA0355 | Hypothetical protein | + | 97769 (0.97) | + | + | |
| 21 | AAGGTGGGTC | 19q13.12 | 40889171 | Int | |||||||
| 152 | AAGGTCTACC | 19q13.33 | 55192036 | I | VRK3 | Vaccinia virus-related kinase 3 | − | 28412 (0.58) | + | + | |
| 21 | AAGGTGAGTT | 19q13.43 | 61779316 | I | Predictedg | − | 9227 (0.17) | + | + | ||
| 20 | 153 | AAGGTGAGGA | 20q13.13 | 50223866 | I | ADNP | Activity-dependent neuroprotector | − | 9083 (0.22) | + | − |
| 22 | 22 | AAGGTCAATT | 22q11.21 | 19609567 | I | CRKL | v-crk oncogene homolog | + | 13299 (0.40) | + | + |
| 153 | AAGGTGAGGA | 22q13.1 | 35834266 | Int | LTR (endogenous retrovirus 1) | ||||||
| 21 | AAGGTTAGTG | 22q13.1 | 36501308 | I | EIF3S6IP | Eukaryotic translation initiation factor 3 | − | 12844 (0.33) | + | + | |
| 22 | AAGGTCATGA | 22q13.33 | 49112299 | Int | Alu | ||||||
| X | 22 | AAGGTCTAAA | Xp11.3 | 41971993 | I | Predictedg | + | 75610 (0.48) | − | − | |
Junction between the 5′ end of the HIV-1 LTR (first five letters) and the host cell DNA.
The host nucleotide number at the junction was determined by using the UCSC Bioinformatics Human Genome Database (July 2003 assembly freeze). Bold type indicates clusters of two integration events within a 1-Mb window (P, 3.9 × 10−5). Italic type indicates clusters of three integration events within a 1-Mb window (P, 2.6 × 10−5).
Nature of the integration site: I, intron; E, exon; Int, intergenic. Overall, 93.2% (69 of 74) of the integration sites were in defined or predicted genes, while 6.8% (5 of 74) were in intergenic regions. Among integrations in genes, 94.2% (65 of 69) were in introns and 5.8% (4 of 69) were in exons.
Transcriptional orientation: +, the host gene and the HIV-1 insert have the same transcriptional orientation; −, the gene and the insert have the opposite orientation. Of genes in transcription units, 49.3% (34 of 69) were in the + orientation and 50.7% (35 of 69) were in the − orientation.
Distance, in nucleotides, between the start site for transcription of the host gene and the HIV-1 integration site. Numbers in parentheses indicate the relative position of the integration site within the gene, with 0 representing the start of transcription and 1 representing the end of the transcript.
For integration sites within known or predicted genes, RT-PCR was carried out with gene- specific primers spanning an intron on total RNA isolated from purified resting or activated (with PHA) CD4+ T cells. +, presence of a PCR product of the predicted size; −, no PCR product under conditions that gave a readily detectable band for a ubiquitously expressed gene (GAPDH) and, for characterized genes, a correct product from cell lines or primary tissues known to express the gene. These included chondrocytes (LOX), kidney cells (ADD3), and mesenchymal stem cells (MEOX1). The expression of TOP2A, MEOX1, and a human mRNA from chromosome 12p11.21 was detected in activated but not resting CD4+ T cells. For integration events in well-characterized (RefSeq) genes, expression of the targeted gene in resting CD4+ T cells was observed for 91.1% (51 of 56) of the genes.
Gene predicted by the Genscan algorithm.
hmRNA, human mRNA from GenBank.
The start site for transcription has not yet been determined.
Among the 74 sites identified, we observed for different patients several clusters of independent integration events that fell within 1 Mb of each other (Table 1, bold and italic type). All of these clusters were in gene-dense regions. Only 5 out of 74 integration events occurred in intergenic regions, and 4 of these were found within 10 kb of RefSeq transcription units. There was no apparent tendency for HIV-1 integration into repetitive elements, despite their frequency within the human genome (Table 1). Despite previous work suggesting that HIV-1 latency involves integration into centromeric DNA (32), no sites were identified in alphoid regions of centromeres.
