Abstract
Three late assembly domain consensus motifs, namely PTAP, PPPY, and LYPXL, have been identified in different retroviruses. They have been shown to interact with the cellular proteins TSG101, Nedd4, and AP2 or AIP, respectively. Human T-cell leukemia virus type 1 (HTLV-1) has a PPPY and a PTAP motif, separated by two amino acids, located at the end of MA, but only the PPPY motif is conserved in the deltaretrovirus group. Like other retroviral peptides carrying the late motif, MA is mono- or di-ubiquitinated. A mutational analysis showed that 90% of PPPY mutant particles were retained in the cell compared to 15% for the wild-type virus. Mutations of the PTAP motif resulted in a 20% decrease in particle release. In single-cycle infectivity assays, the infectious titers of late motif mutants correlated with the amounts of released virus, as determined by an enzyme-linked immunosorbent assay. We observed binding of MA to the WW domains of the Nedd4 family member WWP1 but not to the amino-terminal ubiquitin E2 variant domain of TSG101 in mammalian two-hybrid analyses. The binding to WWP1 was eliminated when the PPPY motif was mutated. However, MA showed binding to TSG101 in the yeast two-hybrid system that was dependent on an intact PTAP motif. A dominant-negative (DN) mutant of WWP1 could inhibit budding of the intact HTLV-1 virus. In contrast, DN TSG101 only affected the release of virus-like particles encoded by Gag expression plasmids. Electron and fluorescent microscopy showed that Gag accumulates in large patches in the membranes of cells expressing viruses with PPPY mutations. Very few tethered immature particles could be detected in these samples, suggesting that budding is impaired at an earlier step than in other retroviruses.
The last stage in the life cycle of retroviruses is budding from the cell accompanied by enveloping of the virus particle with the plasma membrane. These final steps are completed in a similar fashion in other enveloped viruses such as filoviruses and rhabdoviruses. For retroviruses, the Gag precursor protein alone is sufficient for the assembly and release of virus-like particles (VLPs) (14). Three distinct assembly motifs which mediate membrane attachment (M), the interaction of subunits (I), and late assembly functions (L) have been identified in Gag precursor proteins. L domains of several retroviruses have been characterized and found to be responsible for the fission of the plasma membrane stalk connecting the cell and the budding virus (20, 41, 61, 62, 65).
Recent work revealed that the late assembly domains present on the nascent particle recruit components of the cellular vesicle trafficking machinery to achieve membrane fission. The particular cellular component that interacts with the virus protein depends on the sequence of the late domain motif. To date, three different motifs have been well characterized: they are PPXY, P(T/S)AP, and LYPXL. Most lentiviruses carry a P(T/S)AP motif which is located at the end of Gag and interacts with TSG101 (13, 15, 29, 55). TSG101 is a member of the ubiquitin E2 variant (UEV) family, a group of proteins with close homology to bona fide ubiquitin conjugases, except that they lack a cysteine in the catalytic center. TSG101 is the mammalian homologue of the yeast protein vps23p, which is part of ESCRT1 (2, 22). ESCRT (endosomal sorting complex required for transport) complexes assemble proteins that are modified by the addition of one to four ubiquitins at the limiting membranes of multivesicular bodies (MVBs), eventually causing the invagination and budding of vesicles (6, 30). This process is topologically equivalent to virus budding.
The LYPXL motif was first identified in the p9 region of the Gag precursor of equine infectious anemia virus (EIAV). The motif has been shown to interact with the μ-chain of the adapter complex 2 (5, 41, 42). More recently, a variant of the motif was also identified in p6 of human immunodeficiency virus (HIV) and other primate lentiviruses and was shown to recruit the AIP protein connecting ESCRTI and ESCRTIII (50, 56).
The PPXY motif, which is found in the amino-terminal half of the Gag precursors of retroviruses belonging to several genera, interacts in vitro with WW domains found in many proteins. WW domains share relatively little sequence homology except for two conserved tryptophan residues (52). Members of the Nedd4 family of ubiquitin ligases, which contain one to four WW domains, are the most likely in vivo partners for PPXY motifs. While not an integral member of the vesicle protein sorting pathway, Nedd4 has been shown to initiate the downregulation of several cell surface receptor complexes after stimulation. Nedd4 family proteins modify these receptors by low-level ubiquitination and target them via MVBs for lysosomal degradation (reviewed in references 18 and 43). In addition to catalyzing the initial modification at the plasma membrane, they can act at various other sites along the endosomal downregulation pathway, as demonstrated for the yeast orthologue (9, 57). Although different retroviruses use distinct motifs to recruit different components of the endosomal trafficking machinery, the motifs can be interchanged and are independent of location to some extent (27, 36, 64). However, while the sequence of the particular L domain defines the interacting partner, recent studies by Strack et al. (51) have demonstrated that additional elements in the Gag precursor are necessary for complete egress of the HIV type 1 (HIV-1) particle. Indeed, it has become apparent that most viruses have complex or composite L-domain motifs. Ebola virus contains overlapping PTAP and PPXY motifs (PTAPPEY) in its matrix protein that can replace the L domain in HIV-1 and recruit TSG101 (17, 29). Replacing the HIV-1 p6 peptide with this short Ebola virus sequence is sufficient to allow budding of a minimal HIV-1 Gag precursor. Although the PPXY component of the Ebola motif is not necessary in the context of the complete HIV Gag precursor, abolishing it from the minimal precursor prevents the budding of VLPs, indicating that it contributes in that context.
Human T-cell leukemia virus type 1 (HTLV-1) is a member of the deltaretrovirus family (21). It contains the sequence PPPYVEPTAP at the C-terminal end of the MA region. In this report, we show that mutation of the PPPY motif in provirus expression plasmids severely impeded virus budding, while changing the PTAP sequence caused only a modest defect. Additionally, we show that the loss of the PPPY motif also prevented the ubiquitination of MA, while changes in PTAP did not. We demonstrate that wild-type (wt) MA and a PTAP− mutant interacted with Nedd4 family member WWP1 in mammalian two-hybrid assays but that the binding was eliminated by changes in the PPPY motif. The interaction of wt and PPPY− mutant Gag with TSG101 was detected in yeast two-hybrid analyses, but it was absent for PTAP mutants. A dominant-negative (DN) mutant of WWP1 was shown to inhibit HTLV-1 budding. Electron microscopic imaging of cells expressing the PPPY mutant virus revealed that budding was arrested at an earlier stage than that of other retroviruses with late-assembly-domain mutations.
MATERIALS AND METHODS
Plasmids.
pX1MT and pCMVHT1 have been described previously (8, 19, 48). Mutations were generated by overlapping PCRs and were transferred into the proviral clone as NotI-DraIII restriction fragments, which were completely sequenced. pCMVHT-NCY was generated by inserting the enhanced yellow fluorescent protein (EYFP) gene in frame into the gag gene of pCMVHT1 at codon 427. cDNA clones for TSG101 and WWP1 were generated by reverse transcription-PCR using primers based on GenBank sequences NM006292 and NM007013, respectively. Expression clones were generated by using the Gateway system according to the protocols provided by the manufacturer (Invitrogen, Carlsbad, Calif.). The TSG5′ expression clone was obtained from Z. Sun via E. O. Freed. The retroviral expression vector pUCHRatt is a derivative of pHR (31; D. Derse, P. Lloyd, S. Hill, and G. Heidecker, unpublished data) containing the Gateway attA cassette. The plasmids pCMVΔ8.2, pCMV-VSVg, and pHTC-GFPluc have been described previously (8, 31). The components for the mammalian and yeast two-hybrid systems were purchased from Clontech, Palo Alto, Calif. Plasmid DNAs were isolated with Qiagen (Hilden, Germany) columns and protocols.
Cell cultures, transfections, virus preparation, and infections.
