Abstract
Systems cell biology melds high-throughput experimentation with quantitative analysis and modeling to understand many critical processes that contribute to cellular organization and dynamics. Recently, there have been several advances in technology and in the application of modeling approaches that enable the exploration of the dynamic properties of cells. Merging technology and computation offers an opportunity to objectively address unsolved cellular mechanisms, and has revealed emergent properties and helped to gain a more comprehensive and fundamental understanding of cell biology.
Systems cell biology: What it is and what it is not
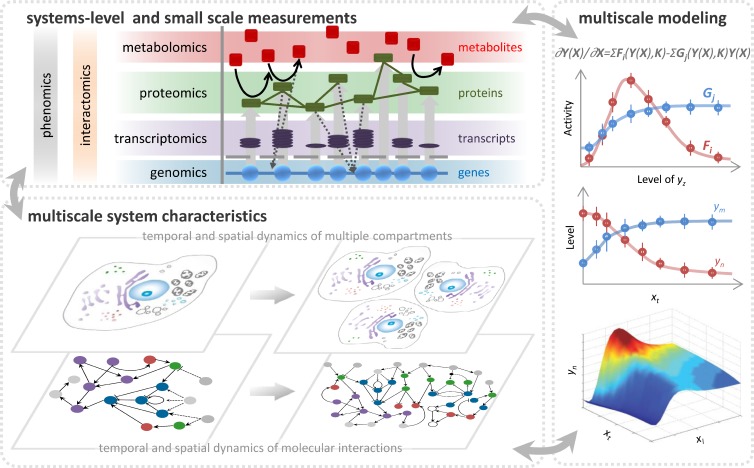
Systems cell biology is the study of the emergent properties of a cell and its component parts using comprehensive and quantitative experimental methods that are interpreted by predictive mathematical and statistical models. Emergent properties result from “the whole being greater than the sum of its parts.” The progression to studying the cell as a system is a natural one for cell biologists who have always sought to meld the biochemical processes of molecules and modules with the spatial and structural features of cells (Alberts, 1998; Hartwell et al., 1999). Thus, understanding cell biology is inherently a multiscale problem, with many levels and hierarchies of cellular organization, compartmentalization, and temporal regulation (Fig. 1). Emergent properties within a cell derive from the interplay of system components arranged in complex motifs such as logic gates, feedback and feed-forward loops, and combinations thereof (Alon, 2007; Tyson and Novák, 2010). This complex interplay leads to behaviors that include switch-like functionality, filtering, signal amplification, oscillations, and multistability. This gives rise to systems-level properties of cells including robustness, hysteresis, modularity, and population heterogeneity. The goal of systems cell biology is therefore to achieve more than a description of the individual components and component properties. It is to achieve an understanding of how information is transmitted and interpreted by the cell. Systems cell biology is also more than simply the acquisition of large amounts of data, or the assembly and visualization of that data into networks, heat maps, and diagrams. It is also not an unbiased replacement for intuition, as many cellular processes can be intuitively explored from a systems perspective. A prime example is the eukaryotic cell cycle, where a rich history of applying nonlinear dynamical systems models that rely, in part, on an intuition about the interactions of key cell cycle regulators has dramatically advanced our understanding of this process (see Ferrell et al., 2011 and the references within).
Figure 1.
Exploring the cell as a system. Systems cell biology incorporates systems-level and small-scale measurements of information flow from all the components of the system and from all the hierarchies of the relevant temporal and spatial scale. Multiscale modeling approaches are used to infer missing parameters and visualize phenotypes as the function of multiscale system characteristics. The process is iterative, and the model is refined until it is predictive.
Systems biology: A glove for every hand?
Systems biology is broadly defined as a framework for conducting quantitative and comprehensive scientific enquiry. This framework facilitates a rigorous analysis of the complexity of biological systems at all levels of cellular organization that contribute to a behavior or phenotype of interest (Kitano, 2002). However, this common definition is rather vague, and this has encouraged skepticism with regard to the ability of systems biology research to achieve the lofty goal of understanding complex biology (Brenner, 2010).
Irrespective of the focus of a study, a systems biology approach often includes several common elements: exploratory data acquisition and visualization, data integration and the formulation of quantitative models, and the testing of these models, along with the hypotheses they generate, with further experimentation (Ideker et al., 2001; Aitchison and Galitski, 2003). These results can then be used to guide iterative cycles of the systems approach that serve to refine the model in question (Fig. 1). Another way to consider this is that systems biology enables the identification of the many ways information can flow and be processed within a biological system (Ideker et al., 2001; Nurse, 2008). To function in this capacity, systems biology requires systems-level data collection.
The omics of systems biology: Exploratory data acquisition and visualization
Systems biology is commonly associated with large-scale “-omics” technologies such as genomics, proteomics, and functional genetics that are used to explore the state of a system under investigation (Short, 2009). However, it is a misapprehension to think that systems biology is only the acquisition of such large-scale datasets. The inclusion of an omic discovery component to the analysis of biological complexity assists in identifying situations where a phenotype is caused by an emergent or unanticipated property of the system (Aitchison and Galitski, 2003). This does not imply that emergent properties are naturally revealed through omics approaches, but rather that through the acquisition of a comprehensive and quantitative dataset such properties can be revealed through mathematical modeling and computational analysis. To be of practical use in discovering these essential components, omics technologies must be quantitative and amenable to high throughput approaches, comprehensible visualization, and statistical approaches. Even when properly executed, experiments often fall short of this goal, and, ideally, computational interpretation of the results can assist in identifying missing variables and influences or measurements that would be more informative. Moreover, computational analyses and modeling strategies can assist in revealing the underlying mechanisms of the system (Fig. 1). From this vantage point, proximal causes and effects can be separated from distal ones and be analyzed further with a cycle of modeling and experimentation.
Next-generation sequencing (NGS): Understanding genetic determinants in cellular systems
For the correct implementation of a great deal of other systems approaches, such as proteomics and functional genetics, a reference genome is an essential starting point and necessary tool for their implementation. NGS, also known as “deep-sequencing,” is helping to redefine our understanding of chromatin structure and organization (Yen et al., 2013), as well as the regulation of transcription (Rhee and Pugh, 2011, 2012) and translation (Ingolia et al., 2009, 2011; Guttman et al., 2013). Although we will avoid giving a thorough overview of the technology and various NGS platforms (which can be found in Koboldt et al., 2013; Mardis, 2013), we highlight recent discoveries that illustrate the need to view the cell from a systems perspective.
A current trend from the rapid rise in genomic sequencing is the inclusion of phylogenetic and comparative genomic analyses in considering mechanistic models of cell biology (Liti et al., 2009; Finnigan et al., 2012; Mast et al., 2014). Addressing the challenge of assigning cell components to a particular function is thus partly alleviated by considering the idiosyncratic origins of the components of a system. Evolutionary analyses of cell biology on a systems scale have benefitted from increased taxon sampling and are enabling tests of the implicit assumptions of molecular cell biology as well as the study of cellular phenomena in model systems. These evolutionary comparisons on a systems level are helping to place the findings from one cellular system within the context of all cellular systems (Elias et al., 2012; Koonin and Mulkidjanian, 2013). Furthermore, it enables the exploration of the origins of cellular complexity and helps restrict the search space of causal mechanisms to those that are congruent with evolutionary theory (Koonin, 2011; Doolittle, 2012; Koumandou et al., 2013). See the JCB review series on evolution (http://jcb.rupress.org/cgi/collection/7).
NGS has also resulted in an increase in the number of individual genomes from a single species (Liti et al., 2009; 1000 Genomes Project Consortium et al., 2010; Koboldt et al., 2013). Phenotypic analysis of related strains of budding yeast showed remarkable differences in response to a variety of stimuli including acclimation to temperature and tolerance to drugs (Liti et al., 2009). From a medical perspective, the intraorganismal comparisons of genome-wide association studies (GWAS) are identifying allelic heterogeneity that has important implications for organismal development and disease diagnosis, progression, and prognosis (Welch et al., 2012). For example, polymorphisms in the FOXO3 locus, a member of the forkhead family of transcription factors with roles in diverse cellular processes (Litvak et al., 2012; Eijkelenboom and Burgering, 2013), are prognostic for the outcome of patients diagnosed with Crohn’s disease (Lee et al., 2013). Importantly, the polymorphisms in FOXO3 are not diagnostic for the disease, and susceptibility is therefore contingent on other factors. In addition to Crohn’s disease, FOXO3 may also affect the severity of prognosis for other autoimmune diseases including rheumatoid arthritis. These results highlight the importance of allelic diversity to cellular function, an underexplored topic that will only be understood from the context of a systems perspective.
