Abstract
The cum1 and cum2 mutations of Arabidopsis thaliana inhibit cucumber mosaic virus (CMV) multiplication. In cum1 and cum2 protoplasts, CMV RNA and the coat protein accumulated to wild-type levels, but the accumulation of the 3a protein of CMV, which is necessary for cell-to-cell movement of the virus, was strongly reduced compared with that in wild-type protoplasts. In cum2 protoplasts, the accumulation of turnip crinkle virus (TCV)-related RNA and proteins was also reduced. Positional cloning demonstrated that CUM1 and CUM2 encode eukaryotic translation initiation factors 4E and 4G, respectively. Unlike most cellular mRNA, the CMV RNA lacks a poly(A) tail, whereas the TCV RNA lacks both a 5′-terminal cap and a poly(A) tail. In vivo translation analyses, using chimeric luciferase mRNA carrying the terminal structures and untranslated sequences of the CMV or TCV RNA, demonstrated that these viral untranslated sequences contain elements that regulate the expression of encoded proteins positively or negatively. The cum1 and cum2 mutations had different effects on the action of these elements, suggesting that the cum1 and cum2 mutations cause inefficient production of CMV 3a protein and that the cum2 mutation affects the production of TCV-encoded proteins.
Most eukaryotic cellular mRNA has a 5′-terminal cap (m7GpppN-) and a poly(A) tail, which lead to increased translation efficiency (11; reviewed in references 12 and 29) and increased mRNA stability (reviewed in reference 32). The cap structure serves as the binding site for eukaryotic translation initiation factor 4F (eIF4F). eIF4F consists of two subunits: eIF4E cap-binding subunit and eIF4G (reviewed in reference 14). In plants, an isoform of eIF4F, designated eIFiso4F, has been identified. eIFiso4F consists of an eIFiso4E cap-binding subunit and eIFiso4G (reviewed in reference 4). In vitro eIF4F promotes the translation of mRNA with stable secondary structures in the 5′ untranslated regions (UTRs) more efficiently than does eIFiso4F (13). Other functional differences between eIF4F and eIFiso4F have also been reported (9, 13, 21, 23). eIF4G and eIFiso4G interact not only with eIF4E and eIFiso4E, respectively, but also with eIF4A, eIF3, and the poly(A)-binding protein. eIF3 recruits the 40S ribosomal subunit, whereas eIF4A is a DEAD-box RNA helicase that unwinds the 5′ UTR of mRNA to facilitate binding of the 40S ribosomal subunit to the 5′ UTR. After binding to the 5′UTR, the 40S ribosomal subunit migrates in a 5′-to-3′ direction to the initiation codon, where the 60S ribosomal subunit binds and translation begins (reviewed in references 4 and 14). Through the association of the poly(A) tail, the poly(A)-binding protein, eIF4G, eIF4E, and the 5′ cap, capped, polyadenylated mRNA is circularized (36; reviewed in reference 29). This mRNA circularization is thought to contribute to synergistic translation enhancement by the cap and poly(A) structures.
The mRNAs of many RNA viruses lack either the 5′ cap, the 3′ poly(A) tail, or both, but they are stable and efficiently translated in the cytoplasm of host cells through mechanisms that have evolved for each virus (reviewed in reference 12). For example, the genomic RNAs of cucumber mosaic virus (CMV), tobacco mosaic virus (TMV), and brome mosaic virus have a 5′ cap but lack a 3′ poly(A) tail, and their 3′ UTRs contain, instead, tRNA-like structures. The 3′ UTRs of the TMV and brome mosaic virus RNAs contain sequence elements that increase translation efficiency (10, 20; reviewed in reference 12). The genomic RNAs of some plant viruses, including barley yellow dwarf virus (BYDV), turnip crinkle virus (TCV), tomato bushy stunt virus, and satellite tobacco necrosis virus, are uncapped and nonpolyadenylated (1, 7, 25, 37). The translation of BYDV RNA depends strongly on a cap-independent translation enhancer (TE) sequence in the 3′ UTR, and eIF4F is involved in this 3′ TE-enhanced translation in vitro (35). The 5′ UTR of the BYDV RNA is also necessary for the 3′ TE to function (35). TCV, tomato bushy stunt virus, and satellite tobacco necrosis virus RNAs also contain cap-independent TEs in their 3′ UTRs, and their 5′ UTRs are also necessary for efficient translation, at least for the TCV and satellite tobacco necrosis virus RNAs (7, 25, 37).
