Abstract
Background & objectives:
Colorectal cancer (CRC) is second only to breast cancer as the leading cause of cancer-related deaths in Malaysia. In the Asia–Pacific area, it is the highest emerging gastrointestinal cancer. The aim of this study was to identify single nucleotide polymorphisms (SNPs) and environmental factors associated with CRC risk in Malaysia from a panel of cancer associated SNPs.
Methods:
In this case-control study, 160 Malaysian subjects were recruited, including both with CRC and controls. A total of 768 SNPs were genotyped and analyzed to distinguish risk and protective alleles. Genotyping was carried out using Illumina's BeadArray platform. Information on blood group, occupation, medical history, family history of cancer, intake of red meat and vegetables, exposure to radiation, smoking and drinking habits, etc was collected. Odds ratio (OR), 95% confidence interval (CI) were calculated.
Results:
A panel of 23 SNPs significantly associated with colorectal cancer risk was identified (P<0.01). Of these, 12 SNPs increased the risk of CRC and 11 reduced the risk. Among the environmental risk factors investigated, high intake of red meat (more than 50% daily proportion) was found to be significantly associated with increased risk of CRC (OR=6.52, 95% CI :1.93 - 2.04, P=0.003). Two SNPs including rs2069521 and rs10046 in genes of cytochrome P450 (CYP) superfamily were found significantly associated with CRC risk. For gene-environment analysis, the A allele of rs2069521 showed a significant association with CRC risk when stratified by red meat intake.
Interpretation & conclusions:
In this preliminary study, a panel of SNPs found to be significantly associated with CRC in Malaysian population, was identified. Also, red meat consumption and lack of physical exercise were risk factors for CRC, while consumption of fruits and vegetables served as protective factor.
Keywords: Cytochrome P450, gastrointestinal carcinoma, gene-environment interaction, genetic polymorphism, risk factors, SNPs
Worldwide, colorectal cancer (CRC) is the third most common cancer and its incidence is increasing in East Asia, including Malaysia. The 2007 report of National Cancer Registry shows a total of 2,246 CRC cases registered nationwide, representing 12.3 per cent of all registered cases1. The age standardized incidence rates among men and women were 20.9 and 16.8 per 100,000 population, respectively between 2003 and 20052. A high incidence of cancers of the colon and rectum is consistently observed in populations with a western type diet including countries of Europe, North America, New Zealand and Australia3. Epidemiological evidence indicates that physical inactivity, high prevalence of obesity, and a diet high in red and processed meats may all contribute to CRC. In addition, fibers may have a protective role as suggested in The European Cancer Prevention Organization Intervention Study4.
Cancer is a complex disease involving numerous genes and pathways acting in synergy to exert varied risk effects. The effect of each individual SNP (single nucleotide polymorphism) may or may not be critical. Better insight into the genetic effects on function as seen by the clinical parameters such as tumour phenotype, extent of its spread is important. To date, numerous microarray studies have recognized the importance of genetic polymorphisms in CRC risk5. However, these microarray studies have been predominantly performed in populations of European countries. The demography of CRC in Asian populations is also different from western populations. For Whites and African-Americans, the incidence of CRC in the proximal colon was significantly higher compared with Asians and Pacific Islanders who had a higher incidence of distal cancers6. The specific differences in anatomic location of the tumour may also suggest genetic variables.
There are two distinct genomic instability pathways leading to CRC. The first pathway is chromosomal instability (CIN) pathway also known as the adenoma-carcinoma sequence7. It is characterized by genetic alteration through chromosomal losses and gains (aneuploidy) in adenomatous polyposis coli (APC), kirsten rat sarcoma viral oncogene (KRAS), Small mothus against decapentaplagic (SMAD) and tumour protein-53 (TP53) genes8,9. Chromosome instability appears to be the most frequent cause for sporadic CRC10. However in the second pathway, microsatellite instability (MSI) is the hallmark of Lynch syndrome [Hereditary non-polyposis colorectal cancer (HNPCC)]. MSI pathway is involved with mutations in mismatch-repair genes (MMR)11,12. The primary function of MMR genes is to eliminate base–base mismatches and insertion–deletion loops that arise because of DNA polymerase slippage during DNA replication. It has been hypothesized that SNPs in MMR genes may change gene expression and activity, hence influencing the effectiveness of cancer treatment and survival of patients13.
Carcinogenesis of CRC arises from the substantial accumulation of genetic and epigenetic alterations. The process is also influenced by environmental risk factors. Most genetic association studies in Asia examined the correlation between the cancer and a single or a small number of polymorphic variants14. Thus the goal of the current study was to screen a larger number of polymorphisms and environmental factor that may contribute to the development of CRC in our local setting. To this end, we analyzed 768 SNPs that are known to be involved in the development of various cancers for their potential role in the development of CRC.
Material & Methods
Study design and participants: Based on microarray sample size calculation (http://bioinformatics.mdanderson.org/MicroarraySampleSize/) with 768 genes (SNPs), 99 % confidence interval and 90 per cent power of study, about 40 samples per group (CRC and control) were considered appropriate. But the sample size was increased to 80 samples per group (CRC and control) to improve the power of the study.
