Abstract
Foamy virus (FV) replication, while related to that of orthoretroviruses, differs at a number of steps. Several of these differences involve the reverse transcriptase (RT). There appear to be fewer RTs present in FV than in orthoretroviruses; we previously proposed that the polymerase of FV RT was more active than orthoretroviral RTs to compensate for the numerical difference. Here we present further characterization of the RT of FV. The polymerase activity of FV RT was greater than that of human immunodeficiency virus type 1 RT in a variety of assays. We also examined the RNase H activity of FV RT, and we propose that FV RT has a basic loop in the RNase H domain. Although the sequence of the basic loop of FV RT is different from the basic loop of either Moloney leukemia virus RNase H or Escherichia coli RNase H, the FV RT basic loop appears to have a similar function.
Foamy viruses (FVs) are retroviruses (subfamily Spumaretrovirinae) but are in some ways distinct from typical retroviruses. The organization of the gag, pol, and env genes in the FV genome is the same as in orthoretroviruses and, like orthoretroviruses, FVs convert their single-stranded RNA genomes into integrated double-stranded DNA proviruses flanked by two long terminal repeats (LTRs), using the enzymes reverse transcriptase (RT) and integrase (IN). However, FVs have distinct features that set them apart from orthoretroviruses. Several of these differences involve either reverse transcription or the RT enzyme itself. In orthoretroviruses, such as human immunodeficiency virus type 1 (HIV-1), RT converts the single-stranded RNA genome into double-stranded DNA after the virion enters the target cell (8). In contrast, a significant portion of FV particles contain double-stranded DNA (39, 41). Inhibitor studies make it clear that it is the virions with DNA genomes that infect the target cells, indicating that FV RT must be active in the cells producing the viral particles (27, 41).
Orthoretroviruses use a single full-length RNA transcript as the mRNA for both the gag and pol genes. Translation of the full-length RNA produces both the Gag and Gag-Pol polyproteins (8). Approximately 20 Gag proteins are produced for every Gag-Pol fusion protein (8). Gag self-assembles to form viral particles; during virus assembly, the Gag portion of the Gag-Pol fusion protein interacts with the Gag proteins. This is how the Pol proteins are incorporated into virions; the resulting virions have approximately 50 to 100 Gag-Pol polyproteins per particle (14, 18, 28, 38). However, in an FV-infected cell, the Pol polyprotein is made independently from Gag; Pol is translated from a spliced message (13, 19, 39). The FV Pol polyprotein is proteolytically processed; however, it undergoes only limited processing. There is a single protease cleavage between RT and IN which releases IN and a protease-RT fusion protein (30). Protease is not cleaved from RT. The mechanism for packaging FV Pol protein into the virion appears to involve interactions of FV Pol with specific sequences in the RNA genome (17). The number of Pol proteins within the virion appears to be significantly lower than for orthoretroviruses (2, 3, 15, 16, 17). FV particles may contain as few as two RT molecules (one RT bound at each specific genomic site) to carry out the conversion of the single-strand RNA genome into double-stranded DNA, compared to 50 to 100 RT proteins in an orthoretrovirus virion. In HIV-1, it takes approximately 20 to 30 enzymatically active RTs to successfully carry out reverse transcription (20).
Since there are relatively few RT proteins in the FV particle compared to that in orthoretroviruses, we proposed that the polymerase of FV RT would be more efficient than orthoretroviral RTs, such as HIV-1 RT, to overcome this numerical disadvantage (31). Retroviral RTs have two enzymatic activities, polymerase and RNase H, which cleaves the RNA strand of an RNA-DNA duplex. These two enzymatic activities are both necessary and sufficient for RT to convert the single-stranded viral RNA genome into double-stranded DNA. We previously reported that the polymerase activity of the RT from the simian FV-chimpanzee (human isolate) (previously designated human FV and now designated prototype FV [PFV]) was more processive than HIV-1 RT using a DNA template (31). We report here additional analysis of the polymerase activity and RNase H activity of PFV RT and compare these activities to the corresponding activities of HIV-1 RT and Moloney leukemia virus (MLV) RT.
MATERIALS AND METHODS
In vitro polymerase assays.
The construction of the FV RT expression clone and the protocol for expressing and purifying the RT have been previously described (31). The protein consists of an inactive protease (which contains the active-site mutation D24A) fused to the FV RT. The expression and purification of HIV-1 RT has been previously described (5, 7). MLV RT was obtained from Invitrogen. The DNA-dependent DNA polymerase assay was done as previously described (5). The primer −47 (New England Biolabs) was 5′-end labeled and then annealed to single-strand M13mp18 DNA (New England Biolabs). The primer was extended with either HIV-1 RT or FV RT as previously described (5, 31). The final concentration of template-primer (T/P) in each reaction mixture was approximately 2.5 nm; the RT was in molar excess (∼85 nM). The cold trap, poly(rC) · oligo(dG), was added in excess relative to RT (300 nM) after the RT was allowed to bind to the labeled T/P. The extension products were suspended in 2× gel loading buffer (Ambion) and heated at 65°C to denature the samples. An alkaline agarose gel was prepared (33), and the samples were loaded. After electrophoresis for 15 h, the gel was neutralized and dried. The products were visualized by exposure to X-ray film.
The RNA-dependent DNA polymerase assay has been previously described (4). A clone containing a T7 RNA polymerase promoter, the HIV-1 polypurine tract (PPT), U3, R, U5, and the HIV-1 primer binding site (PBS) was linearized with NotI. An RNA transcript was generated using the T7 MEGAScript kit (Ambion) that contains the HIV-1 PPT, LTR, and PBS sequences in the same sense as the HIV-1 one LTR wide. The sequences were derived from pNL 4-3 (1). A synthetic DNA oligonucleotide (Biosource) complementary to the PBS sequence (5′-GTCCCTGTTCGGGCGCCA-3′) was 5′-end labeled and then annealed to the RNA template. The primer was extended as previously described (4) in the presence of various nucleocapsid (NC) concentrations (kindly provided by Louis Henderson). The final concentration of the labeled T/P in each reaction mixture was approximately 15 nM; the RT was in molar excess (∼85 nM). The extension products were suspended in 2× gel loading buffer (Ambion), heated to denature the products, and then separated on a 6% polyacrylamide sequencing gel. The products were visualized by exposure to X-ray film.
Strand transfer assay.
The strand transfer assay was similar to that described by Davis et al. (10). The strand transfer primer (5′-GCATCTGGCGCTCGCAAATTTG-3′) was 5′-end labeled and then annealed to the RNA strand transfer template (5′-AGGUGAGUGAGAUGAUAACAAAUUUGCGAGCGCCAGAUGC-3′). The concentration of the T/P was 40 nM in the reaction mixture, and the primer was extended by 170 nM HIV-1 RT or FV RT in the presence of 25 mM Tris (pH 8.0), 75 mM KCl, 10 mM MgCl2, 100 μg of bovine serum albumin/ml, 10 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 25 μM (each) dATP, dCTP, dGTP, and dTTP, 1 U of Superasin (Ambion)/μl, and 400 nM strand transfer acceptor (5′-GAGCTGCTTGAATTCTGCGTACTAGGTGAGTGAGATAACA-3′). The reaction was allowed to proceed for various lengths of time (5, 10, 25, or 45 min) before being terminated by the addition of EDTA and ethanol precipitation. The samples were suspended in 2× gel loading buffer and fractionated on a 15% polyacrylamide sequencing gel. The products were visualized by exposure to X-ray film.