Virus multiplication depends on the host translation system and many other host functions. The loss of one such host factor results in resistance to the virus. The resistance is expected to be genetically recessive in this case. In fact, several wild-type genes corresponding to recessive mutations that confer resistance to viruses have been identified, and it is thought that they are required for viral multiplication (34, 38). For example, recessive genetic traits of independent plant species that confer resistance against potyviruses have turned out to correspond to mutations in genes encoding eIFiso4E or eIF4E (9, 21, 23, 27). Several lines of experimental evidence indicate that eIFiso4E or eIF4E plays an important, but as yet unrevealed, role in potyvirus infection by interacting with VPg, a small polypeptide covalently attached to the 5′ terminus of the potyvirus RNA (30).
Two recessive mutations of Arabidopsis thaliana, cucumovirus multiplication 1 (cum 1) and cum2, each reduce the accumulation of CMV in plants. Since they do not drastically affect the multiplication of CMV RNA in protoplasts, it has been suggested that the cum1 and cum2 mutations inhibit the cell-to-cell movement of CMV (39, 40). Comparison of the multiplications of a crucifer-infecting tobamovirus (TMV-Cg) and TCV, which belong to different genera (Tobamovirus and Carmovirus, respectively), with that of CMV (genus Cucumovirus) revealed that these mutations do not affect the multiplication of TMV-Cg but that the cum2 mutation does affect the multiplication of TCV (39, 40). Here, we report that the CUM1 and CUM2 genes encode eIF4E and eIF4G, respectively.
MATERIALS AND METHODS
Plants and viruses.
A. thaliana (L.) Heynh ecotypes Columbia (Col-0) and Landsberg erecta (Ler) were used as wild-type ecotypes. The cum1 and cum2 mutants are in the Col-0 background (39, 40). CMV-Y, TCV-B, and TMV-Cg were described by Yoshii et al. (39).
Mapping and identification of the cum1 and cum2 mutations.
The cum1 and cum2 mutations were mapped by using 1750 and 1958 F2 plants, respectively, which were generated by genetic crosses between the respective mutants (Col-0 background), and Ler wild-type plants (details are available on request). The CUM genotypes of F2 plants were determined by measuring the accumulation of viral coat proteins (CPs) in 10 to 18 F3 plants for each F2 plant. In some cases, the CUM genotypes were confirmed in the F4 generation. The cleaved amplified polymorphic sequence (CAPS) markers g4539, AG, and GL1; the simple sequence length polymorphism marker nga112; and the restriction fragment length polymorphism marker mi32 have been described by the Arabidopsis Information Resource (http://www.arabidopsis.org). The yeast artificial chromosomes (YACs) EW2E9, EW8D1, and CIC1E8L and the bacterial artificial chromosomes (BACs) F15J5 and F27H5 were obtained from the Arabidopsis Biological Resource Center at Ohio State University. The restriction fragment length polymorphism markers EW2E9L, EW8D1L, and CIC1E8L were prepared from the corresponding YACs by using the thermal asymmetric interlaced-PCR method (22). Restriction digestion with EcoRV, AccII, and AccI was used to generate polymorphic fragments between Col-0 and Ler for EW2E9L, EW8D1L, and CIC1E8L, respectively. The CAPS markers T2O9DraI and F27H5HaeIII were created based on the genomic sequence information for Col-0 and Ler in the Arabidopsis Information Resource. Digestion of PCR products obtained by using primer sets 5′-GGGCCCTATTTCGACATTTT-3′ and 5′-TCGGAAATCGTTCCTGTTTC-3′ for T2O9DraI and 5′-GGCAACATGGAAGCTAGTTCCATTGGTATACC-3′ and 5′-GATGGAAGAAATAGGGCTGTCTCGAAG-3′ for F27H5HaeIII with DraI and HaeIII, respectively, generated polymorphic DNA fragments between Col-0 and Ler.
To find the cum1 and cum2 mutations, 1- to 2-kbp DNA fragments were amplified from the mapped regions of the respective mutant genomic DNA by PCR with specific primers designed from the wild-type (Col-0) sequence in the Arabidopsis Information Resource, purified from gels, and directly sequenced by using a 377 DNA sequencer (Applied Biosystems) and a BigDye terminator sequencing kit (Perkin-Elmer). Information on the primers used to sequence the mutant genomes is available on request. The mutant genome sequences were compared with the wild-type sequence in the database to find base substitutions. The wild-type nucleotides at the candidate mutation sites were confirmed by direct sequencing of relevant DNA fragments that were amplified from the wild-type genomic DNA by PCR.