The study was conducted in Kuala Lumpur, the capital city of Peninsular Malaysia. In April 2010 and July 2011, 80 incident cases with a primary diagnosis of CRC were identified from two different hospitals in Malaysia: 30 (37.5%) from Hospital Sultan Ismail and 50 (62.5%) from Hospital Kuala Lumpur (HKL). The inclusion criteria for selection of cases were: age >18 yr, Malaysian citizens and confirmed diagnosis with CRC stage I - stage IV. The exclusion criteria for cases were: CRC patients with unknown primary cancer site and secondary metastasis site. A total of 80 controls were selected from Hospital Putrajaya. The inclusion criteria for controls were: Malaysians aged more than 18 yr who had never been diagnosed with any cancer and did not have a family history of any cancers. Controls that had a diagnosis of any malignancy and chronic illnesses before or after recruitment were excluded. Control subjects were matched best to case subjects based on their gender, age, ethnicity and smoking status. This study's procedures and protocols were approved by the Medical Research Ethics Committee, National Institute of Health, Malaysia (Research ID: NMRR-10-652-6473).
Control subjects were healthy subjects who accompanied the real patients to the hospitals and volunteered to be our controls. 16 controls were selected from another hospital because our project was for genotyping 12 different types of cancers and we had enrolled 5-6 hospitals of Malaysia. Controls from hospital putrajaya matched perfectly with our cases.
CRC was staged according to TNM (tumour node metastasis) classification system using information from the surgical file and/or pathology report. Early stage CRC was referred as stage I and stage II. Late stage CRC was referred as stage III and stage IV. Demographic data and information about the known risk factors for CRC were collected through face-to-face interviews. The structured questionnaire included questions regarding age, sex, ethnicity, height and weight, blood group, occupation, personal medical history, family history of cancer, red meat and vegetable intake, exposure to radiation, social history including smoking and drinking habit, and for female subjects, obstetrics and gynaecology history. In accordance with Malaysian dietary habit, instead of measuring food groups in number of servings, study subjects were asked to quantify each food group as an average proportion (%) of their total daily food intake. The subjects were specifically asked to provide information in accordance with their dietary habit 5-10 yr prior to the time of recruitment.
SNP selection: Candidate SNPs were identified following three strategies: (i) focused mainly on SNPs that were reported in Asian populations to be associated with 12 types of cancers, including female breast, colorectal, lung, cervical, leukaemia, nasopharyngeal, gastric, prostate, ovarian, liver, thyroid, and oral cancers; (ii) only polymorphisms and mutations reported among East, Southeast, and South Asian populations were included; and (iii) SNPs were only selected if these have been independently reported at least twice and values of reported odds ratio or relative risk were ≤ 0.9 and ≥ 1.1 for protective and risk alleles, respectively. SNPs were identified via literature search and an established dbSNP (The Single Nucleotide Polymorphism database of The National Center for Biotechnology Information; NCBI) http://www.ncbi.nlm.nih.gov/snp/?term=SNP.
Genotyping: Genomic DNA was extracted using QIAamp DNA Blood Mini Kit (Qiagen, GMBH, Hilden, Germany). The concentration and purity of the isolated DNA was determined by a Smart SpecTM plus spectrophotometer (Bio-rad, Inc., USA), and the integrity of the DNA was assessed by 0.8 per cent agarose gel electrophoresis. Genotyping was carried out according to the Illumina Golden Gate Genotyping Assay (Illumina, Inc. San Diego, USA). A custom designed BeadChip (microarray) containing the probes specific for the selected polymorphisms was employed for the study. The Genome Studio software version 2011.1 (Illumina, Inc. San Diego, USA) was used for automated genotype clustering and calling. SNPs that had call rates of less than 93 per cent, SNPs that deviated from Hardy-Weinberg Equilibrium (HWE, P<0.05) and monoallelic SNPs were excluded from further analysis. By using Bayesian model15, GenCall and GenTrain were assigned to measure how well an individual SNP assay clusters and how well an individual sample fits into a given cluster. A confidence score that bound between 0 and 1 for each genotype call correlates with accuracy of the genotype call. Our threshold limit for both scores was 0.25.
Statistical analysis: Statistical analyses were performed using STATA version 10 (STATA, College Station, TX, USA). P≤0.01 was considered significant. Disparities of demographic characteristics among cases and controls group were assessed using the independent t-test. Multivariate methods based on conditional logistic regression were used to calculate odds ratios (OR) and confidence intervals (95% CI) for the significant SNPs associated with CRC risk and the relation of red meat intake by cytochrome P450 genotype. The association between individual SNPs and CRC risk was assessed by comparing genotype frequencies in cases and controls. OR was calculated by using the most frequent homozygous genotype in controls as the reference for dominant model. OR for recessive model was calculated by using the combination of homozygotes and heterozygotes that have a higher frequency in the controls as reference. SNPs with ORs less than 1 were considered to confer protective effect (OR<1); and SNPs with ORs more than 1 (OR>1) were considered to confer risk of developing CRC. For these analyses with SNP genotypes, false discovery rate estimates (q-values) were determined using the adaptive 2-stage procedure defined by Benjamini and colleagues16 and the Excel-based calculator described by Pike17.