Pyrophosphorylysis.
ATP- and NaPPi-dependent pyrophosphorylysis analysis was done as previously described (7). A synthetic DNA oligonucleotide (5′-GTCCCTGTTCGGGCACCA-3′; Biosource) was 5′-end labeled and then annealed to the template (5′-AGTCAGTGTAGACAATCCCTAGCAATGGTGCCCGAACAGGGAC-3′). The 3′ end of the primer was then blocked by the addition of zidovudine monophosphate (AZTMP) (7). After purification of the blocked T/P, the T/P was incubated with HIV-1 RT or FV RT as previously described (7) in the presence of various concentrations of either NaPPi (50, 100, or 200 μM) or ATP (2.0, 5.0, or 10.0 mM) for 10 min. The final concentration of T/P in each reaction mixture was approximately 15 nM; the RT was in molar excess (∼171 nM). The reactions were halted by the addition of EDTA, and the T/P was precipitated by the addition of isopropyl alcohol. The products were fractionated on a 15% polyacrylamide sequencing gel. The total amount of T/P (blocked and unextended plus deblocked and extended) and the amount of full-length product (deblocked and extended to the end of the template) were determined using a PhosphorImager.
RNase H assays.
Several RNA-DNA heteroduplexes were used and are shown below in Fig. 5. The RNA oligonucleotides were obtained from Dharmacon Research, Inc. The RNA oligonucleotides were 5′-end labeled and then annealed to synthetic DNA oligonucleotides by heating and slow cooling. A 0.2 μM concentration of T/P was suspended in 25 mM Tris (pH 8.0), 50 mM NaCl, 5.0 mM MgCl2, 100 μg of bovine serum albumin/ml, 10 mM CHAPS, and 1 U of Superasin (Ambion)/μl. The final reaction volume was 12 μl. The reactions were initiated by the addition of the indicated amount of RT and were incubated at 37°C. Aliquots were removed at the indicated time points, and the reaction was halted by addition of 2× gel loading buffer. The reaction products were fractionated on a 15% polyacrylamide sequencing gel. Products were visualized by exposure to X-ray film.
FIG. 5.
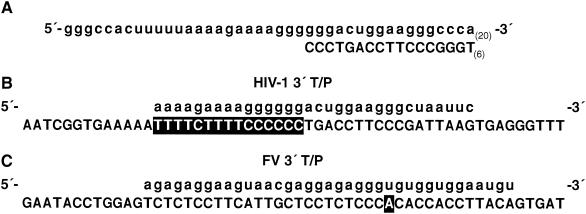
RNA and DNA substrates used in the RNase H assays. (A) A long 5′-end-labeled RNA was annealed to a short DNA oligonucleotide. The RNA sequence is sense strand and is derived from the sequence of the HIV-1 PPT and part of the HIV-1 U3 region (1). The 3′ end of the DNA primer is bound at the polymerase active site, and the RNase H active site of FV RT or HIV-1 RT will cleave the RNA template in the double-stranded region of the substrate. Uncleaved RNA is 60 nt in length. (B) A short 5′-end-labeled RNA annealed to a long DNA. The sequences are also derived from the PPT/U3 boundary region of the HIV-1 genome. The RNA is sense strand. The PPT of HIV-1 is highlighted. Uncleaved RNA is 32 nt in length. The distance from the labeled 5′ end of the RNA to the PPT-U3 junction is 15 nt. (C) A short 5′-end-labeled RNA annealed to a long DNA. The sequences are derived from the PPT/U3 region of FV (24). The 5′ boundary of the FV PPT is not known. The first residue of the U3 region is highlighted. Uncleaved RNA is 37 nt in length. The distance from the labeled 5′ end of the RNA to the PPT/U3 junction is 24 nt.
RESULTS
Polymerase activity.
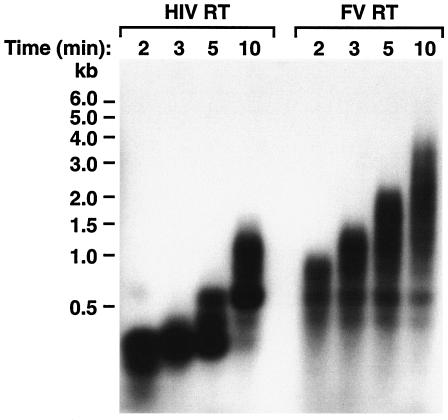
As described in the introduction, PFV RT remains covalently joined to the protease (PR) after proteolytic maturation of the virion (30). We have expressed a PFV PR-RT fusion protein in Escherichia coli in which the PR is inactivated by an amino acid substitution in the active site (D24A). This protein was designated D/A PFV RT and was shown to have significantly greater processivity than HIV-1 RT when DNA was the template (31). The processivity assay measures the lengths of extension products from a single cycle of polymerization, because there is a nonradioactive trap present in the reaction. We measured the lengths of the extension products generated in the absence of an unlabeled trap at various time points. A 5′-end-labeled DNA primer was annealed to single-stranded M13mp18 DNA and extended by either HIV-1 RT or D/A PFV RT, called hereafter FV RT. The M13mp18 DNA used as the template in these experiments was a single-stranded circle 7,249 nucleotides (nt) in length. After 10 min, the majority of extension products generated by HIV-1 RT were less than 1,500 nt in length (Fig. 1); FV RT had synthesized products over 4,000 nt long. The results were similar for other time points (Fig. 1), and the results were reproducible with different batches of FV RT and HIV-1 RT (data not shown). After 15 min, FV RT extended the primer all the way around the circular M13mp18 template and had begun to displace the 5′ end of the primer strand (data not shown). At 15 min, HIV-1 RT had extended less than 4,000 nt (data not shown). Both HIV-1 RT and FV RT were shown to have strand displacement activity (data not shown).
FIG. 1.
DNA-dependent DNA polymerase activity of FV RT and HIV-1 RT. A 5′-end-labeled DNA primer was annealed to a single-stranded M13mp18 DNA template and extended with either HIV-1 RT or FV RT for the lengths of time indicated. Samples were precipitated with ethanol and then fractionated on a 2.0% alkaline agarose gel. After electrophoresis, the gel was neutralized and dried as described in Materials and Methods. The products were visualized by autoradiography. The size marker was 5′-end-labeled 1.0-kb marker from New England Biolabs.
