In spite of global transcriptional inhibition, a decrease in HSF2 expression during mitosis allows for heat shock protein expression and protects cells against proteotoxicity.
Abstract
Unless mitigated, external and physiological stresses are detrimental for cells, especially in mitosis, resulting in chromosomal missegregation, aneuploidy, or apoptosis. Heat shock proteins (Hsps) maintain protein homeostasis and promote cell survival. Hsps are transcriptionally regulated by heat shock factors (HSFs). Of these, HSF1 is the master regulator and HSF2 modulates Hsp expression by interacting with HSF1. Due to global inhibition of transcription in mitosis, including HSF1-mediated expression of Hsps, mitotic cells are highly vulnerable to stress. Here, we show that cells can counteract transcriptional silencing and protect themselves against proteotoxicity in mitosis. We found that the condensed chromatin of HSF2-deficient cells is accessible for HSF1 and RNA polymerase II, allowing stress-inducible Hsp expression. Consequently, HSF2-deficient cells exposed to acute stress display diminished mitotic errors and have a survival advantage. We also show that HSF2 expression declines during mitosis in several but not all human cell lines, which corresponds to the Hsp70 induction and protection against stress-induced mitotic abnormalities and apoptosis.
Introduction
Cells and their proteomes are continuously challenged by various environmental stresses, such as heat and heavy metals, pathophysiological states, or physiological conditions including cell proliferation, which cause acute or chronic stress. To maintain proteostasis, cells respond to stress stimuli by cytoprotective mechanisms, one of which is the evolutionarily well-conserved heat shock response (HSR). The HSR promotes cell survival through robust inducible expression of heat shock proteins (Hsps) that act as molecular chaperones to prevent aggregation of misfolded proteins and facilitate their refolding or degradation (Richter et al., 2010).
Proteotoxic stress, such as acute heat shock, stalls cell cycle progression at the G1/S or G2/M transition (Kühl et al., 2000; Nakai and Ishikawa, 2001), but the exact mechanism remains unknown. In mitosis, a global reduction of transcription, including stress-inducible expression of Hsps, leaves mitotic cells particularly vulnerable to protein damage (Martínez-Balbás et al., 1995). Consequently, many cells subjected to proteotoxic stress undergo apoptosis or mitotic catastrophe, and surviving cells are likely to accumulate mitotic errors, e.g., multipolar spindles and chromosome misalignment (Martínez-Balbás et al., 1995; Hut et al., 2005), resulting in chromosomal instability (CIN).
The stress-induced expression of Hsps is regulated by a family of transcription factors called heat shock factors (HSFs). Of the four mammalian HSFs (HSF1–4), HSF1 is the major stress-responsive factor, required for the inducible expression of, for example, Hsp70 (Åkerfelt et al., 2010). The function of HSF1 is not limited to the expression of Hsps, but it has specific target genes, for example, in cancer, where HSF1 promotes survival and proliferation of highly malignant cells (Dai et al., 2007; Mendillo et al., 2012; Santagata et al., 2013; Vihervaara and Sistonen, 2014). The activity of HSF1 is regulated by a multitude of posttranslational modifications, including phosphorylation, sumoylation, and acetylation (Sarge et al., 1993; Holmberg et al., 2001; Hong et al., 2001; Hietakangas et al., 2003; Guettouche et al., 2005; Westerheide et al., 2009; Anckar and Sistonen, 2011). HSF2 is best known for its role in the developing brain and reproductive organs (Åkerfelt et al., 2010). Evidence for a stress-regulated function of HSF2 is accumulating, as it binds to the promoters of Hsps and modulates the activity of HSF1 through formation of HSF1-HSF2 heterotrimers (Loison et al., 2006; Östling et al., 2007; Sandqvist et al., 2009). HSF2 deficiency has been shown to reduce the temperature at which HSF1 is activated (Shinkawa et al., 2011), but HSF2 alone is a poor activator of Hsp transcription upon stress (Kroeger et al., 1993). Unlike the ubiquitously and constitutively expressed HSF1, HSF2 is a short-lived protein with a tissue- and developmental stage–specific expression pattern (Fiorenza et al., 1995; Björk et al., 2010), and HSF2 activity is mainly regulated by its levels in the cell (Sarge et al., 1991, 1993; Sandqvist et al., 2009; Björk and Sistonen, 2010). As an example, during spermatogenesis, HSF2 is posttranscriptionally regulated by a micro RNA, miR-18, which belongs to the Oncomir-1 cluster (Björk et al., 2010). However, in response to acute stress, the ubiquitin E3 ligase anaphase-promoting complex/cyclosome (APC/C) rapidly ubiquitylates HSF2 and directs it to proteasomal degradation (Ahlskog et al., 2010).
Upon the addition of stress, the inactive HSF1 monomers form homo- or heterotrimers with HSF2 and bind to specific DNA sequences, thus inducing expression of target genes (Sandqvist et al., 2009; Anckar and Sistonen, 2011). In a recent genome-wide study, we compared the target specificity of HSF1 and HSF2 in cycling and mitotic human K562 cells. In stressed cycling cells, HSF1 occupies 1,242 loci, including genes that code for chaperones, transcriptional and translational regulators, and cell cycle determinants, whereas HSF1 is profoundly displaced from mitotic chromatin, and only 35 target loci remain occupied (Vihervaara et al., 2013). This exclusion of HSF1 from chromatin may contribute to the heat sensitivity observed in mitotic cells (Martínez-Balbás et al., 1995). Because HSF2 is capable of binding to >500 loci during mitosis (Vihervaara et al., 2013), we now addressed the functional relevance of HSF2 in the regulation of cell survival in mitotic cells exposed to acute stress. We found that HSF2 levels decline during mitosis in a cell type–specific manner. Our data reveal that in cells where HSF2 was down-regulated, both HSF1 and RNA polymerase II (RNAPII) that are normally displaced from mitotic chromatin were able to access the hsp70 promoter, allowing stress-inducible Hsp70 expression. Intriguingly, HSF2-deficient cells showed diminished mitotic abnormalities and increased survival upon acute heat stress that may provide a protective mechanism to mitotic cells.
Results
HSF2 levels are strictly regulated in mitotic cells
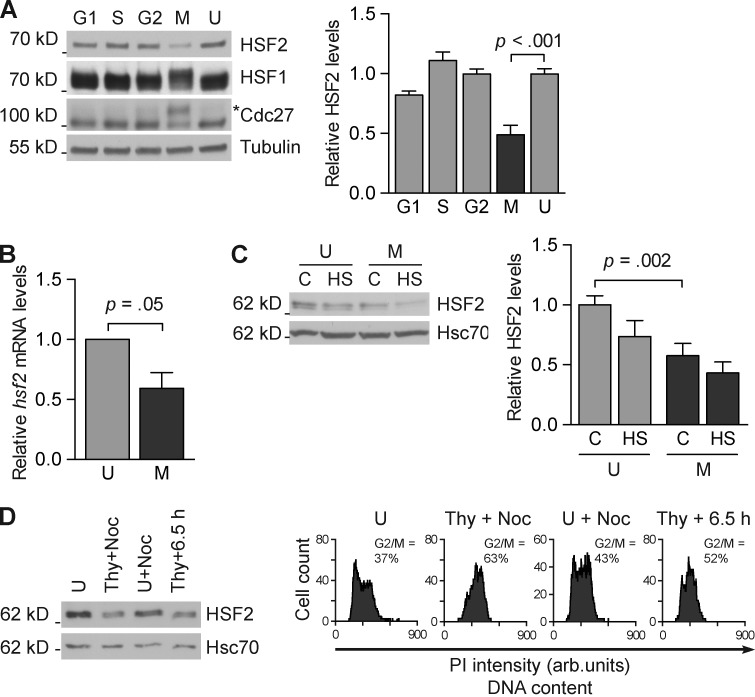
Similar to many other transcription factors, the activity of HSF2 is regulated by its levels in cells (Sandqvist et al., 2009), and prominent changes in HSF2 amounts have been observed both during cell differentiation and upon stress application (Rallu et al., 1997; Mathew et al., 1998; Ahlskog et al., 2010; Björk et al., 2010). Because the DNA-binding activity of HSF2 in mitotic cells has been reported (Xing et al., 2005; Vihervaara et al., 2013), we investigated if there are cell cycle–dependent variations in HSF2 levels. We synchronized K562 cells into G1, S, G2, and M phases and found that both protein and mRNA levels of HSF2 declined in mitosis (Fig. 1, A and B), which is in agreement with previous genome-wide studies showing that hsf2 mRNA is regulated in a cell cycle–dependent manner in HeLa and U2OS cells (Whitfield et al., 2002; Grant et al., 2013). In accordance with our previous results (Ahlskog et al., 2010), HSF2 protein levels were further decreased in response to a 30-min heat shock followed by a 1-h recovery in both unsynchronized and mitotic cells (Fig. 1 C). To confirm that the decrease in HSF2 during mitosis was not due to the experimental procedure, we used different synchronization protocols and observed a decline in HSF2 protein levels in mitosis that correlated with the amount of cells in mitosis (Fig. 1 D). K562 cells that were synchronized by thymidine into S phase, followed by a 4-h release into normal growth medium and a 3-h treatment with nocodazole, showed the lowest levels of HSF2 protein. Cells that were synchronized by thymidine and left to proceed in the cell cycle for 6.5 h, when the cells start entering mitosis, showed a marked decrease in HSF2, whereas unsynchronized cells treated with nocodazole for 3 h had only a minor decrease in HSF2 levels. These results suggest that the expression of HSF2 is delicately regulated during mitosis, clearly implying a function for HSF2 in mitosis.
Figure 1.
HSF2 protein and mRNA levels decrease during mitosis. (A, left) Representative immunoblot analysis of HSF2 protein in unsynchronized K562 cells (U), cells synchronized into mitosis by a thymidine-nocodazole block (M), G1 by releasing mitotic cells for 5 h (G1), S phase by a double-thymidine block (S), or G2 by releasing S phase cells for 5 h (G2). Immunoblotting was performed with the indicated antibodies. Tubulin serves as a loading control. The asterisk denotes phosphorylated Cdc27, which serves as a control indicating that cells are in mitosis. (A, right) Quantification of immunoblots (n = 3) showing HSF2 protein levels related to tubulin. (B) qRT-PCR analysis of hsf2 mRNA in unsynchronized (U) and mitotic (M) K562 cells. Relative quantities of hsf2 mRNA were first normalized to gapdh, and then unsynchronized samples were arbitrarily set to a value of 1 (n = 5). (C, left) Immunoblot analysis of HSF2 protein in unsynchronized (U) and mitotic (M) K562 cells. Cells were left untreated (C) or subjected to heat shock (HS; 30 min at 42°C, 1 h at 37°C). (C, right) Quantifications of blots where HSF2 was related to Hsc70 (n = 4). (D) Representative immunoblot analysis of HSF2 protein in K562 cells (n = 3). Cells were either left unsynchronized (U); treated with thymidine (24 h), followed by, after a 4-h release, the addition of nocodazole for 3 h (Thy-Noc); only treated with nocodazole for 3 h (U+Noc); or released from thymidine for 6.5 h (Thy+6.5 h). (D, right) Flow cytometry analysis of propidium iodide–stained cells. The data shown are from a single representative experiment out of three repeats. For the experiment shown, n = 4,000. All values represent mean + SEM (error bars).