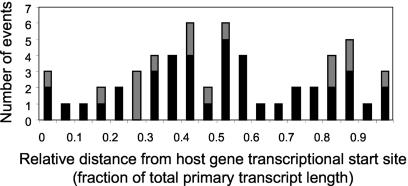
Although there was a striking preference for integration into transcriptional units, the orientation of the HIV-1 genome with respect to the host gene and its location within the host gene appeared to be random (Table 1 and Fig. 3). Unlike the findings for endogenous human retroviruses (59), there was no correlation between the orientation of integrated HIV-1 in vivo and the direction of cellular gene transcription (P > 0.5) (Table 1). In contrast to the findings for murine leukemia virus, which integrates preferentially in the vicinity of transcription start sites (68), there was no significant correlation between HIV-1 integration site and the start site for the transcription of the targeted host gene (P = 0.8955) (Fig. 3). For both RefSeq and non-RefSeq genes, HIV-1 integration sites in resting CD4+ T cells were found along the entire length of the host genes (Fig. 3).
FIG. 3.
Distribution of HIV-1 integration events along the lengths of targeted genes. Results are shown both for genes that are part of the RefSeq database (51) (black bars [bottom portion of bars]) of well-characterized genes and for genes predicted on the basis of sequenced human mRNAs or by sequence analysis (gray bars [top portion of bars]). Each transcript, regardless of its length, was divided into 20 equal parts, and the position of HIV-1 integration within the transcript was plotted by using these relative length units.
Integration sites in infected resting CD4+ T cells are in host genes that are actively expressed.
To determine whether integration sites in resting CD4+ T cells were in host genes that were transcriptionally active, we analyzed the expression status of the targeted host genes in populations of resting CD4+ T cells. Because each integration site is unique and is likely present in only a single cell in any blood sample, it is not possible to determine simultaneously both the nature of the integration site in a given cell and the level of expression of the targeted host gene in that particular cell. However, infected resting CD4+ T cells from patients on HAART express little HIV-1 RNA (7, 28) and are therefore very similar to uninfected resting CD4+ T cells. A recent study by Chun et al. showed that gene expression profiles for resting CD4+ T-cell populations from aviremic patients on HAART are similar to profiles for resting CD4+ T cells from healthy donors (12). Therefore, the expression of targeted host genes was analyzed by RT-PCR with RNA from purified resting CD4+ T cells from HIV-1-negative donors (Fig. 4 and Table 1). Expression patterns in mitogen-activated CD4+ T cells and appropriate control cell lines were also examined.
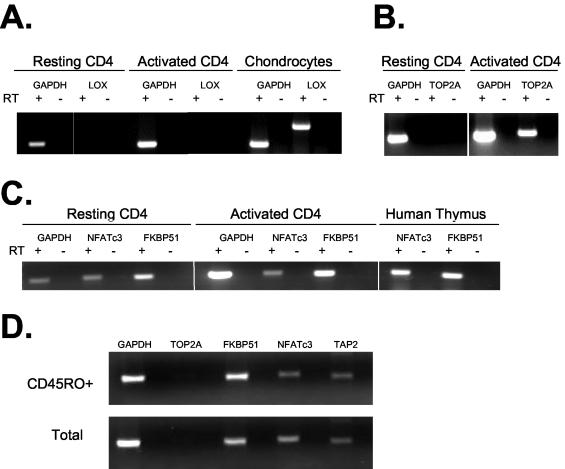
FIG. 4.
Analysis of host gene expression in resting CD4+ T cells. All transcriptional units in which an integration site was identified were analyzed by RT-PCR (Table 1). Representative examples are shown. In all three panels, GAPDH served as a positive control for RT-PCR. RT-PCRs were carried out with (+) or without (−) RT. (A) Patterns of expression of GAPDH and LOX by resting CD4+ T cells, by activated CD4+ T cells, and by chondrocytes. In all instances, RT-PCR signals were dependent on the presence of RT. (B) Expression of TOP2A by mitogen-activated but not resting CD4+ T cells. RT-PCR signals were dependent on the presence of RT. (C) Analysis of NFATc3 and FKBP51 expression in resting and activated CD4+ T cells and in the thymus. RT-PCR signals were dependent on the presence of RT. (D) Patterns of expression of genes targeted for integration in resting CD4+ T cells and in the memory subset of resting CD4+ T cells.
For each of the integration sites in known or predicted genes, primers spanning an intron and producing amplicons of approximately 400 to 500 bases were designed. RT-PCRs were carried out with a standard set of conditions that gave expected results for the characterized genes. In all instances when results were negative with RNA from resting CD4+ T cells, the integrity of the RNA sample was assessed by successful detection of transcripts from ubiquitously expressed genes, such as that for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).In addition, we evaluated the ability of the primers to detect the expression of the relevant gene in activated CD4+ T cells or in positive control cell lines from a tissue known to express the gene.