We examined the ability of the enzymes to extend a 5′-end-labeled DNA primer when RNA was the template. As described in Materials and Methods, the template RNA was transcribed from the HIV-1 LTR using T7 RNA polymerase. The resulting RNA has the same polarity as HIV-1 genomic RNA. A 5′-end-labeled DNA primer complementary to the PBS sequence in the HIV-1 genome was annealed to the RNA template and extended with either HIV-1 RT or FV RT in the presence of various concentrations of HIV-1 NC (kindly provided by Louis Henderson). In the orthoretrovirus virion, the RNA genome is coated by NC. It has been suggested that NC may aid in strand transfer, probably by stimulating nucleic acid annealing, and may help RT pass through regions of secondary structure by destabilizing hairpin structures in the RNA (reference 8 and references therein). However, it is also possible that NC may hinder processivity, particularly in regions without significant secondary structure, because RT must displace NC from the RNA as it is polymerizing. In our experiments, extension of the primer to the end of the template strand would result in a full-length product of 680 nt. As shown in Fig. 2, in a 10-min reaction FV RT extended the primer to the end of the RNA template, while the largest HIV-1 RT products synthesized were 300 nt. In the absence of NC, the two RTs behaved somewhat differently when they encountered RNA secondary structure (0.0 μM NC lane). After incorporating approximately 95 nt, the enzymes reached the 3′ end of a secondary structure designated the poly(A) hairpin (9) and, after incorporating approximately 142 nt, the 3′ end of the TAR hairpin (for review, see reference 8). HIV-1 RT has a tendency to pause at such hairpins and, when paused, has a greater probability to disassociate from the T/P (8). FV RT also paused at the base of hairpins; however, the paused products were 1 or 2 nt shorter for FV RT than for HIV-1 RT (Fig. 2). Similar results were obtained in experiments done with different batches of HIV-1 RT and FV RT (data not shown).
FIG. 2.
RNA-dependent DNA polymerase activity of FV RT and HIV-1 RT. The clone PPT-PBS (An) has been previously described (4), and a brief description is given in Materials and Methods. The RNA template has the same sequence as the HIV-1 RNA genome and contains sequences from PPT, U3, R, U5, and PBS. A DNA primer complementary to the PBS was 5′-end labeled and annealed to the RNA. The primer was extended as described in Materials and Methods. NC was added to give the concentrations indicated before RT was added to the reaction mixture. The 3′ end of the poly(A) hairpin is approximately 95 nt from the 5′ end of the primer, while the 3′ end of the TAR hairpin is approximately 142 nt from the 5′ end of the primer.
The enzymes also differed in their ability to extend the primer in the presence of HIV-1 NC. At the lowest concentration of NC where, on average, there was one NC bound every 70 nt, no differences were seen in the products produced by either RT compared to those produced in the absence of NC (Fig. 2). At the highest concentration of HIV-1 NC (one NC molecule bound for every 7 nt of RNA template), the amount of extended product for HIV-1 RT was greatly reduced (Fig. 2). It appears that there was a significant inhibition of the initiation of polymerization by HIV-1, presumably because NC bound to the T/P and interfered with the binding of HIV-1 RT. However, for FV RT, the amount of full-length product was somewhat higher in the presence of HIV-1 NC than in the absence of HIV-1 NC. This suggests FV RT binds strongly to the T/P and can effectively compete with HIV-1 NC. Similar results were obtained with different batches of HIV-1 RT (data not shown).
Pyrophosphorylysis.
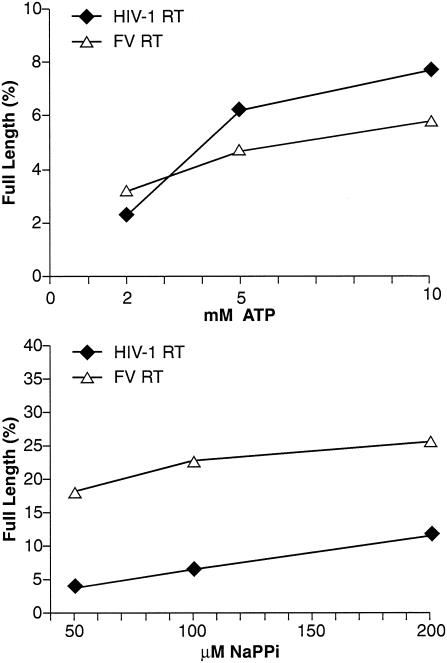
Polymerization is the addition of a deoxynucleoside triphosphate (dNTP) (or dNTP analog) to a primer. This increases the length of the primer by 1 nt, with the concomitant release of a pyrophosphate (PPi). The reverse of the polymerase reaction is pyrophosphorylysis: the primer is shortened by 1 nt and PPi is added to the released nucleotide, generating a dNTP (or dNTP analog). Thus, polymerase activity and PPi pyrophosphorylysis activity should be related. The resistance of HIV-1 to AZT is the result of enhanced pyrophosphorylysis. However, for AZT-resistant HIV-1 RT, the biologically relevant pyrophosphate donor appears to be ATP rather than PPi (e.g., references 7, 25, and 26 and references therein). The basic reaction is similar to PPi pyrophosphorylysis; however, when ATP is used as the pyrophosphate donor, a dinucleoside tetraphosphate is the excision product. FV replication in tissue culture is sensitive to AZT (27, 31, 32, 34, 41, 42). We measured the ability of FV RT to catalyze pyrophosphorylysis with either ATP or NaPPi as a pyrophosphate donor. Pyrophosphorylysis was measured as the ability to remove an AZT nucleotide analog blocking the 3′ end of a primer, which then allowed the primer to be extended to the end of the template strand, generating a full-length product. As shown in Fig. 3, FV RT was more efficient at removing an AZTMP from the end of the primer than was HIV-1 RT when PPi was the pyrophosphate donor. Since this type of pyrophosphorylysis is the reverse of DNA synthesis, the higher level of PPi-dependent pyrophosphorylysis matches the higher polymerase activity of FV RT than of HIV-1 RT (31) (see above).
FIG. 3.
NaPPi- and ATP-dependent pyrophosphorylysis. As described in Materials and Methods, a DNA primer was 5′-end labeled and then annealed to a DNA template. AZTMP was incorporated at the 3′ end of the primer by polymerization with HIV-1 RT in the presence of AZT triphosphate. After purification, the T/P was incubated with either HIV-1 RT or FV RT in the presence of dNTPs and either ATP or NaPPi as the pyrophosphate donor. Pyrophosphorylysis was measured as the ability of the RT to excise the AZT group blocking the primer and extend the primer to the end of the template strand. The amount of total label in the sample and the amount of label in the full-length product were determined with a phosphorimager, and the data are expressed as a percentage of the full-length product. Experiments were done in duplicate, and the results were quite reproducible. Data from a representative experiment are shown.