HSF2 represses stress-inducible Hsp70 expression during mitosis
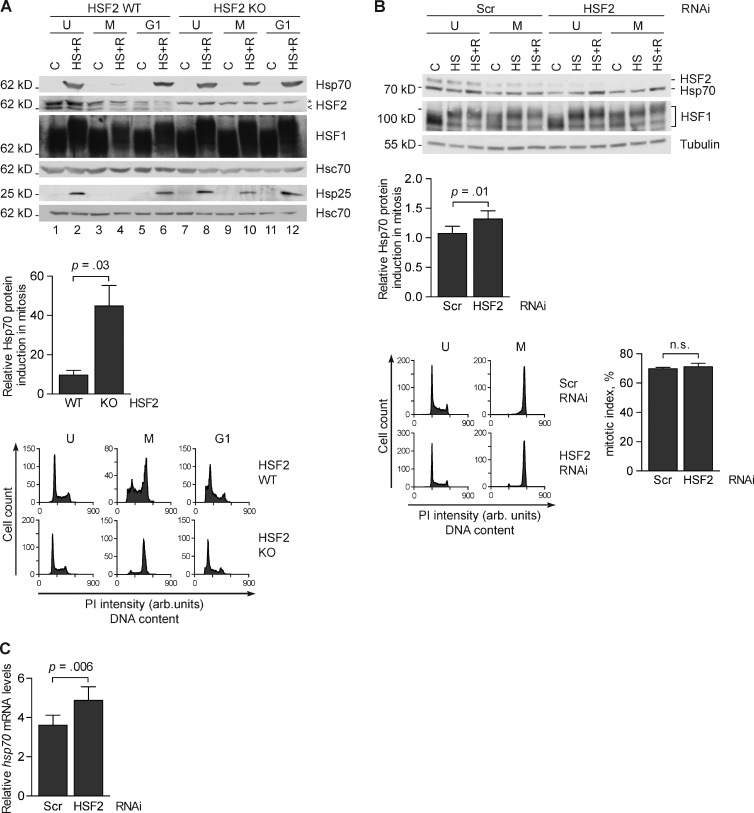
Because HSF2 has been shown to modulate the HSR in interphase cells (Östling et al., 2007; Shinkawa et al., 2011), we investigated the impact of HSF2 on the HSR in mitotic cells using hsf2+/+ (HSF2 wild type [WT]) and hsf2−/− (HSF2 knockout [KO]) immortalized mouse embryonic fibroblasts (MEFs). Hsp70 is one of the classical heat-induced chaperones and a shared target for HSF1 and HSF2 (Östling et al., 2007; Vihervaara et al., 2013), and was chosen as a marker for the HSR. Unsynchronized, mitotic, or G1 phase cells were left untreated or heat treated for 1 h at 43°C, followed by a 3-h recovery at 37°C. Cells were synchronized into mitosis by a thymidine-nocodazole block, and mitotic cells were harvested by mitotic shake-off. Unsynchronized MEFs responded to heat shock by elevated expression of Hsp70 (Fig. 2 A, lanes 2 and 8). In mitosis, the stress-inducible expression of Hsps was repressed in HSF2 WT MEFs (Fig. 2 A, lane 4), which is in accordance with the previously reported mitosis-specific inhibition of transcription (Martínez-Balbás et al., 1995). When cells proceeded to G1 phase, the stress-inducible Hsp expression was restored (Fig. 2 A, lane 6). Surprisingly, in heat-shocked HSF2 KO MEFs, Hsp70 was prominently increased during mitosis (Fig. 2 A, lane 10). Another stress-inducible Hsp, Hsp25, was similarly up-regulated by heat shock in the absence of HSF2 (Fig. 2 A). To determine whether the effect of HSF2 is conserved in human cells, HSF2 was transiently down-regulated with a specific shRNA (Fig. 2 B). We confirmed that down-regulation of HSF2 did not affect entry into mitosis by calculating the mitotic index of cells synchronized into mitosis, and found an equal amount of mitotic cells in both Scrambled and HSF2 shRNA-transfected K562 cells (Fig. 2 B, bottom right). In mitotic K562 cells, depletion of HSF2 led to increased hsp70 mRNA expression upon a 30-min heat shock at 42°C compared with Scrambled control shRNA-transfected cells (Fig. 2 C), and the Hsp70 protein levels increased correspondingly after a 1-h recovery at 37°C (Fig. 2 B). Because the cell population studied was not purely mitotic (Fig. 2 B, bottom right), our results could have been influenced by the residual nonmitotic cells. However, previous studies using unsynchronized populations, with >90% of cells in interphase, have demonstrated that HSF2 positively modulates Hsp70 and Hsp25 (Östling et al., 2007), which reinforces the finding that the negative effect of HSF2 on Hsp70 induction originates from the mitotic cell population. Together, these results suggest that HSF2 functions as a repressor of the HSR during mitosis in both murine and human cells.
Figure 2.
HSF2 represses the HSR in mitotic cells. (A, top) Representative immunoblot (n = 3) of Hsp70 and Hsp25 proteins upon heat shock in HSF2 WT and HSF2 KO MEFs at different phases of the cell cycle. Unsynchronized (U), mitotic (M), or G1-phase (G1) cells were left untreated (C) or heat-treated (HS+R, 1 h at 43°C, 3 h at 37°C). Cells were synchronized into mitosis by a thymidine-nocodazole block, and mitotic cells were harvested by mitotic shake-off. G1 phase cells were obtained by releasing mitotic cells into normal growth media for 3 h. Immunoblotting was performed with the indicated antibodies. <, HSF2; *, an unspecific band. Hsc70 serves as a loading control. (A, middle) Quantifications of Hsp70 protein induction in mitotic cells (n = 3). Hsp70 was normalized to Hsc70 and then related to the respective untreated mitotic samples, arbitrarily set to a value of 1. (A, bottom) Representative flow cytometry analyses of propidium iodide–stained cells. The data shown are from a single representative experiment out of three repeats. For the experiment shown, n = 10,000. (B, top) A representative immunoblot of K562 cells transfected with the indicated shRNAs and then either left unsynchronized (U) or synchronized into mitosis (M) by a thymidine-nocodazole block. Cells were treated with a 30-min heat shock at 42°C (HS) or a 30-min heat shock followed by a 1-h recovery at 37°C (HS+R). Samples were analyzed with the indicated antibodies and tubulin was used as a loading control. (B, middle) Quantifications of Hsp70 protein induction in mitotic cells (n = 5). Hsp70 was normalized to tubulin and analyzed as in A. (B, bottom left) Flow cytometry analyses of propidium iodide–stained cells. The data shown are from a single representative experiment out of five repeats. For the experiment shown, n = 10,000. (B, bottom right) The mitotic index was determined based on chromosome morphology of cells DNA stained with Hoechst. 80 cells per treatment from three independent experiments were counted. (C) qRT-PCR analysis of hsp70 mRNA induced by heat shock (30 min at 42°C) in mitotic K562 cells. Cells were transfected and synchronized as in B. Relative quantities of hsp70 mRNA were first normalized to gapdh and then calculated from the respective untreated samples, arbitrarily set to a value of 1. Values represent mean + SEM (error bars; n = 5). n.s., not specific.
Stress-inducible binding of HSF1 to chromatin is enhanced in mitotic cells depleted of HSF2
HSF1 is the major stress-responsive factor responsible for inducing the expression of Hsps. HSF2, however, is not capable of inducing Hsps alone, but only in conjunction with active HSF1 (Östling et al., 2007). Upon heat shock, HSF1 forms homotrimers, or heterotrimerizes with HSF2, and is posttranslationally modified by phosphorylation and sumoylation for maximal transactivation capacity (Sarge et al., 1991, 1993; Hietakangas et al., 2006; Sandqvist et al., 2009; Anckar and Sistonen, 2011). To elucidate whether the striking repressive effect of HSF2 on Hsp70 transcription in mitosis is caused by changes in HSF1, we analyzed activation of HSF1 in mitotic K562 cells transfected with either HSF2-specific shRNA or Scrambled control shRNA. As judged by the migration pattern of HSF1 on immunoblots (Sarge et al., 1993), HSF1 was phosphorylated upon heat shock in both unsynchronized and mitotic cells independently of HSF2 levels (Fig. 2 B), which suggests that HSF1 is activated upon stress in mitosis. To investigate whether the stress-induced interaction of HSF1 and HSF2 that was originally observed in unsynchronized cells (Alastalo et al., 2003) also occurred in mitosis, we performed coimmunoprecipitation. Despite the fact that the total amount of HSF2 was lower in mitotic cells, the HSF1–HSF2 interaction was similar to that in unsynchronized cells (Fig. 3 A).
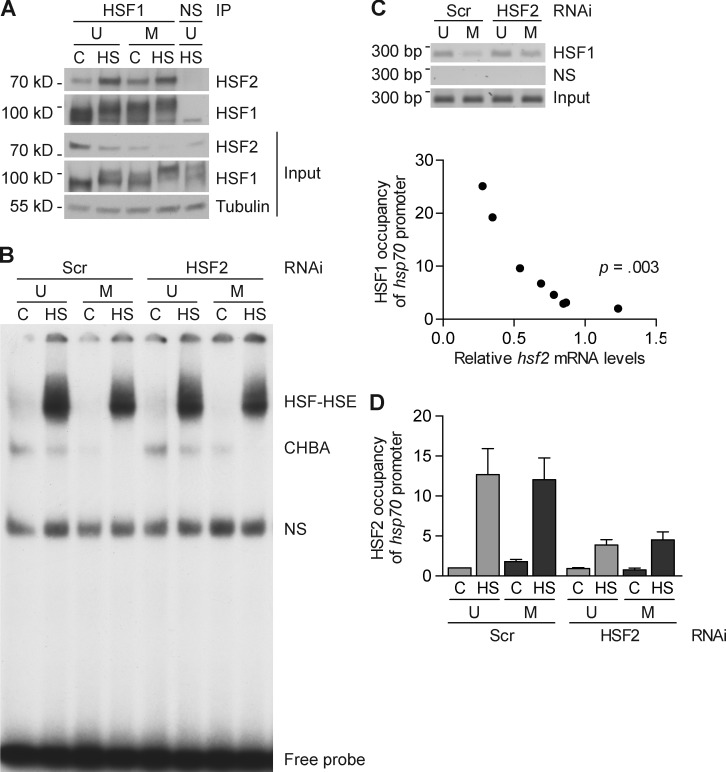
Figure 3.
HSF2 interferes with the stress-inducible binding of HSF1 to the hsp70 promoter in mitotic cells. (A) Representative immunoblot analysis of coprecipitated HSF1 and HSF2 (n = 3). Lysates from K562 cells, either unsynchronized (U) or synchronized into mitosis by a thymidine-nocodazole block (M), were left untreated (C) or heat shocked (HS; 30 min at 42°C), and immunoprecipitated with an anti-HSF1 antibody or normal rabbit IgG (NS). Input: whole-cell lysates were analyzed by immunoblotting with the indicated antibodies. (B) Representative EMSA analysis of HSF DNA-binding activity (n = 3). K562 cells were transfected with indicated shRNAs and either left unsynchronized (U) or synchronized into mitosis (M) as in A before they were left untreated or subjected to a 30-min heat shock at 42°C. Whole cell lysates were analyzed by EMSA, using a 32P-labeled probe corresponding to the proximal HSE from the human hsp70 promoter. HSF-HSE indicates the specific DNA-binding complex, CHBA indicates the constitutive HSE-binding activity, and NS indicates nonspecific DNA–protein complexes. (C, top) ChIP analysis of stress-induced (30 min at 42°C) recruitment of HSF1 to the hsp70 promoter in K562 cells transfected and synchronized as in B. NS indicates nonspecific DNA binding. Input represents 10% of the material used in the ChIP assay. (C, bottom) There is an inverse relationship between HSF1 binding and hsf2 mRNA levels. qPCR quantification of HSF1 occupancy of hsp70 promoter and hsf2 mRNA levels of individual samples from heat-treated mitotic samples, transfected with either Scrambled or HSF2-specific shRNA, was plotted and the correlation was calculated (n = 8). (D) qPCR quantification of HSF2 occupancy at the hsp70 promoter in K562 cells transfected, synchronized, and treated as in B (n = 3). Values represent mean + SEM (error bars).