The role of NGS in providing high-resolution data on the transcriptome of cells also has mechanistic relevance for cell biology. Deep sequencing of RNA (RNaseq) provides absolute transcription levels of both annotated and unannotated regions of the genome. The result of its application has revealed a wealth of unanticipated complexity in transcript heterogeneity, including novel splice variants, alternative start and stop sites, the lengths of 5′ and 3′ untranslated regions, and the dynamic expression of bicistronic transcripts (Pelechano et al., 2013; Gupta et al., 2014; Pelechano et al., 2014). Transcription of the genome is also much more pervasive and ubiquitous than previously thought (Djebali et al., 2012). Use of NGS technology in combination with novel processing steps is just beginning to vastly redefine our understanding of transcriptional regulation and complexity (Mudge et al., 2013). For example, a recent survey of the yeast transcriptome identified 1.88 million unique mRNA transcript reads (Pelechano et al., 2013). From an organism originally characterized as having 5,885 genes (Goffeau et al., 1996), this is a staggering amount of diversity at the mRNA level. To avoid the biases of isoform analysis that result from enrichment strategies to sequence only the 5′ or 3′ end of individual mature mRNA molecules, a novel intramolecular ligation step after mRNA isolation allowed joint sequencing of both ends of a single mRNA isoform (Pelechano et al., 2013). Consistent with a functional relevance for at least some of this diversity, rather than a result of stochastic transcription initiation or termination, isoform variation was demonstrated to be responsive to changes in growth conditions. These new layers of transcriptional complexity will be of use in refining our understanding of the regulation and plasticity of a cell’s transcriptional response to environmental perturbations.
The ability to precisely map protein–nucleic acid interactions in a quantitative way is perhaps the most demonstrative example of the advancement and refinement of NGS technology. Protein–DNA or protein–RNA purification strategies in combination with exonuclease treatment before deep sequencing of the protected fragments provides the ability to map such interactions at a genome-wide scale to within single nucleotide resolution (Ingolia et al., 2009; Rhee and Pugh, 2011). These high-resolution genome-wide studies enable the comprehensive study and direct visualization of chromatin remodeling dynamics (Yen et al., 2013), identification of transcription factor binding sites (Rhee and Pugh, 2011), assembly of RNA polymerase pre-initiation complexes (Rhee and Pugh, 2012), and the profiling of ribosome occupancy of mRNA (Ingolia et al., 2009; Ingolia et al., 2011; Guttman et al., 2013). When visualized globally, the noisy signals from individual genes are smoothed and universal mechanisms are revealed. Aligning DNA sequences bound to RNA polymerase II pre-initiation complexes and viewing them at a genome scale has provided a unifying view of many regulatory mechanisms governing transcription. One striking revelation was the presence of degenerate TATA-like elements at previously characterized “TATA-less” promoters in yeast (Rhee and Pugh, 2012). Assembling genome-wide maps for an ensemble of RNA polymerase II–associated general transcription factors has also revealed consequences for deviations from the TATA consensus sequence, including increased reliance on nucleosome positioning for proper assembly (Rhee and Pugh, 2012). The fate of both coding and noncoding RNA has been mapped by immunoprecipitations of mRNA-binding proteins (Tuck and Tollervey, 2013). Sorting the ribonucleoprotein complexes using clustering approaches allowed for the identification and classification of several mRNP subclasses with implications for the importance of 3′ processing events in biogenesis, localization, and turnover (Tuck and Tollervey, 2013).
From genomics to proteomics
Despite the advancements in NGS, gene expression and mRNA levels are not very good proxies for protein levels or function in cells. Regulatory mechanisms exist at each stage of a protein’s life cycle: synthesis, folding, targeting, integration into distinct compartments and complexes, activity, stability, and degradation (Vogel and Marcotte, 2012). Measuring the half-life of proteins on a global scale has revealed complexity in protein turnover in a cell type–dependent manner (Claydon and Beynon, 2012). The constituents of protein complexes measured typically have similar turnover rates, although there are exceptions (for examples see Price et al., 2010). In addition, translation and proteolysis not only regulate the synthesis and degradation of proteins, but also serve to buffer intracellular amino acids levels, and must therefore receive regulatory inputs from several sources (Vogel and Marcotte, 2012). Profiling the association of ribosomes with mRNA provides one measure of the rate of protein synthesis (Ingolia et al., 2011). This technique was recently complemented with a proteomic analysis of protein longevity using isotope pulse labeling combined with shotgun tandem mass spectrometry (MS) to measure both the translation of new protein and the longevity of old protein in rat liver and brain cells (Toyama et al., 2013). In addition to discovering 37 long-lived proteins, the combined approach of ribosome profiling and semiquantitative MS revealed that despite the longevity of these proteins, all were pervasively translated. In several cases, discrepancies in the longevity of members of histone and nuclear pore complexes suggest mechanisms regulating the turnover and assembly of these complexes (Toyama et al., 2013).
Information on the pathways and function of a protein is often derived from knowledge of the proteins with which it interacts. Protein–protein interactions (PPIs) range from stable molecular machines of defined stoichiometries and functions to transient interactions whose mechanisms of dynamics are poorly defined. Areas of outstanding interest in proteomics research therefore concern the composition and stoichiometry of protein complexes, the interconnectivity and presence of shared components of different protein complexes, and identification of sites of posttranslational modifications. While detectable at the genomic and transcriptional level, the functional consequence of variation from alternative splicing, allelic variations, and point mutations often plays out in the altered activity or binding capacity of the encoded proteins. For example, one possible phenotype caused by the reduced expression or enhanced turnover of a protein may have more to do with the effect this has on that protein’s binding partners.
Quantitative, sensitive, and reproducible proteomics approaches
Recently, alternative operation modalities using certain types of mass spectrometers are making MS-based proteomics studies quantitative and reproducible, with attomole sensitivity (Doerr, 2013; Marx, 2013; Picotti et al., 2013a). Accumulated data on the fragmentation properties and chromatographic behavior of peptides has enabled the development of targeted and data-independent proteomics approaches (Farrah et al., 2012). In targeted proteomics, e.g., selective reaction monitoring (SRM), the mass spectrometer is tuned to selectively monitor predefined pairs of precursor and product ion masses of unique proteins. This approach has been greatly enabled by the availability of genomic data, inexpensive de novo peptide synthesis techniques, and large-scale peptide reference maps (Ackermann et al., 2008; Farrah et al., 2012; Holman et al., 2012; Picotti et al., 2013b). Multiplexing the assay by retuning the filter allows one to keep a quantitative tally for several hundred proteins in a single experiment.
Targeted proteomics enable an interrogation of the dynamics of PPI networks. Importantly, the focus of proteomic studies can move beyond the technicalities of coverage depth or reproducibility, and allows one to pursue interrogation of the kinetic properties of protein complexes. For example, by adapting an affinity purification strategy to SRM-MS, Bisson et al. (2011) identified 90 reproducible interactors of GRB2, an important hub in growth factor signaling, and mapped the binding site of each protein to one of three characterized protein binding domains within GRB2. Thus, with a single experiment, detailed and quantitative data for 90 PPIs were collected. The dynamics of GRB2 signaling hub complexes and their association with different receptor tyrosine kinases (RTKs) to form signaling scaffolds were then measured against a battery of different growth factor receptor stimulants (Bisson et al., 2011). These experiments revealed stimulation-specific GRB2 complexes that displayed unique temporal kinetics of assembly and disassembly (Bisson et al., 2011). Similarly, the consequences of the temporal kinetics of RTK scaffold assembly were explored with the epidermal growth factor receptor (EGFR) signaling scaffold protein Shc1 (Zheng et al., 2013). The dynamics of Shc1 phosphorylation at six residues and its association with 41 binding partners was followed over multiple time points after activation of EGFR by EGF. Analysis of the results revealed a dynamic network of phosphorylation-dependent regulated recruitment and assembly of three distinct signaling complexes (Zheng et al., 2013). Therefore, SRM-MS offers the ability to explore the dynamic properties of protein networks that are essential for mechanistic understanding of biological function. In addition to studying signaling cascade kinetics, SRM-MS has also been successfully applied to the interrogation of 464 known and putative RNA polymerase II–associated general transcription factors and used to probe them for DNA binding capacity (Mirzaei et al., 2013).
Refinements of MS methodologies have also allowed for the development of quantitative data-independent approaches. For example, the systematic fragmentation of precursor ions independently of ion count was applied to a proteomic analysis of the principle of polydispersity (Jung et al., 2013). Polydispersity is a population phenomenon of proteins owing to their localization to one or more organelles that have nonuniform properties leading, for example, to a collection of different sedimentation coefficients (De Duve et al., 1960; de Duve, 1964). The cosedimentation profile of proteins from cytosolic and organellar fractions of yeast grown under different nutrient conditions enabled a comprehensive look at the dynamics of protein movement between the cytosol and organelles such as mitochondria and peroxisomes. This data-independent acquisition protocol improved the dynamic range of protein identification by over an order of magnitude from the classic shotgun MS/MS approach (Yi et al., 2002; Marelli et al., 2004; Jung et al., 2013). Remarkably, this approach revealed that as many as ∼1,200 proteins, a substantial portion of the yeast proteome, shift their relative distributions between the cytosol and an organellar fraction in response to changes in nutrient conditions (Jung et al., 2013).