When the pyrophosphate donor was ATP, the products of the forward reaction (polymerization) were not the same as the substrates for the excision reaction (ATP-dependent pyrophosphorylysis). Wild-type HIV-1 RT is able to bind ATP and use it as a pyrophosphate donor in an excision reaction (7, 25, 26). As shown in Fig. 3, despite the greater polymerase activity of FV RT compared to HIV-1 RT, the level of pyrophosphorylysis with ATP as the pyrophosphate donor was slightly lower for FV RT than for HIV-1 RT. This suggests that the ability of HIV-1 RT to bind ATP and use it in an excision reaction may be fortuitous; FV RT, despite its higher level of polymerase and PPi-mediated pyrophosphorylysis activity, is no better than HIV-1 RT in carrying out ATP-mediated pyrophosphorylysis. Since FV requires high polymerase activity to generate infectious virus, its modest ability to excise AZT from a blocked primer using ATP as the pyrophosphate donor has implications for the development of AZT resistance (see Discussion).
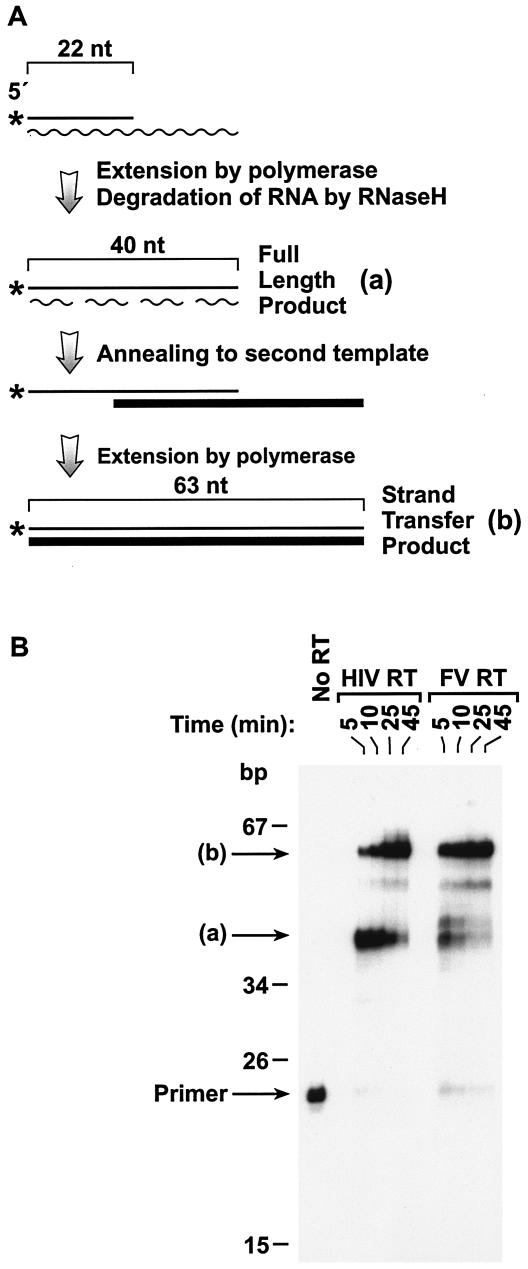
Strand transfer.
One of the key reactions RT must carry out is transfer between templates (strand transfer) (Fig. 4A). The first strand transfer in reverse transcription depends on the polymerase activity and on a second enzymatic activity, that of RNase H, which degrades RNA only when it is part of an RNA-DNA heteroduplex (8). We generated a model substrate for strand transfer by annealing a 5′-end-labeled DNA primer to an RNA template (Materials and Methods). The sequence of the donor strand was chosen to avoid any problems with hairpin DNA synthesis; experiments in which the acceptor strand was omitted from the reaction clearly showed no hairpin DNA was synthesized (data not shown). The DNA primer can be extended to the end of the RNA template to produce a full-length product. RNase H activity degrades the RNA template, allowing the extended DNA primer to anneal to a DNA template present in the reaction. After this annealing occurs, the primer can be extended to yield a larger strand transfer product (Fig. 4B). In the reaction, the full-length product, which depends only on polymerase activity, accumulates first. After RNase H degrades the RNA template and the primer anneals to the second template, the strand transfer product, whose synthesis is dependent on the enzymatic activities of both polymerase and RNase H, begins to appear. If the original RNA template is not degraded (RNase H−), the primer cannot be extended beyond the end of the original RNA template and only the full-length product will be generated.
FIG. 4.
Strand transfer by FV RT and HIV-1 RT. (A) Schematic of the strand transfer assay. The 5′-end-labeled DNA primer (22 nt in length) is annealed to an RNA template (wavy line). Extension of the primer by the polymerase to the end of the RNA template will give a full-length product (designated as a) that is 40 nt in length. The RNA strand in the RNA-DNA duplex is then degraded by the RNase H activity of the RT, allowing the transfer of the DNA strand to a new template (thick line). Further extension of the labeled DNA to the end of the second template will give a strand transfer product (designated as b) that is 63 nt in size. (B) Strand transfer assay. As described, a DNA primer was 5′-end labeled and annealed to an RNA template. The primer was extended by either FV RT or HIV-1 RT for the indicated lengths of time in the presence of dNTPs and a second DNA oligonucleotide, designated the acceptor template. Extension of the primer to the end of the RNA template generates the full-length product (labeled a). Extension to the end of the acceptor template yields the strand transfer product (labeled b). The time necessary to generate the full-length product is dependent only on the polymerase activity of the RT, while that for the strand transfer product is dependent on both the polymerase and the RNase H activities of the enzyme.
As shown in Fig. 4B, HIV-1 RT was able to extend almost all of the primer to the end of the RNA template in 5 min, and some strand transfer product was produced. By 45 min, most of the full-length product had been extended to produce the strand transfer product. In contrast, after 5 min, the majority of the DNA synthesized by FV RT was strand transfer product. This result can be explained in part by the higher polymerase activity of FV RT compared to HIV-1 RT. However, it also suggests that FV RT has a relatively high RNase H activity, since the starting RNA template must be degraded before the DNA template can be annealed and the strand transfer product synthesized.
RNase H activity.