Recently, we reported that both HSF1 and HSF2 access mitotic chromatin; however, HSF1 occupation of DNA is markedly reduced, whereas HSF2 binds a multitude of target loci (Vihervaara et al., 2013). We therefore examined whether the absence of HSF2 altered HSF1 DNA-binding capacity in mitosis, which could explain the more pronounced induction of Hsps (Fig. 2). Upon activation, trimeric HSFs bind target promoters containing heat shock elements (HSEs), and both HSF1 and HSF2 recognize the same sequence, i.e., inverted repeats of the pentameric sequence nGAAn, but with different affinity (Sakurai and Enoki, 2010; Vihervaara et al., 2013). To examine HSE-binding activity, we exposed unsynchronized and mitotic K562 cells to a 30-min heat shock at 42°C and performed a gel mobility shift assay (EMSA). In accordance with earlier studies showing that the in vitro DNA-binding activity of both HSF1 and HSF2 is intact in mitosis (Martínez-Balbás et al., 1995; Vihervaara et al., 2013), no major difference in heat-induced HSF-HSE formation was observed between unsynchronized and mitotic cells (Fig. 3 B). Furthermore, HSF2 depletion did not affect the HSF–HSE complex formation, which indicates that HSF2 does not alter the in vitro DNA-binding capacity of HSF1 (Fig. 3 B).
The mitotic chromatin is strongly condensed to allow for sister chromatid separation, resulting in DNA that is less accessible to transcription factors, including HSF1 (Martínez-Balbás et al., 1995; Vihervaara et al., 2013). Using chromatin immunoprecipitation (ChIP) on samples from heat-treated K562 cells transfected with either Scrambled control or HSF2-specific shRNA, we determined the DNA-binding properties of HSF1 in the silenced mitotic chromatin environment. Consistent with our previous study (Vihervaara et al., 2013), the heat-inducible binding of HSF1 to the hsp70 promoter was reduced in mitotic cells when compared with unsynchronized cells (Fig. 3 C, top). Intriguingly, in mitotic cells, HSF2 down-regulation resulted in a more pronounced HSF1 occupancy of the hsp70 promoter than in Scrambled control (Fig. 3 C, top), which suggested that HSF1 is allowed to access the mitotic chromatin in the absence of HSF2. Next, we performed quantitative HSF1 ChIP in heat-shocked mitotic cells and related HSF1 occupancy to hsf2 mRNA expression in the corresponding samples. HSF1 binding to the hsp70 promoter was inversely correlated with hsf2 mRNA levels (Fig. 3 C, bottom). ChIP analysis of HSF2 revealed that HSF2 occupancy of the hsp70 promoter slightly increased in mitosis under nonstressed conditions, whereas HSF2 binding was equally induced in unsynchronized and mitotic cells upon heat shock (30 min at 42°C; Fig. 3 D), which is in agreement with previous studies showing that HSF2 avidly interacts with the hsp70 promoter in mitosis (Xing et al., 2005; Vihervaara et al., 2013). Intriguingly, in cells where HSF2 was down-regulated, the residual HSF2 bound to the hsp70 promoter in a stress-inducible fashion. We conclude that HSF2 interferes with the accessibility of HSF1 to mitotic chromatin, possibly by competing with HSF1 for binding to the hsp70 promoter.
HSF2 affects the chromatin environment of the hsp70 promoter in mitotic cells
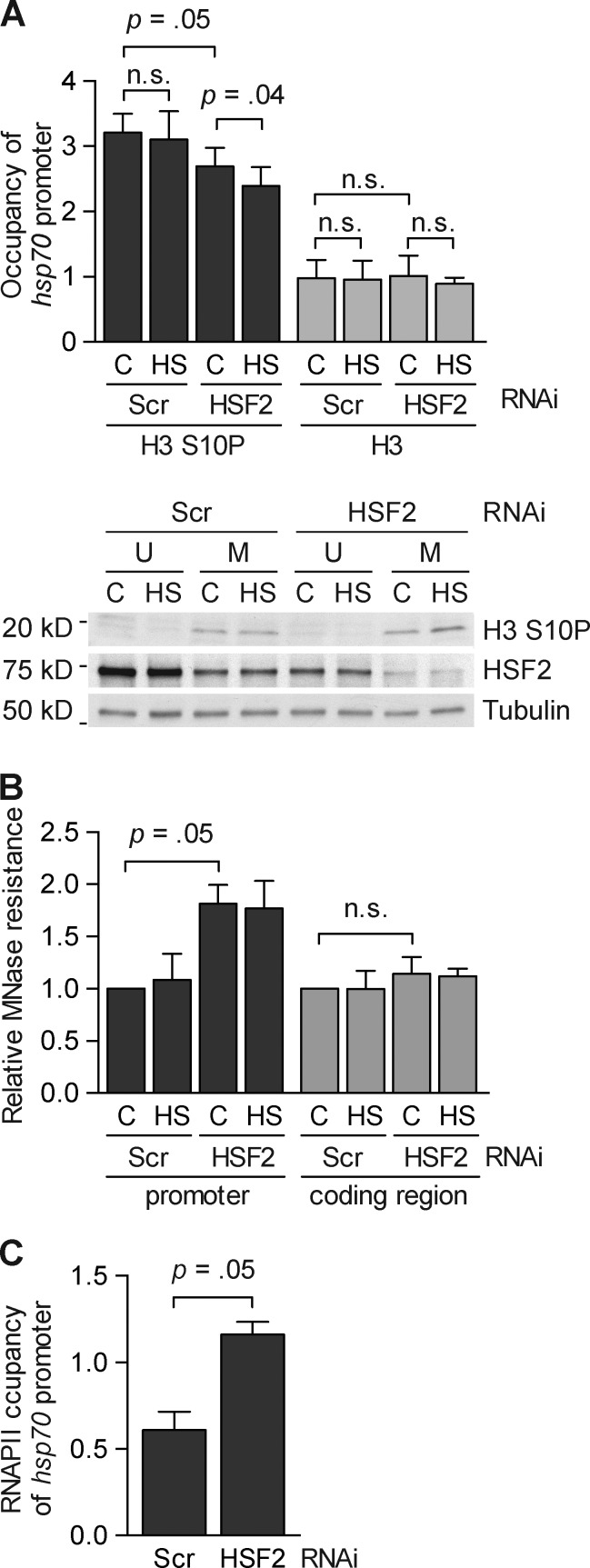
How does HSF2 exert its inhibitory effect on the accessibility of HSF1 to the hsp70 promoter? HSF2 has previously been shown to be required for correct chromatin compaction during spermatogenesis (Åkerfelt et al., 2008). These earlier results, combined with the observation that the DNA-binding capacity of HSF1 in vitro, but not in vivo, was preserved in mitosis (Fig. 3), led us to reason that HSF2 could affect the chromatin environment in mitotic cells. At the onset of mitosis, the chromatin environment is dramatically changed: RNA polymerases and transcription factors are displaced from chromatin, posttranslational modifications of the histone tails are altered, and the chromatin is condensed (Martínez-Balbás et al., 1995; Sif et al., 1998; Christova and Oelgeschläger, 2002; Chen et al., 2005). Phosphorylation of serine 10 on histone H3 (H3 S10P) is associated with a repressive chromatin environment (Sawicka and Seiser, 2012). Mitotic cells display elevated H3 S10P levels on the hsp70 promoter (Valls et al., 2005). Using ChIP with an H3 S10P antibody, we examined the impact of HSF2 on the levels of H3 S10P at the hsp70 promoter. In mitotic control cells, the occupancy of H3 S10P was 3.2-fold higher than in unsynchronized cells, whereas less H3 S10P was bound (2.7-fold) in HSF2-depleted cells (Fig. 4 A, top). Upon heat shock (30 min at 42°C), H3 S10P further decreased at the hsp70 promoter in cells where HSF2 was down-regulated (Fig. 4 A, top). Because no differences in the total histone H3 levels were detected (Fig. 4 A, top), the decline in phosphorylation of histone H3 upon HSF2 down-regulation was not caused by a decreased histone H3 occupancy on the hsp70 promoter. To investigate whether the HSF2 levels had an impact on the global amount of H3 S10P in mitosis, we analyzed H3 S10P levels in HSF2-depleted cells by immunoblotting, and found no significant differences compared with the Scrambled control cells (Fig. 4 A, bottom).
Figure 4.
HSF2 affects chromatin organization at the hsp70 promoter during mitosis. (A, top) Occupation of phosphorylated histone H3 (H3 S10P) and total histone H3 (H3) at the hsp70 promoter in nonstressed (C) and heat-shocked (HS; 30 min at 42°C) mitotic K562 cells was analyzed by ChIP and quantified with qPCR (n = 4). Cells were transfected with the indicated shRNA constructs and synchronized into mitosis by a thymidine-nocodazole block. The qPCR values of the IP were normalized to the input values and related to the Scrambled unsynchronized control that was arbitrarily set to a value of 1. (A, bottom) Representative immunoblot (n = 4) of H3 S10P levels in K562 cells. Immunoblotting was performed with the indicated antibodies. (B) MNase sensitivity analysis of the hsp70 promoter (−377 to −229 bp) and coding region (+1,286 to +1,397 bp) in mitotic K562 cells as measured by qPCR (n = 4). Cells were transfected, synchronized, and treated as in A. The qPCR values of the MNase-treated samples were related to input and to the Scrambled control sample that was arbitrarily set to a value of 1. (C) RNAPII occupation of the hsp70 promoter in nonstressed mitotic cells. K562 cells were transfected, synchronized and analyzed as in A, analyzed by ChIP, and quantified with qPCR (n = 4). All values represent mean + SEM (error bars). n.s., not specific.
We further investigated the chromatin structure using a micrococcal nuclease (MNase) DNA accessibility assay in mitotic K562 cells transfected with either Scrambled control or HSF2 shRNA. In mitotic HSF2-depleted K562 cells, we found an enhanced MNase resistance of the hsp70 promoter, but not the coding region (Fig. 4 B), which indicates proteins binding to the hsp70 promoter in the absence of HSF2. In interphase cells, RNAPII is associated with the hsp70 promoter as a paused polymerase (Brown et al., 1996; Adelman and Lis, 2012), and its recruitment to the chromatin increases MNase resistance (Weber et al., 2010). However, during mitosis RNAPII is displaced from the chromatin, including the hsp70 promoter (Parsons and Spencer, 1997; Christova and Oelgeschläger, 2002). Accordingly, a ChIP analysis of K562 cells revealed that RNAPII at the hsp70 promoter decreased in mitotic Scrambled control cells compared with unsynchronized cells (Fig. 4 C). We reasoned that in the presence of HSF2, RNAPII could be prevented from accessing the promoter in mitotic cells, and that in the absence of HSF2, RNAPII binding would be facilitated. Indeed, down-regulation of HSF2 led to increased RNAPII occupancy of the hsp70 promoter in nonstressed mitotic cells to a level similar to unsynchronized cells (Fig. 4 C). Collectively, our results indicate that HSF2 modulates the chromatin environment and prevents HSF1 and RNAPII from binding to and transactivating the hsp70 promoter during mitosis.