The goal of an unbiased data-independent approach is to combine the benefits of increased sensitivity and quantitative capacity of targeted approaches with the discovery component found in data-dependent approaches (Gillet et al., 2012). As with targeted SRM-MS, a priori information from preassembled spectral libraries can be used by targeted data-mining algorithms to identify protein-specific peptide fragment ion traces in complex fragment ion spectra (Gillet et al., 2012). With specialized mass spectrometers, a complete record of the proteins contained in a sample can be recorded by implementing comprehensive and systematic acquisition protocols that produce time-resolved and mass-segmented complex spectral ion maps. One such promising approach is sequential window acquisition of all theoretical spectra (SWATH-MS), which refers to the way the mass spectrometer is operated to collect these comprehensive proteomics data (Collins et al., 2013). In a proof-of-principle experiment, SWATH-MS of affinity purified 14-3-3β, an abundant cytosolic scaffold protein, consistently identified 1,967 interacting proteins and quantified the dynamic changes of 567 members of the promiscuous 14-3-3β scaffold interactome after stimulation of the insulin–PI3K–AKT pathway (Collins et al., 2013). In a complementary study, the interactome data generated by SWATH-MS was used to track changes to PPI networks induced by chemical inhibitors or allelic variations linked to disease pathologies (Lambert et al., 2013). Retaining the discovery component of traditional MS, experiments conducted with data-independent approaches, such as SWATH-MS, ensure an accurate measurement of the effect of biological perturbations on the study of cellular mechanisms. Also, the comprehensive spectral data generated serve as a reliable digital record of a protein sample, and ensure data integrity. These spectral maps can assist in experiment optimization or in comparing protocols or results between laboratories, or can be used for reassessment of samples to look for features that might have been initially missed or deemed unimportant. Excitingly, targeted and data-independent MS combined with cross-linking agents is an emerging approach to improve the detection and measurement of transient PPIs and for discovering the dynamic rearrangements within protein complexes (Gingras et al., 2007; Politis et al., 2014).
Systematically deciphering the genotype-to-phenotype paradigm
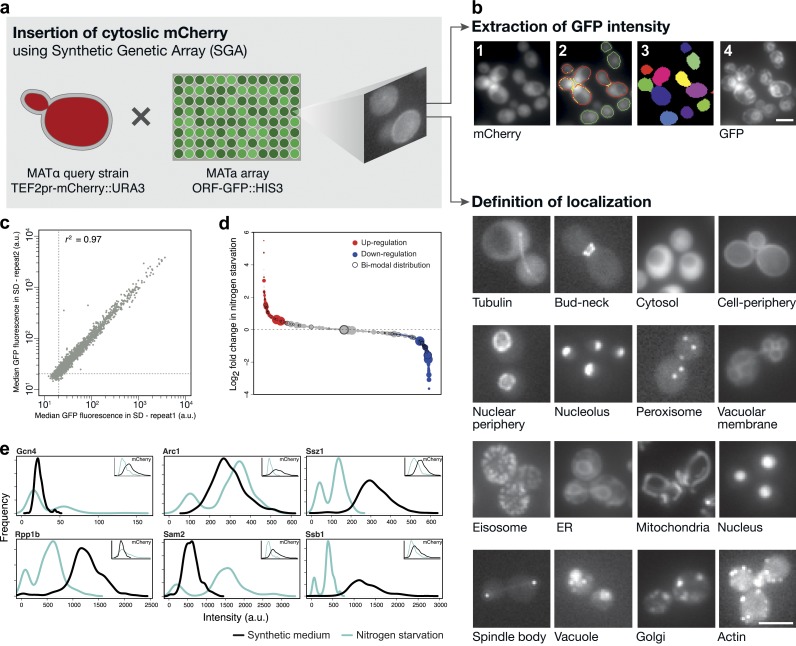
Functional genomic studies pursue a mechanistic explanation for the cause and effect relationship between genotype and phenotype (Fig. 2). At a systems level, the cause and effect of genetic perturbations are typically considered from a network perspective. In organisms that are easy to manipulate genetically, i.e., Saccharomyces cerevisiae, functional genetics have been automated using robotics-assisted synthetic genetic array (SGA) methodology and measurements of colony size as a function of cellular fitness for a phenotype (Tong et al., 2001; Schuldiner et al., 2005; Tong and Boone, 2006; Roguev et al., 2008). The first compilation of a global genetic map was composed of genetic interaction profiles that covered 75% of all genes in yeast (Costanzo et al., 2010). These initial studies have revealed that the genetic interaction profile of one allele, against a genomic collection of other alleles, comprises a unique phenotypic signature that can be used to deduce uncharacterized functions and to order sets of genes within novel functional pathways (Beltrao et al., 2010; Costanzo et al., 2010; Baryshnikova et al., 2013). Such global genetic interaction networks are assembled by systematically measuring the degree of epistasis that pairs of genetic alleles impart on each other. The strength of epistasis of one allele against another cannot be assumed to scale linearly across a systematic array of all alleles in a genome. However, the systematic assembly of epistatic interactions between an allele of one gene against alleles in all other genes has successfully revealed the modularity of protein complexes as well as the cooperativity and redundancy that exists between known biological pathways and processes (Baryshnikova et al., 2013). For example, comparing the genetic interaction network profiles with networks identified by chemical–genetic perturbations can help predict the cellular targets of chemical compounds (Hillenmeyer et al., 2008, 2010; Costanzo et al., 2010; Lee et al., 2014). These functional genetics studies also highlight the challenge of pleiotropy for determining gene function with a reductionist approach. The unbiased, systematic, and quantitative characterization of genetic interaction networks has inverted the reductionist paradigm in defining a process-centric model of gene function to a component-centric model (Weissman, 2010). For example, a compilation of 53 point mutation alleles of yeast RNA polymerase II was used to assemble and systematically interrogate the functional characteristics of each of its subdomains (Braberg et al., 2013). This detailed analysis allowed a high-resolution dissection of coordinated RNA polymerase II activities in transcriptional regulation, including the rate of transcription, splicing events, and start site selection (Braberg et al., 2013). Phenotypic screening is also not limited to cellular growth or fitness. For example, SGA technology has been coupled to an automated microscopy platform to allow systematic interrogation of spindle pole body assembly and microtubule dynamics in yeast (Vizeacoumar et al., 2010; Breker et al., 2013; Fig. 2). The combination of high-content screening and SGA technology has also been used to study peroxisome dynamics (Saleem et al., 2010; Cohen et al., 2014).
Figure 2.
High-content microscopy screening combined with SGA technology. (a) SGA methods are used to integrate fluorescent markers into a yeast library. Depicted is the combining of two fluorescent markers, TEF2pr-mCherry, and a library where each yeast open reading frame has been C-terminally tagged with GFP. (b) Automated microscopy and image processing techniques enable the extraction of relevant information such as protein localization and intensity, which is a proxy for abundance. (c–e) Statistical methods can be used to normalize the quantified data and group proteins and classify them in relation to a particular cellular response. Reproduced from Breker et al. (2013). Bars, 5 µm.
One exciting application of functional genetics is identifying novel drug candidates for cancer (Kuiken and Beijersbergen, 2010). Here, the idea is to search for pathways and genetic interactions that are relevant in the context of a particular cancer or infection and target these pathways and genes for therapeutic intervention. For example, synthetic lethal interactors of oncogenic MYC have been identified through systematic siRNA screens of “druggable” genes, a collection of the human genome whose protein products are known or considered likely to bind with high affinity to known small molecules (Cheng et al., 2007; Toyoshima et al., 2012). This strategy ensures that sensitivity to the drug only occurs in the presence of oncogenic MYC and therefore is applicable in cases where targeting the oncogene itself is not practical or feasible. It also greatly expands the number of druggable targets for a given disease. Similar strategies for certain infectious diseases, where a virus or bacterial pathogen usurps the role of host cellular machinery, seem possible and are another potential application of functional genomics and systems cell biology.
Systems analysis using public databases: Modeling guides experimentation
Publicly available databases of genetic expression data, proteomics data, functional genomic screens, and automated microscopy data repositories are available to provide the inputs necessary for large-scale systems analysis to initiate systems-level interrogations. In many cases, hypotheses formed from the evaluation of a systems dataset are easily addressed with targeted and more traditional approaches to cell biology. They may also serve as a guide for choosing the right type of systems approach to invest for use in further study.