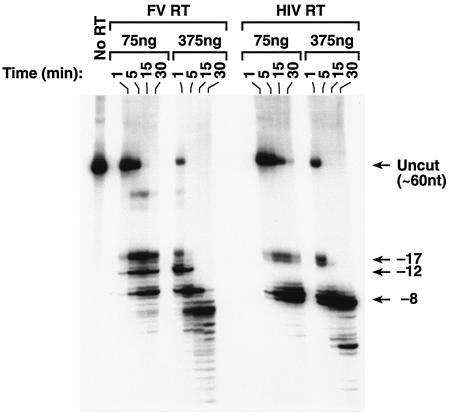
When HIV-1 RT binds an RNA-DNA T/P such as is shown in Fig. 5A, the 3′ end of the DNA primer is located at the polymerase active site and the RNase H active site contacts the RNA template approximately 17 to 18 nt from the polymerase active site (for review, see reference 8). When RNase H cleaves the RNA strand, the initial cleavages occur approximately 17 nt from the polymerase active site. These cleavages have been designated the −17 family of cleavages (8). HIV-1 RT then alters its interactions with the T/P so that the RNase H domain cleaves the RNA strand approximately 8 nt from the 3′ end of the primer (−8 cleavages). We compared the RNase H activity of FV RT to HIV-1 RT using model substrates. For the initial assay, RNase H activity was measured by annealing a 5′-end-labeled RNA to a smaller DNA primer (Fig. 5A). Because the RNase H activity of FV RT had not been previously measured, two concentrations of enzyme were tested (Fig. 6). The labeled RNA substrate was 60 nt in length. As expected, HIV-1 RT produced both the −17 and the −8 family of cleavages (Fig. 6). The RNase H of FV RT cleaved the substrate at sites similar to the −17 and −8 cleavages made by HIV-1 RT (Fig. 6). FV RT also made a third family of cleavages between the −17 and −8 sites, which we have designated −12 (Fig. 6). At high concentrations of FV RT (375 ng), products smaller than 8 nt in length were also detectable; such small products were not seen in the HIV-1 RT digests. There was also a small amount of a larger cleavage product that was detected only in the FV RT reactions that was less than 60 nt in length but was significantly larger than the products generated by the −17 family of cleavages. This product appears to result from cleavages in the poly(A) region of the template RNA; however, the origin of these cleavages is not clear. It should also be noted that HIV-1 RT made two predominant cleavages in both the −17 and the −8 positions, while FV RT has only one predominant cleavage in the −17 and −8 positions; there was one predominant cleavage at the −12 position as well. Although there are similarities, the RNase H cleavage patterns of HIV-1 RT and FV RT are different. To try to understand the basis of these differences, we compared the RNase H sequence of FV RT to MLV RT and to HIV-1 RT (Fig. 7A).
FIG. 6.
RNase H activity of HIV-1 RT and FV RT. The labeled RNA-DNA substrate shown in Fig. 5A was incubated with either HIV-1 RT or FV RT for the indicated lengths of time. Because FV RT had not been previously analyzed in this system, two concentrations of enzyme were tested. The size of intact RNA is 60 nt, as shown in the no-RT lane. The RNA fragments derived from the −17, −12, and −8 families of cleavages are shown.
FIG. 7.
(A) Sequence alignment of the basic loop region. The alignment is based on previous reports (6). A catalytically important aspartic acid residue was chosen as the starting point (amino acids [aa] 498 in HIV-1 RT, aa 583 in MLV RT, aa 669 in FV RT, and aa 70 in E. coli RNase H) and is highlighted. The basic loops for MLV RT and E. coli RNase H are shaded; the RNase H domain HIV-1 RT does not have a basic loop. The basic amino acids in the putative FV RT basic loop are also shaded. (B) Previously reported sequence of a section of the FV RNase H domain (24). (C) The FV RNase H sequence as determined in this study.
The RNase H domain of MLV RT has a basic loop similar to the basic loop of E. coli RNase H (Fig. 7A) (6, 23, 37). This basic loop has a role in binding the nucleic acid substrate (6, 23, 37). The RNase H of HIV-1 RT does not have a related basic loop. The sequence of the RNase H domain of FV RT was compared to the other RT sequences using the RNase H active site amino acids to guide the alignment. The basic loop region of the MLV RNase H domain and E. coli RNase H is shown in Fig. 7A. The most striking feature was the presence of three basic amino acids (-RRR- in the MLV RNase H and -KKR- in E. coli RNase H) near the N terminus of the loop. There are other basic amino acids in the basic loop, but the amino acid positions are not as well conserved between the two proteins. In contrast, the FV RNase H domain only has one basic amino acid at this location (Fig. 7A). The RNase H of FV does have a group of three lysine residues that are in positions similar to two lysine residues in E. coli RNase H (Fig. 7A). The alignment suggests that FV RT has a basic loop, but it appears to be somewhat different than the basic loop in MLV RT and E. coli RNase H.
When sequencing was done in the FV RNase H domain, differences were noted between our sequence (Fig. 7C) and the published sequence (Fig. 7B, Medline loci HSU21247 and NC_001736). Our sequence has one more guanosine at codon 723 and one less adenosine residue at codon 734. In all, 11 amino acids are different in our sequence than in the published sequence, although none of the amino acids appears to be part of the RNase H active site. Our FV RT was derived from the human spumaretrovirus 13 molecular clone, which is infectious (24).
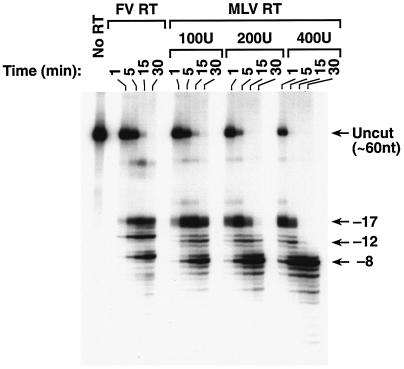
We compared the RNase H activities of FV and MLV RTs. As shown in Fig. 8, MLV RT also had three regions of RNase H cleavage rather than the two seen for HIV-1 RT. In a previous report, a slightly different substrate was used, and the three families of cleavages were not as well resolved (6). Although both RTs make three families of cleavages, there were clear differences in the cleavages made by the FV and MLV RTs. FV RT made one predominant cleavage at each of the three positions. MLV RT made two primary cleavages at the −17 position; one matched the position cleaved by FV RT, while the other was 1 nt closer to the polymerase active site. MLV RT made one predominant cleavage at the −12 and −8 locations, but both products appeared to be 1 nt closer to the polymerase active site than the cleavages made by FV RT (Fig. 8). For both MLV RT and FV RT, it appears that the −17 cleavages are made first, followed by the −12 and −8 cleavages. Taken together, the data suggest that the basic loop of MLV RT has a role in the −12 cleavages and that the basic region of the FV RT RNase H domain also functions as a basic loop, leading to −12 cleavages (see Discussion).
FIG. 8.
RNase H activity of MLV RT and FV RT. The labeled RNA-DNA substrate shown in Fig. 5A was incubated with either MLV RT or FV RT for the indicated lengths of time. Several concentrations of MLV RT were used to simplify the comparison. The RNA products that derive from the −17, −12, and −8 families of cleavages are indicated.
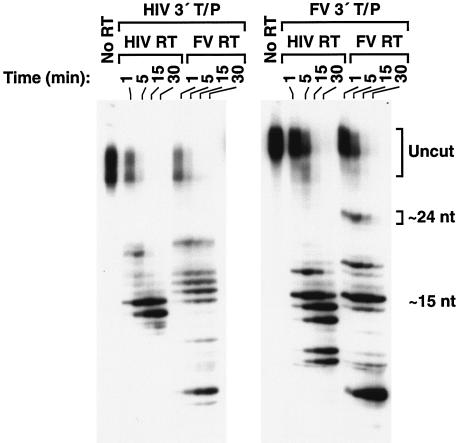
We compared RNase H activities of HIV-1 RT and FV RT with a different set of substrates in which a short 5′-end-labeled RNA was annealed to a longer DNA. The sequences were derived from the PPT/U3 sequences of either the HIV-1 genome (Fig. 5B) or the FV genome (Fig. 5C). While the cleavage that defines the 5′ end of the FV PPT has not yet been determined, the boundary between the PPT and U3 is known. The labeled RNA spans the PPT/U3 boundary in both substrates. As seen in Fig. 9, the enzymes cleaved the substrates differently. When HIV-1-derived sequences were used, HIV-1 RT produced two major cleavages, which were located near the junction between the HIV-1 PPT/U3 sequences (Fig. 5B and 9). FV RT produced a family of products resulting from cleavages within the 5′ end of HIV-1 U3; the products were larger than those produced by HIV-1 RT (Fig. 5B and 9). There were also smaller products resulting from cleavages within the HIV-1 PPT region. When FV-derived sequences were used as the substrate, FV RT produced a large cleavage product that corresponded to a cleavage near the FV PPT/U3 junction and smaller products derived from cleavages within the FV PPT. In contrast, HIV-1 RT produced a series of products resulting from cleavages within the FV PPT region and did not have the larger cleavage product resulting from a cleavage at the FV PPT/U3 junction (Fig. 5C and 9).