Results
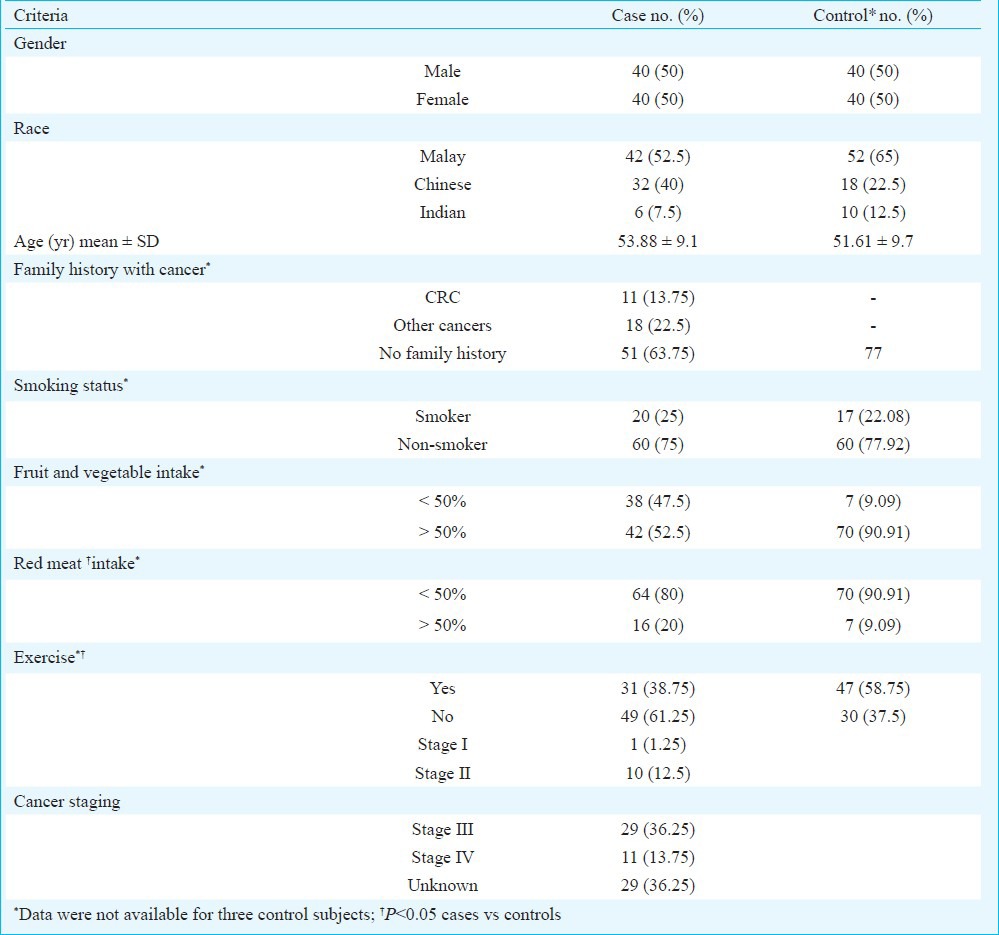
Demographic and lifestyle characteristics of the CRC patients and control subjects are shown in Table I. The mean age for the cancer patients and the controls were 53.88 ± 9.1 and 51.61 ± 9.7 years, respectively. No significant differences were observed in age, gender, ethnicity and smoking status. Among the cancer subjects, 36.25 per cent had family history with cancers. This included first and second degree relatives; 13.75 per cent of the cases had early stages of CRC; and 50 per cent of the overall CRC cases had advanced stages of CRC. Red meat, fruits and vegetable consumption and exercise were significantly associated with colorectal cancer risk relative to controls (P<0.05, data not shown). Conditional logistic regression model showed that a diet with red meat comprising > 50 per cent of total daily dietary intake increased the risk of CRC development [OR=6.52, 95% CI : 93-2.04, P=0.003; meanwhile, high fruits and vegetables intake (at least 50% daily food intake) and physical exercise at least once a week served as protective factors against CRC.
Table I.