B5, 293T, and HeLa cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, penicillin, and streptomycin. Transfections were done essentially as previously described (19). Six to 18 μg of plasmid DNA was transfected into 5 × 106 293T cells by calcium phosphate precipitation in BES [N,N-bis(2-hydroxyethyl)-2-aminoethane sulfonic acid]-buffered saline. Some experiments were done on a smaller scale with 5 × 105 293T cells, 2 μg of plasmid DNA, and Lipofectamine 2000 (Invitrogen). HeLa cells were transfected by using Fugene (Roche). The medium was changed at 8 to 10 h posttransfection. Supernatants were harvested 24 h later and filtered through 0.45-μm-pore-size filters. For immunoblot analyses, viruses were pelleted through a 20% glycerol cushion at 100,000 × g for 90 min. Cells were washed with phosphate-buffered saline (PBS), lysed at room temperature in TBST (Tris-buffered saline with 1% NP-40 and a protease inhibitor cocktail [Sigma]) for 15 min, and then kept on ice. Lysates were cleared at 20,000 × g for 15 min. Single-cycle infection assays were performed as previously described (8).
Immunoprecipitation, Western blot analysis, and ELISA.
Virus pellets were lysed in 2% sodium dodecyl sulfate-100 mM dithiothreitol for 10 min at 70°C. Lysates were diluted 30-fold with TBS, precleared with normal rabbit antibodies and protein A/G-agarose (Santa Cruz Biotechnologies, Inc.), and then precipitated with a rabbit anti-MA (HTLV-1) or rabbit anti-ubiquitin (Dako, Glostrup, Denmark) antibody in the presence of protein A/G-agarose (Santa Cruz Biotechnologies, Inc.). Precipitates were washed three times in TBST and denatured in sample buffer. Denaturing gel electrophoresis was carried out in 4 to 12% NUPAGE gels in morpholineethanesulfonic acid (MES) or morpholinepropanesulfonic acid (MOPS) buffer according to the manufacturer's directions (Invitrogen). Gels were transferred to Immobilon membranes (Millipore, Bedford, Mass.) and blocked in TBS with 3% dried milk. Probing was done in the same solution with mouse anti-p19 (Zeptomatrix, Buffalo, N.Y.) and mouse anti-ubiquitin (Santa Cruz Biotechnologies, Inc.) monoclonal antibodies. Washing and exposure to the appropriate horseradish peroxidase-linked secondary antibody (Cell Signaling) were done in TBS-0.2% Tween. Blots were developed with ECL+ (Amersham, Piscataway, N.J.) and exposed to X-ray film or analyzed with the STORM system (Molecular Dynamics). Enzyme-linked immunosorbent assay (ELISA) determinations were carried out with filtered tissue culture supernatants and cell extracts by using anti-HTLV-1 p19 ELISA kits from Zeptomatrix according to the manufacturer's protocol.
Mammalian and yeast two-hybrid experiments.
Plasmids from the Matchmaker II or mammalian Matchmaker system were obtained from Clontech. Inserts were generated by PCRs using primers containing the appropriate restriction sites. Transfections for mammalian Matchmaker experiments were done in triplicate in 24-well plates seeded with 293T cells. Luciferase assays were performed by using a kit (Promega, Madison, Wis.) according to the manufacturer's directions. For yeast two-hybrid analyses, plasmids were transfected into competent AH209 cells from Clontech according to the manufacturer's protocol. Growth on SD medium (Invitrogen, Carlsbad, Calif.) without leucine, tryptophan, histidine, and adenine was used as evidence of an interaction, and blue staining of replicated colonies was done in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). In both systems, Gag proteins were expressed as DNA-binding-domain fusions while the TSG101 or WWP1 sequences were fused to the activation domain.
Immunofluorescence microscopy.
For immunofluorescence microscopy analyses, HeLa cells were seeded on glass coverslips. After transfection and growth, cells were fixed in 3.5% paraformaldehyde (Sigma) in PBS for 15 min. After being washed with PBS, the cells were permeabilized and blocked with 0.25% saponin (Sigma) in PBS containing 3% normal goat serum. Samples were incubated with antibodies and washed in the same solution. Samples were mounted in Prolong (Molecular Probes, Eugene, Oreg.) and inspected under a T2000S microscope (Nikon). Images were captured with a Coolsnap Fx digital camera (Roper Scientific).
TEM and IEM.
Detailed procedures for standard thin-section transmission electron microscopy (TEM) and postembedding immunoelectron microscopy (IEM) were described previously (54). Modified procedures were performed as follows. For TEM, cultured cells were scraped, centrifuged at 150 × g, and fixed in 2% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4) overnight at 4°C, followed by 1% osmium fixation for 1 h at room temperature. Cell pellets were dehydrated in graded ethanol (35, 50, 70, 95, and 100%) and propylene oxide (100%). Cell pellets were infiltrated overnight with a 1:1 mixture of propylene oxide and epoxy resin, embedded in pure resin, and cured at 55°C for 48 h. The cured block was thin sectioned, mounted on naked 200-mesh copper grids, and stained in uranyl acetate and lead citrate. For IEM, cultured cells were collected as described above and fixed in 4% paraformaldehyde in PBS for 2 h. Cell pellets were rinsed in cold PBS, dehydrated in cold graded ethanol, infiltrated with a 1:1 mixture of 100% cold ethanol and LR White resin, and then incubated in pure LR White resin overnight at 4°C. Cell pellets were embedded in pure LR White resin and cured at 55°C for 24 h. The cured block was thin sectioned and mounted on naked 200-mesh nickel grids. The grids were incubated for 30 min in PBS containing 10% (wt/vol) normal goat serum, 0.1% (wt/vol) bovine serum albumin (BSA), and 0.05% Tween 20 in order to block nonspecific binding sites. The primary antibody incubation was carried out in serial dilutions made in PBS-BSA buffer overnight at 4°C. The grids were washed in a PBS-BSA-Tween 20 solution for 2 h and incubated with a 1:100 dilution of immunogold-conjugated secondary antibody for 1 h. The grids were washed as described above and stained in uranyl acetate and lead citrate. Digital images were obtained with an electron microscope equipped with a digital camera system.
RESULTS
The PPPY sequence is the dominant late assembly motif in HTLV-1.
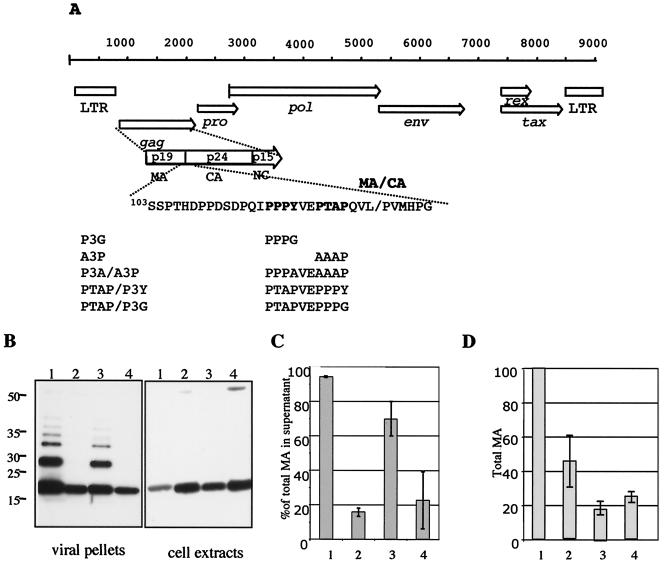
To determine whether the PPPY or the PTAP motif, found at the end of the HTLV-1 MA protein, contributes to the late assembly function, we mutated the elements individually or in combination (Fig. 1A). Previous work by other groups demonstrated that the mutation of PTAP to AAAP (A3P) or PPXY to PPPG or PPPA (P3G or P3A) eliminated late assembly functions in other viruses (25, 35, 59, 62). These mutations were introduced into the infectious molecular clone of HTLV-1 (pX1MT) to give pX1MT-P3G and pX1MT-A3P or the double mutant pX1MTP3A/A3P. The virus mutants were transfected into 293T cells; virus particles in the supernatants were harvested and concentrated by centrifugation through a glycerol cushion. Cell extracts were prepared by nonionic detergent lysis. The same fraction of the total supernatant and the total cell extract was loaded into each lane of the immunoblot gels and was probed with an anti-MA antibody while adjusting to keep the total amount of MA constant (Fig. 1B). Most of the MA protein was found in virus particles, with only a small proportion retained in cells transfected with the wt provirus (lanes 1). Very little budding occurred with the pX1MT-P3G (lanes 2) or double mutant (lanes 4) constructs. In contrast, the pX1MT-A3P construct (lanes 3) released virus into the supernatant with only a slightly lower efficiency than the wt. Interestingly, the Gag precursor that was retained in the cells was significantly processed, with only a slight decrease in processing for late domain mutants compared to the wt.