FIG. 9.
RNase H activity of HIV-1 RT and FV RT. The 5′-end-labeled RNA-DNA substrates were derived from HIV-1 genome sequences (42) (Fig. 5B) or FV genome sequences (24) (Fig. 5C) around the PPT/U3 boundaries. The substrates were incubated with HIV-1 RT or FV RT for the indicated lengths of time. The distance from the 5′-end label to the HIV PPT-U3 junction is approximately 15 nt, while the distance from the 5′-end label to the FV PPT-U3 junction is approximately 24 nt.
DISCUSSION
As described in the introduction, orthoretroviruses, such as HIV-1, have approximately 50 to 100 Pol proteins per particle (14, 18, 28, 38). In contrast, there are relatively few FV RT molecules in the virion to carry out the conversion of the single-strand FV RNA genome into a double-strand DNA (16, 17). We previously proposed that the RT of FV would need to be more efficient than HIV-1 RT to overcome this numerical disadvantage, and initial characterization of the polymerase activity of FV RT, using a DNA template, showed that FV RT was more processive HIV-1 RT (31). Here, we report additional analysis of the polymerase activity of FV RT and a characterization of the RNase H activity of FV RT.
In polymerase assays, using either RNA or DNA as the template, FV RT produced longer extension products than did HIV-1 RT. The longer products could be the result of two mechanisms: either FV RT catalyzes the polymerization reaction faster than does HIV-1 RT, or FV RT binds more tightly to the T/P than HIV-1 RT and disassociates less frequently. These mechanisms are not mutually exclusive, and it appears that both are involved in efficient polymerization by FV RT. Evidence for greater polymerization activity was found in the DNA-dependent DNA polymerase assay (Fig. 1), the RNA-dependent DNA polymerase assay (Fig. 2), the PPi-dependent pyrophosphorylysis assay (Fig. 3), and the strand-transfer assay (Fig. 4). In all the assays, FV RT and the HIV-1 RT were present at molar excess compared to the T/P. The ability of the enzyme to remain bound to the T/P should not be a critical factor in these assays, since there is an excess of RT ready to bind to any free primer end and begin polymerization. In some respects, this is similar to orthoretrovirus particles, where 50 to 100 RT molecules are available to initiate DNA synthesis from the two tRNAs bound to the dimeric RNA genome. In all assays, FV RT produced larger products than HIV-1 RT, suggesting FV RT polymerizes more efficiently than HIV-1 RT.
The polymerase data also show that FV RT and HIV-1 RT interact with the extended template strand differently. In our previous experiments, FV RT produced a slightly different pattern of products than did HIV-1 RT when DNA was the template (31). We report here that there are subtle differences when RNA is the template (Fig. 2). RTs tend to pause at sites where there are secondary structures. The RNA template used in these experiments was derived from the LTR of HIV-1 and contained the poly(A) hairpin and the TAR hairpin (8). FV RT appeared to pause at a location slightly further away from hairpins than did HIV-1 RT (Fig. 2). One possibility is that the fingers and palm subdomains of FV RT protrude further 5′ of the polymerase active site than do the fingers and palm of HIV-1 RT. This would cause FV RT to encounter a secondary structure sooner than HIV-1 RT, and the pause site would produce a slightly smaller extension product.
There are also differences in the effects of HIV-1 NC on HIV-1 RT and FV RT. As described in the introduction, depending on the amount of secondary structure in the template, NC can help or hurt polymerization by RT. In our assay, high concentrations of NC impaired the ability of HIV-1 RT to extend the primer on an unstructured template. The NC on the RNA template has to be displaced by HIV-1 RT as it is polymerizing, and high concentrations appear to be detrimental to polymerization in our assay. The overall band pattern, however, of the products generated by HIV-1 RT did not appear to be greatly affected by the addition of HIV-1 NC (Fig. 2). This suggests that NC did not completely destabilize the hairpins in the RNA we used. In contrast, HIV-1 NC had little effect on the quantity of DNA synthesized by FV RT; the presence of HIV-1 NC may increase the efficiency of FV RT polymerase. FV Gag protein is minimally processed by FV PR (30) and, therefore, FV does not have a free NC, as do HIV-1 and other orthoretroviruses (8). However, there is a glycine-arginine motif near the C terminus of the FV Gag protein that has nucleic acid binding activity (40); whether this binding activity would be similar to that of HIV-1 NC and increase the efficiency of polymerization by FV RT is not known. We believe that the effects of HIV-1 NC on polymerization by HIV-1 RT and FV RT reflect how well the different RTs bind to the T/P and that FV RT is better able to compete with HIV-1 NC for the nucleic acid substrate.