This approach was recently validated for a globally predictive environmental and gene regulatory influence network (EGRIN) model of peroxisome biogenesis in yeast (Danziger et al., 2014). The predictive capacity of the model was subsequently verified in a gene-by-gene focused study of the top candidates to more accurately assess activator or repressor function. This layered and iterative approach added an additional regulatory circuit composed of genes previously not associated with regulating peroxisome biogenesis and integrated them into a model containing a well-studied regulatory circuit. The virtuous cycle of model refinement and the explanatory power of the mechanistically predictive model aptly demonstrate the promise of systems biology to improving our understanding of cellular mechanisms.
An important outcome and aim of systems cell biology will need to be the continued creation and curation of high-quality repositories for systems-level data that ensure accessibility and ease of use for the entire biological community (Hakenberg et al., 2004; Stark et al., 2006; Kowald and Schmeier, 2011; Chatr-aryamontri et al., 2013).
Modeling cellular systems
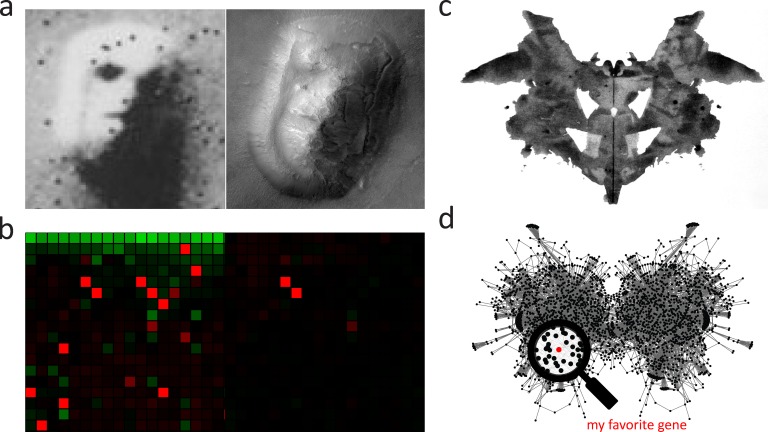
The spectrum of modeling approaches span from conceptual to mechanistic and from focused to broad (Aldridge et al., 2006). It is beyond the scope of this review to cover the plethora of modeling approaches that exist for cell biology (see Meier-Schellersheim et al., 2009; Chen et al., 2010; Ferrell et al., 2011; Ratushny et al., 2011a; Mogilner et al., 2012; Lander, 2013). However, within a systems biology paradigm, modeling forms a central part of a cycle that includes the interpretation and integration of existing and new data, the formation of new hypotheses, and the exploration of relative parameters that aid in designing new experiments to test the model (Fig. 1). Modeling brings objectivity and minimizes the phenomenon of pareidolia, the illusion or misperception of perceiving a vague or obscure stimulus as clear and distinct, in the complex patterns found in systems biology data (Fig. 3).
Figure 3.
Common pitfalls in systems cell biology include pareidolia and experimenter bias. (a) Two satellite photos of an elevated area of land in the Cydonia region of Mars. The image on the left gives the illusion of a face due to the perspective of the image and its relatively low resolution. This illusion is missing from the higher-resolution image on the right. Images captured by the National Aeronautics and Space Administration (NASA). (b) Results from an SGA experiment before (left) and after (right) statistical normalization of the data reflect the pareidolia phenomenon. (c) Rorschach blot 01 is the first in a series of ink blots used in psychological tests that evaluate a subject’s interpretation of ambiguous images for clues about their personality and mental state. Image generated by Hermann Rorschach. (d) A network diagram based on the yeast regulome (Payne and Garrels, 1997) is subject to a similar bias when a familiar node of interest is focused upon.
The goal of modeling is to not merely imitate biological behavior but to simulate perturbations to the system in order to provide quantitative and reliable predictions of function. However, the relationship between any particular model and a set of observations is rarely unique; the number of possible models for a given system is too large without a theory to focus the search space (Brenner, 2010). Therefore, the pairing of a modeling approach with a biological system is important, as each modeling method has individual requirements, limitations, and predictive power. The utility of any given model is in its ability to focus experiments that are predicted to be most informative to the biological area of interest. This is critical given the vast potential of solutions imparted by evolution. Models sharpen questions (Matessi and Karlin, 1984).
Combining modeling with experimentation often leads to new insights synergistically. For example, global monitoring of the GINS complex combined with a very simple model of its movements revealed a surprisingly uniform progression of replication across the genome (Sekedat et al., 2010). The GINS complex is essential for establishing the DNA replication fork that is central to chromosome replication (Labib and Gambus, 2007). Time-resolved chromatin immunoprecipitation (ChIP)-chip experiments were compared with simulations that recapitulated the observed dynamics using an iterative model that relies on reliable assumptions of the distribution of start times, replication velocity, efficiency of initiation, and pausing. The combination of systems data acquisition and accurate models that simulated the data was then used to study firing efficiencies at several replication origins and to study the effect of highly transcribed transfer RNA (tRNA) genes on replication fork arrest (Sekedat et al., 2010).
Qualitative models that use pictures and diagrams with connecting arrows to propose mechanisms are likely the most common and most familiar type of models used by cell biologists. Challenges arise when such models are too abstract, when they depict mechanisms that operate outside of the scale of study, or when an attempt is made to incorporate many different types of experimental observations made under different time frames, conditions, or scales. Formalizing these qualitative models into more mechanistic and multiscale models is an essential step in systems cell biology. For example, we have studied the mechanisms of peroxisome regulation and biogenesis by integrating various global systems datasets to build both kinetic models and genome-wide statistical models (Smith et al., 2007, 2011a,b; Ratushny et al., 2008, 2012; Danziger et al., 2014). These and other studies have revealed the coordination of peroxisome dynamics with other cellular processes. Focusing on peroxisome biogenesis, transcription was shown to control peroxisomal metabolism and peroxisome import and fission machineries, but not components of de novo peroxisome biogenesis. This suggests the utility of transcriptional regulatory data in informing models of regulated peroxisome biogenesis (for review see Smith and Aitchison, 2013).
Models also help to explore the features and topologies of large networks that are useful for studying emergent systems properties. Research into the universality of network structure has revealed several shared characteristics including the small-world phenomenon; that is, molecular networks are like social networks, separated by only a handful of connections (Milgram, 1967; Watts and Strogatz, 1998; Barzel and Barabási, 2013). However, at this level many networks fall prey to the “hair-ball” syndrome and can become unintelligible. Furthermore, the ontological assignments provided in these large-scale networks are oftentimes myopic because they are assigned based on partially characterized phenomena and ignore the unknowns. One solution to this problem is to systematically infer ontological features from the data itself (Dutkowski et al., 2013). By repeating this process in combination with the integration of new data into repositories, we can refine the ontologies that reflect the system characteristics of individual cellular components.
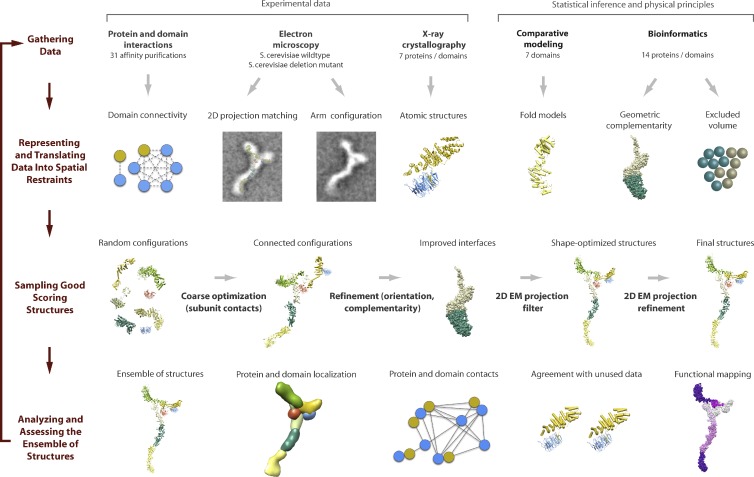
Bringing the leverage of systems biology tools from the level of large networks to the level of the molecules and macromolecular complexes populating these networks is a frontier where both progress and challenges exist. Here, the central challenge is to equate the structural elements of a protein encoded in the genome with the functional capabilities that are phenotypically observed. This is confounded by the modularity evidenced in cells as well as the observed fact that many proteins participate in multiple different complexes. Efforts to map the structure–function relationship with the subcomponents of the nuclear pore complex (NPC) help to illustrate this point (Rout et al., 2000; Hetzer and Wente, 2009; Aitchison and Rout, 2012; Fig. 4). Structural, biochemical, and genetic evidence has revealed a modular NPC with eightfold symmetry (Alber et al., 2007; Hoelz et al., 2011). Forming the outer rings of the NPC is the Nup84 complex, a heptameric modular structure composed of Nup133, Nup120, Nup145c, Nup85, Nup84, Seh1, and Sec13. Seh1 and Sec13 are also components of the Seh1-associated complex and the COPII vesicle-coating complex (Barlowe et al., 1994; Stagg et al., 2006), and this complicates the matter of assigning specific functions to these proteins within the NPC. To determine the subunit arrangement and morphology of the Nup84 complex, an extensive domain-mapping proteomics approach was used to identify contact points within the subcomplex, as well as between the Nup84 complex and the rest of the NPC (Fernandez-Martinez et al., 2012). In addition, negative stain electron microscopy was used to obtain structural information on the different truncated forms of the complex. These data were then translated into spatial restraints and integrated with existing structural data for individual components to build a density map for the Nup84 complex and its arrangement within the NPC (Fig. 4). This process of data integration in combination with modeling, iteration, and refinement, while specific to a portion of a much larger nuclear pore complex, is a specific example of how to systematically explore cellular function.