A decrease in HSF2 levels protects mitotic cells against acute heat stress
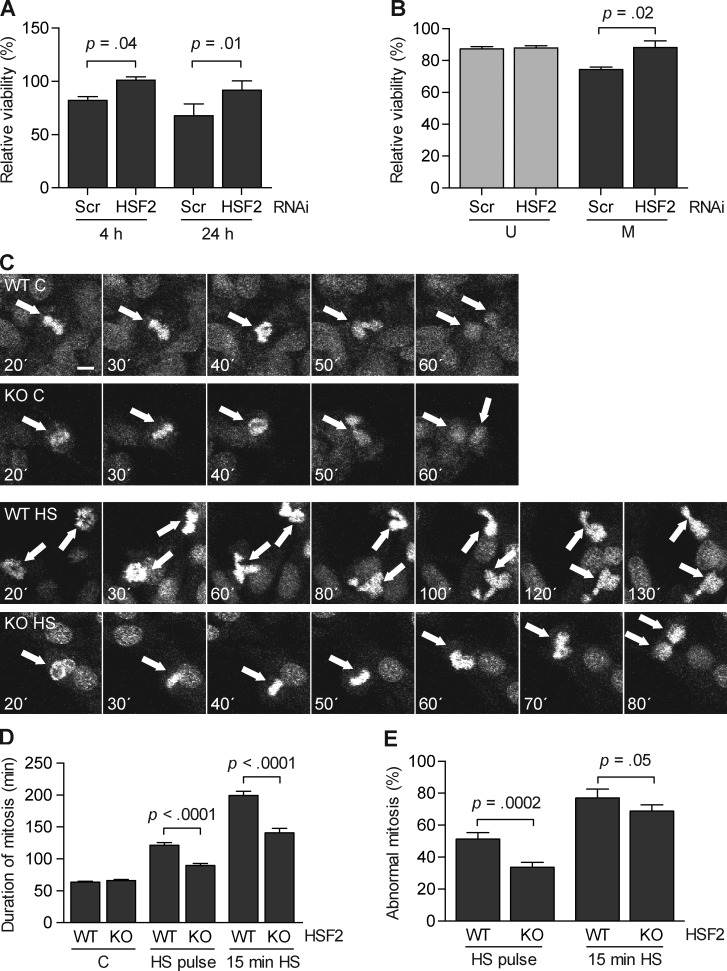
Heat stress impairs the function of the microtubule-organizing centers, centrosomes, which are essential in ensuring the fidelity of chromosome segregation (Debec and Marcaillou, 1997; Nakahata et al., 2002; Hut et al., 2005). Interestingly, overexpression of Hsp70 before mitosis protects cells from heat-induced mitotic abnormalities by chaperoning the centrosomes (Hut et al., 2005). HSF2-deficient cells were capable of inducing Hsp70 during mitosis (Fig. 2), which could pose a survival advantage upon heat stress. To test this hypothesis, we synchronized K562 cells with down-regulated HSF2, subjected the cells to a 30-min heat shock at 42°C, and counted them after a recovery for 4 or 24 h. The results showed a mean 20% increase in viability in HSF2-depleted mitotic cells compared with the corresponding Scrambled control cells (Fig. 5 A). To confirm these results, the impact of HSF2 was assessed using the MTS survival assay, which measures cell metabolism. In unsynchronized HeLa cells, HSF2 down-regulation had no effect on survival upon a 30-min heat shock at 43°C, whereas HSF2-depleted mitotic HeLa cells survived heat shock better than their Scrambled shRNA-treated counterparts (Fig. 5 B), thereby supporting our hypothesis.
Figure 5.
Diminished HSF2 levels promote cell survival after heat shock in mitosis. (A) K562 cells transfected with the indicated shRNA constructs were synchronized into mitosis by a thymidine-nocodazole block and exposed to heat shock (30 min at 42°C). Cells were counted in triplicate after 4 h and 24 h of recovery at 37°C. The number of cells was related to their untreated counterparts (n = 4). (B) HeLa cells transfected as in A were left unsynchronized (U) or synchronized into mitosis and collected by mitotic shake-off (M) before being heat-treated (30 min at 43°C) or left untreated. Survival was measured by an MTS assay after a 2-h recovery at 37°C. Values from the heat-treated samples were related to their respective control (n = 4). (C) Selected still frames from time-lapse microscopy of DRAQ5-stained HSF2 WT and HSF2 KO MEFs that were either left untreated (C) or subjected to a pulse of heat shock at 43°C (HS; for details see Materials and methods). Time is given as minutes after heat shock. Arrows denote mitotic cells. Bar, 10 µm. See Videos 1–4. (D) Duration of mitosis was measured from nuclear envelope breakdown to cytokinesis in untreated (C) or heat-treated (pulse at 43°C or 15 min at 43°C) HSF2 WT and HSF2 KO MEFs. At least 20 cells per treatment from four individual experiments were monitored. (E) Ratio of abnormal mitosis in cells from D. Abnormal mitosis was measured as the percentage of cells with defects in chromosome segregation, such as daughter cells forming micronuclei, chromatin decondensing without division, formation of more than two daughter cells, or apoptosis. All values represent mean + SEM (error bars).
To understand the mechanism underlying increased survival in the absence of HSF2, we used time-lapse microscopy to monitor the progression of unsynchronized cells through mitosis in HSF2 WT and KO MEFs that were either untreated or exposed to heat shock. Time-lapse microscopy revealed that heat-treated HSF2-deficient MEFs progressed through mitosis faster than HSF2 WT MEFs. In HSF2 WT MEFs, the mean duration of mitosis was 64 min in normal conditions, and the duration of mitosis was prominently prolonged to 122 min after a pulse of heat shock, and to 199 min after a 15-min heat shock (Fig. 5, C and D; and Videos 1–4). The corresponding results in HSF2 KO MEFs were 66 min under normal conditions, 91 min after a heat shock pulse, and 140 min after a 15-min heat shock (Fig. 5, C and D). The mean number of HSF2 WT ( = 24 cells) and KO ( = 20 cells) MEFs entering mitosis within 3 h of heat shock did not significantly differ, which indicates that the absence of HSF2 did not affect the G2/M checkpoint (data are from three experiments).
Because a delayed exit from mitosis can cause CIN and apoptosis (Sotillo et al., 2007; Orth et al., 2012), we investigated whether the prolonged mitosis, as observed upon heat shock, was accompanied by an HSF2-dependent increase in mitotic errors. Cells with defects in chromosome segregation, such as daughter cells forming micronuclei, chromatin decondensing without division, formation of more than two daughter cells, or apoptosis, were scored as mitotic errors. A pulse of heat shock caused mitotic errors in 52% of the HSF2 WT MEFs, in contrast to only 34% in HSF2 KO MEFs (Fig. 5 E). Similarly, after a 15-min heat shock, HSF2 KO MEFs displayed fewer abnormalities (69%) during mitosis than their WT counterparts (77%, Fig. 5 E). These results suggest that decreased levels of HSF2 provide protection against prolonged mitosis and mitotic errors during acute stress.
HSF2 expression declines during mitosis in several but not all human cell lines
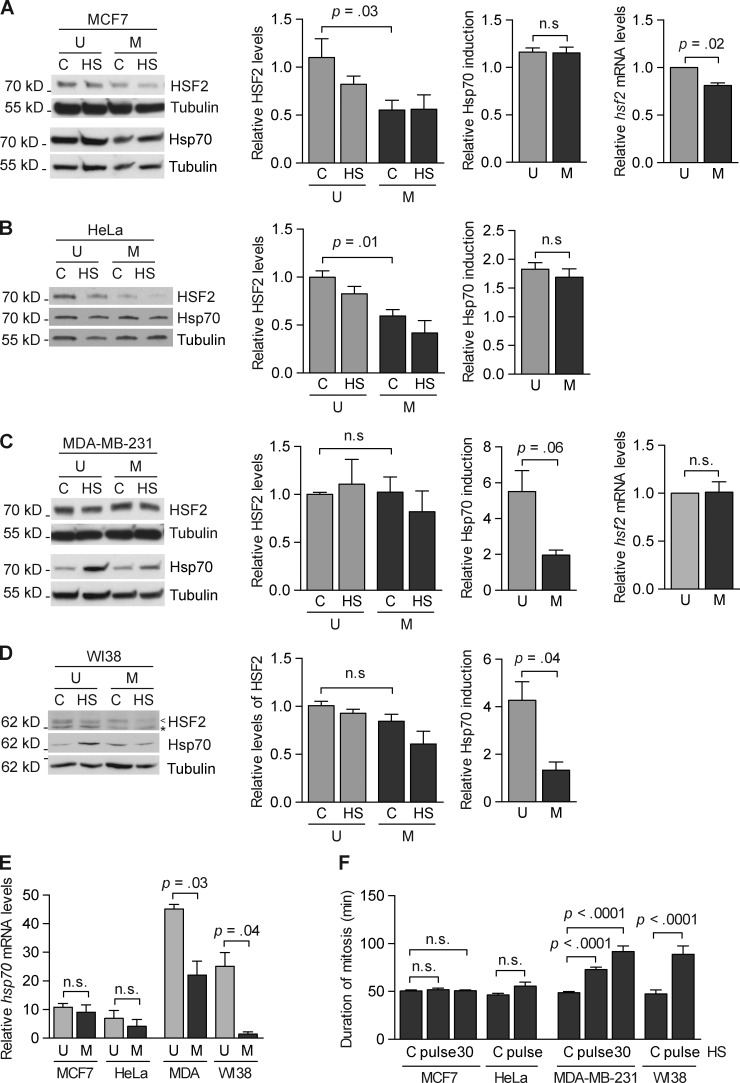
Decreased levels of HSF2 increase cell survival and protect mitotic cells against prolonged mitosis and mitotic errors upon proteotoxic stress (Fig. 5). To determine the significance of this finding, we investigated whether the mitosis-dependent decrease in HSF2 levels, as observed in K562 cells (Fig. 1, A–C), also occurred in other human cells. For this purpose, we synchronized MCF7, HeLa, MDA-MB-231, and WI38 cells into mitosis with a thymidine-nocodazole block and collected mitotic cells by mitotic shake-off. HSF2 protein levels were analyzed by immunoblotting, and a 40–45% decrease in HSF2 protein levels was observed in untreated mitotic MCF7 and HeLa cells compared with unsynchronized cells (Fig. 6, A and B). In contrast, mitotic MDA-MB-231 and WI38 cells did not display a significant change in their HSF2 protein levels (Fig. 6, C and D). Next, we examined the hsf2 mRNA levels in MCF7 and MDA-MB-231 cells and found a consistent, albeit mild (19%) decrease in hsf2 mRNA levels of mitotic MCF7 cells (Fig. 6 A). In accordance with the constant HSF2 protein levels, hsf2 mRNA expression remained unchanged in mitotic MDA-MB-231 cells (Fig. 6 C), which suggests that HSF2 expression is regulated on both protein and mRNA levels during mitosis in a cell type–specific manner. This observation is in line with previous genome-wide studies showing that hsf2 mRNA levels decrease during mitosis in certain cell lines, i.e., HeLa and U2OS, but not in HaCaT immortalized keratinocytes or primary foreskin fibroblasts (Whitfield et al., 2002; Bar-Joseph et al., 2008; Peña-Diaz et al., 2013; Grant et al., 2013).