Pyrophosphorylysis, which has also been called the excision reaction, has been well characterized as a mechanism of resistance to AZT in mutant variants of HIV-1 RT (references 7, 25, and 26 and references therein). If AZTMP has been incorporated at the 3′ terminus of the primer strand, HIV-1 RT can use a pyrophosphate donor to excise the AZTMP, freeing the end of the primer for elongation. Several studies have indicated that, for HIV-1 RT, the in vivo pyrophosphate donor is probably ATP rather than PPi (7, 25, 26). Wild-type HIV-1 RT has the ability to use ATP to excise AZTMP from the primer (with moderate efficiency), indicating that wild-type HIV-1 RT can bind ATP, albeit relatively inefficiently. The presence of AZT resistance mutations, most notably T215Y/F, increases the ability of HIV-1 RT to bind ATP, thus increasing the ability of the mutant enzyme to excise the AZTMP (7, 25, 26). We suggest that ATP-dependent AZTMP excision became a preferred pathway for AZT resistance in HIV-1 RT precisely because wild-type HIV-1 RT has a nascent ability to bind ATP. This could allow for the evolution of resistance via excision: the AZT resistance mutations improve the ATP binding ability of HIV-1 RT and increase the efficiency of the excision reaction. FV is also sensitive to AZT, and it has proven difficult to isolate FV that is resistant to AZT (C. R. Stenbak and M. L. Linial, unpublished results). When we compared the ability of FV RT and HIV-1 RT to excise AZTMP using ATP as the pyrophosphate donor, FV RT was slightly less efficient than HIV-1 RT, even though FV RT has greater polymerase activity. Our data suggest that FV RT can also bind ATP and use it for pyrophosphorylysis, but at a slightly lower level than HIV-1 RT. What is more important than the absolute level of ATP-dependent excision is the comparison of the rate of the forward (polymerase) reaction and the rate of the excision reaction for FV RT. Resistance to AZT requires that the excision of AZT be more efficient, in relative terms, than its incorporation during polymerization. Because FV RT is a very efficient polymerase, resistance via pyrophosphorylysis requires very efficient excision. We previously described a mutation in FV RT (V313 M) that retained a relatively high level of polymerase activity (50% of wild-type FV RT). However, viruses containing the mutant RT replicated poorly and were unable to generate significant levels of full-length DNA products (31). In a similar manner, the modest level of ATP-dependent AZT excision present in wild-type FV RT is probably too low to remove the AZTMP from the end of the primer quickly enough to compensate for the efficient incorporation of AZTTP by FV RT. We propose that the basal level of ATP-dependent AZT excision of wild-type FV RT would need to be significantly higher to favor this mechanism for the development of AZT resistance. These data reinforce the idea that, in vivo, it is unlikely that PPi is the pyrophosphate donor. FV RT is more efficient than HIV-1 RT in excising AZTMP using PPi. This efficient PPi-dependent excision is related to the greater polymerase activity of FV RT (PPi-mediated pyrophosphorylysis is the reverse of the polymerase reaction). However, this efficient PPi-dependent pyrophosphorylysis is not sufficient to give AZT resistance within the cell; FV is sensitive to AZT (27, 31, 32, 34, 41, 42).
There are differences in the cleavages made by the RNase H of FV RT compared to that of HIV-1 RT. The RNase H of HIV-1 RT produces two families of cleavages: the −17 family and the −8 family. With the same T/P substrate, FV RT produces three families of cleavages. These three families of cleavages are similar to the three cleavages made by MLV RT. We suggest that the most likely explanation for the difference between the cleavages made by HIV-1 RT and FV and MLV RT is the presence of an extra nucleic acid binding region, the basic loop, in the RNase H domain of FV RT and MLV RT that is not present in HIV-1 RT (Fig. 7A). However, it is clear from the sequence comparison that the basic loop of FV RT differs considerably from the basic loop of MLV RT (Fig. 7A). The basic loop in MLV RT is important for the polymerase and RNase H activities (6, 23, 37), and it is probable that the basic loop of FV RT also contributes to efficient binding of the nucleic acid substrate. As discussed above, if only a few FV RT proteins were present in the viral particle, tight binding of the nucleic acid substrate would appear to be essential for the proper conversion of the RNA genome into double-stranded DNA.
Others have shown, using RNA-DNA substrates in which the 5′-end-labeled RNA is shorter than the DNA it is annealed to, that the RNase H activity of HIV-1 RT progressively cleaves toward the labeled 5′ end of the RNA (11, 12, 22, 29). We designed similar substrates in which the DNA contains the entire PPT region and part of the U3 region of either HIV-1 or FV. The RNA spans the PPT-U3 boundary. The crystal structure of HIV-1 RT bound to an RNA-DNA substrate containing the PPT region of HIV-1 has distortions within the PPT, which may help to direct HIV-1 RNase H cleavage to the PPT-U3 boundary (35). Distortions may be present in the PPT even in the absence of HIV-1 RT (21). As the RNase H activity of either HIV-1 or FV RT progressively degrades toward the labeled 5′ end of the RNA, it will encounter the PPT. We examined the pattern of cleavages made by HIV-1 RT and FV RT RNase H on the PPT substrates derived from the FV and HIV genomes and asked whether the enzymes were able to specifically cleave at the U3/PPT boundary. Each RT was able to recognize its own PPT with reasonably good specificity; however, neither enzyme cleaved the noncognate PPT appropriately. It is clear from these data that FV RT and HIV-1 RT interact with the same nucleic acid substrates in different ways, and it is likely that the overall structures of the RNA-DNA heteroduplexes that contain the cognate PPT sequences play a major role in determining where the RNase H of the two enzymes cleaves the RNA templates.
A question that is harder to answer is how the overall RNase H activity of FV RT compares to that of HIV-1 RT. A part of the problem is that the answer will depend on the nature of the substrate(s) used to compare the activities. As described above, we believe that the polymerase of FV RT is more active than the polymerase of HIV-1 RT, because it is likely that there are only a few molecules of FV RT present in the virion. RNase H activity is also required for retroviral replication; however, it would appear that HIV-1 virions have a significant excess of RNase H activity (3). Given this fact, is the overall RNase H activity of FV RT significantly greater than that of HIV-1 RT? From the assays we have done, the RNase H activity of FV RT appears to be only slightly higher than that measured for HIV-1 RT. This conclusion is based on the relative rates of cleavage of a standard RNA-DNA template (Fig. 6). The difference between FV and HIV-1 RT RNase H activities does not appear to be as large as the difference between the respective polymerase activities. The idea that there is a significant excess of RNase H activity in HIV-1 RT (and HIV-1 virions) is also supported by the observation that the RNase H activity of HIV-2 RT is much lower than the RNase H activity of HIV-1 RT (36). For this reason, it may not be surprising that the level of RNase H activity of FV RT is not significantly higher than that of HIV-1 RT.