Examples of this analysis are shown in Fig. 4. Under the conditions used, RNA for the ubiquitously expressed enzyme (GAPDH) was readily detectable in resting CD4+ T cells, while RNA for the tissue-specific enzyme lysyl oxidase (LOX) was not detected (Fig. 4A). LOX is an amine oxidase important for cross-linking collagen and elastin in the extracellular matrix. The LOX gene is expressed by fibrogenic cells, and there is no evidence for expression in lymphocytes (36). Activated CD4+ T cells also failed to express LOX, but expression was readily detected in a chondrocyte cell line, confirming the ability of the RT-PCR to detect LOX RNA when present (Fig. 4A). As expected, resting CD4+ T cells did not express genes that are expressed only in cycling cells, such as the gene for topoisomerase II (TOP2A), an enzyme that is involved in DNA replication. As shown in Fig. 4B, the expression of the TOP2A gene was not detected in purified resting CD4+ T cells but was readily detected in activated CD4+ T cells. Thus, the RT-PCR conditions used gave the results expected for the characterized genes.
With this approach, the expression of targeted host genes was analyzed for all of the integration sites for which there was sufficient information on intron-exon structure to permit the design of primers (n = 68). The results are shown in Table 1. The most important finding was that a high proportion of the genes in which integration events were found were actively transcribed in resting CD4+ T cells from healthy donors. For the 68 genes analyzed, expression in resting CD4+ T cells was clearly detectable for 55 (81%). For five genes for which expression was not detected, the integration site was located in a putative gene that was predicted by the Genscan algorithm but for which no human mRNA had yet been described. It is therefore possible that the absence of expression reflected deficiencies in the gene prediction program. When the analysis is restricted to the 56 integration sites in molecularly characterized genes that are part of the RefSeq database (36), the results are even more striking. Expression in resting CD4+ T cells was observed for 50 of 56 genes (89%). Some examples are shown in Fig. 4C. RNAs for both nuclear factor of activated T cells (NFATc3) and FK506-binding protein 51 (FKBP51) were expressed in resting CD4+ T cells as well as activated CD4+ T cells (Fig. 4C). NFATc3 is a member of the nuclear factors of activated T cells family of transcription factors, which are present in the cytosol of resting lymphocytes (22) and which are translocated to the nucleus following dephosphorylation by calcineurin upon T-cell activation. FKBP51 is a widely expressed member of the FK506-binding protein family (2). Two of the genes in which integration sites were found (those for TOP2A and mesenchyme homeobox 1 [MEOX1]) were expressed in activated but not resting CD4+ T cells; a single gene (that for the activity-dependent neuroprotector [ADNP]) was expressed in resting but not activated cells. Overall, 91% (51 of 56) of the targeted genes were expressed in resting CD4+ T cells (Table 1). Patterns of gene expression were also analyzed by using cDNA arrays (data not shown). In general, the results were similar, but because RT-PCR is more sensitive and reliable, results from the RT-PCR analysis are presented. Taken together, our results suggest that in infected resting CD4+ T cells, HIV-1 genomes reside predominantly in genes that are actively transcribed in resting CD4+ T cells.
Previous studies suggested that in resting CD4+ T cells, the majority of integrated HIV-1 DNA is present in cells of the memory subset (6, 14). We therefore confirmed that the expression of the genes in Table 1 was similar for the memory subset of CD4+ T cells. During the purification of resting cells, the memory subset was enriched to >95% by depletion of cells expressing the RA isoform of CD45 and sorting for cells expressing the RO isoform. The memory subset was shown to have a pattern of expression of the relevant genes very similar to that of the entire resting CD4+ T-cell population. For example, as shown in Fig. 4D, both populations of cells expressed FKBP51, NFATc3, and TAP2 but not TOP2A.
Incorporation of HIV-1 sequences into transcripts of host genes.