Distributions of selected demographic characteristics and potential risk factors of colorectal cancer among cancer cases and controls
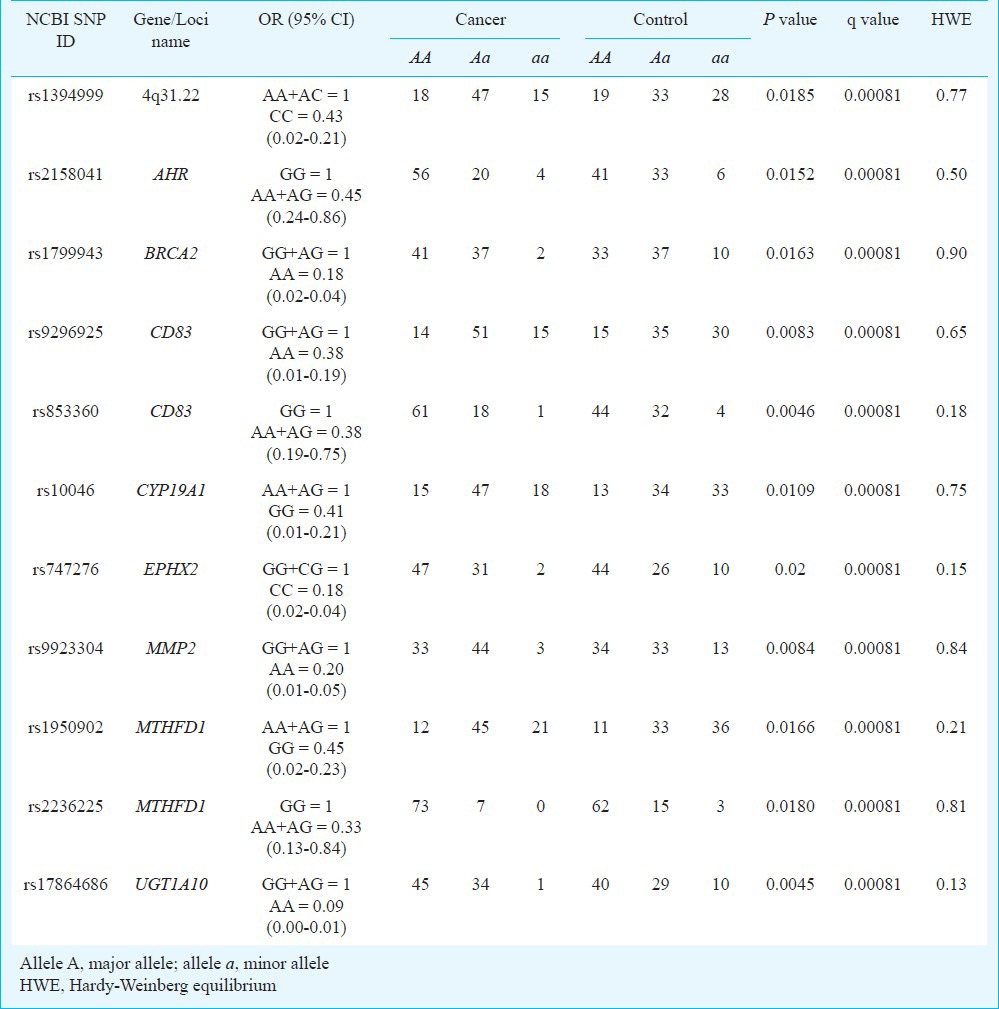
Genotyping was successful for all the 160 samples. For each SNP, the average call rate was 99.4 per cent. SNPs that departed from Hardy-Weinberg equilibrium (P<0.05) were excluded from the analysis. Based on dominant and recessive models, 12 SNPs were found to increase the risk of CRC, and 11 SNPs reduced the risk of CRC (Tables II, III, P<0.01). Among these, a SNP located in genes of cytochrome P450 (CYP) superfamily, rs2069521 of CYP1A2 has been shown to yield the highest significant value with adjusted P value (q-value) 0.00011. rs10046 in CYP19A1 was found to be protective against CRC risk among Malaysian population [OR=0.41 (95% CI: 0.01-0.21), q<0.001].
Table II.
Risk allele frequencies and genotype distribution in patients with colorectal cancer and controls
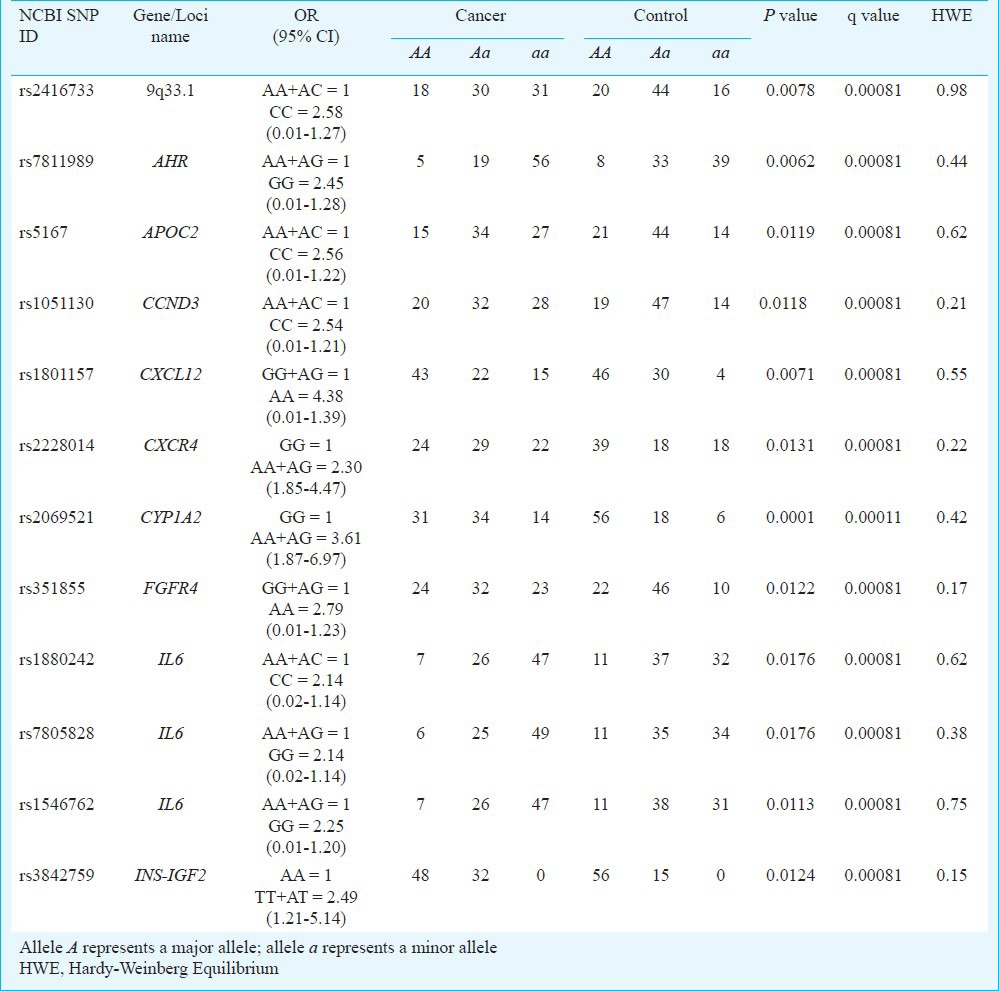
Table III.
Protective allele frequencies and genotype distribution in patients with colorectal cancer and controls
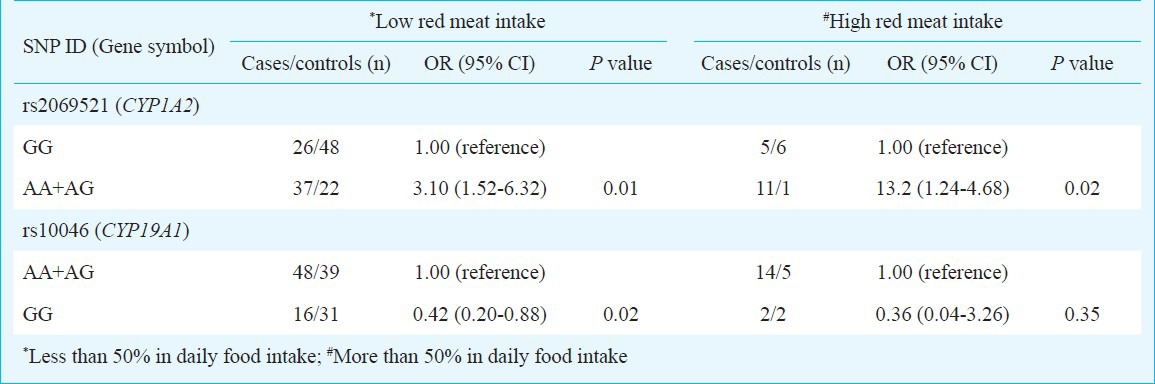
The potential gene-environment interaction of the aforementioned SNPs with red meat consumption and CRC susceptibility was investigated. In Table IV, the association of rs2069521 and rs10046 with CRC risk was stratified according to red meat intake. A significant association (P<0.05) was found between the SNP rs2069521 of CYP1A2 and red meat intake. Among