Complementation analysis of the cum1 and cum2 mutations.
The T-DNA clone pRK-1 was constructed by subcloning the 6.5-kb XbaI fragment from the BAC F15J5 into the pCLD04541 vector (GenBank accession no. AF184978). The T-DNA clone pKT-1 was constructed by subcloning the 15-kb SpeI fragment from the BAC F27H5 into the pCLD04542 vector. T-DNA clones were electroporated into Agrobacterium tumefaciens C58C1 (pGV2260), and these strains were used to transform A. thaliana plants by the vacuum infiltration method (2). To confirm that the entire T-DNA fragments were integrated into the mutant genomes, the junction regions between each insert DNA and the cloning site were amplified with the specific primer sets.
Determination of the CUM1 and CUM2 mRNA sequences.
The entire sequences of the CUM1 and CUM2 mRNAs were determined by 5′ and 3′ rapid amplification of cDNA ends with a SMART PCR cDNA synthesis kit (Clontech) and a CAP site cDNA kit (Nippon Gene), followed by DNA sequencing.
Antisera and quantification of viral proteins.
cDNA fragments corresponding to subregions of the CMV-Y 1a protein (amino acids 473 to 748) and the TMV-Cg 130,000-molecular-weight (130K) protein (amino acids 643 to 1103) were cloned into the pGEX5X-1 (Amersham-Pharmacia) and pET16b (Novagen) vectors, respectively. For CMV-Y 3a, TMV-Cg 30K, and TCV-B p28, the entire coding regions were cloned into the pET3b (Novagen), pGEX6P-1 (Amersham-Pharmacia), and pET-32c (Novagen) vectors, respectively. Expression, protein purification, and production of rabbit antiserum for the TMV-Cg 130K fusion protein were performed as described for the anti-TMV-L 130K protein serum by Hagiwara et al. (16). The other fusion proteins were expressed in Escherichia coli BL21 strains (Novagen) as inclusion bodies, purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, eluted from polyacrylamide gel slices, and injected into rabbits. Rabbit antiserum against TMV-Cg CP was prepared by using gel-purified CP from virus particles. Rabbit antisera against CMV-Y 2a, the CMV-D virion, and the TCV-M virion were obtained from C. Masuta, (American Type Culture Collection, Manassas, Va.) and A. E. Simon, respectively. Mouse antiserum against TCV p8 was obtained from T. J. Morris. Immunoblot analysis was performed with the ECL Plus Western Blotting System (Amersham-Pharmacia) according to the manufacturer's instructions.
In vivo translation assay.
DNA fragments corresponding to the 5′ UTRs of RNA3 and RNA4 were PCR amplified from an infectious clone for CMV-Y RNA3 obtained from C. Masuta, using primer sets 5′-GGGGTACCGTAATCTAACCACCTGTGTGT-3′-5′-CATGCCATGGAACCTTGGAAAGCCATGCCTCG-3′ and 5′-GGGGTACCGTTATTGTCTACTGACTATATAGA-3′-5′-CATGCCATGGATTCAGATTTGTCCATGACTCGA-3′, respectively. The PCR products were digested with KpnI and NcoI (sites are underlined in the primer sequences) and subcloned between the corresponding restriction sites of the pSP-luc+ vector (Promega) to obtain pMP-5′ and pCP-5′. From the pSP-luc+ vector, the AatII-XbaI fragment containing the SP6 promoter, the 5′ UTR, and the luc+ coding region was transferred to the corresponding AatII-XbaI region of the pSP64Poly(A) vector (Promega) to obtain pV-luc+. DNA fragments corresponding to the 3′ UTRs of CMV-Y RNA3 and RNA4 were amplified from the infectious clone of CMV-Y RNA3 by PCR with the forward primers 5′-GCTCTAGACACTTTGGTGCGTATTAGTAT-3′ and 5′-GCTCTAGATTCCGTGTTCCCAGAATCCTCCCTCCGATC-3′, respectively, and the reverse primer 5′-CCGGAATTCTGGTCTCCTTTTGGAGGCCCCCACGAA-3′. The fragments were digested with XbaI and EcoRI (sites are underlined in the primer sequences) and were cloned between the corresponding restriction sites of the pSP-luc+ vector to create pMP-3′ and pCP-3′. The XbaI-EcoRI fragments of pMP-3′ or pCP-3′ containing the 3′ UTR sequences were introduced into pMP-5′ or pCP-5′ by using the corresponding restriction sites to create pMP-5′+3′, pCP-5′+3′, or pMP-5′+CP-3′.