FIG. 1.
The PPPY motif is responsible for virus export. (A) Genetic map of HTLV-1 showing the major open reading frames. The processed peptides of Gag are indicated in the expanded view of the Gag precursor protein. The late assembly motif and its surrounding sequence are shown; below, the mutations used in this study are indicated. pX1MT-PTAP/P3Y and pX1MT-PTAP/P3G contain an extra proline codon. We also tested the construct without the additional proline and saw no difference (data not shown). (B) Western blots of proteins from viral pellets (left) and cellular extracts (right) probed with a mouse monoclonal anti-MA antibody followed by a horseradish peroxidase-conjugated anti-mouse antibody. Blots were developed with ECL+ and exposed to X-ray film. Pellets and extracts were obtained from 293T cell cultures transiently transfected with pX1MT (lanes 1), pX1MT-P3G (lanes 2), pX1MT-A3P (lanes 3), and pX1MT-P3A/A3P (lanes 4). The positions of molecular weight markers are given on the left. (C) ELISA determination of the percentages of MA in the supernatant compared to the total MA in the culture. The results of three experiments are summarized. Sample designations are the same as for panel B. (D) Relative total amounts of MA in each culture expressed as percentages of MA expressed by the pX1MT construct. The results of three experiments are summarized. Sample designations are the same as for panel B. Error bars in panels C and D indicate standard errors.
Virus particle release was quantified by measuring the amount of MA in supernatants and cell extracts by p19 (MA) ELISA. The summarized results of three experiments are depicted in Fig. 1C. For the wt virus, 95% ± 0.2% of the total MA protein was in the culture supernatant. In contrast, only 17% ± 2.2% or 22% ± 15% of the total MA was released by cells transfected with the P3G or P3A/A3P mutant, respectively. The A3P mutant consistently had a significantly less severe phenotype (70% ± 10%). Mutation of the late motifs, especially the PTAP sequence, resulted in a drop in steady-state levels of the corresponding Gag protein (Fig. 1D).
Phenotypes of L-domain mutants are independent of Gag expression levels or proteolytic processing.
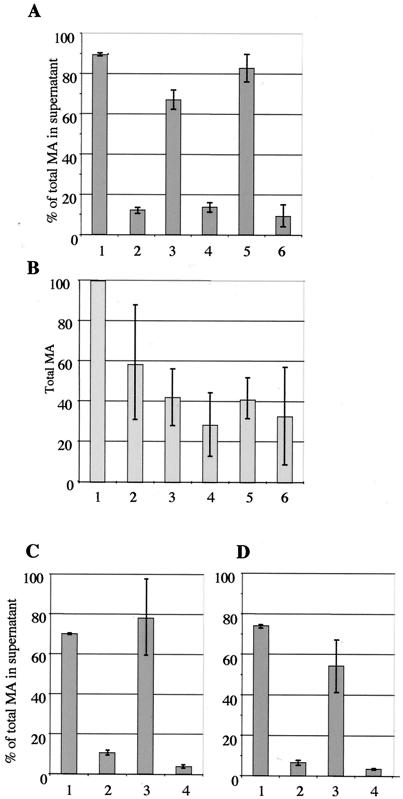
Studies of the late domain mutations in other retroviruses have shown that high levels of Gag expression can overcome the block in budding (for a review, see reference 12); indeed, the late assembly function of the HIV-1 p6 peptide was initially overlooked due to the large amounts of virus produced in the experimental setting. To test whether HTLV-1 late assembly domain mutants could be complemented by higher expression levels of Gag, we transferred the mutations into the provirus expression plasmid pCMVHT1, in which the 5′ long terminal repeat (LTR) was replaced with the cytomegalovirus (CMV) immediate-early promoter. These constructs directed the expression of viral proteins in transfected cells to levels that were about 50-fold higher than those in pX1MT. We used ELISA quantitation to analyze the budding efficiency of virions after transfection into 293T cells. Figure 2A summarizes the ELISA results of several experiments. Consistently, pCMVHT-P3G had a pronounced budding defect comparable to that of pX1MT-P3G; only about 12% ± 1.2% of the total MA protein in the culture was found in the supernatant, compared with about 89.4% ± 1% for pCMVHT1. Again, the construct with the A3P mutation budded relatively efficiently (67% ± 4.5%). To test whether the relative positions of the motifs affected their ability to function as late domains, we changed the PPPYVEPTAP sequence to PTAPVEPPPY. The Gag protein with this mutation was able to form virions as well as the wt (82% ± 6.6%). However, after further mutation of the sequence to PTAPEVP3G, the level of virus particle release dropped to that of the P3G mutant (9% ± 5%). These data indicate that PTAP alone cannot function as a late assembly motif in this context. Cultures transfected with the CMVHT-1 late domain mutants attained lower steady-state levels of Gag protein than those expressing the wt construct (Fig. 2B).
FIG. 2.
PPPY defect cannot be complemented by overexpression or stabilization of Gag precursor. (A) ELISA determination of the percentages of total MA reactive protein present in the supernatants of 293T cultures transfected with pCMVHT1 (bar 1), pCMVHT1-P3G (bar 2), pCMVHT1-A3P (bar 3), pCMVHT1-P3A/A3P (bar 4), pCMVHT1-PTAPVEP3Y (bar 5), and pCMVHT1-PTAPVEP3G (bar 6). The average and standard deviations of five independent experiments are shown. (B) Relative total amounts of MA in each culture expressed as percentages of MA expressed by the pCMVHT-1 construct. The results of three experiments are summarized. Sample designations are the same as for panel A. (C) Percentages of total MA in the supernatants of 293T cell cultures transfected with pCMVHT1-PR− (bar 1), pCMVHT1-PR−-P3G (bar 2), pCMVHT1-PR−-A3P (bar 3), and pCMVHT1-PR−-P3A/A3P (bar 4). (D) Percentages of total MA in the supernatants of 293T cell cultures transfected with pCMVHT1-NCYΔX (bar 1), pCMVHT1-NCYΔX-P3G (bar 2), pCMVHT1-NCYΔX-A3P (bar 3), and pCMVHT1-NCYΔX-P3A/A3P (bar 4).
We next asked whether stabilizing Pr55 Gag by incorporating an inactivating mutation in the protease gene could facilitate the egress of late domain mutant virions. This phenomenon has been observed in other retroviral systems, including the closely related bovine leukemia virus (BLV) (20, 59). Figure 2C shows that pCMVHT1-PR− P3G exhibited an L-domain mutant phenotype, while no defect was discernible for the pCMVHT1-PR−-carried viruses. Similar results were obtained when we fused the gene for EYFP to the end of the Gag gene (Fig. 2D). The fused gene was inserted into a proviral construct with a deletion in the envelope gene, giving pCMV-NCYΔX. The sequences appended to the end of gag disrupted the frameshift signal necessary for the translation of the protease gene, and the construct consequently expressed an 82-kDa Gag-EYFP fusion protein. While the expression levels of Gag-EYFP were about 10-fold lower than those of Gag from pCMVHT1 or pCMVHT1-PR− constructs, the distributions of MA between the supernatant and cells for the wt and mutant viruses were comparable.
Released particles are infectious.