Figure 6.
HSF2 expression declines during mitosis in a cell type–specific manner, thus protecting cells against prolonged mitosis. (A–D) Representative immunoblots of HSF2 protein in unsynchronized (U) and mitotic (M) MCF7, HeLa, MDA-MB-231, and WI38 cells. Cells were synchronized into mitosis by a thymidine-nocodazole block and collected by mitotic shake-off. Cells were left untreated (C) or subjected to heat shock (HS; 30 min at 42°C, 1 h at 37°C). Immunoblotting was performed with the indicated antibodies. Tubulin serves as a loading control. <, HSF2; *, unspecific band. (left middle panels) Quantification of immunoblots (n = 3) showing HSF2 protein related to tubulin. (right middle panels) Quantification of immunoblots (n = 3) showing Hsp70 induction in unsynchronized and mitotic cells. Samples were analyzed as in Fig. 2 B. (A and C, right) qRT-PCR analysis of hsf2 mRNA in unsynchronized (U) and mitotic (M) cells (n = 3). Relative quantities of hsf2 mRNA were analyzed as in Fig. 1 B. (E) qRT-PCR analysis of hsp70 mRNA induction upon a 30-min heat shock at 42°C in unsynchronized (U) and mitotic (M) MCF7, HeLa, MDA-MB-231 (MDA), and WI38 cells. Relative quantities of hsp70 mRNA were analyzed as in Fig. 2 C. (n = 3). (F) Duration of mitosis was measured from rounding up of cells until the formation of two daughter cells in untreated (C) or heat-treated (pulse at 42°C or 30 min at 42°C) cells. At least 20 cells per treatment from five (MCF7 and MDA-MB-231) or two (HeLa and WI38) individual experiments were monitored. All values represent mean + SEM (error bars). n.s., not specific.
To examine if the decrease in HSF2 levels altered the stress-induced Hsp70 expression, we subjected the unsynchronized and mitotic cells to heat shock (30 min at 42°C, 1 h at 37°C). In accordance with our hypothesis, the hsp70 mRNA and protein induction in mitotic MCF7 and HeLa cells with decreased HSF2 expression was similar in both unsynchronized and mitotic cells (Fig. 6, A, B, and E). In mitotic MDA-MB-231 and WI38 cells with unaltered HSF2 levels, the heat-induced hsp70 mRNA and protein expression was significantly reduced compared with that in unsynchronized cells (Fig. 6, C–E).
By using IncuCyte live cell imaging technology, we investigated whether the mitosis-specific regulation of HSF2 influenced mitosis in these different cell lines. The length of mitosis was calculated from the rounding up of the cells until two daughter cells were formed in unsynchronized cells. In untreated cells, the length of mitosis varied between a mean of 46 min in HeLa cells and 50 min in MCF7 cells (Fig. 6 F). In MCF7 and HeLa cells, neither a 42°C heat shock pulse nor a 30-min heat shock affected the length of mitosis. In MDA-MB-231 cells, a 42°C heat shock prominently prolonged mitosis from 49 min in control cells to 73 min in heat pulse–treated cells, and it was nearly doubled (92 min) upon a 30-min heat shock. In WI38 cells, a pulse of heat shock caused a robust 1.9-fold increase in the mean length of mitosis (89 min) compared with the untreated cells (47 min, Fig. 6 F). Collectively, these results demonstrate that cells where HSF2 levels decrease during mitosis are better protected against heat-induced prolonged mitosis compared with cells whose HSF2 levels are constant throughout the cell cycle. Because the length of mitosis correlates with the amount of mitotic errors (Fig. 5, D and E), these results suggest that cells with decreased mitotic HSF2 levels are subjected to fewer mitotic errors in response to acute heat stress.
Discussion
Although the HSR, and especially the fundamental functions of HSF1 therein, have been thoroughly studied in unsynchronized cells, little is known about HSR regulation in mitotic cells. Mitosis aims at faithful chromosome segregation and maintenance of genome stability. Any errors can influence cell viability and/or the risk of triggering uncontrolled proliferation. For the chromosomes to be separated properly, the chromatin is condensed, gene expression is repressed, and nuclear structures are dissolved (Delcuve et al., 2008). Considering these major changes, it is evident why proteotoxic stress in combination with a repressed HSR during these important events can be detrimental to the individual cell and the whole organism. In the present study, we uncover an inhibitory effect for HSF2 on stress-inducible transcription in mitotic cells and reveal a mechanism underlying this HSF2-dependent regulation. Surprisingly, HSF2, which has been shown to contribute to the HSR in interphase cells by positively modulating Hsp expression (Östling et al., 2007), functions as a repressor of transcription in mitosis. Even more intriguing is our finding that a decline in HSF2 levels enables inducible gene expression in the otherwise transcriptionally silenced mitotic cells.
Although it is considered a dogma in cell biology, it is not mechanistically understood how transcription is repressed during mitosis. It could be due to the condensed chromatin environment or through inhibitory posttranslational modifications, e.g., phosphorylation of DNA-bound proteins such as histones, chromatin remodeling complexes, and other regulatory proteins that decrease accessibility or prevent the DNA-binding activity of transcription factors and RNA polymerases (Gottesfeld and Forbes, 1997; Chen et al., 2005). It has previously been shown that the mitotic hsp70 promoter remains sensitive to DNase I treatment, which indicates that it remains open and not affected by chromatin remodeling complexes (Martínez-Balbás et al., 1995; Michelotti et al., 1997). It has been suggested that this “bookmarking” of the promoter is executed by HSF2, which remains bound to the hsp70 promoter throughout mitosis (Fig. 3 D; Xing et al., 2005; Vihervaara et al., 2013). Xing et al. (2005) have suggested that HSF2 down-regulation leads to increased chromatin compaction of the hsp70 promoter in mitotic cells. The results obtained in our study did not point to differences in the nucleosome composition of the hsp70 promoter in the absence of HSF2. The total histone H3 levels at the hsp70 promoter are similar in both unsynchronized and mitotic cells, independently of HSF2 levels (Fig. 4 A). Moreover, we propose that the increased MNase resistance, which was observed upon HSF2 down-regulation, could be caused by enhanced accessibility of RNAPII and other DNA-binding proteins to the hsp70 promoter (Fig. 4 C). Despite the open chromatin structure, both RNAPII and HSF1 are excluded from the hsp70 promoter during mitosis (Martínez-Balbás et al., 1995). In the absence of HSF2, the hsp70 promoter is also accessible to both HSF1 and RNAPII during mitosis (Figs. 3 C and 4 C), enabling Hsp70 expression, and thereby protecting cells against heat-induced mitotic errors. We show a reduced occupancy of the repressive histone marker, H3 S10P, at the hsp70 promoter in HSF2-depleted mitotic cells (Fig. 4 C). This reduction is small but consistent and significant, and it is worth noticing that a more robust change in H3 S10P could have adverse effects on cells, given earlier reports showing that a considerable reduction in the chromatin histone H3 S10P levels impairs chromosome segregation (Wei et al., 1999; Sawicka and Seiser, 2012). It has previously been shown that the phosphorylation of an adjacent threonine residue (H3 T3) functions as a switch, which inhibits the DNA binding of TFIID in mitotic cells, thereby inhibiting transcription (Varier et al., 2010). It is plausible that the small change in phosphorylation status observed in our experiments (Fig. 4 A) could similarly function as a switch that increases the accessibility of the hsp70 promoter to HSF1 and RNAPII (Figs. 3 C and 4 C). Alternatively, the change in the ratio between HSF1 and HSF2, due to HSF2 decline, might affect HSF1 DNA-binding activity, e.g., via HSF2 acting as a competitive inhibitor, or by affecting trimer composition.
For protecting themselves against the proteotoxic stress to which all cells, particularly cancer cells, are continuously subjected (Solimini et al., 2007; Luo et al., 2009; Richter et al., 2010), it would be beneficial for cells to also induce Hsp70 in mitosis. Here we describe a novel mechanism by which cells with reduced levels of HSF2 are able to express Hsp70 in mitosis. We show that certain cells, such as K562, MCF7, and HeLa cells, decrease their HSF2 levels during mitosis, whereas MDA-MB-231 and WI38 cells do not (Fig. 6). Accordingly, only the mitotic cells that contain diminished HSF2 levels display stress-inducible Hsp70 expression during mitosis, almost to the same extent as in unsynchronized cells (Fig. 6 E). It is possible that the constitutively high Hsp70 levels observed in K562, MCF7, and HeLa cells might promote Hsp70 induction in mitosis (Figs. 2 B and 6, A and B). Intriguingly, when exposed to acute heat stress, cells with decreased amounts of HSF2 are capable of completing mitosis faster than cells with constant HSF2 levels (Fig. 6 F). Delicate regulation of HSF2 expression is crucial for cells, and as previously shown, HSF2 is a short-lived protein that can be regulated both posttranscriptionally by miR-18 and posttranslationally through ubiquitylation by APC/C (Ahlskog et al., 2010; Björk et al., 2010). APC/CCdc20 is active from prometaphase to anaphase when it mediates the proteasomal degradation of several proteins, e.g., cyclin A and cyclin B (Pines, 2011). Because the mitotically active APC/CCdc20 is able to ubiquitylate HSF2 in vitro (Ahlskog et al., 2010), APC/C could contribute to the declined HSF2 protein levels. The decrease in hsf2 mRNA levels observed in mitosis facilitates reduced HSF2 protein levels. This decrease is only observed in some, but not all, cell lines. Intriguingly, several of the cell lines with stable mitotic HSF2 are nontumorigenic, including WI38, HaCaT, and primary foreskin fibroblasts (Fig. 6; Whitfield et al., 2002; Bar-Joseph et al., 2008; Grant et al., 2013; Peña-Diaz et al., 2013). The upstream signaling that mediates mitosis-specific HSF2 decrease in certain cells needs to be explored in future studies.