Figure 4.
Integrative biology combining multiple experimental approaches with statistical inference and modeling approaches. Structural data on the Nup84 complex was derived from domain mapping, negative stain electron microscopy, and x-ray crystallography experiments and integrated with comparative data modeling and bioinformatics methods. The resulting structural models were then overlaid with data from functional mapping experiments. The process is refined by iteration and results in a set of possible structural configurations from which inferences can be made regarding the dynamics of this subcomplex within the much larger nuclear pore complex. Reproduced from Fernandez-Martinez et al. (2012).
Challenges
Notwithstanding our expanded potential to map and quantify molecular components, processes, and functions of biological systems with advanced technologies, our understanding of many parts of these systems is far from complete. There are several major challenges that remain to be addressed in order to effectively model, systematically explore, and predictably control biological systems.
First, high-throughput experimental measurements often uncover intricate relationships between hundreds or thousands of molecular components. This simple fact dramatically increases the number of parameters that need to be included in corresponding models, which in turn necessitates a deluge of new experiments and system interrogations for validation of these parameters. It is important to develop modeling and analytical approaches for rational formulation and parameterization of mathematical models and optimal experimental design. This is especially critical for analysis of combinatorial regulations to avoid an explosion in the number of parameters using algorithms for rational reduction of the model complexity (Bongard and Lipson, 2007; Likhoshvai and Ratushny, 2007). It is also important to develop methods for linking genome-scale models (Bonneau et al., 2007; Danziger et al., 2014) with meso- (Martin et al., 1990; Hoffmann et al., 2002; Ratushny et al., 2008; Ashall et al., 2009; Pang et al., 2013) and small-scale (Brandman et al., 2005; Tsai et al., 2008; Ratushny et al., 2012; Wurtmann et al., 2014) models of relevant subsystems for effective and systematic exploration of the underlying detailed mechanisms. Such methods and models (Karr et al., 2012) are essential for the investigation of transient responses of biological systems when their complexity is maximally exposed due to the presence of nonlinear cooperative and synergistic effects, along with feedback and feed-forward regulatory mechanisms (Alon, 2007; Bennett et al., 2008; Ratushny et al., 2008, 2011b; Ashall et al., 2009; Litvak et al., 2009).
This issue is compounded by the fact that many systems techniques take measurements of populations of cells and molecules rather than of single cells and molecules. Averaging a signal over the population can obscure important phenomena such as phase variation and the processing and response to stochasticity. Techniques that enable the measurement of the genome, transcriptome, metabolome, or proteome of single cells are being developed (Rubakhin et al., 2011; Giesen et al., 2014; Grün et al., 2014; Klemm et al., 2014). The ability to make quantitative measurements of many different molecules repeated on the same cell over time and to multiply this process for many cells would pave the way for understanding biological variability.
Second, the heterogeneity of experimental data and the difficulties in obtaining relevant high-quality data for particular conditions challenges the building cycle of predictive models. Often, available experimental data are not sufficient to fully inform molecular mechanisms of the vast majority of biological systems, and researchers struggle with “black” or “gray” box problems, trying to investigate partially known or poorly understood molecular subsystems. Thus, it is important to develop modeling approaches that allow rational selection of the most appropriate level of detail in the model and match its complexity with the complexity of available experimental data and prior knowledge about the biological system (Likhoshvai and Ratushny, 2007; Bongard and Lipson, 2007; Schmidt and Lipson, 2009; Ratushny et al., 2011a).
Third, molecular processes of many biological systems inherently occur on multiple scales in time and space. Temporally, they span from very fast processes (e.g., formation of molecular complexes and signaling) to relatively slow processes (e.g., organelle biogenesis and cell division). Spatially, biological systems are multicompartmental structures, and frequently many biomolecular processes within any given compartment are inhomogeneous (Mogilner et al., 2012). Integration and exploration of biological processes at multiple levels simultaneously or, in contrast, constructing multiscale models, are fundamental challenges. Furthermore, the nature of biological processes is very diverse and can be viewed/modeled as preferably discrete and stochastic, or, conversely, relatively continuous and deterministic. Therefore, it is crucial to develop flexible hybrid modeling approaches that simultaneously and effectively span the various processes in cells, from the molecular to the morphological. These approaches should integrate temporal and spatial multiscale properties of biological systems and allow a reversible cross-scale flow of information within a single model.
How the cell manages the processing, storage, and transmission of information across multiple scales remains an exceptional challenge (Nurse and Hayles, 2011). In particular, discovering the extent to which different systems mechanisms are responsible for cellular function, and how systems motifs can be combined to bring about new phenotypes, are exciting avenues of pursuit. The future of cell biology as outlined here will increasingly come to rely on systems approaches. It is an exciting time to be pursuing the new biology of the 21st century.
Acknowledgments
The authors thank members of the Aitchison laboratory for critical comments on the manuscript and for discussion. We also thank the anonymous reviewers, even Reviewer 3, who in this case was phenomenal.
Research in the Aitchison laboratory is supported by grants P50 GM076547, U54 GM103511, and U01 GM098256 from the National Institutes of Health.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- MS
- mass spectrometry
- NGS
- next-generation sequencing
- PPI
- protein–protein interaction
- SGA
- synthetic genetic array
- SRM
- selective reaction monitoring
- SWATH
- sequential window acquisition of all theoretical spectra
References
- 1000 Genomes Project Consortium, Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., and McVean G.A.. 2010. A map of human genome variation from population-scale sequencing. Nature. 467:1061–1073 (published erratum appears in Nature. 2011. 473:544) 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann, B.L., Berna M.J., Eckstein J.A., Ott L.W., and Chaudhary A.K.. 2008. Current applications of liquid chromatography/mass spectrometry in pharmaceutical discovery after a decade of innovation. Annu. Rev. Anal. Chem. (Palo Alto Calif). 1:357–396 10.1146/annurev.anchem.1.031207.112855 [DOI] [PubMed] [Google Scholar]
- Aitchison, J.D., and Galitski T.. 2003. Inventories to insights. J. Cell Biol. 161:465–469 10.1083/jcb.200302041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison, J.D., and Rout M.P.. 2012. The yeast nuclear pore complex and transport through it. Genetics. 190:855–883 10.1534/genetics.111.127803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber, F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007. Determining the architectures of macromolecular assemblies. Nature. 450:683–694 10.1038/nature06404 [DOI] [PubMed] [Google Scholar]
- Alberts, B.1998. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 92:291–294 10.1016/S0092-8674(00)80922-8 [DOI] [PubMed] [Google Scholar]
- Aldridge, B.B., Burke J.M., Lauffenburger D.A., and Sorger P.K.. 2006. Physicochemical modelling of cell signalling pathways. Nat. Cell Biol. 8:1195–1203 10.1038/ncb1497 [DOI] [PubMed] [Google Scholar]
- Alon, U.2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8:450–461 10.1038/nrg2102 [DOI] [PubMed] [Google Scholar]
- Ashall, L., Horton C.A., Nelson D.E., Paszek P., Harper C.V., Sillitoe K., Ryan S., Spiller D.G., Unitt J.F., Broomhead D.S., et al. 2009. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science. 324:242–246 10.1126/science.1164860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., and Schekman R.. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 77:895–907 10.1016/0092-8674(94)90138-4 [DOI] [PubMed] [Google Scholar]
- Baryshnikova, A., Costanzo M., Myers C.L., Andrews B., and Boone C.. 2013. Genetic interaction networks: toward an understanding of heritability. Annu. Rev. Genomics Hum. Genet. 14:111–133 10.1146/annurev-genom-082509-141730 [DOI] [PubMed] [Google Scholar]