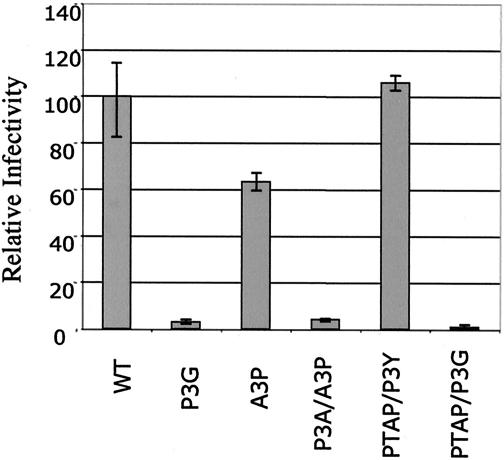
To assess the quality of virus particles produced by wt and late domain mutant CMVHT-1 constructs, we used a single-cycle infection assay that was recently established in our laboratory (8). The CMVHT-1 series of wt and late domain plasmids were used as packaging constructs and were cotransfected into 293T cells with a luciferase-expressing reporter plasmid carrying the HTLV-1 LTRs and packaging signal (pHTC-GFPLuc) and pCMV-VSV to provide the vesicular stomatitis virus glycoprotein envelope. Supernatants were filtered and used to infect B5 cells, a rhesus monkey lung fibroblast line previously shown to be susceptible to HTLV-1 infection but that is very difficult to transfect. B5 cell extracts were assayed after 48 h of infection. The results (Fig. 3) show that the infectious activity of a given supernatant correlated with the amount of viral protein (as determined by MA ELISA) released into the supernatant. The specific infectivities of particles derived from constructs with P3G mutations were lower than those of the other constructs. However, a constant amount of MA in the supernatants of all cultures is probably not in the form of VLPs. This material constitutes a larger fraction of the total MA in the supernatants of cultures expressing PPPY− Gag and lowers the apparent specific activities of these preparations.
FIG. 3.
Late domain mutations do not affect the infectivity of the virus in single-cycle assays. wt and mutant pCMVHT-1-bearing viruses were cotransfected with pHTC-GFPluc and pCMV-VSVg into 293T cells. After 2 days, filtered supernatants were transferred to B5 cells. The luciferase activity was assayed at 2 days postinfection. The infectivity of the mutants is expressed relative to that of pCMVHT1, which was set to 100%.
Immunofluorescence and electron microscopic analyses of budding HTLV-1 particles.
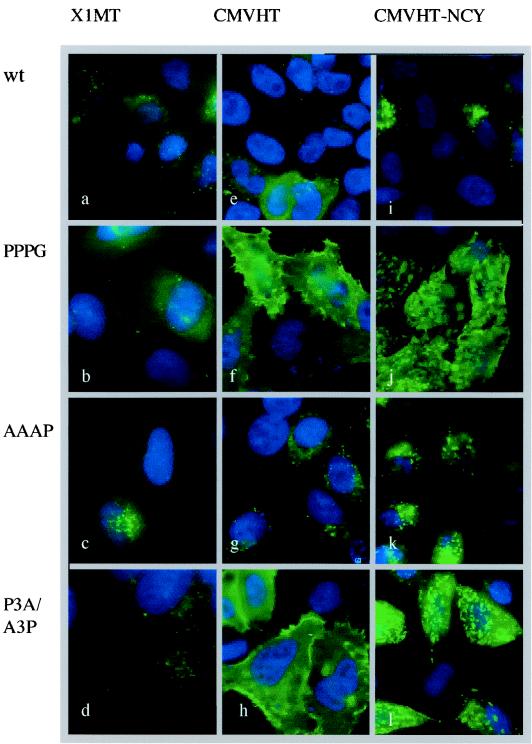
The imaging of MA by immunofluorescence microscopy demonstrated an increased retention of Gag in cells transfected with the P3G and P3A/A3P mutants compared to those transfected with a wt or A3P construct (Fig. 4). In HeLa cells transfected with pX1MT or the A3P mutant and stained with an anti-MA antibody, only a few intracellular speckles were visible (Fig. 4a and c). In contrast, cells expressing the P3G or P3A/A3P mutant virus displayed a considerable amount of anti-MA reactive material which was visible on the cell surface in small bright patches (Fig. 4b and d). The enhanced retention of Gag with the P3G mutation was even more pronounced when the pCMVHT series of constructs was used, since the higher level of expression made visualization easier. Again, only small amounts of MA, mostly located in the cytoplasm in a speckled pattern, were visible in cells expressing wt and A3P constructs (Fig. 4e and g). Cells expressing P3G and P3A/A3P mutants were brightly decorated uniformly on the plasma membrane (Fig. 4f and h). In cells transfected with the pCMVHT-NCYΔX-P3G and -P3A/A3P constructs, the Gag protein accumulated in large patches.
FIG. 4.
Immunofluorescence microscopy reveals retention of L-domain mutant Gag in plasma membrane. HeLa cells were transfected with HTLV-1 expression plasmids as indicated at the top encoding the L domains indicated on the left. The cells expressing the pX1MT and pCMVHT1 constructs were stained with a monoclonal anti-MA antibody, followed by anti-mouse immunoglobulin G-Alexa-488 (Molecular Probes). All samples were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) to visualize the nuclei (Molecular Probes).
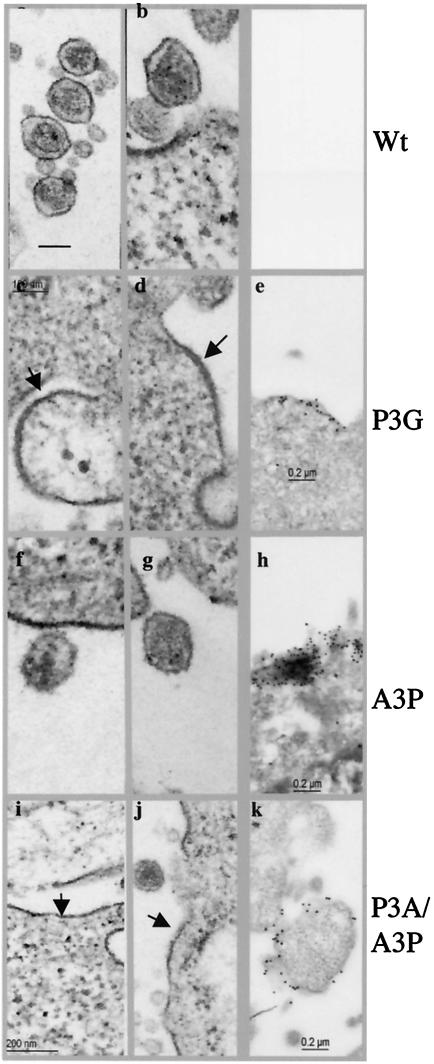
To visualize the morphology of the budding viruses, we used TEM and IEM. Very few virus particles at any stage of budding could be detected in cells transfected with the wt pX1MT or pCMVHT plasmid (Fig. 5a and b). This finding is perhaps not unexpected when one considers that very little Gag protein stays associated with cells, as measured by ELISA. TEM showed that the mutants with changes in the PPPY motif caused a thickening of the membranes in large patches, with the characteristic appearance seen when Gag is bound to the inner leaf of the plasma membrane (Fig. 5c, d, i, and j). This was verified by IEM, which showed these areas of the membrane decorated with gold beads (Fig. 5 e and k). Since only very few nascent budding viruses were observed for PPPY− mutants, the HTLV-1 late domain mutants appear to be arrested at an early stage of morphogenesis. This is in agreement with results reported by Le Blanc et al. for HTLV (25) but in contrast to observations for most other retroviruses, in which mutation of the late domain motif affects the very last step of pinching off, leaving the budded viruses tethered to the cell or to each other, depending on the cell type (7).
FIG. 5.