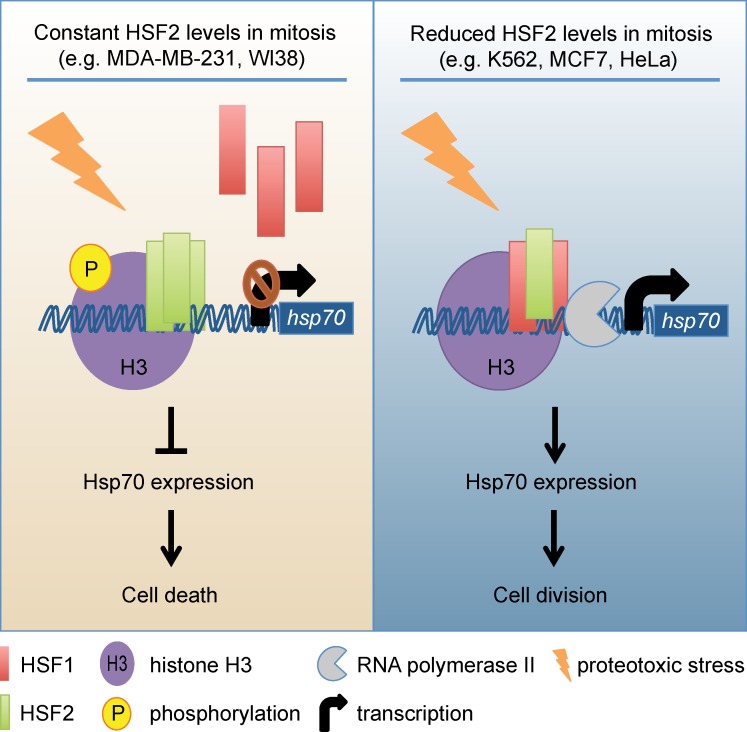
In the light of our results, we propose a model (Fig. 7) where mammalian cells with decreased HSF2 levels (e.g., MCF7, K562, and HeLa) are better protected against proteotoxic insults than cells where HSF2 does not decline during mitosis (e.g., MDA-MB-231 and WI38). Repression of HSF2 expression leads to changes in the chromatin environment, as demonstrated here with the hsp70 promoter, thereby making the otherwise silenced target promoter accessible to RNAPII and HSF1. The HSF1-mediated increase in Hsp70 expression prevents mitotic defects under stress conditions, thereby providing a survival advantage upon exposure to detrimental stimuli that would jeopardize proteostasis during this sensitive phase of the cell cycle. There is rapidly accumulating evidence for HSF1 playing a key regulatory role in many cancer types (Dai et al., 2007; Santagata et al., 2011; Mendillo et al., 2012; Ciocca et al., 2013; Santagata et al., 2013). Our results showing that HSF2 represses HSF1-inducible Hsp70 expression in mitosis, accompanied by an increase in mitotic defects, might give new insights into the function of HSFs in malignant transformation. Mitotic errors cause CIN, which is a hallmark of cancer as low levels of CIN promote oncogenesis, whereas high levels of CIN are associated with tumor suppressor functions (Weaver et al., 2007; Pfau and Amon, 2012). The decreased viability of cells is due to apoptosis, or G1 arrest, caused by severe mitotic errors or prolonged mitosis (Uetake and Sluder, 2010; Orth et al., 2012). Cells with only modest mitotic problems survive and propagate CIN (Weaver et al., 2007; Schvartzman et al., 2010). According to the working model (Fig. 7), we hypothesize that loss of HSF2 in mitosis might protect cells against excessive mitotic problems that would result in cell death, but instead leads to accumulation of minor mitotic errors. Thus, the mitosis-associated decrease in HSF2 levels, as reported here, could contribute to malignant transformation of cancer cells. In the forthcoming studies, it will be important to establish the mechanisms by which HSF2 expression is regulated in mitosis of healthy and cancer cells.
Figure 7.
A model for transcriptional regulation in mitosis. (left) In mitotic cells, e.g., MDA-MB-231 and WI38 cells, where HSF2 is present, HSF1 and RNAPII are displaced from the hsp70 promoter, which prevents stress-induced expression of Hsp70 and ultimately leads to mitotic problems and cell death. (right) In cells, e.g., K562, MCF7, and HeLa cells, where HSF2 levels are dramatically reduced during mitosis, the hsp70 promoter is accessible to RNAPII and HSF1, thus enabling Hsp70 expression and cell survival upon proteotoxic stress.
Materials and methods
Cell culture and heat treatments
Human K562 erythroleukemia cells and MDA-MB-231 breast adenocarcinoma cells were cultured in RPMI medium (Sigma-Aldrich) supplemented with 10% FCS, 2 mM l-glutamine, streptomycin, and penicillin. Human HeLa cells, MCF7 breast adenocarcinoma cells, and WI38 lung fibroblasts were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FCS, 2 mM l-glutamine, streptomycin, and penicillin. MEFs from HSF2-deficient and wild-type mice were derived from HSF2 KO and WT mouse embryos. The HSF2 KO mice strains were generated using gene targeting in mouse embryonic stem cells and harboring an hsf2 gene where exon 5 has been replaced by the β-geo gene (Kallio et al., 2002). MEFs were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FCS, 2 mM l-glutamine, 10 mM nonessential amino acids, 1.2 mM sodium pyruvate, streptomycin, and penicillin. All cells were maintained at 5% CO2 in a humidified 37°C incubator. Heat shock treatments were performed in a 42°C (K562, MDA-MB-231, HeLa, MCF7, and WI38) or 43°C (MEFs) water bath for indicated times.
Synchronization of cells
Synchronization of K562, HeLa, MDA-MB-231, MCF7, and WI38 cells was modified from a previously described protocol (Whitfield et al., 2000). Cells were treated for 20 h with 2 mM thymidine (Sigma-Aldrich), washed twice with PBS (Sigma-Aldrich), and released into complete media. After 6 h, nocodazole (Fluka) was added (100 ng/ml) for 12 h. Mitotic HeLa, MDA-MB-231, MCF7, and WI38 cells were collected by mitotic shake-off, and all K562 cells were collected. K562 cells were synchronized into G1 phase by releasing mitotic cells for 5 h, into S phase by a double thymidine block (after incubation with 2 mM thymidine for 16 h, cells were washed and released into complete media for 8 h, followed by another 16-h of incubation with 2 mM thymidine), and into G2 by releasing S phase cells for 5 h. MEFs were synchronized into mitosis by adding 2 mM thymidine for 16 h before cells were released into complete media for 2 h before incubation with nocodazole (300 ng/ml) for 3 h. Mitotic cells were collected by mitotic shake-off. MEF cells were synchronized into G1 phase by releasing mitotic cells into complete media for 3 h. Inhibitors were removed 10 min before heat shock treatment by washing twice in complete media. The mitotic index was monitored by flow cytometry by FACSCalibur (BD). Cells were fixed in 70% ethanol before staining with propidium iodide (40 µg/ml; Sigma-Aldrich). The flow cytometry profiles were analyzed using FlowJo X10 software. Live cells were gated and the histogram function was plotted in the GraphPad Prism software.
Depletion of HSF2 by RNAi using shRNA
HSF2 was down-regulated using pSUPER vector-encoded sequences as described previously (Östling et al., 2007). In brief, the sequence specific for HSF2 (5′-CAGGCGAGTACAACAGCAT-3′), or a scrambled sequence (5′-GCGCGCTTTGTAGGATTCG-3′) was used. HeLa or K562 cells were transfected with the shRNA vectors by electroporation (975 µF, 230 mV), and cells were incubated 72 h before harvesting. Synchronization was initiated 24 h after transfection.
Mitotic index
Cells synchronized into mitosis by a thymidine-nocodazole block were allowed to attach to poly-l-lysine–coated cover glasses for 30 min, after which they were DNA stained with Hoechst. The cover glasses were mounted and images were acquired using a microscope (LSM 780; Carl Zeiss). The mitotic index was determined based on the morphology and intensity of the stained nuclei.
Immunoprecipitation (IP) and Western blot analysis
The co-IP was performed as described previously (Ahlskog et al., 2010). In brief, samples were lysed in lysis buffer (25 mM Hepes, pH 8.0, 100 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 20 mM β-glycerophosphate, 20 mM para-nitrophenylphosphate, 100 µM sodium orthovanadate, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 1× Complete mini-protease inhibitor cocktail [Roche Diagnostics]). Samples (500 µg) were precleared with a 50% protein G–Sepharose slurry before incubation with the HSF1 antibody (SPA-901; Enzo Life Sciences) for 30 min at 4°C. Samples were then incubated with a 50% slurry of Protein G–Sepharose for 2 h at 4°C and washed with TEG buffer (20 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 10% glycerol) containing 150 mM NaCl and 0.1% Triton X-100. Western blotting was performed by running samples on a Mini-protean TGX gel (4–15%; Bio-Rad Laboratories). Proteins were transferred to nitrocellulose membrane (Protran nitrocellulose), and upon immunoblotting, visualized by enhanced chemiluminescence. Immunoblotting was performed with antibodies against HSF1 (rabbit, SPA-901, 1:5,000; Enzo Life Sciences), HSF2 (rat, 3E2, 1:500; EMD Millipore), HSF2 (rabbit, 1:5,000; Östling et al., 2007), Hsp70 (mouse, SPA-810, 1:5,000; Enzo Life Sciences), Hsp27 (mouse, SPA-803, 1:5,000; Enzo Life Sciences), Hsp25 (rabbit, SPA-801, 1:5,000; Enzo Life Sciences), Cdc27 (mouse, 610454, 1,5000; BD), and Histone H3 phospho-S10 (mouse, ab14955, 1:1,000; Abcam). Antibodies against Hsc70 (rat, SPA-815, 1:5,000; Enzo Life Sciences) and β-tubulin (mouse, AA2, 1:1,000; Sigma-Aldrich) were used as loading controls. The rabbit polyclonal anti-HSF2 antibody was generated by immunizing rabbits with recombinant full-length mouse HSF2 containing a His tag (Östling et al., 2007). Quantifications were done using ImageJ (National Institutes of Health) and the densitometry values of HSF2 were normalized to the values of Hsc70 or β-tubulin. A mean of all the unsynchronized control samples was calculated, and all values were compared with that mean. Statistics were calculated using repeated measures analysis of variance (ANOVA), and unsynchronized and mitotic control samples were compared with a post hoc test.
Quantitative RT-PCR (qRT-PCR)
RNA was isolated using an RNeasy kit (QIAGEN). Of each sample, 1 µg of RNA was treated with RQ1 DNaseI (Promega) and reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories). Kapa probe fast ABI prism qPCR kit (Kapa Biosystems) was used for the qPCR reaction. The following probes and primers were used: hsp70.1 probe, 5′-FAMTTACACACCTGCTCCAGCTCCTTCCTCTTTAMRA-3′, 5′-GCCGAGAAGGACGAGTTTGA-3′, and 5′-CCTGGTACAGTCCGCTGATGA-3′; hsf2 probe #36 (Universal Probe Library, Roche), 5′-GGAGGAAACCCACACTAACG-3′ and 5′-ATCGTTGCTCATCCAAGACC-3′; gapdh probe, 5′-FAMACCAGGCGCCCAATACGACCAATAMRA-3′, 5′-GTTCGACAGTCAGCCGCATC-3′, and 5′-GGAATTTGCCATGGGTGGA-3′. Relative quantities of hsf2 and hsp70 were normalized against their respective gapdh, and fold inductions were determined after the control was arbitrarily set to a value of 1. The results were analyzed using SDS 2.3 and RQ Manager software (Applied Biosystems). Statistics were calculated using a paired two-tailed Student’s t test for hsp70 mRNA induction and a one-sample t test for the hsf2 mRNA expression.
EMSA
EMSA analysis was performed as described previously (Östling et al., 2007). In brief, whole cell extracts (12 µg of total protein) were incubated with a 32P-labeled oligonucleotide corresponding to the proximal HSE of the human hsp70.1 promoter. Complexes were separated on a native 4% polyacrylamide gel and detected with a light-sensitive film.