the subjects with low red meat consumption (<50% daily dietary intake), AA and AG genotypes of rs2069521 conferred cancer risk to the Malaysian population (OR=3.10 versus 3.61). However, the genotypes conferred an increased risk of CRC by four-fold among subjects with a high red meat consumption diet (≥ 50% daily dietary intake; OR=13.2; Table IV). This demonstrated that the allele A of rs2069521 might allow CYP1A2 to functionally increase the activation of oxidizing carcinogens, resulting in an increased rate of carcinogenic DNA damage. This effect was further compounded by unhealthy high red meat diet, the source of CYP1A2 substrates.
Table IV.
Odds ratio for interactions between cytochrome P450 genotypes and colorectal cancer risk stratified by red meat intake
Discussion
The present study was conducted to explore the association between cancer-related polymorphisms and environmental factors with the risk of CRC among Malaysian population. A total of 23 SNPs were identified in association with CRC risk (P<0.01, q<0.001). Among these 23 SNPs, two have been reported previously in colorectal cancer association studies in Asian populations, including rs747276 and rs180115718,19. Environmental risk factors such as dietary red meat intake and physical inactivity were identified as major risk factors for CRC, consistent with previously published studies20,21. Our results also showed that the risk conferred by SNP rs2069521 of CYP1A2 was compounded by high daily consumption of red meat.