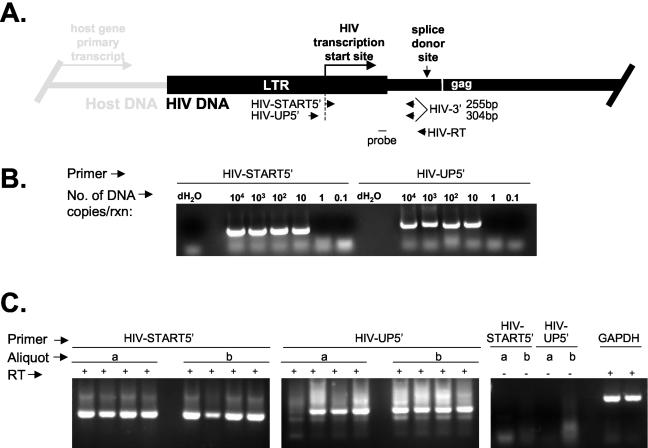
The finding that HIV-1 integration sites in resting CD4+ T cells are located predominantly within introns of genes that are actively expressed in resting CD4+ T cells predicts that the HIV-1 genome should be included within the primary transcripts of the targeted host genes and then degraded along with the remainder of the relevant introns following splicing. Thus, RNA species derived from host promoters and containing HIV-1 sequences may be present in resting CD4+ T cells, although these cells are not permissive for high-level expression from the HIV-1 LTR. To test this hypothesis, RT-PCR experiments were designed to detect low levels of transcripts containing HIV-1 sequences upstream of the HIV-1 transcription start site and to distinguish them from the low levels of bona fide HIV-1 transcripts produced in infected resting CD4+ T cells (Fig. 5A). RNA from resting CD4+ T cells was reverse transcribed with the gene-specific primer HIV-RT, which binds upstream of the first HIV-1 splice site. The resulting cDNA was amplified with two primer sets that share a 3′ primer. One primer set (HIV-UP5′ and HIV-3′) amplifies only HIV-1 sequences produced as a result of transcription of the targeted host gene reading through the HIV-1 genome that is inserted into the gene. Because the forward primer is located upstream of the transcription start site (nt 456 of the LTR) and the reverse primer is located downstream of the LTR, only RNA species initiating upstream of the HIV-1 transcription start site are amplified. The other primer set (HIV-START5′ and HIV-3′) is able to amplify these transcription products in addition to any HIV-1 transcripts that have initiated at the HIV-1 LTR. To prevent amplification from HIV-1 DNA, isolated RNA was treated with DNase before RT-PCR. In addition, control reactions from which RT was omitted were included in each experiment and were invariably negative. In control experiments, both primer sets were shown to have comparable sensitivities on a plasmid template (Fig. 5B).
FIG. 5.
Detection of transcripts containing HIV-1 sequences in resting CD4+ T lymphocytes from patients on HAART. (A) RT-PCR strategy for detecting HIV-1 transcripts initiating at the HIV-1 LTR and transcripts initiating upstream of the transcription start site. The positions of the two forward PCR primers (HIV-START5′ and HIV-UP5′) relative to the transcription start site are indicated. (B) Sensitivity of heminested PCR analysis with, as a template, 10-fold serial dilutions of a plasmid (pLAI) containing the entire HIV-1 genome. The forward primer was either HIV-START5′ or HIV-UP5′. For the first reaction (rxn), the reverse primer was HIV-RT. For the nested reaction, the reverse primer was HIV-3′. dH2O, distilled H2O. (C) Analysis of transcripts from two independent aliquots (a and b) of 106 sorted, resting CD4+ T cells from a representative patient. (Left panel) Transcripts were amplified in quadruplicate reactions with forward primer HIV-START5′. (Middle panel) Transcripts were amplified in quadruplicate reactions with forward primer HIV-UP5′. (Right panel) Control reactions lacking RT were performed with the indicated forward primers. GAPDH served as a positive control for RNA isolation and RT-PCRs for each aliquot.
The presence of both types of transcripts in highly purified populations of resting CD4+ T cells from patients on HAART and showing stable suppression of viremia to below the limit of detection was analyzed. Single-round, semiquantitative RT-PCR demonstrated that RNAs containing HIV-1 sequences upstream of the transcription start site were present, but at levels that were even lower than the very low levels of transcripts initiating at the HIV-1 transcription start site (data not shown). This finding may reflect the rapid turnover of the HIV-1 sequences contained within the introns of other genes as well as the fact that only half of the integration sites were in the same transcriptional orientation as the host gene (Table 1). For these reasons, a more sensitive heminested PCR approach was used to detect low levels of short-lived RNA species (Fig. 5C). Reverse primers HIV-RT and HIV-3′ were used in the first and second rounds, respectively. Two independent aliquots of RNA from 106 purified resting CD4+ T cells were analyzed in quadruplicate with each primer set. Both types of transcripts could be readily detected as bands of the expected sizes. The nature of the PCR products was confirmed by Southern blot hybridization with an internal probe (Fig. 5A) and by direct cloning and sequencing. With this approach, transcripts originating from upstream of the HIV-1 LTR could be detected in purified resting CD4+ T cells from each of six patients studied. Control reactions that lacked RT were invariably negative (Fig. 5C, right panel). Taken together, these results demonstrate that RNA molecules containing HIV-1 LTR sequences upstream of the HIV-1 transcription start site can be detected in resting CD4+ T cells, consistent with the active transcription of host genes carrying the integrated HIV-1 genome.