ChIP
ChIP was performed as described previously (Östling et al., 2007). In brief, K562 cells were cross-linked immediately after treatment with a final concentration of 1% formaldehyde, followed by quenching in 125 mM glycine. After lysis, chromatin was sonicated using Bioruptor. Samples were precleared using a 50% slurry of protein G–Sepharose beads blocked with bovine serum albumin. Immunoprecipitation was performed overnight at 4°C using antibodies against HSF1 (rabbit, SPA-901; Enzo Life Sciences), HSF2 (rabbit; Östling et al., 2007), Histone H3 (rabbit, ab1791; Abcam), Histone H3 phospho-S10 (mouse, ab14955; Abcam), RNAPII (mouse, MMS-129R; Covance), or rabbit IgG (sc-2027; Santa Cruz Biotechnology) as a nonspecific (NS) antibody. Upon washing of immunocomplexes, proteins and RNA were digested using proteinase K and RNase A, and cross-links were reversed by incubating samples overnight at 65°C. DNA was purified with phenol–chloroform, and PCR was performed on 5% of each sample using pure Taq Ready-to-go PCR beads (GE Healthcare) and primers for human hsp70.1 (5′-CCATGGAGACCAACACCCT-3′; 5′-CCCTGGGCTTTTATAAGTCG-3′; Östling et al., 2007). The input lanes represent 10% of the material used in the ChIP assay. Quantitative PCR analysis was performed using an ABI Prism 7900HT system (Applied Biosystems), a Kapa probe fast ABI prism qPCR kit (Kapa Biosystems), and primers and probe for the human hsp70 promoter (hsp70.1 F, 5′-CTGGCCTCTGATTGGTCCAA-3′; hsp70.1 R, 5′-CACGGAGACCCGCCTTTT-3′; 5′-FAM-CGGGAGGCGAAACCCCTGGAA-BHQ-3′). IP samples were normalized to values obtained for input before fold enrichment was determined by arbitrarily setting Scrambled unsynchronized control to a value of 1. For the statistical analysis, GraphPad Prism software was used, a linear regression was calculated for the correlation, and a two-tailed paired Student’s t test was used for column statistics.
Cell viability assay
Unsynchronized or synchronized cells were treated with heat shock at the indicated times. Cell viability was determined either by manually counting cells using a Bürker chamber or by the MTS-based CellTiter 96 Aqueous One Solution Reagent (Promega). At least three technical replicates were used for each biological sample. Values were obtained by relating the treated cells with an untreated counterpart. A paired two-tailed Student’s t test was used to calculate statistics.
Time-lapse imaging
Images for time-lapse imaging were acquired every 10 min using a confocal laser microscope (Leica SP5; Leica), using the LAS AF software with the Matrix extension (Leica), enabling imaging of multiple positions simultaneously. Cells (10,000 cells/well) were plated in normal growth medium into a glass-bottom 96-well plate (Greiner) and stained with 2.5 µM DRAQ5 (Biostatus Limited). Imaging was performed in a heated (37°C) chamber with a 5% CO2 atmosphere. Cells were imaged in four optical planes (5 µm each). Heat shock was performed in two different ways. For a pulse of heat shock, the temperature of the medium was increased to 43°C and was then left to cool off. The longer heat shock was performed by submerging the plate for 15 min in a 43°C water bath. Time-lapse recordings were started within 20 min of heat shock. Images were collected using a dry 20×, 0.70 NA, HC Plan-Apochromat PH2 objective lens (Leica) and a 633-nm laser for DRAQ5 staining and bright field images. Data were analyzed using the ImageJ software. Maximum intensity projections were made from the stacks. Time from nuclear envelope breakdown until telophase was scored manually in blind. Progression through mitosis was scored as normal when DNA aligned in a metaphase plate and two identical daughter cells were formed. Mitosis was scored as abnormal when there were lagging chromosomes, when the daughter cells formed micronuclei, when chromatin was decondensed without division, or when more than two daughter cells were formed (Hut et al., 2005). Statistical analysis was performed on a minimum of 20 cells per treatment from four independent experiments. For the length of mitosis, a two-tailed independent Student’s t test was used.
Time-lapse imaging using IncuCyte
Time-lapse imaging of MCF7, MDA-MB-231, HeLa, and WI38 cells was performed in an incubator outfitted with an IncuCyte ZOOM microscope (Essen BioScience) equipped with a 20× dry objective lens (0.45 NA, S Plan Fluor, DIC N1, ELWD; Nikon). Cells (30,000 cells/well) were plated in normal growth medium into a 12-well plate. After 24 h, the unsynchronized cells were either left untreated, or treated with a pulse of heat (42°C) or a 30-min heat shock at 42°C. Imaging was started within 10 min of heat shock. Phase-contrast images were acquired every 10 min. Images were organized into a stack using ImageJ software. Length of mitosis was calculated from when the cells rounded up until the formation of two daughter cells or the onset of apoptosis. Data were analyzed manually in blind. Statistical analysis was performed on 20 cells from five independent repeats for MCF7 and MDA-MB-231, and two independent repeats for HeLa and WI38 cells. A two-tailed independent Student’s t test was used to calculate statistics.
MNase assay
MNase assay was modified from a previously described protocol (Krishnakumar and Kraus, 2010). Proteins and DNA were cross-linked by incubating cells for 10 min at 37°C with 1% paraformaldehyde, after which 125 mM glycine was added for 5 min at 4°C. Cell pellets were washed and resuspended in TM2 buffer (10 mM Tris, pH 7.5, 2 mM MgCl2, 1 mM DTT, and 1× Roche’s protease inhibitor cocktail). Cells were permeabilized with 1.5% NP-40 and resuspended in MNase buffer (10 mM Tris, pH 7.5, 2 mM MgCl2, 2 mM CaCl2, 1 mM DTT, and 1× protease inhibitor cocktail [Roche]). Samples were divided into two aliquots: one was digested with MNase and the other was sonicated. Samples were incubated with MNase (New England Biolabs, Inc.) at a final concentration of 6.3 U/µl for 10 min at 37°C, after which the reaction was stopped (5% SDS and 50 mM EGTA), 0.2 M NaCl was added, and samples were incubated at 65°C overnight. Samples were treated with proteinase K (50 µg/ml) and RNase A (6 µg/ml). DNA was purified with phenol–chloroform. DNA (20 ng) was analyzed using qPCR as described for ChIP. In addition to primers and a probe against the human hsp70 promoter as described in the ChIP section, the following primers and probes were used: hsp70 promoter (hsp70.1 F2, 5′-CTGGCCTCTGATTGGTCCAA-3′; hsp70.1 R2, 5′-CACGGAGACCCGCCTTTT-3′; 5′-FAM-CGGGAGGCGAAACCCCTGGAA-BHQ-3′), and hsp70 coding region (hsp70.1 CR F, 5′-AGCTGCTGCAGGACTTCTTC-3′; hsp70.1 CR R, 5′-GACTTGTCCCCCATCAGGA-3′; Roche universal probe library probe #70). The enrichment of MNase-digested DNA was related to sonicated genomic DNA. Values were compared with the Scrambled control, which was arbitrarily set to a value of 1. Statistics were calculated using a one-sample t test where the mean MNase resistance of HSF2 down-regulated cells was compared with a value of 1.
Online supplemental material
Video 1 shows time-lapse imaging of an HSF2 WT MEF going through mitosis under nonstressed conditions. Video 2 shows time-lapse imaging of a HSF2 KO MEF going through mitosis under nonstressed conditions. Video 3 shows time-lapse imaging of two HSF2 WT MEFs displaying mitotic abnormalities after being subjected to a pulse of heat shock at 43°C. Video 4 shows time-lapse imaging of an HSF2 KO MEF going through mitosis after being subjected to a pulse of heat shock at 43°C. All videos are associated with Fig. 5 C. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201402002/DC1.
Supplementary Material
Acknowledgments
We thank Anna Moa Westerlund-Rönnberg for ideas and expert technical help in the initial phase of this study. We are grateful to all members of the Sistonen laboratory for stimulating discussions and critical comments on the manuscript, and to the Cell Imaging Core at Turku Centre for Biotechnology for technical assistance.
The research was funded by the Academy of Finland (L. Sistonen, J.K. Björk, and M.J. Kallio), the Sigrid Jusélius Foundation, Finnish Cancer Organizations (L. Sistonen), Magnus Ehrnrooth Foundation (L. Sistonen and P. Roos-Mattjus), Åbo Akademi University Foundation (L. Sistonen, A.N. Elsing, J.K. Björk, and P. Roos-Mattjus), the Tor, Joe and Pentti Borg Foundation (J.K. Björk and P. Roos-Mattjus), Medicinska understödsföreningen Liv och Hälsa r.f. (A.N. Elsing and P. Roos-Mattjus), the Turku Graduate School of Biomedical Sciences, the Swedish Cultural Foundation, the Finnish Cultural Foundation (A.N. Elsing), and Victoriastiftelsen (C. Aspelin).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- ChIP
- chromatin immunoprecipitation
- CIN
- chromosomal instability
- H3 S10P
- phosphorylation of serine 10 on histone H3
- HSE
- heat shock element
- HSF
- heat shock factor
- Hsp
- heat shock protein
- HSR
- heat shock response
- IP
- immunoprecipitation
- KO
- knockout
- MEF
- mouse embryonic fibroblast
- MNase
- micrococcal nuclease
- RNAPII
- RNA polymerase II
- WT
- wild type
References
- Adelman, K., and Lis J.T.. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13:720–731 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlskog, J.K., Björk J.K., Elsing A.N., Aspelin C., Kallio M., Roos-Mattjus P., and Sistonen L.. 2010. Anaphase-promoting complex/cyclosome participates in the acute response to protein-damaging stress. Mol. Cell. Biol. 30:5608–5620 10.1128/MCB.01506-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerfelt, M., Henriksson E., Laiho A., Vihervaara A., Rautoma K., Kotaja N., and Sistonen L.. 2008. Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc. Natl. Acad. Sci. USA. 105:11224–11229 10.1073/pnas.0800620105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerfelt, M., Morimoto R.I., and Sistonen L.. 2010. Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11:545–555 10.1038/nrm2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alastalo, T.P., Hellesuo M., Sandqvist A., Hietakangas V., Kallio M., and Sistonen L.. 2003. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J. Cell Sci. 116:3557–3570 10.1242/jcs.00671 [DOI] [PubMed] [Google Scholar]
- Anckar, J., and Sistonen L.. 2011. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80:1089–1115 10.1146/annurev-biochem-060809-095203 [DOI] [PubMed] [Google Scholar]
- Bar-Joseph, Z., Siegfried Z., Brandeis M., Brors B., Lu Y., Eils R., Dynlacht B.D., and Simon I.. 2008. Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc. Natl. Acad. Sci. USA. 105:955–960 10.1073/pnas.0704723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk, J.K., and Sistonen L.. 2010. Regulation of the members of the mammalian heat shock factor family. FEBS J. 277:4126–4139 10.1111/j.1742-4658.2010.07828.x [DOI] [PubMed] [Google Scholar]
- Björk, J.K., Sandqvist A., Elsing A.N., Kotaja N., and Sistonen L.. 2010. miR-18, a member of Oncomir-1, targets heat shock transcription factor 2 in spermatogenesis. Development. 137:3177–3184 10.1242/dev.050955 [DOI] [PubMed] [Google Scholar]
- Brown, S.A., Imbalzano A.N., and Kingston R.E.. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 10:1479–1490 10.1101/gad.10.12.1479 [DOI] [PubMed] [Google Scholar]
- Chen, D., Dundr M., Wang C., Leung A., Lamond A., Misteli T., and Huang S.. 2005. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 168:41–54 10.1083/jcb.200407182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova, R., and Oelgeschläger T.. 2002. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 4:79–82 10.1038/ncb733 [DOI] [PubMed] [Google Scholar]
- Ciocca, D.R., Arrigo A.P., and Calderwood S.K.. 2013. Heat shock proteins and heat shock factor 1 in carcinogenesis and tumor development: an update. Arch. Toxicol. 87:19–48 10.1007/s00204-012-0918-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, C., Whitesell L., Rogers A.B., and Lindquist S.. 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 130:1005–1018 10.1016/j.cell.2007.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debec, A., and Marcaillou C.. 1997. Structural alterations of the mitotic apparatus induced by the heat shock response in Drosophila cells. Biol. Cell. 89:67–78 10.1016/S0248-4900(99)80082-3 [DOI] [PubMed] [Google Scholar]