In Malaysia, Ramadas et al22 have reported that red meat intake affects the risk of colorectal adenoma among Malaysian population. The heme iron in red meat has been proposed to promote CRC, yet the mechanism is still poorly understood. A meta-analysis of prospective cohort studies of colon cancer suggests that catalytic effect of heme iron may result in the formation of carcinogenic N-nitroso compounds (NOC) and the end products of lipid peroxidation, lead to the development of adenoma and carcinoma23.
The SNPs identified in the present study are located on various genes involved in many biochemical pathways, including DNA repair, cell cycle regulation, xenobiotic metabolic process, inflammatory response, angiogenesis and others. Genes involved in DNA repair and cell cycle regulation, such as aryl hydrocarbon receptor (AHR); breast cancer 2, early onset (BRCA2) and excision repair cross-complementing rodent repair deficiency, complementation group 6 (ERCC6) play important roles in maintaining the stability of the human genome. Genetic polymorphisms within these genes may cause the protein expressed to be malfunctioned. This may result in inefficient repair of DNA damaged by the exposure to radiation and chemicals, and thus may contribute to abnormal cellular functioning. As a consequence, it may lead to tumour development, and eventually colorectal cancer.
In addition, genes that are involved in the chemokine activity and inflammatory pathway were also highly represented in our SNPs including chemokine (C-X-C motif) ligand 12 (CXCL12); chemokine (C-X-C motif) receptor 4 (CXCR4); interleukin 6 (IL6) and CD83 molecule (CD83). Our data showed the polymorphisms in CXCL12 (rs1801157) and CXCR4 (rs2228014) genes were significantly associated with CRC susceptibility. The effect of rs1801157 (CXCL12) in colorectal cancer risk has been investigated in Swedish population and the carrying rate of A allele of rs1801157 was found to be higher in colon cancer patients compared to rectal cancer patients24. The molecular CXCR4 pathway has also been shown to be associated with advanced colorectal cancer25. The frequency of GA/AA genotypes of rs2228014 (Ile142Ile) has been shown to be significantly (P<0.001) higher in CRC patients with lymph node metastasis than in CRC patients without lymph node metastasis26. The study has also suggested that rs2228014 polymorphism may increase the expression of SDF-1a mRNA and may be used as a predictive marker of lymph node metastasis in CRC27.
The chemoattraction of leukocytes to inflammatory sites is predominantly involved with chemokines and cytokines. These partially mediate the interaction of leukocytes between tumour cells and stromal cells in tumour initiation and progression. Several evidences have shown pro-inflammatory cytokines regulate cancer cell growth and thereby contribute to tumour promotion and progression28,29. Among these, IL-6 seems to take a center stage in human colorectal cancer development. People with chronic inflammation of the large bowel such as ulcerative colitis, familial adenomatous polyposis have a greater likelihood of developing CRC. The rs1800795 polymorphism of the IL6 gene has been one of the most commonly studied polymorphisms and it has been reported that individuals with the C allele in rs1800795 have reduced risk of developing colorectal cancer29. Enjuanes et al29 have recently described an association of a polymorphism of the IL-6 promoter (rs1546762) with an increased risk for giant cell arteritis.
The mechanism of action of fibroblast growth factors (FGFs) is not clearly described compared to the function of vascular endothelial growth factor (VEGF) in the process of tumour angiogenesis. A significant association between FGFR4 polymorphism (rs351855) and prostate cancer has been identified by Liwei et al30 using a total of 2618 cases and 2305 controls (a meta-analysis study). Current results suggest that role of rs351855 risk variant-expressing cells (T allele) is to either sustain cell–cell adhesion or loose cell–cell adhesion and invade as single cells in promoting tumor expansion31.
The gene for CYP1A2 has been mapped on chromosome 15q24.1 and shares a bidirectional promoter with another cytochrome P450 gene, CYP1A132. CYP1A2 is highly polymorphic and to date, more than 200 SNPs have been deposited in the NCBI database and frequency of these SNPs varies by ethnicity. Polymorphisms in CYP1A2 gene have been studied extensively to determine their potential mechanisms associated with CRC risk33,34. However, none of these have discussed CYP1A2 rs2069521 association with CRC risk. In our study, with high daily red meat consumption, CYP1A2 rs2069521 increased the risk of CRC by four-fold as compared with unstratified population. Increased activity of CYP1A2 has been shown to increase the likelihood of CRC development35. Yet the mechanism by which the variant allele affects the enzymatic activity is still not clear. Our findings demonstrated an example of gene-environment interaction leading to increased cancer risk.
However, these results should be interpreted with caution, as there are limitations in this study. The study sample was small, thus prone to the effects of various biases. These include selection bias as the subjects were not randomized and not equally stratified. There was an element of recall bias as the information collected was solely questionnaire based.
In conclusion, the present study analyzed 768 SNPs in an attempt to investigate association of these variations with colorectal cancer. The results of our study suggest that dietary intake of red meat may be associated with the development of colorectal cancer. As no research studies have been published on CRC risk related to genetic-environmental exposure in Malaysia, our findings serve as a preliminary analysis study. Additionally, our investigations indicated that red meat consumption and the lack of physical exercise were risk factors for colorectal cancer; and increased consumption of fruits and vegetables served as protective factor. The identified polymorphisms may provide insightful link for further genetic or epidemiological studies with an emphasis on functional aspects.