DISCUSSION
We have analyzed sites of HIV-1 integration in vivo in highly purified resting CD4+ T cells from the peripheral blood of patients on HAART. This cell population is of particular interest because, despite the presence of cells carrying integrated HIV-1 DNA (13, 14, 16), there is no detectable virus production without some form of stimulation (12, 14, 16, 28). It was therefore of interest to determine whether the nonproductive nature of the infection of resting CD4+ T cells in vivo is related to the nature of the integration site. HIV-1 integration sites in transformed cell lines infected in vitro with HIV-1 were previously analyzed (32, 54, 68). Transcriptional units were favored sites of integration in two studies (54, 68), but when cells were selected for reversibly nonproductive viral gene expression, integration into regions of heterochromatin was favored (32).
In our in vivo analysis of integration sites in resting CD4+ T cells, we found that integration into transcriptional units was strongly favored (93%). Typically, HIV-1 integration sites were within introns of the relevant genes. Moreover, many of the genes found to contain integration sites were actively expressed in resting CD4+ T cells from healthy donors (91%). This result was obtained by direct RT-PCR analysis of each gene and did not rely solely on cDNA array analysis. The finding that HIV-1 genomes reside within introns of genes actively expressed in resting CD4+ T cells predicts that HIV-1 sequences may be included in the primary transcripts of the relevant host genes. This hypothesis is supported by the results of RT-PCR analysis with primers that can amplify only templates containing HIV-1 sequences upstream of the HIV-1 transcription start site. We conclude that for the majority of resting CD4+ T cells with integrated HIV-1 DNA, the absence of virus production cannot be attributed to integration into chromosomal regions that are intrinsically repressive for transcription. Furthermore, our results show that the presence of integrated HIV-1 DNA within an actively transcribed host gene is not sufficient for productive infection of resting CD4+ T cells. Mechanisms unrelated to the nature of the integration site must account for the nonproductive nature of the infection in resting CD4+ T cells.
One potential explanation for the nonproductive nature of the infection in resting CD4+ T cells is that these cells carry defective HIV-1 genomes. The in vivo error rate for HIV-1 RT is estimated to be 3.4 × 10−5 substitutions/nucleotide/cycle. For a genome of 9.6 × 103 nt, there is only a 1 in 3 probability (0.326) that each newly arising HIV-1 genome will contain a new mutation, and only a fraction of these mutant genomes will be defective for replication. However, there may be some selection for defective genomes in vivo. In some of the activated CD4+ T cells that become infected, new mutations arising during reverse transcription may adversely affect virus gene expression. As a result, the cells will not express viral proteins and will not die from viral cytopathic effects or host cytolytic effector mechanisms. In principle, such cells will have a greater chance of surviving long enough to revert back to a resting state. Thus, resting CD4+ T cells carrying defective genomes may accumulate.
Nevertheless, many studies have confirmed that replication-competent virus can persist in a latent form in resting CD4+ T cells for long periods of time, even in patients who show complete suppression of detectable viremia while on HAART (3, 13, 14, 16, 21, 67). Potential mechanisms for HIV-1 latency include proviral integration into chromosomal sites that are or that become repressive for transcription (26, 32, 33), the absence in resting CD4+ T cells of activation-dependent host transcriptional activators necessary for HIV-1 gene expression (5, 17, 45, 64), premature termination of HIV-1 transcripts due to the absence of HIV-1 Tat and Tat-associated host factors (1, 25, 29-31, 35), and the failure to export unspliced HIV-1 RNA due to insufficient levels of Rev (43, 50). Because only a small fraction of the resting CD4+ T cells with integrated HIV-1 DNA can produce replication-competent virus following cellular activation, our data do not exclude the possibility that HIV-1 latency involves integration into chromosomal regions that are or that become repressive for transcription (26, 32, 33). Although we did not observe integration into regions of heterochromatin such as the alphoid repeat regions of centromeres, it remains possible that rare integrations into such sites occur in vivo and that such integrations are associated with latency.