Phenotypes of late domain mutants by electron microscopy. HeLa cells were transfected with the indicated pCMVHT-1 constructs. Arrows indicate a thickening of the plasma membrane due to the retention of viral proteins. (a to d, f, g, i, and j) TEM images; (e, h, and k) IEM images. Staining was done with a monoclonal anti-MA antibody followed by anti-mouse immunoglobulin G-gold (5-nm diameter).
HTLV late domain interacts with WW domains of WWP1 and with the amino-terminal region of TSG101.
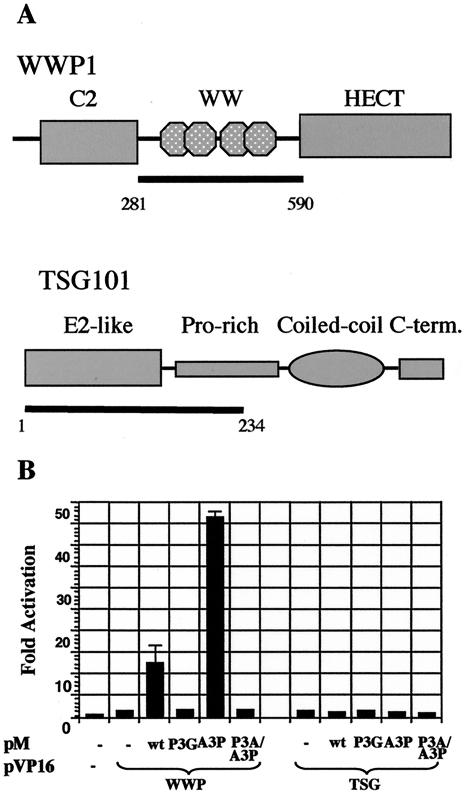
PPXY motifs have been shown to interact with WW domains, so named because they contain two conserved tryptophan residues (52). Several papers have reported an interaction of other PPPY-type L domains with the WW domains of the Nedd4 family of ubiquitin ligases (16, 17, 24, 28, 53, 63). We used the mammalian two-hybrid system to investigate the ability of MA to bind to WWP1, the human homologue of the chicken Nedd4 family member LDI-1 identified by Kikonyogo et al. as a partner of the Rous sarcoma virus (RSV) late motif (24). The WW domains carrying the sequence (nucleotides 841 to 1744) of a cDNA clone of WWP1 were inserted into the activation domain-expressing plasmid pVP16 (Fig. 6A). The plasmid encoding the DNA binding partner (pMp19*) contained the sequence for codons 5 to 154 of the Gag precursor (i.e., the fusion product extended 25 residues into the CA part of the precursor). A positive interaction was detected when the pMp19* plasmid encoded either a wt or A3P version of the HTLV-1 late domain, but not when the PPPY motif was altered. Interestingly, the interaction observed with the A3P mutant was consistently three- to fourfold higher than that observed with the wt construct. These experiments were performed several times in 293T as well as HeLa cells, with reproducible levels of interaction.
FIG. 6.
HTLV-1 late motif interacts with WW domains of WWP1. (A) Schematic representation of major domains of WWP1 (top) and TSG101 (bottom). The regions inserted into the activation domain fusion plasmids of the mammalian and yeast two-hybrid systems (pVP16 and pAS-2, respectively) are indicated by the bars. (B) pM-p19+ and its late domain mutant versions expresses the Gal4 DNA-binding domain fused to residues 5 to 154 of Gag. The reporter plasmid pGL5-Luc expresses the luciferase gene under the control of five copies of the Gal4 consensus binding site. Representative results from one of three experiments are shown. Activation is expressed as the fold induction over that seen with the pM-p19+/pVP16 combination.
We also performed similar experiments with the UEV domain of TSG101, which has been shown to be the interacting partner of PTAP-type late assembly motifs (6, 13, 39). Amino acid residues 1 to 238 of TSG101 were expressed as an activation domain fusion. We did not detect any interaction of the TSG101 construct with any of the late domain-DNA binding domain fusions in the mammalian two-hybrid system. However, since attempts to detect an interaction between HIV-Gag and TSG101 failed, we performed a yeast two-hybrid analysis. The HTLV-1 and HIV-1 full-length Gag proteins, as well as MA and p6, were expressed as Gal4 DNA binding fusions, and the WW domain of WWP1 and the TSG101 UEV domain were expressed as Gal4 activation domain fusions. As Table 1 shows, with this system we could easily detect an interaction between the amino-terminal half of TSG101 and HTLV-1 MA or full-length Gag as well as the binding of HIV-1 full-length Gag and p6 to the TSG101 UEV domain. Only HTLV-1 Gag with an intact PTAP motif was able to interact with TSG101 UEV in this assay.
TABLE 1.
Yeast two-hybrid interaction between HTLV Gag proteins and TSG101
| DNA-binding domain plasmid | Interaction with activation domain plasmida
|
||
|---|---|---|---|
| No insert | TSG101-UEV | WW domains (WWP1) | |
| HIV-Gag | − | ++ | − |
| HIV-p6 | − | + | − |
| HTLV-Gag | − | ++++ | ± |
| HTLV-MA | − | ++++ | ± |
| HTLV-MA-P3G | − | +++ | ND |
| HTLV-MA-A3P | − | − | ND |
| HTLV-MA-P3AA3P | − | − | ND |
+, interaction is present, as evidenced by the growth of blue colonies in the presence of X-Gal on medium without leucine, tryptophan, histidine, and adenine. The number of plus symbols indicates the level of activity in the β-galactosidase assay of replicated colonies. −, no growth and no interaction; ND, not done.
DN WWP1 and TSG101 inhibit HTLV-1 budding.
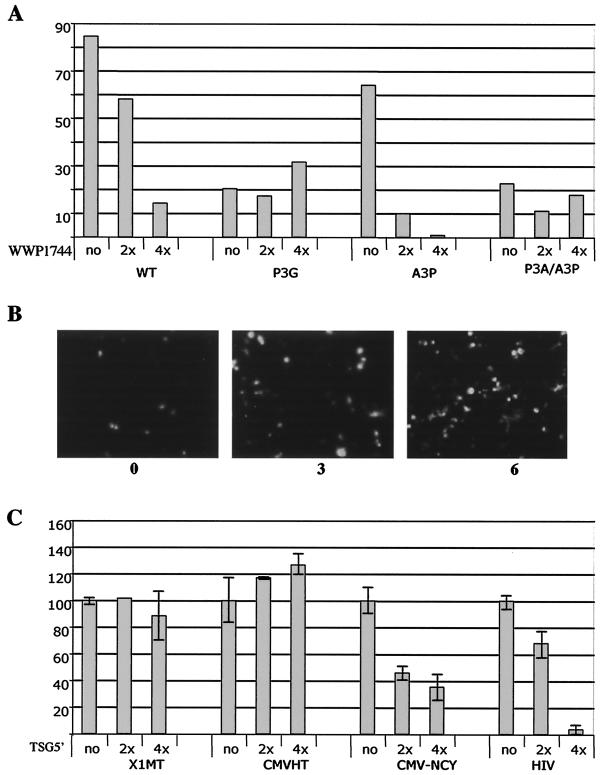
The mammalian two-hybrid experiment indicated that the Nedd4 family member WWP1 interacted with HTLV-1 Gag. To explore this point further, we generated an expression plasmid producing a truncated, hemagglutinin-tagged version of WWP1 encoding everything but the HECT domain (pUCHR-WWP1744). The equivalent fragment of the homologous avian virus protein has been shown to function as a DN inhibitor of RSV budding (24, 38). The cotransfection of 293FT cells with pX1MT-wt or pX1MT-A3P and increasing amounts of the WWP1-1744 expression plasmid resulted in a dose-dependent decrease in virus budding, as determined by ELISA (Fig. 7A). In contrast, pX1MT-P3G or the double mutant was not affected by WWP1-1744 expression. The inhibitory effect of WWP1-1744 on HTLV-1 budding was also visualized by fluorescence microscopy of cells transfected with the pCMVHT-NCYΔX series of late domain mutants and increasing amounts of pUCHR-WWP1744. When the Gag protein had the wt or A3P L motif, increasing amounts of cotransfected pCMV-WWP1744 resulted in a higher level of retention of Gag in the cells, as revealed by the increased numbers of bright cells. Figure 7B shows 293T cells transfected with pCMVHT-NCY and increasing amounts of pCMV-WWP1744. This effect was not discernible for cells expressing Gag with the P3G mutation.