References
- 1.Ariffin OZ, Saleha IT. Kuala Lumpur (Malaysia): Ministry of Health; 2011. Malaysia cancer statistics – Data and figure 2007. [Google Scholar]
- 2.Lim GCC, Rampal S, Halimah Y, editors. Kuala Lumpur (Malaysia): National Cancer Registry; 2008. Cancer incidence in peninsular Malaysia, 2003 - 2005; pp. 1–176. [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Bonithon-Kopp C, Kronborg O, Giacosa A, Rath U, Faivre J. Calcium and fibre supplementation in prevention of colorectal adenoma recurrence: a randomised intervention trial. European Cancer Prevention Organisation Study Group. Lancet. 2000;356:1300–6. doi: 10.1016/s0140-6736(00)02813-0. [DOI] [PubMed] [Google Scholar]
- 5.Abuli A, Fernandez-Rozadilla C, Giraldez MD, Munoz J, Gonzalo V, Bessa X, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. A two-phase case-control study for colorectal cancer genetic susceptibility: candidate genes from chromosomal regions 9q22 and 3q22. Br J Cancer. 2011;105:870–5. doi: 10.1038/bjc.2011.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu X, Chen VW, Martin J, Roffers S, Groves FD, Correa CN, et al. Subsite-specific colorectal cancer incidence rates and stage distributions among Asians and Pacific Islanders in the United States, 1995 to 1999. Cancer Epidemiol Biomarkers Prev. 2004;13:1215–22. [PubMed] [Google Scholar]
- 7.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 8.Sameer AS, Chowdri NA, Syeed N, Banday MZ, Shah ZA, Siddiqi MA. SMAD4--molecular gladiator of the TGF-beta signaling is trampled upon by mutational insufficiency in colorectal carcinoma of Kashmiri population: an analysis with relation to KRAS proto-oncogene. BMC Cancer. 2010;10:300. doi: 10.1186/1471-2407-10-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sameer AS, ul Rehman S, Pandith AA, Syeed N, Shah ZA, Chowdhri NA, et al. Molecular gate keepers succumb to gene aberrations in colorectal cancer in Kashmiri population, revealing a high incidence area. Saudi J Gastroenterol. 2009;15:244–52. doi: 10.4103/1319-3767.56102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worthley DL, Leggett BA. Colorectal cancer: molecular features and clinical opportunities. Clin Biochem Rev. 2010;31:31–8. [PMC free article] [PubMed] [Google Scholar]
- 11.Nissar S, Lone TA, Banday MZ, Rasool R, Chowdri NA, Parray FQ, et al. Arg399Gln polymorphism of XRCC1 gene and risk of colorectal cancer in Kashmir: a case control study. Oncol Lett. 2013;5:959–63. doi: 10.3892/ol.2013.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sameer AS, Nissar S, Qadri Q, Alam S, Baba SM, Siddiqi MA. Role of CYP2E1 genotypes in susceptibility to colorectal cancer in the Kashmiri population. Hum Genomics. 2011;5:530–7. doi: 10.1186/1479-7364-5-6-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wijnen JT, Brohet RM, van Eijk R, Jagmohan-Changur S, Middeldorp A, Tops CM, et al. Chromosome 8q23.3 and 11q23.1 variants modify colorectal cancer risk in Lynch syndrome. Gastroenterology. 2009;136:131–7. doi: 10.1053/j.gastro.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 14.Guo X, Zhang L, Wu M, Wang N, Liu Y, Er L, et al. Association of the DNMT3B polymorphism with colorectal adenomatous polyps and adenocarcinoma. Mol Biol Rep. 2010;37:219–25. doi: 10.1007/s11033-009-9626-z. [DOI] [PubMed] [Google Scholar]
- 15.Morris AP. Direct analysis of unphased SNP genotype data in population-based association studies via Bayesian partition modelling of haplotypes. Genet Epidemiol. 2005;29:91–107. doi: 10.1002/gepi.20080. [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93:491–507. [Google Scholar]
- 17.Pike N. Using false discovery rates for multiple comparisons in ecology and evolution. Methods Ecol Evol. 2011;2:278–82. [Google Scholar]
- 18.Ikeda S, Sasazuki S, Natsukawa S, Shaura K, Koizumi Y, Kasuga Y, et al. Screening of 214 single nucleotide polymorphisms in 44 candidate cancer susceptibility genes: a case-control study on gastric and colorectal cancers in the Japanese population. Am J Gastroenterol. 2008;103:1476–87. doi: 10.1111/j.1572-0241.2008.01810.x. [DOI] [PubMed] [Google Scholar]