The nature of the HIV-1 integration sites in vivo can provide insights into the accessibility of different chromosomal regions and the targeting mechanisms used in the integration process. Because direct infection of resting CD4+ T cells does not lead to integration due to blocks at the level of entry (48, 52), reverse transcription (49, 69), and nuclear import (9, 38), it is most likely that resting CD4+ T cells with integrated HIV-1 DNA arise when activated CD4+ T cells become infected and then revert back to a resting state (3). According to this model, the characteristics of the integration sites in vivo should reflect host DNA accessibility and viral targeting characteristics operative in activated CD4+ T cells. Our results are consistent with this model in that most targeted genes were actively expressed in both activated and resting CD4+ T cells. This pattern is strikingly different from that observed for some other retroviruses and retroelements. Avian leukosis virus integration is directed away from actively transcribed genes (66). Murine leukemia virus tends to integrate near transcription start sites (68) and not throughout the targeted gene, as observed here. The Ty1-4 retrotransposons of Saccharomyces cerevisiae integrate upstream of genes transcribed by RNA polymerase III (4). In contrast to the random transcriptional orientation of HIV-1 in resting CD4+ T cells, endogenous human retroviruses tend to have an antisense orientation (59). Despite widely different patterns of integration adopted by various retroelements, the pattern of HIV-1 integration observed in resting CD4+ T cells is essentially the same as that observed in extensive studies of HIV-1 integration into proliferating cell lines in vitro (54, 68). This finding reinforces the notion that the nonproductive nature of the infection in resting CD4+ T cells with integrated HIV-1 DNA is not the result of the nature of the integration site.
It is not yet possible to determine how the present results bear on the critical issue of HIV-1 latency. Such a determination will require the development of methods for simultaneously determining the integration site and the replication competence of individual proviruses. Although difficult, this is an important goal. The capacity of replication-competent HIV-1 genomes to persist in a latent state in resting CD4+ T cells for many years, even in patients on HAART, represents a major barrier to HIV-1 eradication (13, 20, 21, 57, 62, 67). If the general patterns observed here for integrated proviruses in resting CD4+ T cells also apply to the subset of proviruses that are replication competent, then the latent reservoir may be more accessible to therapeutic intervention than previously thought.
Acknowledgments
We thank members of the Siliciano laboratory for many helpful discussions and Mike Paradise for help in recruiting patients.
This work was supported by NIH grant AI43222, a grant from the Johns Hopkins Center for AIDS Research, the Doris Duke Charitable Foundation, and the Howard Hughes Medical Institute.
REFERENCES
- 1.Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baughman, G., G. J. Wiederrecht, F. Chang, M. M. Martin, and S. Bourgeois. 1997. Tissue distribution and abundance of human FKBP51, an [sic] FK506-binding protein that can mediate calcineurin inhibition. Biochem. Biophys. Res. Commun. 232:437-443. [DOI] [PubMed] [Google Scholar]
- 3.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 4.Boeke, J. D., and S. E. Devine. 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93:1087-1089. [DOI] [PubMed] [Google Scholar]
- 5.Bohnlein, E., J. W. Lowenthal, M. Siekevitz, D. W. Ballard, B. R. Franza, and W. C. Greene. 1988. The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-alpha gene and type 1 HIV. Cell 53:827-836. [DOI] [PubMed] [Google Scholar]
- 6.Brenchley, J. M., B. J. Hill, D. R. Ambrozak, D. A. Price, F. J. Guenaga, J. P. Casazza, J. Kuruppu, J. Yazdani, S. A. Migueles, M. Connors, M. Roederer, D. C. Douek, and R. A. Koup. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J. Virol. 78:1160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks, D. G., D. H. Hamer, P. A. Arlen, L. Gao, G. Bristol, C. M. Kitchen, E. A. Berger, and J. A. Zack. 2003. Molecular characterization, reactivation, and depletion of latent HIV. Immunity 19:413-423. [DOI] [PubMed] [Google Scholar]