- Barzel, B., and Barabási A.-L.. 2013. Universality in network dynamics. Nat. Phys. 9:750 10.1038/nphys2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao, P., Cagney G., and Krogan N.J.. 2010. Quantitative genetic interactions reveal biological modularity. Cell. 141:739–745 10.1016/j.cell.2010.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.R., Pang W.L., Ostroff N.A., Baumgartner B.L., Nayak S., Tsimring L.S., and Hasty J.. 2008. Metabolic gene regulation in a dynamically changing environment. Nature. 454:1119–1122 10.1038/nature07211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson, N., James D.A., Ivosev G., Tate S.A., Bonner R., Taylor L., and Pawson T.. 2011. Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat. Biotechnol. 29:653–658 10.1038/nbt.1905 [DOI] [PubMed] [Google Scholar]
- Bongard, J., and Lipson H.. 2007. Automated reverse engineering of nonlinear dynamical systems. Proc. Natl. Acad. Sci. USA. 104:9943–9948 10.1073/pnas.0609476104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau, R., Facciotti M.T., Reiss D.J., Schmid A.K., Pan M., Kaur A., Thorsson V., Shannon P., Johnson M.H., Bare J.C., et al. 2007. A predictive model for transcriptional control of physiology in a free living cell. Cell. 131:1354–1365 10.1016/j.cell.2007.10.053 [DOI] [PubMed] [Google Scholar]
- Braberg, H., Jin H., Moehle E.A., Chan Y.A., Wang S., Shales M., Benschop J.J., Morris J.H., Qiu C., Hu F., et al. 2013. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 154:775–788 10.1016/j.cell.2013.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman, O., Ferrell J.E. Jr, Li R., and Meyer T.. 2005. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 310:496–498 10.1126/science.1113834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breker, M., Gymrek M., and Schuldiner M.. 2013. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 200:839–850 10.1083/jcb.201301120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S.2010. Sequences and consequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:207–212 10.1098/rstb.2009.0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-aryamontri, A., Breitkreutz B.-J., Heinicke S., Boucher L., Winter A., Stark C., Nixon J., Ramage L., Kolas N., O’Donnell L., et al. 2013. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 41:D816–D823 10.1093/nar/gks1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W.W., Niepel M., and Sorger P.K.. 2010. Classic and contemporary approaches to modeling biochemical reactions. Genes Dev. 24:1861–1875 10.1101/gad.1945410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, A.C., Coleman R.G., Smyth K.T., Cao Q., Soulard P., Caffrey D.R., Salzberg A.C., and Huang E.S.. 2007. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 25:71–75 10.1038/nbt1273 [DOI] [PubMed] [Google Scholar]
- Claydon, A.J., and Beynon R.. 2012. Proteome dynamics: revisiting turnover with a global perspective. Mol. Cell. Proteomics. 11:1551–1565 10.1074/mcp.O112.022186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, Y., Klug Y.A., Dimitrov L., Erez Z., Chuartzman S.G., Elinger D., Yofe I., Soliman K., Gärtner J., Thoms S., et al. 2014. Peroxisomes are juxtaposed to strategic sites on mitochondria. Mol. Biosyst. 10:1742–1748 10.1039/c4mb00001c [DOI] [PubMed] [Google Scholar]
- Collins, B.C., Gillet L.C., Rosenberger G., Röst H.L., Vichalkovski A., Gstaiger M., and Aebersold R.. 2013. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods. 10:1246–1253 10.1038/nmeth.2703 [DOI] [PubMed] [Google Scholar]
- Costanzo, M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L.Y., Toufighi K., Mostafavi S., et al. 2010. The genetic landscape of a cell. Science. 327:425–431 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danziger, S.A., Ratushny A.V., Smith J.J., Saleem R.A., Wan Y., Arens C.E., Armstrong A.M., Sitko K., Chen W.-M., Chiang J.-H., et al. 2014. Molecular mechanisms of system responses to novel stimuli are predictable from public data. Nucleic Acids Res. 42:1442–1460 10.1093/nar/gkt938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve, C.1964. Principles of tissue fractionation. J. Theor. Biol. 6:33–59 10.1016/0022-5193(64)90065-7 [DOI] [PubMed] [Google Scholar]
- De Duve, C., Beaufay H., Jacques P., Rahman-Li Y., Sellinger O.Z., Wattiaux R., and De Coninck S.. 1960. Intracellular localization of catalase and of some oxidases in rat liver. Biochim. Biophys. Acta. 40:186–187 10.1016/0006-3002(60)91338-X [DOI] [PubMed] [Google Scholar]
- Djebali, S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. 2012. Landscape of transcription in human cells. Nature. 489:101–108 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerr, A.2013. Mass spectrometry-based targeted proteomics. Nat. Methods. 10:23 10.1038/nmeth.2286 [DOI] [PubMed] [Google Scholar]
- Doolittle, W.F.2012. Evolutionary biology: A ratchet for protein complexity. Nature. 481:270–271 [DOI] [PubMed] [Google Scholar]
- Dutkowski, J., Kramer M., Surma M.A., Balakrishnan R., Cherry J.M., Krogan N.J., and Ideker T.. 2013. A gene ontology inferred from molecular networks. Nat. Biotechnol. 31:38–45 10.1038/nbt.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelenboom, A., and Burgering B.M.T.. 2013. FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 14:83–97 10.1038/nrm3507 [DOI] [PubMed] [Google Scholar]
- Elias, M., Brighouse A., Gabernet-Castello C., Field M.C., and Dacks J.B.. 2012. Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases. J. Cell Sci. 125:2500–2508 10.1242/jcs.101378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrah, T., Deutsch E.W., Kreisberg R., Sun Z., Campbell D.S., Mendoza L., Kusebauch U., Brusniak M.-Y., Hüttenhain R., Schiess R., et al. 2012. PASSEL: the PeptideAtlas SRMexperiment library. Proteomics. 12:1170–1175 10.1002/pmic.201100515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez, J., Phillips J., Sekedat M.D., Diaz-Avalos R., Velazquez-Muriel J., Franke J.D., Williams R., Stokes D.L., Chait B.T., Sali A., and Rout M.P.. 2012. Structure-function mapping of a heptameric module in the nuclear pore complex. J. Cell Biol. 196:419–434 10.1083/jcb.201109008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell, J.E. Jr, Tsai T.Y.-C., and Yang Q.. 2011. Modeling the cell cycle: why do certain circuits oscillate? Cell. 144:874–885 10.1016/j.cell.2011.03.006 [DOI] [PubMed] [Google Scholar]
- Finnigan, G.C., Hanson-Smith V., Stevens T.H., and Thornton J.W.. 2012. Evolution of increased complexity in a molecular machine. Nature. 481:360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesen, C., Wang H.A.O., Schapiro D., Zivanovic N., Jacobs A., Hattendorf B., Schüffler P.J., Grolimund D., Buhmann J.M., Brandt S., et al. 2014. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods. 11:417–422 10.1038/nmeth.2869 [DOI] [PubMed] [Google Scholar]
- Gillet, L.C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., and Aebersold R.. 2012. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 11:O111.016717 10.1074/mcp.O111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.-C., Gstaiger M., Raught B., and Aebersold R.. 2007. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8:645–654 10.1038/nrm2208 [DOI] [PubMed] [Google Scholar]
- Goffeau, A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., et al. 1996. Life with 6000 genes. Science. 274:546–567 10.1126/science.274.5287.546 [DOI] [PubMed] [Google Scholar]
- Grün, D., Kester L., and van Oudenaarden A.. 2014. Validation of noise models for single-cell transcriptomics. Nat. Methods. 11:637–640 10.1038/nmeth.2930 [DOI] [PubMed] [Google Scholar]
- Gupta, I., Clauder-Münster S., Klaus B., Järvelin A.I., Aiyar R.S., Benes V., Wilkening S., Huber W., Pelechano V., and Steinmetz L.M.. 2014. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol. Syst. Biol. 10:719 10.1002/msb.135068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman, M., Russell P., Ingolia N.T., Weissman J.S., and Lander E.S.. 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 154:240–251 10.1016/j.cell.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakenberg, J., Schmeier S., Kowald A., Klipp E., and Leser U.. 2004. Finding kinetic parameters using text mining. OMICS. 8:131–152 10.1089/1536231041388366 [DOI] [PubMed] [Google Scholar]
- Hartwell, L.H., Hopfield J.J., Leibler S., and Murray A.W.. 1999. From molecular to modular cell biology. Nature. 402(6761Suppl):C47–C52 10.1038/35011540 [DOI] [PubMed] [Google Scholar]
- Hetzer, M.W., and Wente S.R.. 2009. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell. 17:606–616 10.1016/j.devcel.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer, M.E., Fung E., Wildenhain J., Pierce S.E., Hoon S., Lee W., Proctor M., St. Onge R.P., Tyers M., Koller D., et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 320:362–365 10.1126/science.1150021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer, M.E., Ericson E., Davis R.W., Nislow C., Koller D., and Giaever G.. 2010. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol. 11:R30 10.1186/gb-2010-11-3-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz, A., Debler E.W., and Blobel G.. 2011. The structure of the nuclear pore complex. Annu. Rev. Biochem. 80:613–643 10.1146/annurev-biochem-060109-151030 [DOI] [PubMed] [Google Scholar]