cDNA fragments of the 5′ and 3′ portions of the TCV-B genomic RNA (nucleotides 1 to 90 and 2247 to 4050 of the sequence under GenBank accession no. M22445 [5]) were amplified from TCV-B virion RNA by reverse transcription-PCR with the specific primer sets 5′-CGGGGTACCGGTAATCTGCAAATCCCTGCA-3′-5′-CATGCCATGGCTGTGTTGAGTGTGTGTAGAAG-3′and 5′-CCGCTCGAGGGTTCGGCTACACTCCAGA-3′-5′-GATCCCCGGGCAGGCCCCCCCCCCGCG-3′, respectively. The PCR products were digested with KpnI plus NcoI and XhoI plus SmaI (the sites are underlined in the primer sequences) and inserted between the corresponding restriction sites of the pV-luc+ and pGEM7Zf(+) vectors to obtain p28/88-5′ and pTCV-B1, respectively. The pTCV-B1 sequence differed from the sequence under GenBank accession no. M22445 at nine positions (details are available on request). A DNA fragment corresponding to the 5′ UTR of TCV subgenomic RNA2 (nucleotides 2605 to 2769) was amplified by PCR from pTCV-B1 by using the primer set 5′-CGGGGTACCGGTAATATATGCTTTCTACAACTCTCTC-3′ and 5′-CATGCCATGGACTTCCGGACTCTAGGATCATT-3′. The PCR product was digested with KpnI and NcoI (the sites are underlined in the primer sequences) and cloned between the corresponding restriction sites of pV-luc+ to obtain p38-5′. DNA fragments corresponding to the 3′ UTR of the TCV genomic RNA (nucleotides 2390 to 4050) and subgenomic RNA2 (nucleotides 3798 to 4050) were prepared by PCR with the forward primers 5′-GCTCTAGAGCGACAGCGACGCAACAG-3′ and 5′-GCTCTAGATACGGTAATAGTGTAGTCTTCTCATCT-3′, respectively, and the M13 reverse primer of the sequence 5′-CAGGAAACAGCTATGAC-3′. PCR products were digested with XbaI (the site is underlined in the primer sequences) and SmaI (present in the multicloning site) and inserted between corresponding restriction sites in the pV-luc+ vector to obtain p28/88-3′ and p38-3′. The XbaI-SmaI TCV 3′ UTR fragments were also inserted between the corresponding restriction sites of p28/88-5′ and p38-5′ to obtain p28/88-5′+3′ and p38-5′+3′, respectively. The Renilla luciferase (RLUC) plasmid, a derivative of pSP64Poly(A), was described by Chiba et al. (6).
Uncapped and capped RNAs were synthesized from linearized plasmids by using the RiboMax (Promega) and AmpliCap (Epicentre Technologies, Madison, Wis.) kits, respectively. Preparation of protoplasts and RNA transfection were carried out essentially as described by Yoshii et al. (39). Briefly, 1.0 × 106 protoplasts were electroporated with 20 pmol each of firefly luciferase (FLUC) and RLUC RNAs at 475 V/cm with a capacitance of 1,000 μF. Protoplasts were cultured at 23°C for 6 h in darkness and harvested. FLUC and RLUC activities were measured by using a dual-luciferase reporter assay system (Promega). RLUC activity per milligram of protein in cum1 protoplasts was significantly lower than that in wild-type protoplasts (P < 0.05 by Student's paired t test; approximately 70% of the wild-type level), while RLUC activity in cum2 protoplasts was similar to that in wild-type protoplasts. Each electroporation was repeated at least three times with protoplasts that were prepared independently.