- 8.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl. Acad. Sci. USA 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukrinsky, M. I., T. L. Stanwick, M. P. Dempsey, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo, K. H., B. Vissel, A. Nagy, E. Earle, and P. Kalitsis. 1991. A survey of the genomic distribution of á satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res. 19:1179-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun, T. W., J. S. Justement, R. A. Lempicki, J. Yang, G. Dennis, Jr., C. W. Hallahan, C. Sanford, P. Pandya, S. Liu, M. McLaughlin, L. A. Ehler, S. Moir, and A. S. Fauci. 2003. Gene expression and viral production in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proc. Natl. Acad. Sci. USA 100:1908-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. M. Mican, M. Baseler, Lloyd, A. L., M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun, T.-W., L. Carruth, D. Finzi, X. Shen, J. A. Digiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y.-H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantitation of latent tissue reservoirs and total body load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 15.Chun, T.-W., D. Engel, S. B. Mizell, L. A. Ehler, and A. S. Fauci. 1998. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 188:83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun, T.-W., D. Finzi, J. Margolick, K. Chadwich, D. Schwartz, and R. F. Siliciano. 1995. Fate of HIV-1-infected T cells in vivo: rates of transition to stable latency. Nat. Med. 1:1284-1290. [DOI] [PubMed] [Google Scholar]
- 17.Duh, E. J., W. J. Maury, T. M. Folks, A. S. Fauci, and A. B. Rabson. 1989. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc. Natl. Acad. Sci. USA 86:5974-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckstein, D. A., M. L. Penn, Y. D. Korin, D. D. Scripture-Adams, J. A. Zack, J. F. Kreisberg, M. Roederer, M. P. Sherman, P. S. Chin, and M. A. Goldsmith. 2001. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity 15:671-682. [DOI] [PubMed] [Google Scholar]
- 19.Festenstein, R., S. N. Pagakis, K. Hiragami, D. Lyon, A. Verreault, B. Sekkali, and D. Kioussis. 2003. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299:719-721. [DOI] [PubMed] [Google Scholar]
- 20.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 21.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 22.Flanagan, W. M., B. Corthesy, R. J. Bram, and G. R. Crabtree. 1991. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature 352:803-807. [DOI] [PubMed] [Google Scholar]
- 23.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganesh, L., E. Burstein, A. Guha-Niyogi, M. K. Louder, J. R. Mascola, L. W. Klomp, C. Wijmenga, C. S. Duckett, and G. J. Nabel. 2003. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature 426:853-857. [DOI] [PubMed] [Google Scholar]
- 25.Ghose, R., L. Y. Liou, C. H. Herrmann, and A. P. Rice. 2001. Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4+ T lymphocytes by combination of cytokines. J. Virol. 75:11336-11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, G., L. Ylisastigui, and D. M. Margolis. 2002. The regulation of HIV-1 gene expression: the emerging role of chromatin. DNA Cell Biol. 21:697-705. [DOI] [PubMed] [Google Scholar]
- 27.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermankova, M., J. D. Siliciano, Y. Zhou, D. Monie, K. Chadwich, J. B. Margolick, T. C. Quinn, and R. F. Siliciano. 2003. Analysis of HIV-1 gene expression in latently infected resting CD4+ T lymphocytes in vivo. J. Virol. 77:7383-7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrmann, C. H., R. G. Carroll, P. Wei, K. A. Jones, and A. P. Rice. 1998. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 72:9881-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, K. A., and B. M. Peterlin. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63:713-743. [DOI] [PubMed] [Google Scholar]
- 32.Jordan, A., D. Bisgrove, and E. Verdin. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaech, S. M., E. J. Wherry, and R. Ahmed. 2002. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2:251-262. [DOI] [PubMed] [Google Scholar]
- 35.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 36.Kim, Y., C. D. Boyd, and K. Csiszar. 1995. A new gene with sequence and structural similarity to the gene encoding human lysyl oxidase. J. Biol. Chem. 270:7176-7182. [DOI] [PubMed] [Google Scholar]
- 37.Korin, Y. D., D. G. Brooks, S. Brown, A. Korotzer, and J. A. Zack. 2002. Effects of prostratin on T-cell activation and human immunodeficiency virus latency. J. Virol. 76:8118-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korin, Y. D., and J. A. Zack. 1999. Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes. J. Virol. 73:6526-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, P. A. Marx, F. McCutchan, J. W. Mellors, and S. Wolinsky. 2001. HIV sequence compendium 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 40.Kulkosky, J., D. M. Culnan, J. Roman, G. Dornadula, M. Schnell, M. R. Boyd, and R. J. Pomerantz. 2001. Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98:3006-3015. [DOI] [PubMed] [Google Scholar]
- 41.Kulkosky, J., R. A. Katz, and A. M. Skalka. 1990. Terminal nucleotides of the preintegrative linear form of HIV-1 DNA deduced from the sequence of circular DNA junctions. J. Acquir. Immune Defic. Syndr. 3:852-858. [PubMed] [Google Scholar]
- 42.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. Fitzhugh, et al. 2001. Intitial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 43.Malim, M. H., and B. R. Cullen. 1991. HIV-1 structural gene expression requires the binding of multiple rev monomers to the viral RRE: implications for HIV-1 latency. Cell 65:241-248. [DOI] [PubMed] [Google Scholar]