- Hoffmann, A., Levchenko A., Scott M.L., and Baltimore D.. 2002. The IκB-NF-κB signaling module: temporal control and selective gene activation. Science. 298:1241–1245 10.1126/science.1071914 [DOI] [PubMed] [Google Scholar]
- Holman, S.W., Sims P.F.G., and Eyers C.E.. 2012. The use of selected reaction monitoring in quantitative proteomics. Bioanalysis. 4:1763–1786 10.4155/bio.12.126 [DOI] [PubMed] [Google Scholar]
- Ideker, T., Galitski T., and Hood L.. 2001. A new approach to decoding life: systems biology. Annu. Rev. Genomics Hum. Genet. 2:343–372 10.1146/annurev.genom.2.1.343 [DOI] [PubMed] [Google Scholar]
- Ingolia, N.T., Ghaemmaghami S., Newman J.R.S., and Weissman J.S.. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324:218–223 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia, N.T., Lareau L.F., and Weissman J.S.. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 147:789–802 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, S., Smith J.J., von Haller P.D., Dilworth D.J., Sitko K.A., Miller L.R., Saleem R.A., Goodlett D.R., and Aitchison J.D.. 2013. Global analysis of condition-specific subcellular protein distribution and abundance. Mol. Cell. Proteomics. 12:1421–1435 10.1074/mcp.O112.019166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr, J.R., Sanghvi J.C., Macklin D.N., Gutschow M.V., Jacobs J.M., Bolival B. Jr, Assad-Garcia N., Glass J.I., and Covert M.W.. 2012. A whole-cell computational model predicts phenotype from genotype. Cell. 150:389–401 10.1016/j.cell.2012.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano, H.2002. Systems biology: a brief overview. Science. 295:1662–1664 10.1126/science.1069492 [DOI] [PubMed] [Google Scholar]
- Klemm, S., Semrau S., Wiebrands K., Mooijman D., Faddah D.A., Jaenisch R., and van Oudenaarden A.. 2014. Transcriptional profiling of cells sorted by RNA abundance. Nat. Methods. 11:549–551 10.1038/nmeth.2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt, D.C., Steinberg K.M., Larson D.E., Wilson R.K., and Mardis E.R.. 2013. The next-generation sequencing revolution and its impact on genomics. Cell. 155:27–38 10.1016/j.cell.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V.2011. The Logic of Chance. First edition FT Press Service, Upper Saddle River, NJ: 516 pp [Google Scholar]
- Koonin, E.V., and Mulkidjanian A.Y.. 2013. Evolution of cell division: from shear mechanics to complex molecular machineries. Cell. 152:942–944 10.1016/j.cell.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Koumandou, V.L., Wickstead B., Ginger M.L., van der Giezen M., Dacks J.B., and Field M.C.. 2013. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit. Rev. Biochem. Mol. Biol. 48:373–396 10.3109/10409238.2013.821444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowald, A., and Schmeier S.. 2011. Text mining for systems modeling. Methods Mol. Biol. 696:305–318 10.1007/978-1-60761-987-1_19 [DOI] [PubMed] [Google Scholar]
- Kuiken, H.J., and Beijersbergen R.L.. 2010. Exploration of synthetic lethal interactions as cancer drug targets. Future Oncol. 6:1789–1802 10.2217/fon.10.131 [DOI] [PubMed] [Google Scholar]
- Labib, K., and Gambus A.. 2007. A key role for the GINS complex at DNA replication forks. Trends Cell Biol. 17:271–278 10.1016/j.tcb.2007.04.002 [DOI] [PubMed] [Google Scholar]
- Lambert, J.-P., Ivosev G., Couzens A.L., Larsen B., Taipale M., Lin Z.-Y., Zhong Q., Lindquist S., Vidal M., Aebersold R., et al. 2013. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat. Methods. 10:1239–1245 10.1038/nmeth.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, A.D.2013. How cells know where they are. Science. 339:923–927 10.1126/science.1224186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.C., Espéli M., Anderson C.A., Linterman M.A., Pocock J.M., Williams N.J., Roberts R., Viatte S., Fu B., Peshu N., et al. UK IBD Genetics Consortium. 2013. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 155:57–69 10.1016/j.cell.2013.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, A.Y., St. Onge R.P., Proctor M.J., Wallace I.M., Nile A.H., Spagnuolo P.A., Jitkova Y., Gronda M., Wu Y., Kim M.K., et al. 2014. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science. 344:208–211 10.1126/science.1250217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likhoshvai, V., and Ratushny A.. 2007. Generalized hill function method for modeling molecular processes. J. Bioinform. Comput. Biol. 5:521–531 10.1142/S0219720007002837 [DOI] [PubMed] [Google Scholar]
- Liti, G., Carter D.M., Moses A.M., Warringer J., Parts L., James S.A., Davey R.P., Roberts I.N., Burt A., Koufopanou V., et al. 2009. Population genomics of domestic and wild yeasts. Nature. 458:337–341 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak, V., Ramsey S.A., Rust A.G., Zak D.E., Kennedy K.A., Lampano A.E., Nykter M., Shmulevich I., and Aderem A.. 2009. Function of C/EBPδ in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat. Immunol. 10:437–443 10.1038/ni.1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak, V., Ratushny A.V., Lampano A.E., Schmitz F., Huang A.C., Raman A., Rust A.G., Bergthaler A., Aitchison J.D., and Aderem A.. 2012. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature. 490:421–425 10.1038/nature11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis, E.R.2013. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. (Palo Alto Calif.). 6:287–303 10.1146/annurev-anchem-062012-092628 [DOI] [PubMed] [Google Scholar]
- Marelli, M., Smith J.J., Jung S., Yi E., Nesvizhskii A.I., Christmas R.H., Saleem R.A., Tam Y.Y.C., Fagarasanu A., Goodlett D.R., et al. 2004. Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J. Cell Biol. 167:1099–1112 10.1083/jcb.200404119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, R.J., Kusel J.R., and Pennington A.J.. 1990. Surface properties of membrane vesicles prepared from muscle cells of Ascaris suum. J. Parasitol. 76:340–348 10.2307/3282663 [DOI] [PubMed] [Google Scholar]
- Marx, V.2013. Targeted proteomics. Nat. Methods. 10:19–22 10.1038/nmeth.2285 [DOI] [PubMed] [Google Scholar]
- Mast, F.D., Barlow L.D., Rachubinski R.A., and Dacks J.B.. 2014. Evolutionary mechanisms for establishing eukaryotic cellular complexity. Trends Cell Biol. 24:435–442 10.1016/j.tcb.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Matessi, C., and Karlin S.. 1984. On the evolution of altruism by kin selection. Proc. Natl. Acad. Sci. USA. 81:1754–1758 10.1073/pnas.81.6.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Schellersheim, M., Fraser I.D.C., and Klauschen F.. 2009. Multiscale modeling for biologists. Wiley Interdiscip. Rev. Syst. Biol. Med. 1:4–14 10.1002/wsbm.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgram, S.1967. The small world problem. Psychol. Today. 2:60–67 [Google Scholar]
- Mirzaei, H., Knijnenburg T.A., Kim B., Robinson M., Picotti P., Carter G.W., Li S., Dilworth D.J., Eng J.K., Aitchison J.D., et al. 2013. Systematic measurement of transcription factor-DNA interactions by targeted mass spectrometry identifies candidate gene regulatory proteins. Proc. Natl. Acad. Sci. USA. 110:3645–3650 10.1073/pnas.1216918110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner, A., Allard J., and Wollman R.. 2012. Cell polarity: quantitative modeling as a tool in cell biology. Science. 336:175–179 10.1126/science.1216380 [DOI] [PubMed] [Google Scholar]
- Mudge, J.M., Frankish A., and Harrow J.. 2013. Functional transcriptomics in the post-ENCODE era. Genome Res. 23:1961–1973 10.1101/gr.161315.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse, P.2008. Life, logic and information. Nature. 454:424–426 10.1038/454424a [DOI] [PubMed] [Google Scholar]
- Nurse, P., and Hayles J.. 2011. The cell in an era of systems biology. Cell. 144:850–854 10.1016/j.cell.2011.02.045 [DOI] [PubMed] [Google Scholar]
- Pang, W.L., Kaur A., Ratushny A.V., Cvetkovic A., Kumar S., Pan M., Arkin A.P., Aitchison J.D., Adams M.W.W., and Baliga N.S.. 2013. Metallochaperones regulate intracellular copper levels. PLOS Comput. Biol. 9:e1002880 10.1371/journal.pcbi.1002880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, W.E., and Garrels J.I.. 1997. Yeast Protein database (YPD): a database for the complete proteome of Saccharomyces cerevisiae. Nucleic Acids Res. 25:57–62 10.1093/nar/25.1.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano, V., Wei W., and Steinmetz L.M.. 2013. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 497:127–131 10.1038/nature12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano, V., Wei W., Jakob P., and Steinmetz L.M.. 2014. Genome-wide identification of transcript start and end sites by transcript isoform sequencing. Nat. Protoc. 9:1740–1759 10.1038/nprot.2014.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti, P., Bodenmiller B., and Aebersold R.. 2013a. Proteomics meets the scientific method. Nat. Methods. 10:24–27 10.1038/nmeth.2291 [DOI] [PubMed] [Google Scholar]