FIG. 7.
DN WWP1 and DN TSG101 inhibit egress of virus or VLPs, respectively. (A) Two micrograms of wt or mutant pX1MT was transfected with 6 or 12 μg of pUCHRattWWP1744 or an empty vector into 2 × 106 293T cells. The amounts of MA in the supernatants and cell extracts were determined by p19 MA ELISA. The graph shows the percentages of total MA detected in the supernatant. (B) Fluorescence microscopy of cultures transfected with pCMVHT-NCYΔX alone or with a three- or sixfold excess of pUCHRattWWP1744. The increase in fluorescence indicates the larger amounts of Gag protein retained in the cells in the presence of DN WWP1. (C) Two hundred nanograms of pX1MT, pCMVHT, pCMV-NCYΔX, or pCMVΔ8.2 was transfected with 0, 400, or 800 ng of pTSG5′. DNA concentrations in the transfection reactions were kept constant with pCMVpA.
The coexpression of pTSG5′, a DN mutant of TSG101, did not interfere with HTLV-1 budding when we used the full-length proviral construct pX1MT or pCMVHT (Fig. 7C). However, TSG5′ had a negative effect on the budding efficiency of pCMV-NCYΔX proportional to the added amount of the DN construct. The reason for the discrepancy in the effects of TSG5′ on the constructs is unclear at this point. It is probably not a function of the Gag expression level: we performed this experiment in 12-well plates and the total yield of MA per well was about 10 ng for the pCMV-NCYΔX construct, compared to 2 ng for pX1MT and 100 ng for pCMVHT. The other obvious differences were the defect in env in pCMV-NCYΔX and the inability of cells with this construct to process the Gag precursor.
MA is ubiquitinated.
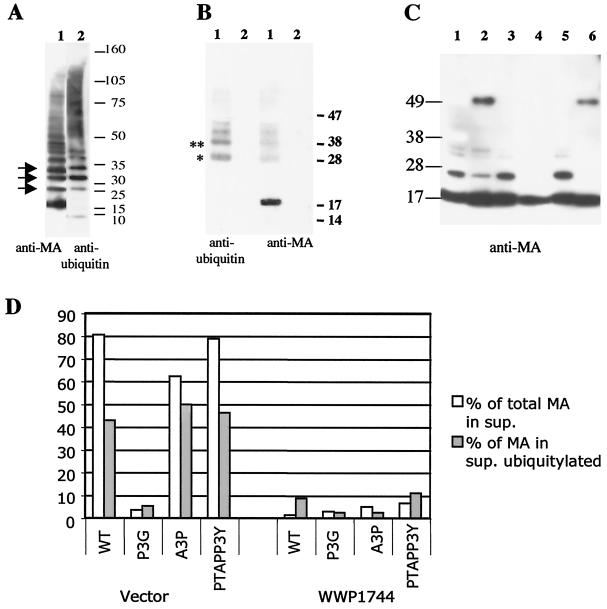
Previous work by several groups has shown that a small fraction of the late-motif-containing Gag peptide, as well as the Gag precursor of some viruses, is mono- or diubiquitinated in virions (32, 33, 37, 49). Western blots of virion proteins showed several bands after anti-MA probing that migrated with mobilities that cannot be assigned to any partially processed Gag precursors (for an example, see Fig. 1). Gag precursors expressed in the absence of protease also showed additional bands of reduced mobility (data not shown). To test whether these MA-epitope-containing peptides were modified by ubiquitination, we sequentially probed a Western blot of virions separated by density (produced by MT2 cells) with rabbit anti-ubiquitin and mouse anti-MA antibodies. Figure 8A shows that the HTLV-1 preparation contained free ubiquitin as well as several high-molecular-weight peptides that reacted with the anti-ubiquitin antibody and that several bands on the anti-ubiquitin blot comigrated with bands on the anti-MA blot. Interestingly, we did not observe ubiquitinated MA in cellular extracts (Fig. 1B).
FIG. 8.
MA is ubiquitinated in a WWP1-dependent fashion. (A) Immunoblot analysis of density-separated virus from HTLV-1-infected MT2 cell line with an anti-MA antibody (left) and an anti-ubiquitin antibody (right). Arrows indicate comigrating bands. (B) Pelleted supernatants from 293T cell cultures transfected with pCMVHT1 (lanes 1) or left untransfected (lanes 2) were denatured and immunoprecipitated with a rabbit anti-MA antibody (see Materials and Methods). The right panel was probed with a mouse monoclonal anti-ubiquitin antibody. After stripping, the blot was reprobed with a mouse monoclonal anti-MA antibody (left). The positions of molecular weight markers are given on the right. * and **, predicted positions for mono- and di-ubiquitinated MA, respectively. (C) Immunoblot analysis with 2 ng of the respective matrix protein (according to ELISA) loaded per lane. The mutants used were pCMVHT (lane 1), pCMVHT-P3G (lane 2), pCMVHT-A3P (lane 3), pCMVHT-P3AA3P (lane 4), pCMVHT-PTAP/P3Y (lane 5), and pCMVHT-PTAP/P3G (lane 6). The blot was probed with an anti-MA monoclonal antibody and developed with ECL+ and autoradiography. (D) 293T cells were transfected with 2 μg of the indicated pCMVHT1 constructs plus 15 μg of pUCHR or pUCHR-WWP1744. The fractions of MA in the supernatants were determined by ELISA and are shown as open bars. The fractions of MA from the supernatants that were ubiquitinated are shown as solid bars and were established as described in the text.
To verify that MA was indeed ubiquitinated, we disrupted the pellets from supernatants of untransfected 293T cells or cells transfected with pX1MT with 2% sodium dodecyl sulfate under reducing conditions to minimize protein-protein interactions. The solutions were diluted 30-fold and precipitated with a rabbit anti-MA antibody. An immunoblot analysis of the precipitates with a mouse monoclonal anti-ubiquitin antibody revealed ubiquitinated proteins in the pX1MT lane (lane 1) that migrated with the same mobility as proteins that were visible when the stripped blot was reprobed with a monoclonal anti-MA antibody. No signal was visible on either blot for the supernatants from untransfected cells (Fig. 8B).
To address the dependence of ubiquitination on the presence of a PPPY motif, we compared the MA proteins from wt and mutant viruses by Western blotting after loading equal amounts of MA, as determined by an ELISA. Figure 8C shows that the virions produced by constructs with the PPPG mutations contained significantly less mono- and diubiquitinated MA than those produced by constructs with the intact PPPY motif.
To examine whether the expression of WWP1744 affects ubiquitination, we cotransfected 293T cells with pCMVHT1 or late domain mutant derivatives and an excess of pUCHRWWP1744 or empty pUCHR vector. We quantitated the ratio of MA in the supernatant to that in the cell extract by ELISA and analyzed the matrix protein in virus particles by immunoblotting after adjusting the sample volumes to ensure that there was sufficient material in all lanes. The blots were developed with ECL+ and analyzed by fluorescence imaging. We determined the amount of MA in each sample that was ubiquitinated. Figure 8D shows that only about 5% of the PPPG or double mutant MA was ubiquitinated, while about 40% of the protein with a PPPY motif was modified. The coexpression of DN WWP1 not only drastically lowered the amount of MA protein in the supernatants of cultures transfected with pCMVHT1, but it also prevented ubiquitination of the residual MA protein. The amount of MA in the supernatant of cells transfected with pCMHT-1 and pUCHRWWP1744 was very similar to that produced after transfection with pCMVHT-P3G with or without the DN WWP1 construct.