To determine the genotype of the cum1 locus, the 400-bp PCR fragment, amplified from the genomic DNA by using primers 5′-TTGGGGAAGTTCCTTGCGACCCG-3′ and 5′-AGTCATCACGAAAGTGTTTGCTCAAAT-3′, was digested with MnlI. The fragment derived from the wild-type (Col-0) genome was cleaved into 258-, 94-, 37-, and 11-bp fragments, while the fragment from the cum1 genome was cleaved into 258-, 105-, and 37-bp fragments. To determine the genotype of the cum2 locus, the 1,445-bp PCR fragment, amplified from the genomic DNA by using primers 5′-CCTCCTAAAGTTTGCAGACCTGTG-3′ and 5′-TGTCATCAAATCGAATGTCCTGATTACT-3′, was digested with HaeIII. The fragment derived from the wild-type (Col-0) genome was cleaved into 1,213- and 232-bp fragments, while the fragment from the cum2 genome was not digested with HaeIII.
Nucleotide sequence accession numbers.
The nucleotide sequences of the CUM1 and CUM2 mRNAs have been deposited in the GenBank database (accession no. AB107249 and AB107250).
RESULTS
Map-based cloning of CUM1 and CUM2.
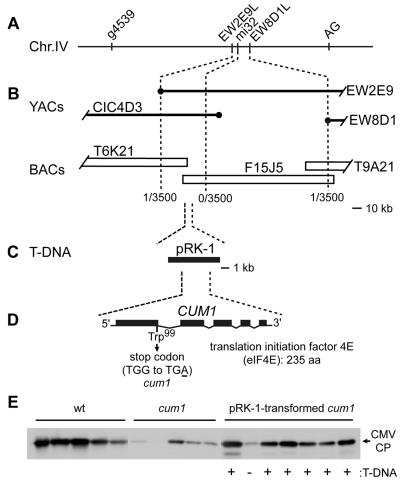
The cum1 mutation was previously mapped between CAPS markers g4539 and AG on A. thaliana chromosome IV (39). Further fine mapping analyses located the cum1 mutation within a 65-kb region between DNA markers EW8D1L and EW2E9L (Fig. 1A and B). Determination of an 11-kb sequences within the 65-kb region of the cum1 genome and comparison with the corresponding wild-type sequences identified a single-base substitution of G to A within the coding region of the At.EIF4E1 (At4g18040) gene encoding eIF4E (26) (GenBank accession no. AB107250). This base substitution changes the 99th tryptophan codon to a nonsense codon (Fig. 1D).
FIG. 1.
Map-based cloning of CUM1. (A) Map around the CUM1 locus on chromosome IV (not drawn to scale). DNA markers are indicated above the chromosome. (B) YAC and BAC contiguous DNA sequences encompassing the CUM1 locus. Circles represent the YAC end probes used for mapping. The numbers of chromatids recombined between the marker and the CUM1 locus per the numbers of chromatids examined are shown. (C) Position of the T-DNA clone pRK-1, which was tested for complementation ability. (D) Exon-intron structure of the CUM1 gene, which encodes eIF4E, and location of the cum1-1 mutation. Filled boxes represent exons, and diagonal lines represent introns. aa, amino acids. (E) Complementation of the cum1-1 mutation by the T-DNA clone pRK-1. T2 plants derived from a single representative pRK-1-transformed cum1-1 mutant plant, wild-type Col-0 plants (wt), and cum1-1 mutant plants were inoculated with CMV-Y. At 48 hpi, inoculated leaves were collected for immunoblotting analysis to assess the accumulation of the CMV-Y CP. Each lane represents one independent plant. The position of the CMV-Y CP is indicated to the right. The presence (+) or absence (−) of the T-DNA insert in each T2 plant is indicated at the bottom.
To confirm that the G-to-A substitution resulted in the cum1 phenotype, the T-DNA clone pRK-1, which harbors the complete At.EIF4E1 gene (Fig. 1C), was stably transformed into the cum1 mutant. Restoration of CMV CP accumulation was observed in inoculated leaves of pRK-1-transformed cum1 mutants (Fig. 1E). No such restoration was observed in cum1 plants transformed with a wild-type genomic DNA fragment that overlaps the pRK-1 insert but does not contain the complete At.EIF4E1 gene (data not shown). The cum1 mutation was also complemented by the introduction of At.EIF4E1 cDNA under the control of the cauliflower mosaic virus 35S RNA promoter (data not shown). These results demonstrate that CUM1 encodes eIF4E.