- Delcuve, G.P., He S., and Davie J.R.. 2008. Mitotic partitioning of transcription factors. J. Cell. Biochem. 105:1–8 10.1002/jcb.21806 [DOI] [PubMed] [Google Scholar]
- Fiorenza, M.T., Farkas T., Dissing M., Kolding D., and Zimarino V.. 1995. Complex expression of murine heat shock transcription factors. Nucleic Acids Res. 23:467–474 10.1093/nar/23.3.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld, J.M., and Forbes D.J.. 1997. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22:197–202 10.1016/S0968-0004(97)01045-1 [DOI] [PubMed] [Google Scholar]
- Grant, G.D., Brooks L. III, Zhang X., Mahoney J.M., Martyanov V., Wood T.A., Sherlock G., Cheng C., and Whitfield M.L.. 2013. Identification of cell cycle-regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol. Biol. Cell. 24:3634–3650 10.1091/mbc.E13-05-0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guettouche, T., Boellmann F., Lane W.S., and Voellmy R.. 2005. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 6:4 10.1186/1471-2091-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas, V., Ahlskog J.K., Jakobsson A.M., Hellesuo M., Sahlberg N.M., Holmberg C.I., Mikhailov A., Palvimo J.J., Pirkkala L., and Sistonen L.. 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 23:2953–2968 10.1128/MCB.23.8.2953-2968.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas, V., Anckar J., Blomster H.A., Fujimoto M., Palvimo J.J., Nakai A., and Sistonen L.. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA. 103:45–50 10.1073/pnas.0503698102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg, C.I., Hietakangas V., Mikhailov A., Rantanen J.O., Kallio M., Meinander A., Hellman J., Morrice N., MacKintosh C., Morimoto R.I., et al. 2001. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 20:3800–3810 10.1093/emboj/20.14.3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y., Rogers R., Matunis M.J., Mayhew C.N., Goodson M.L., Park-Sarge O.K., and Sarge K.D.. 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276:40263–40267 (published erratum appears in J. Biol. Chem. 277:26708) 10.1074/jbc.M104714200 [DOI] [PubMed] [Google Scholar]
- Hut, H.M.J., Kampinga H.H., and Sibon O.C.M.. 2005. Hsp70 protects mitotic cells against heat-induced centrosome damage and division abnormalities. Mol. Biol. Cell. 16:3776–3785 10.1091/mbc.E05-01-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio, M., Chang Y., Manuel M., Alastalo T.-P., Rallu M., Gitton Y., Pirkkala L., Loones M.-T., Paslaru L., Larney S., et al. 2002. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 21:2591–2601 10.1093/emboj/21.11.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar, R., and Kraus W.L.. 2010. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell. 39:736–749 10.1016/j.molcel.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger, P.E., Sarge K.D., and Morimoto R.I.. 1993. Mouse heat shock transcription factors 1 and 2 prefer a trimeric binding site but interact differently with the HSP70 heat shock element. Mol. Cell. Biol. 13:3370–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl, N.M., Kunz J., and Rensing L.. 2000. Heat shock-induced arrests in different cell cycle phases of rat C6-glioma cells are attenuated in heat shock-primed thermotolerant cells. Cell Prolif. 33:147–166 10.1046/j.1365-2184.2000.00175.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loison, F., Debure L., Nizard P., le Goff P., Michel D., and le Dréan Y.. 2006. Up-regulation of the clusterin gene after proteotoxic stress: implication of HSF1-HSF2 heterocomplexes. Biochem. J. 395:223–231 10.1042/BJ20051190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J., Solimini N.L., and Elledge S.J.. 2009. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 136:823–837 10.1016/j.cell.2009.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Balbás, M.A., Dey A., Rabindran S.K., Ozato K., and Wu C.. 1995. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 83:29–38 10.1016/0092-8674(95)90231-7 [DOI] [PubMed] [Google Scholar]
- Mathew, A., Mathur S.K., and Morimoto R.I.. 1998. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 18:5091–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo, M.L., Santagata S., Koeva M., Bell G.W., Hu R., Tamimi R.M., Fraenkel E., Ince T.A., Whitesell L., and Lindquist S.. 2012. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 150:549–562 10.1016/j.cell.2012.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti, E.F., Sanford S., and Levens D.. 1997. Marking of active genes on mitotic chromosomes. Nature. 388:895–899 10.1038/42282 [DOI] [PubMed] [Google Scholar]
- Nakahata, K., Miyakoda M., Suzuki K., Kodama S., and Watanabe M.. 2002. Heat shock induces centrosomal dysfunction, and causes non-apoptotic mitotic catastrophe in human tumour cells. Int. J. Hyperthermia. 18:332–343 10.1080/02656730210129736 [DOI] [PubMed] [Google Scholar]
- Nakai, A., and Ishikawa T.. 2001. Cell cycle transition under stress conditions controlled by vertebrate heat shock factors. EMBO J. 20:2885–2895 10.1093/emboj/20.11.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth, J.D., Loewer A., Lahav G., and Mitchison T.J.. 2012. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol. Biol. Cell. 23:567–576 10.1091/mbc.E11-09-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östling, P., Björk J.K., Roos-Mattjus P., Mezger V., and Sistonen L.. 2007. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J. Biol. Chem. 282:7077–7086 10.1074/jbc.M607556200 [DOI] [PubMed] [Google Scholar]
- Parsons, G.G., and Spencer C.A.. 1997. Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol. Cell. Biol. 17:5791–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Diaz, J., Hegre S.A., Anderssen E., Aas P.A., Mjelle R., Gilfillan G.D., Lyle R., Drabløs F., Krokan H.E., and Sætrom P.. 2013. Transcription profiling during the cell cycle shows that a subset of Polycomb-targeted genes is upregulated during DNA replication. Nucleic Acids Res. 41:2846–2856 10.1093/nar/gks1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfau, S.J., and Amon A.. 2012. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 13:515–527 10.1038/embor.2012.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines, J.2011. Cubism and the cell cycle: the many faces of the APC/C. Nat. Rev. Mol. Cell Biol. 12:427–438 10.1038/nrm3132 [DOI] [PubMed] [Google Scholar]
- Rallu, M., Loones M., Lallemand Y., Morimoto R., Morange M., and Mezger V.. 1997. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc. Natl. Acad. Sci. USA. 94:2392–2397 10.1073/pnas.94.6.2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, K., Haslbeck M., and Buchner J.. 2010. The heat shock response: life on the verge of death. Mol. Cell. 40:253–266 10.1016/j.molcel.2010.10.006 [DOI] [PubMed] [Google Scholar]
- Sakurai, H., and Enoki Y.. 2010. Novel aspects of heat shock factors: DNA recognition, chromatin modulation and gene expression. FEBS J. 277:4140–4149 10.1111/j.1742-4658.2010.07829.x [DOI] [PubMed] [Google Scholar]
- Sandqvist, A., Björk J.K., Åkerfelt M., Chitikova Z., Grichine A., Vourc’h C., Jolly C., Salminen T.A., Nymalm Y., and Sistonen L.. 2009. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol. Biol. Cell. 20:1340–1347 10.1091/mbc.E08-08-0864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata, S., Hu R., Lin N.U., Mendillo M.L., Collins L.C., Hankinson S.E., Schnitt S.J., Whitesell L., Tamimi R.M., Lindquist S., and Ince T.A.. 2011. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. USA. 108:18378–18383 10.1073/pnas.1115031108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata, S., Mendillo M.L., Tang Y.-C., Subramanian A., Perley C.C., Roche S.P., Wong B., Narayan R., Kwon H., Koeva M., et al. 2013. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 341:1238303 10.1126/science.1238303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge, K.D., Zimarino V., Holm K., Wu C., and Morimoto R.I.. 1991. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA-binding ability. Genes Dev. 5:1902–1911 10.1101/gad.5.10.1902 [DOI] [PubMed] [Google Scholar]
- Sarge, K.D., Murphy S.P., and Morimoto R.I.. 1993. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 13:1392–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka, A., and Seiser C.. 2012. Histone H3 phosphorylation - a versatile chromatin modification for different occasions. Biochimie. 94:2193–2201 10.1016/j.biochi.2012.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman, J.-M., Sotillo R., and Benezra R.. 2010. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer. 10:102–115 10.1038/nrc2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkawa, T., Tan K., Fujimoto M., Hayashida N., Yamamoto K., Takaki E., Takii R., Prakasam R., Inouye S., Mezger V., and Nakai A.. 2011. Heat shock factor 2 is required for maintaining proteostasis against febrile-range thermal stress and polyglutamine aggregation. Mol. Biol. Cell. 22:3571–3583 10.1091/mbc.E11-04-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sif, S., Stukenberg P.T., Kirschner M.W., and Kingston R.E.. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842–2851 10.1101/gad.12.18.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solimini, N.L., Luo J., and Elledge S.J.. 2007. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 130:986–988 10.1016/j.cell.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Sotillo, R., Hernando E., Díaz-Rodríguez E., Teruya-Feldstein J., Cordón-Cardo C., Lowe S.W., and Benezra R.. 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 11:9–23 10.1016/j.ccr.2006.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake, Y., and Sluder G.. 2010. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 20:1666–1671 10.1016/j.cub.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls, E., Sánchez-Molina S., and Martínez-Balbás M.A.. 2005. Role of histone modifications in marking and activating genes through mitosis. J. Biol. Chem. 280:42592–42600 10.1074/jbc.M507407200 [DOI] [PubMed] [Google Scholar]
- Varier, R.A., Outchkourov N.S., de Graaf P., van Schaik F.M., Ensing H.J.L., Wang F., Higgins J.M., Kops G.J., and Timmers H.T.. 2010. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO J. 29:3967–3978 10.1038/emboj.2010.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara, A., and Sistonen L.. 2014. HSF1 at a glance. J. Cell Sci. 127:261–266 10.1242/jcs.132605 [DOI] [PubMed] [Google Scholar]
- Vihervaara, A., Sergelius C., Vasara J., Blom M.A.H., Elsing A.N., Roos-Mattjus P., and Sistonen L.. 2013. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc. Natl. Acad. Sci. USA. 110:E3388–E3397 10.1073/pnas.1305275110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, B.A., Silk A.D., Montagna C., Verdier-Pinard P., and Cleveland D.W.. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 11:25–36 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Weber, C.M., Henikoff J.G., and Henikoff S.. 2010. H2A.Z nucleosomes enriched over active genes are homotypic. Nat. Struct. Mol. Biol. 17:1500–1507 10.1038/nsmb.1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., Yu L., Bowen J., Gorovsky M.A., and Allis C.D.. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 97:99–109 10.1016/S0092-8674(00)80718-7 [DOI] [PubMed] [Google Scholar]
- Westerheide, S.D., Anckar J., Stevens S.M. Jr, Sistonen L., and Morimoto R.I.. 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 323:1063–1066 10.1126/science.1165946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, M.L., Zheng L.X., Baldwin A., Ohta T., Hurt M.M., and Marzluff W.F.. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188–4198 10.1128/MCB.20.12.4188-4198.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., and Botstein D.. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 13:1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, H., Wilkerson D.C., Mayhew C.N., Lubert E.J., Skaggs H.S., Goodson M.L., Hong Y., Park-Sarge O.K., and Sarge K.D.. 2005. Mechanism of hsp70i gene bookmarking. Science. 307:421–423 10.1126/science.1106478 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.