DISCUSSION
This report shows that the PPPY tetrapeptide, located close to the end of the MA sequence in the Gag precursor, functions as the major late assembly domain in HTLV-1. Mutation of the sequence to P3G severely diminished the ability of the virus to bud from cells. These data are in agreement with results reported by Le Blanc et al., who showed that the mutation of any residue in PPPY caused budding defects in HTLV-1 (25). In contrast with the case for the HIV-1 system, in which the overexpression of Gag or the prevention of proteolytic processing of the precursor can largely compensate for an impaired L-domain function, no such complementation was observed for HTLV-1. Interestingly, Wang et al. (59) recently reported that for BLV, a close relative of HTLV-1, the budding of VLPs with late domain mutations could be rescued by the inactivation of protease.
We also addressed the role of the PTAP motif located just two residues distal to PPPY and found that its mutation had only modest effects on the budding ability of the infectious virus. This was in contrast to reports by Bouamr et al. (4) and Wang et al. (58), who reported more severe defects for the PTAP mutant and observed an additive effect for the double mutant. In our hands, the quantitative difference between the effect of the single mutation in PPPY and that of the double mutation was not significant. In both reports, the changes introduced into the PTAP motif (ATAA and LIRL, respectively) were more severe than the changes in our study. We had initially changed the motif to PTAA and had also observed more severe effects. However, the overall amount of Gag produced by this mutant dropped to about 1% that of the wt (data not shown). This was the case irrespective of whether we introduced the mutation into the pX1MT, pCMVHT, or even pCMVHT-Pro− background. This finding suggested that the defect was one of stability and not a processing defect. Indeed, even the constructs with the A3P mutation produced, in general, only about 20 to 40% of the wt levels of Gag protein when equal amounts of plasmid DNA were transfected.
It was not completely unexpected that PPPY was the predominant contributor to the late assembly function, as the PTAP motif is not conserved in HTLV-2 and BLV, which are close relatives of HTLV-1 in the deltaretrovirus group. We were, however, surprised to find that the PTAP motif could not replace PPPY, as exchangeability is considered one of the hallmarks of late assembly domains (36, 42). While the PPPY motif functioned equally well when the two motifs were switched, suggesting that the exact position was not important, the PTAP motif alone did not promote significant virus release in either position. Our data are in contrast to those published by Wang et al. (58). They reported that the switch of the two motifs alone totally abolished the budding of VLPs. We had initially generated two different versions of the construct, one encoding PTAPVEPPPY and one with an additional proline following the composite motif. Our thinking had been that the proline might be necessary for maintaining the structure of the Gag precursor and the cleavage site, which is only three residues downstream. However, very little difference was observed between the phenotypes of the PTAPVEPPPY and PTAPVEPPPYP constructs, which budded with wt efficiency, although the overall expression levels were reduced to about 60% that of the wt.
Redundancy in late motifs is not unique to HTLV-1; Mason-Pfizer monkey virus (M-PMV) also has a double motif, PPPYNKATPSAP, in p24/p16. Also, similar to the deltaretroviruses, M-PMV and simian retrovirus 1 have both motifs, while other betaretroviruses, such as simian retrovirus 2 and squirrel monkey retrovirus, only carry the PPPY motif. Yasuda and Hunter reported that deletion of the PPPY motif abolished the budding of M-PMV, while mutations of PSAP had no effect, indicating that PPPY alone is the late assembly motif (62). However, more recent work by Gottwein et al. (15a) showed that the PSAP motif contributes to the budding of M-PMV to about the same extent that we observed with the HTLV-1 system. As mentioned in the introduction, Ebola virus has overlapping motifs (PTAPPEY) in the N terminus of VP40, both of which have been demonstrated to contribute to late assembly events. Furthermore, there is evidence that TSG101 as well as Nedd4 proteins can interact with VP40 (17, 28, 53). Recent work by several laboratories has shown that HIV p6 also carries additional adapter sequences to connect to the MVB budding machinery (30, 50, 56). The interaction was mapped to a LYP-L motif distal to the PTAP sequence and to AIP1, which links ESCRTI and ESCRTIII complexes.
We demonstrated that MA can interact with the Nedd4 family member WWP1, but not with TSG101, based on mammalian two-hybrid experiments. However, we obtained virtually reciprocal results when we used the yeast two-hybrid system. The interaction between HTLV-1 MA or full-length Gag and TSG101 was stronger than that between HIV-1 Gag and TSG101, while only a very weak interaction between WWP1 and Gag was seen in the yeast system. One possible explanation is that the expression of HIV Gag and full-length HTLV-1 Gag as DNA-binding-domain fusions in the mammalian two-hybrid system may not have been at a high enough level to show an interaction; however, it was not possible to assay for an interaction by using reverse constructs, as TSG101 fused to the Gal4 DNA-binding domain was highly transactivating on its own (data not shown). This finding indicates that there are other binding components for TSG101 in the nucleus which may have obscured the sites from interacting with Gag. The interaction of Gag with the WW domains was abolished by mutations in PPPY in the mammalian system, while binding to TSG101 in yeast was dependent on the PTAP sequence.
The Nedd4 family member WWP1 is a likely interaction partner for the HTLV-1 late motif. This protein is the human orthologue of the LDI-1 protein identified by Kikonyogo et al. in a chicken cDNA expression library screen using the RSV late assembly motif peptide as a probe (24). Nedd4 family members are ubiquitin ligases consisting of a Ca2+- and lipid-binding C2 domain, two to four WW domains, and a HECT (homologous to E6-AP carboxy-terminal) domain that constitutes the actual ubiquitin ligase (10, 18). Members of this family regulate the stability of various transmembrane proteins, directing them to be degraded via either the proteasomal or the endosomal pathway by poly- or mono- to tetra-ubiquitination, respectively (44). The best studied Nedd4 substrate in mammals is the transport channel protein EnaC that controls the Na+ balance of extracellular fluids and maintains blood pressure (43). In addition to this and other transporter proteins, several transmembrane receptors are downregulated in a ubiquitination-dependent manner through the endosomal pathway, eventually being deposited in vesicles in MVBs followed by degradation in the lysosome.
Whether WWP1 is the true cellular partner for HTLV-1 has yet to be established, as we have not assayed any other candidates. Yasuda et al. recently identified Bul1 as the Nedd4 family member that interacts with the M-PMV late assembly domain (63), while Timmins et al. showed binding of Ebola virus VP40 to the third WW domain of Nedd4 (53). In our experiments, a mutant WWP1 with a HECT domain deletion showed a DN effect on wt and PTAP mutant virus budding but did not further reduce the budding of the P3G mutant. This finding indicates that WWP1 can act in the budding pathway of HTLV-1.
The interaction between the PTAP motif found in most primate lentiviruses and the UEV domain of TSG101 is well established (6, 13, 15, 29, 55), and the structure of the complex has even been resolved (39). The role of TSG101 in HTLV-1 budding was perplexing in our hands. Like others (4), we observed that DN TSG5′ diminished the release of VLPs when it was coexpressed with constructs encoding only Gag structural proteins. However, we saw no effect on the budding of viruses when we used complete proviral constructs. We are currently addressing the role of other viral proteins in this context.
As in other viruses (32, 33, 37, 49), the Gag precursor of HTLV-1 is ubiquitinated during budding, as evidenced by the fact that a fraction of MA in the virions can be precipitated with an anti-ubiquitin antibody. Ubiquitination was independent of processing, as the modification was found on the precursor in VLPs with an inactive protease. The fraction of total MA which was ubiquitinated was higher than that in other viruses. We observed between 10 and 40% of the wt MA migrating with apparent molecular masses of 25 to 40 kDa, consistent with mono- and di-ubiquitination, compared to the 2 to 5% values reported for HIV-1 and murine leukemia virus (MLV) (32). Ubiquitination was tightly associated with virus egress, as very little ubiquitin was present on cell-associated Gag proteins. Ubiquitination on MA or Pr55 Gag was greatly reduced if the protein carried the P3G mutation. This finding and the observation that the expression of DN WWP1 also prevented the modification of wt Gag suggested that a Nedd4 family member is the ubiquitin ligase responsible for the modification. At this point, our results do not allow us to decide whether ubiquitination is necessary or occurs more as a bystander effect due to interaction with a ubiquitin ligase. This question has not been answered with other retroviral systems either. Most studies on the role of ubiquitination in virus budding were done in the HIV system (33, 34, 46, 49, 52). TSG101 binding to ubiquitinated p6 was shown to be 10-fold stronger than that to p6 alone (13), and the sequestration of ubiquitin into polyubiquitinated proteins by inhibiting proteasomal degradation prevented virus budding (46); however, mutations of lysines, the substrates of ubiquitin ligases, in p6 of HIV or p12 of MLV had little effect on virus egress (32).