- Picotti, P., Clément-Ziza M., Lam H., Campbell D.S., Schmidt A., Deutsch E.W., Röst H., Sun Z., Rinner O., Reiter L., et al. 2013b. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 494:266–270 10.1038/nature11835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis, A., Stengel F., Hall Z., Hernández H., Leitner A., Walzthoeni T., Robinson C.V., and Aebersold R.. 2014. A mass spectrometry-based hybrid method for structural modeling of protein complexes. Nat. Methods. 11:403–406 10.1038/nmeth.2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, J.C., Guan S., Burlingame A., Prusiner S.B., and Ghaemmaghami S.. 2010. Analysis of proteome dynamics in the mouse brain. Proc. Natl. Acad. Sci. USA. 107:14508–14513 10.1073/pnas.1006551107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny, A.V., Ramsey S.A., Roda O., Wan Y., Smith J.J., and Aitchison J.D.. 2008. Control of transcriptional variability by overlapping feed-forward regulatory motifs. Biophys. J. 95:3715–3723 10.1529/biophysj.108.134064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny, A.V., Ramsey S.A., and Aitchison J.D.. 2011a. Mathematical modeling of biomolecular network dynamics. Methods Mol. Biol. 781:415–433 10.1007/978-1-61779-276-2_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny, A.V., Shmulevich I., and Aitchison J.D.. 2011b. Trade-off between responsiveness and noise suppression in biomolecular system responses to environmental cues. PLOS Comput. Biol. 7:e1002091 10.1371/journal.pcbi.1002091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratushny, A.V., Saleem R.A., Sitko K., Ramsey S.A., and Aitchison J.D.. 2012. Asymmetric positive feedback loops reliably control biological responses. Mol. Syst. Biol. 8:577 10.1038/msb.2012.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, H.S., and Pugh B.F.. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 147:1408–1419 10.1016/j.cell.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, H.S., and Pugh B.F.. 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 483:295–301 10.1038/nature10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev, A., Bandyopadhyay S., Zofall M., Zhang K., Fischer T., Collins S.R., Qu H., Shales M., Park H.-O., Hayles J., et al. 2008. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 322:405–410 10.1126/science.1162609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., and Chait B.T.. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–652 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin, S.S., Romanova E.V., Nemes P., and Sweedler J.V.. 2011. Profiling metabolites and peptides in single cells. Nat. Methods. 8:S20–S29 10.1038/nmeth.1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem, R.A., Long-O’Donnell R., Dilworth D.J., Armstrong A.M., Jamakhandi A.P., Wan Y., Knijnenburg T.A., Niemistö A., Boyle J., Rachubinski R.A., et al. 2010. Genome-wide analysis of effectors of peroxisome biogenesis. PLoS ONE. 5:e11953 10.1371/journal.pone.0011953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M., and Lipson H.. 2009. Distilling free-form natural laws from experimental data. Science. 324:81–85 10.1126/science.1165893 [DOI] [PubMed] [Google Scholar]
- Schuldiner, M., Collins S.R., Thompson N.J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J.F., et al. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 123:507–519 10.1016/j.cell.2005.08.031 [DOI] [PubMed] [Google Scholar]
- Sekedat, M.D., Fenyö D., Rogers R.S., Tackett A.J., Aitchison J.D., and Chait B.T.. 2010. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol. Syst. Biol. 6:353 10.1038/msb.2010.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short, B.2009. Cell biologists expand their networks. J. Cell Biol. 186:305–311 10.1083/jcb.200907093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.J., and Aitchison J.D.. 2013. Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14:803–817 10.1038/nrm3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.J., Ramsey S.A., Marelli M., Marzolf B., Hwang D., Saleem R.A., Rachubinski R.A., and Aitchison J.D.. 2007. Transcriptional responses to fatty acid are coordinated by combinatorial control. Mol. Syst. Biol. 3:115 10.1038/msb4100157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.J., Miller L.R., Kreisberg R., Vazquez L., Wan Y., and Aitchison J.D.. 2011a. Environment-responsive transcription factors bind subtelomeric elements and regulate gene silencing. Mol. Syst. Biol. 7:455 10.1038/msb.2010.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.J., Saleem R.A., and Aitchison J.D.. 2011b. Statistical analysis of dynamic transcriptional regulatory network structure. Methods Mol. Biol. 781:337–352 10.1007/978-1-61779-276-2_16 [DOI] [PubMed] [Google Scholar]
- Stagg, S.M., Gürkan C., Fowler D.M., LaPointe P., Foss T.R., Potter C.S., Carragher B., and Balch W.E.. 2006. Structure of the Sec13/31 COPII coat cage. Nature. 439:234–238 10.1038/nature04339 [DOI] [PubMed] [Google Scholar]
- Stark, C., Breitkreutz B.-J., Reguly T., Boucher L., Breitkreutz A., and Tyers M.. 2006. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34:D535–D539 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H.Y., and Boone C.. 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313:171–192 [DOI] [PubMed] [Google Scholar]
- Tong, A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Pagé N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Toyama, B.H., Savas J.N., Park S.K., Harris M.S., Ingolia N.T., Yates J.R. III, and Hetzer M.W.. 2013. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 154:971–982 10.1016/j.cell.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima, M., Howie H.L., Imakura M., Walsh R.M., Annis J.E., Chang A.N., Frazier J., Chau B.N., Loboda A., Linsley P.S., et al. 2012. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc. Natl. Acad. Sci. USA. 109:9545–9550 10.1073/pnas.1121119109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, T.Y.-C., Choi Y.S., Ma W., Pomerening J.R., Tang C., and Ferrell J.E. Jr. 2008. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science. 321:126–129 10.1126/science.1156951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck, A.C., and Tollervey D.. 2013. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 154:996–1009 10.1016/j.cell.2013.07.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson, J.J., and Novák B.. 2010. Functional motifs in biochemical reaction networks. Annu. Rev. Phys. Chem. 61:219–240 10.1146/annurev.physchem.012809.103457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar, F.J., van Dyk N., Vizeacoumar F.S., Cheung V., Li J., Sydorskyy Y., Case N., Li Z., Datti A., Nislow C., et al. 2010. Integrating high-throughput genetic interaction mapping and high-content screening to explore yeast spindle morphogenesis. J. Cell Biol. 188:69–81 10.1083/jcb.200909013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, C., and Marcotte E.M.. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13:227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, D.J., and Strogatz S.H.. 1998. Collective dynamics of ‘small-world’ networks. Nature. 393:440–442 10.1038/30918 [DOI] [PubMed] [Google Scholar]
- Weissman, J.S.2010. The epistemology of cell biology. Mol. Biol. Cell. 21:3825 10.1091/mbc.E10-04-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, J.S., Ley T.J., Link D.C., Miller C.A., Larson D.E., Koboldt D.C., Wartman L.D., Lamprecht T.L., Liu F., Xia J., et al. 2012. The origin and evolution of mutations in acute myeloid leukemia. Cell. 150:264–278 10.1016/j.cell.2012.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtmann, E.J., Ratushny A.V., Pan M., Beer K.D., Aitchison J.D., and Baliga N.S.. 2014. An evolutionarily conserved RNase-based mechanism for repression of transcriptional positive autoregulation. Mol. Microbiol. 92:369–382 10.1111/mmi.12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, K., Vinayachandran V., and Pugh B.F.. 2013. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 154:1246–1256 10.1016/j.cell.2013.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, E.C., Marelli M., Lee H., Purvine S.O., Aebersold R., Aitchison J.D., and Goodlett D.R.. 2002. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis. 23:3205–3216 [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Zhang C., Croucher D.R., Soliman M.A., St-Denis N., Pasculescu A., Taylor L., Tate S.A., Hardy W.R., Colwill K., et al. 2013. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 499:166–171 10.1038/nature12308 [DOI] [PMC free article] [PubMed] [Google Scholar]