- 44.Margolick, J. B., D. J. Volkman, T. M. Folks, and A. S. Fauci. 1987. Amplification of HTLV-III/LAV infection by antigen-induced activation of T cells and direct suppression by virus of lymphocyte blastogenic responses. J. Immunol. 138:1719-1723. [PubMed] [Google Scholar]
- 45.Nabel, G., and D. Baltimore. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711-713. [DOI] [PubMed] [Google Scholar]
- 46.Pauza, C. D. 1990. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology 179:886-889. [DOI] [PubMed] [Google Scholar]
- 47.Persaud, D., T. Pierson, C. Ruff, D. Finzi, K. R. Chadwick, J. B. Margolick, A. Ruff, N. Hutton, S. Ray, and R. F. Siliciano. 2000. A stable latent reservoir for HIV-1 in resting CD4(+) T lymphocytes in infected children. J. Clin. Investig. 105:995-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierson, T., T. L. Hoffman, J. Blankson, D. Finzi, K. Chadwich, J. B. Margolick, C. Buck, J. D. Siliciano, R. W. Doms, and R. F. Siliciano. 2000. Characterization of chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type 1. J. Virol. 74:7824-7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pierson, T. C., Y. Zhou, T. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pomerantz, R. J., D. Trono, M. B. Feinberg, and D. Baltimore. 1990. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell 61:1271-1276. [DOI] [PubMed] [Google Scholar]
- 51.Pruitt, K. D., T. Tatusova, and D. R. Maglott. 2003. NCBI Reference Sequence project: update and current status. Nucleic Acids Res. 31:34-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rabin, R. L., M. K. Park, F. Liao, R. Swofford, D. Stephany, and J. M. Farber. 1999. Chemokine receptor responses on T cells are achieved through regulation of both receptor expression and signaling. J. Immunol. 162:3840-3850. [PubMed] [Google Scholar]
- 53.Riethman, H. C., Z. Xiang, S. Paul, E. Morse, X.-L. Hu, J. Flint, H.-C. Chi, D. L. Grady, and R. K. Moyzis. 2001. Integration of telomere sequences with the draft human genome sequence. Nature 409:948-951. [DOI] [PubMed] [Google Scholar]
- 54.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 55.Schueler, M. G., A. W. Higgins, M. K. Rudd, K. Gustashaw, and H. F. Willard. 2001. Genomic and genetic definition of a functional centromere. Science 294:109-115. [DOI] [PubMed] [Google Scholar]
- 56.Scripture-Adams, D. D., D. G. Brooks, Y. D. Korin, and J. A. Zack. 2002. Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J. Virol. 76:13077-13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siliciano, J. D., J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwich, J. B. Margolick, C. Kovacs, S. J. Gange, and R. F. Siliciano. 2003. Long term follow-up studies confirm the extraordinary stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727-728. [DOI] [PubMed] [Google Scholar]
- 58.Smale, S. T. 2003. The establishment and maintenance of lymphocyte identity through gene silencing. Nat. Immunol. 4:607-615. [DOI] [PubMed] [Google Scholar]
- 59.Smit, A. F. 1999. Interspersed repeats and other momentos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9:657-663. [DOI] [PubMed] [Google Scholar]
- 60.Smith, J. S., S. Kim, and M. J. Roth. 1990. Analysis of long terminal repeat circle junctions of human immunodeficiency virus type 1. J. Virol. 64:6286-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spina, C. A., J. C. Guatelli, and D. D. Richman. 1995. Establishment of a stable, inducible form of human immunodeficiency virus type 1 DNA in quiescent CD4 lymphocytes in vitro. J. Virol. 69:2977-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strain, M. C., H. F. Gunthard, D. V. Havlir, C. C. Ignacio, D. M. Smith, A. J. Leigh-Brown, T. R. Macaranas, R. Y. Lam, O. A. Daly, M. Fischer, M. Opravil, H. Levine, L. Bacheler, C. A. Spina, D. D. Richman, and J. K. Wong. 2003. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc. Natl. Acad. Sci. USA 100:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swingler, S., B. Brichacek, J. M. Jacque, C. Ulich, J. Zhou, and M. Stevenson. 2003. HIV-1 Nef intersects the macrophage CD40L signalling pathway to promote resting-cell infection. Nature 424:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tong-Starksen, S. E., P. A. Luciw, and B. M. Peterlin. 1987. Human immunodeficiency virus long terminal repeat responds to T-cell activation signals. Proc. Natl. Acad. Sci. USA 84:6845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weidhaas, J. B., E. L. Angelichio, S. Fenner, and J. M. Coffin. 2000. Relationship between retroviral DNA integration and gene expression. J. Virol. 74:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 68.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 69.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Y. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 70.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, Z., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353-1357. [DOI] [PubMed] [Google Scholar]