The most puzzling result to us was that mutation of the PPPY motif not only prevented fission to release the budded virus but also arrested virion morphogenesis. This was also reported by Le Blanc et al. (25). Cells expressing the P3G mutant virus displayed hardly any protrusions indicative of nascent virus particles on the cell surface. Instead, immunofluorescence and electron microscopy showed Gag proteins forming large sheets in the plasma membrane. This not only contrasted with the case for viruses using PTAP or LYPXL as late assembly motifs, but also was different from that for viruses with PPPY motifs, including the close relative BLV (59). In all of these cases, electron microscopy revealed that late domain mutants formed immature virus particles tethered to the cell or to each other by thin stalks of plasma membrane, indicating that only the very last step was inhibited. Given our current knowledge of the endosomal pathway for protein downregulation into MVBs and of its recruitment by enveloped viruses for virus egress (for reviews, see references 11, 12, 23, 26, and 40), it is actually the progression of the other PPPY motif viruses to the tethered state that is surprising rather than the early arrest of HTLV-1. The vacuolar protein sorting (VPS) class E pathway is best established in Saccharomyces cerevisiae, but extensive parallels have been identified in mammalian cells. The first step is mono- to tetra-ubiquitination, which hands the proteins—generally transmembrane receptors—the entry ticket to the MVB pathway and causes clustering into clathrin-coated pits, which bud into the cytoplasm (47, 60). The vesicles subsequently are recruited by ESCRTI to fuse with the late endosomal compartment (2, 22, 45). TSG101 is thought to be the ubiquitin-recognizing component of ESCRT1. The binding of ESCRT1 activates ESCRT2 and -3 to form an invagination of the membrane (1, 3). The release of the vesicle into the lumen of the endosome occurs concomitantly with de-ubiquitination to recycle the tag and disassembly of the ESCRT complexes catalyzed by the AAA-ATPase Vps4. It seems obvious at which points it would make sense for the different late assembly domains to accelerate the early arrest of HTLV-1, as the absence of the ubiquitin tag would interfere with all subsequent steps. However, this model also predicts that the inactivation of downstream components of the pathway should interfere with budding. Our results showed that a DN TSG101 mutation had no effect on the release of HTLV-1 virions. This finding is supported by recent reports which demonstrated the same results for the budding of MLV as well as EIAV. On the other hand, viruses with any of the three types of late domains can be inhibited by DN Vps4, suggesting that they utilize the same final step and that shortcuts along the way are possible (13, 30). How HTLV-1 MVBs enter into the MVB pathway and what cellular factors are involved in virion morphogenesis are under investigation.
Acknowledgments
We thank L. Henderson for stimulating discussions and for the rabbit anti-MA antibody. We thank Z. Sun for pTSG5′.
This publication was funded in part by federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248-258. [DOI] [PubMed] [Google Scholar]
- 4.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C., F. Li, and R. C. Montelaro. 2002. Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection. J. Virol. 75:9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2001. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derse, D., S. A. Hill, P. A. Lloyd, H.-K. Chung, and B. A. Morse. 2001. Examining human T-lymphotropic virus type 1 infection and replication by cell-free infection with recombinant virus vectors. J. Virol. 75:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn, R., and L. Hicke. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276:25974-25981. [DOI] [PubMed] [Google Scholar]
- 10.Flasza, M., P. Gorman, R. Roylance, A. E. Canfield, and M. Baron. 2002. Alternative splicing determines the domain structure of WWP1, a Nedd4 family protein. Biochem. Biophys. Res. Commun. 290:431-437. [DOI] [PubMed] [Google Scholar]
- 11.Freed, E. O. 2003. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 11:56-59. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 15.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvey, K. F., and S. Kumar. 1999. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 9:166-169. [DOI] [PubMed] [Google Scholar]
- 19.Heidecker, G., S. Hill, P. A. Lloyd, and D. Derse. 2002. A novel protease processing site in the transframe protein of human T-cell leukemia virus type 1 PR76(gag-pro) defines the N terminus of RT. J. Virol. 76:13101-13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6 Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hull, R. 2001. Classifying reverse transcribing elements: a proposal and a challenge to the ICTV. Arch. Virol. 146:2255-2261. [DOI] [PubMed] [Google Scholar]
- 22.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 23.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell. Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 24.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Blanc, I., M. C. Prevost, M. C. Dokhelar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemmon, S. K., and L. M. Traub. 2000. Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12:457-466. [DOI] [PubMed] [Google Scholar]
- 27.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2002. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Struct. Biol. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 32.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 33.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott, D. E., L. V. Coren, R. C. Sowder, J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77:3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otte, L., U. Wiedemann, B. Schlegel, J. R. Pires, M. Beyermann, P. Schmieder, G. Krause, R. Volkmer-Engert, J. Schneider-Mergener, and H. Oschkinat. 2003. WW domain sequence activity relationships identified using ligand recognition propensities of 42 WW domains. Protein Sci. 12:491-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patnaik, A., V. Chau, and J. W. Wills. 2001. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patnaik, A., and J. W. Wills. 2002. In vivo interference of Rous sarcoma virus budding by cis expression of a WW domain. J. Virol. 76:2789-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 40.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 41.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rotin, D., V. Kanelis, and L. Schild. 2001. Trafficking and cell surface stability of ENaC. Am. J. Physiol. 281:F391-F399. [DOI] [PubMed] [Google Scholar]
- 44.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 45.Sato, T. K., M. Overduin, and S. D. Emr. 2001. Location, location, location: membrane targeting directed by PX domains. Science 294:1881-1885. [DOI] [PubMed] [Google Scholar]
- 46.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with Gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shih, S. C., D. J. Katzmann, J. D. Schnell, M. Sutanto, S. D. Emr, and L. Hicke. 2002. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4:389-393. [DOI] [PubMed] [Google Scholar]
- 48.Shuh, M., S. A. Hill, and D. Derse. 1999. Defective and wild-type human T-cell leukemia virus type I proviruses: characterization of gene products and trans-interactions between proviruses. Virology 262:442-451. [DOI] [PubMed] [Google Scholar]
- 49.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2001. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 51.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sudol, M., and T. Hunter. 2000. NeW wrinkles for an old domain. Cell 103:1001-1004. [DOI] [PubMed] [Google Scholar]
- 53.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493-502. [DOI] [PubMed] [Google Scholar]
- 54.Tobin, G. J., K. Nagashima, and M. A. Gonda. 1996. Immunologic and ultrastructural characterization of HIV pseudovirions containing Gag and Env precursor proteins engineered in insect cells. Methods 10:208-218. [DOI] [PubMed] [Google Scholar]
- 55.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 57.Wang, G., J. M. McCaffery, B. Wendland, S. Dupre, R. Haguenauer-Tsapis, and J. M. Huibregtse. 2001. Localization of the Rsp5p ubiquitin-protein ligase at multiple sites within the endocytic pathway. Mol. Cell. Biol. 21:3564-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, H., N. J. Machesky, and L. M. Mansky. 2004. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J. Virol. 78:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, H., K. M. Norris, and L. M. Mansky. 2002. Analysis of bovine leukemia virus Gag membrane targeting and late domain function. J. Virol. 76:8485-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wendland, B., and S. D. Emr. 1998. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 141:71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]