- 19.Chan DS, Lau R, Aune D, Vieira R, Greenwood DC, Kampman E, et al. Red and processed meat and colorectal cancer incidence: meta-analysis of prospective studies. PLoS One. 2011;6:e20456. doi: 10.1371/journal.pone.0020456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saebo M, Skjelbred CF, Brekke Li K, Bowitz Lothe IM, Hagen PC, Johnsen E, et al. CYP1A2 164 A-->C polymorphism, cigarette smoking, consumption of well-done red meat and risk of developing colorectal adenomas and carcinomas. Anticancer Res. 2008;28:2289–95. [PubMed] [Google Scholar]
- 21.Takachi R, Tsubono Y, Baba K, Inoue M, Sasazuki S, Iwasaki M, et al. Japan Public Health Center-Based Prospective Study Group. Red meat intake may increase the risk of colon cancer in Japanese, a population with relatively low red meat consumption. Asia Pac J Clin Nutr. 2011;20:603–12. [PubMed] [Google Scholar]
- 22.Ramadas A, Kandiah M. Food intake and colorectal adenomas: a case-control study in Malaysia. Asian Pac J Cancer Prev. 2009;10:925–32. [PubMed] [Google Scholar]
- 23.Dimberg J, Hugander A, Lofgren S, Wagsater D. Polymorphism and circulating levels of the chemokine CXCL12 in colorectal cancer patients. Int J Mol Med. 2007;19:11–5. [PubMed] [Google Scholar]
- 24.Wang SC, Lin JK, Wang HS, Yang SH, Li AF, Chang SC. Nuclear expression of CXCR4 is associated with advanced colorectal cancer. Int J Colorectal Dis. 2010;25:1185–91. doi: 10.1007/s00384-010-0999-1. [DOI] [PubMed] [Google Scholar]
- 25.Chang SC, Lin PC, Yang SH, Wang HS, Li AF, Lin JK. SDF-1alpha G801A polymorphism predicts lymph node metastasis in stage T3 colorectal cancer. Ann Surg Oncol. 2009;16:2323–30. doi: 10.1245/s10434-009-0501-x. [DOI] [PubMed] [Google Scholar]
- 26.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H, Ouyang W, Huang C. Inflammation, a key event in cancer development. Mol Cancer Res. 2006;4:221–33. doi: 10.1158/1541-7786.MCR-05-0261. [DOI] [PubMed] [Google Scholar]
- 28.Theodoropoulos G, Papaconstantinou I, Felekouras E, Nikiteas N, Karakitsos P, Panoussopoulos D, et al. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol. 2006;12:5037–43. doi: 10.3748/wjg.v12.i31.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enjuanes A, Benavente Y, Hernandez-Rodriguez J, Queralt C, Yague J, Jares P, et al. Association of NOS2 and potential effect of VEGF, IL6, CCL2 and IL1RN polymorphisms and haplotypes on susceptibility to GCA--a simultaneous study of 130 potentially functional SNPs in 14 candidate genes. Rheumatology. 2012;51:841–51. doi: 10.1093/rheumatology/ker429. [DOI] [PubMed] [Google Scholar]
- 30.Liwei L, Chunyu L, Jie L, Ruifa H. Association between fibroblast growth factor receptor-4 gene polymorphism and risk of prostate cancer: a meta-analysis. Urol Int. 2011;87:159–64. doi: 10.1159/000329069. [DOI] [PubMed] [Google Scholar]
- 31.Rowe RG, Weiss SJ. Navigating ECM barriers at the invasive front: the cancer cell-stroma interface. Annu Rev Cell Dev Biol. 2009;25:567–95. doi: 10.1146/annurev.cellbio.24.110707.175315. [DOI] [PubMed] [Google Scholar]
- 32.Jorge-Nebert LF, Jiang Z, Chakraborty R, Watson J, Jin L, McGarvey ST, et al. Analysis of human CYP1A1 and CYP1A2 genes and their shared bidirectional promoter in eight world populations. Hum Mutat. 2010;31:27–40. doi: 10.1002/humu.21132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi M, Otani T, Iwasaki M, Natsukawa S, Shaura K, Koizumi Y, et al. Association between dietary heterocyclic amine levels, genetic polymorphisms of NAT2, CYP1A1, and CYP1A2 and risk of colorectal cancer: a hospital-based case-control study in Japan. Scand J Gastroenterol. 2009;44:952–9. doi: 10.1080/00365520902964721. [DOI] [PubMed] [Google Scholar]
- 34.Bethke L, Webb E, Sellick G, Rudd M, Penegar S, Withey L, et al. Polymorphisms in the cytochrome P450 genes CYP1A2, CYP1B1, CYP3A4, CYP3A5, CYP11A1, CYP17A1, CYP19A1 and colorectal cancer risk. BMC Cancer. 2007;7:123. doi: 10.1186/1471-2407-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sachse C, Bhambra U, Smith G, Lightfoot TJ, Barrett JH, Scollay J, et al. Colorectal Cancer Study Group. Polymorphisms in the cytochrome P450 CYP1A2 gene (CYP1A2) in colorectal cancer patients and controls: allele frequencies, linkage disequilibrium and influence on caffeine metabolism. Br J Clin Pharmacol. 2003;55:68–76. doi: 10.1046/j.1365-2125.2003.01733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