Acknowledgments
Work at FHCRC was supported by grant CA-18282 to M.L.L. C.R.S. was partially supported by NCI training grant CA-09229. Work in S.H.H.'s laboratory was supported by the National Cancer Institute and NIGMS.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, D. N., and M. L. Linial. 1998. The roles of Pol and Env in the assembly pathway of human foamy virus. J. Virol. 72:3658-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, D. N., and M. L. Linial. 1999. Proteolytic activity, the carboxy terminus of Gag, and the primer binding site are not required for Pol incorporation into foamy virus particles. J. Virol. 73:6387-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer, P. L., J. Lisziewicz, F. Lori, and S. H. Hughes. 1999. Analysis of amino acid insertion mutations in the fingers subdomain of HIV-1 reverse transcriptase. J. Mol. Biol. 286:995-1008. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, P. L., H.-Q. Gao, P. K. Clark, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. YADD mutants of human immunodeficiency type 1 and Moloney murine leukemia virus reverse transcriptase are resistant to lamivudine triphosphate (3TCTP) in vitro. J. Virol. 75:6321-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, P. L., H.-Q. Gao, P. Frank, P. K. Clark, and S. H. Hughes. 2001. The basic loop of the RNase H domain of MLV RT is important both for RNase H and polymerase activity. Virology 282:206-213. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2002. Nucleoside analog resistance caused by insertions in the fingers of human immunodeficiency virus type 1 reverse transcriptase involves ATP-mediated excision. J. Virol. 76:9143-9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J. M., S. H. Hughes, and H. E. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 9.Das, A. T., B. Klaver, B. I. F. Klasens, J. L. B. van Wamel, and B. Berkhout. 1997. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 71:2346-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, W. R., S. Gabbara, D. Hupe, and J. A. Peliska. 1998. Actinomycin D inhibition of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase and nucleocapsid protein. Biochemistry 37:14213-14221. [DOI] [PubMed] [Google Scholar]
- 11.DeStefano, J. J., L. M. Mallaber, P. J. Fay, and R. A. Bambara. 1993. Determinants of the RNase H cleavage specificity of human immunodeficiency virus reverse transcriptase. Nucleic Acids Res. 21:4330-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeStefano, J. J., J. V. Cristofaro, S. Derebail, W. P. Bohlayer, and M. J. Fitzgerald-Heath. 2001. Physical mapping of HIV reverse transcriptase to the 5′ end of RNA primers. J. Biol. Chem. 276:32515-32521. [DOI] [PubMed] [Google Scholar]
- 13.Enssle, J., I. Jordan, B. Mauer, and A. Rethwilm. 1996. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc. Natl. Acad. Sci. USA 93:4137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayman, M. J. 1978. Viral polyproteins in chick embryo fibroblasts infected with avian sarcoma leukosis viruses. Virology 85:241-252. [DOI] [PubMed] [Google Scholar]
- 15.Heinkelein, M., M. Schmidt, N. Fischer, A. Moebes, D. Lindemann, J. Enssle, and A. Rethwilm. 1998. Characterization of a cis-acting sequence in the pol region required to transfer human foamy virus vectors. J. Virol. 72:6307-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinkelein, M., J. Thurow, M. Dressler, H. Imrich, D. Neumann-Haefelin, M. O. McClure, and A. Rethwilm. 2000. Complex effects of deletions in the 5′ untranslated region of primate foamy virus on viral gene expression and RNA packaging. J. Virol. 74:3141-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinkelein, M., C. Leurs, M. Rammling, K. Peters, H. Hanenberg, and A. Rethwilm. 2002. Pregenomic RNA is required for efficient incorporation of Pol polyprotein into foamy virus capsids. J. Virol. 76:10069-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamjoon, G. A., R. B. Naso, and R. B. Arlinghaus. 1977. Further characterization of intracellular precursor polyproteins of Rauscher leukemia virus. Virology 78:11-34. [DOI] [PubMed] [Google Scholar]
- 19.Jordan, I., J. Enssle, E. Guttler, B. Mauer, and A. Rethwilm. 1996. Expression of human foamy virus reverse transcriptase involves a spliced pol mRNA. Virology 224:314-319. [DOI] [PubMed] [Google Scholar]
- 20.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvaratskhelia, M., S. R. Budihas, and S. F. Le Grice. 2002. Pre-existing distortions in nucleic acid structure aid polypurine tract selection by HIV-1 reverse transcriptase. J. Biol. Chem. 277:16689-16696. [DOI] [PubMed] [Google Scholar]
- 22.Lener, D., S. R. Budihas, and S. F. J. LeGrice. 2002. Mutating conserved residues in the ribonuclease H domain of Ty3 reverse transcriptase affects specialized cleavage events. J. Biol. Chem. 277:26486-26495. [DOI] [PubMed] [Google Scholar]
- 23.Lim, D., M. Orlova, and S. P. Goff. 2002. Mutations of the RNase H C helix of the Moloney murine leukemia virus reverse transcriptase reveal defects in polypurine tract recognition. J. Virol. 76:8360-8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Löchelt, M., H. Zentgraf, and R. M. Flügel. 1991. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology 184:43-54. [DOI] [PubMed] [Google Scholar]
- 25.Mas, A., M. Parera, C. Briones, V. Soriano, M. A. Martínez, E. Domingo, and L. Menéndez-Arias. 2000. Role of a dipeptide insertion between codons 69 and 70 of HIV-1 reverse transcriptase in the mechanism of AZT resistance. EMBO J. 19:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer, P. R., S. E. Matsuura, D. Zonarich, R. R. Chopra, E. Pendarvis, H. Z. Bazmi, J. W. Mellors, and W. A. Scott. 2003. Relationship between 3′-azido-3′-deoxythymidine resistance and primer unblocking activity in foscarnet-resistant mutants of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 77:6127-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moebes, A., J. Enssle, P. D. Bieniasz, M. Heinkelein, D. Lindemann, D. Bock, M. O. McClure, and A. Rethwilm. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oppermann, H. J., J. M. Bishop, H. E. Varmus, and L. Levintow. 1977. A joint product of the genes gag and pol of avian sarcoma virus: a possible precursor of reverse transcriptase. Cell 12:993-1005. [DOI] [PubMed] [Google Scholar]
- 29.Palaniappan, C., J. K. Kim, M. Wisniewski, P. J. Fay, and R. A. Bambara. 1998. Control of initiation of viral plus strand DNA synthesis by HIV reverse transcriptase. J. Biol. Chem. 273:3808-3816. [DOI] [PubMed] [Google Scholar]
- 30.Pfrepper, K. I., H. R. Rackwitz, M. Schnolzer, H. Heid, M. Löchelt, and F. M. Flügel. 1998. Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J. Virol. 72:7648-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinke, C. S., P. L. Boyer, D. Sullivan, S. H. Hughes, and M. L. Linial. 2002. Mutation of the catalytic domain of the foamy virus reverse transcriptase leads to loss of processivity and infectivity. J. Virol. 76:7560-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenblum, L. L., G. Patton, A. R. Grigg, A. J. Frater, D. Cain, O. Erlwein, C. L. Hill, J. R. Clarke, and M. O. McClure. 2001. Differential susceptibility of retroviruses to nucleoside analogues. Antivir. Chem. Chemother. 12:91-97. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 34.Santillana-Hayat, M., J. Valla, M. Canivet, J. Peries, and J. M. Molina. 1996. Inhibition of the in vitro infectivity and cytopathic effect of human foamy virus by dideoxynucleosides. AIDS Res. Hum. Retrovir. 12:1485-1490. [DOI] [PubMed] [Google Scholar]
- 35.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA/DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaharabany, M., and A. Hizi. 1992. The catalytic functions of chimeric reverse transcriptases of human immunodeficiency viruses type 1 and type 2. J. Biol. Chem. 267:3674-3678. [PubMed] [Google Scholar]
- 37.Telesnitsky, A., S. W. Blain, and S. P. Goff. 1992. Defects in Moloney murine leukemia virus replication caused by a reverse transcriptase mutation modeled on the structure of Escherichia coli RNase H. J. Virol. 66:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogt, V. M., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication—a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]
- 40.Yu, S. F., K. Edelmann, R. K. Strong, A. Moebes, A. Rethwilm, and M. L. Linial. 1996. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 70:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, S. F., M. D. Sullivan, and M. L. Linial. 1999. Evidence that the human foamy virus genome is DNA. J. Virol. 73:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yvon-Groussin, A., P. Mugnier, P. Bertin, M. Grandadam, H. Agut, J. M. Huraux, and V. Calvez. 2001. Efficacy of dideoxynucleosides against human foamy virus and relationship to its reverse transcriptase amino acid sequence and structure. J. Virol. 75:7184-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]