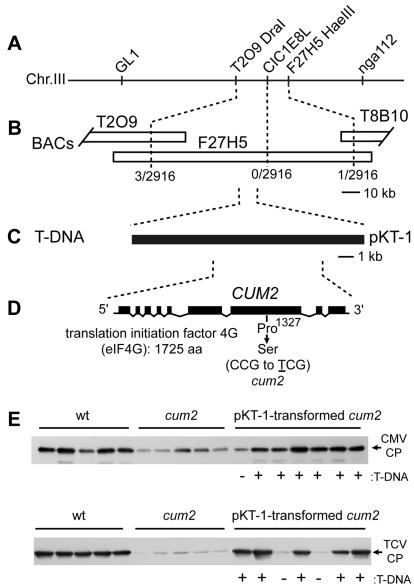
The cum2 mutation was previously mapped between DNA markers GL1 and nga112 on A. thaliana chromosome III (40). Further fine mapping analyses located the cum2 mutation in an 80-kb region between DNA markers T2O9DraI and F27H5HaeIII (Fig. 2A and B). Determination of a 45-kb sequence within the 80-kb region of the cum2 genome and comparison with the corresponding wild-type sequences identified a single-base substitution of C to T within the coding region of the gene encoding eIF4G (At3g60240) (Fig. 2D). Reverse transcription-PCR and 5′ and 3′ rapid amplification of cDNA ends showed that the eIF4G gene contains nine exons and the corresponding mRNA contains 5,451 nucleotides [with the exception of a poly(A) tail] and an open reading frame that encodes a polypeptide of 1,725 amino acids (Fig. 2D). In the cum2 mutant, the 1,327th proline codon is replaced with a serine codon.
FIG. 2.
Map-based cloning of CUM2. (A) Map around the CUM2 locus on chromosome III (not drawn to scale). DNA markers are indicated above the chromosome. (B) BAC contiguous DNA sequence that covers the CUM2 locus. Open boxes represent BAC clones. The numbers of chromatids recombined between the marker and the CUM2 locus per the numbers of chromatids examined are shown below the BACs. (C) Position of the T-DNA clone pKT-1, which was tested for complementation ability. (D) Exon-intron structure of the CUM2 gene, which encodes eIF4G, and location of the cum2-1 mutation. Filled boxes represent exons, and diagonal lines represent introns. aa, amino acids. (E) Complementation of the cum2-1 mutation by the T-DNA clone pKT-1. T2 plants derived from a single, representative pKT-1-transformed cum2-1 mutant plant, the wild-type Col-0 plants (wt), and cum1-1 mutant plants were inoculated with CMV-Y or TCV-B. At 48 hpi, inoculated leaves were collected for immunoblotting analysis to assess the accumulation of the CMV-Y and TCV-B CPs. Each lane represents one independent plant. The positions of the CMV-Y and TCV-B CPs are indicated to the right. The presence (+) or absence (−) of the T-DNA insert in each T2 plant is indicated at the bottom.
To confirm that the C-to-T substitution results in the cum2 phenotype, the T-DNA clone pKT-1, which harbors the complete eIF4G gene (Fig. 2C), was stably transformed into the cum2 mutant. Inoculation of leaves of pKT-1-transformed cum2 mutants with CMV or TCV led to accumulation of the CPs of these viruses to wild-type levels (Fig. 2E). Accumulation of the CPs was not observed in cum2 plants that were transformed with a wild-type genomic DNA fragment that overlaps the pKT-1 insert but does not contain the complete eIF4G gene (data not shown). These results suggest that the C-to-T mutation in the eIF4G gene inhibits the multiplication of both CMV and TCV. Similarly, plants that are homozygous for T-DNA insertions in the eIF4G gene display reduced multiplication of CMV and TCV, as in the cum2 mutant (data not shown).
Reduced accumulation of CMV 3a movement protein in mutant protoplasts.
CMV accumulation is delayed in cum1 and cum2 mutant leaves (39, 40). In protoplasts of these mutants, however, the accumulation patterns of CMV-related RNA and its CP are similar to those in wild-type Col-0 protoplasts (39, 40). To understand how the cum1 and cum2 mutations delay CMV accumulation in plants, we further investigated the effect of these mutations on CMV-encoded protein accumulation.
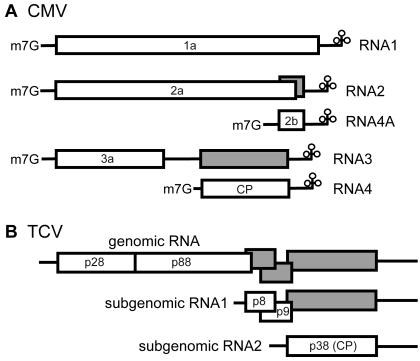
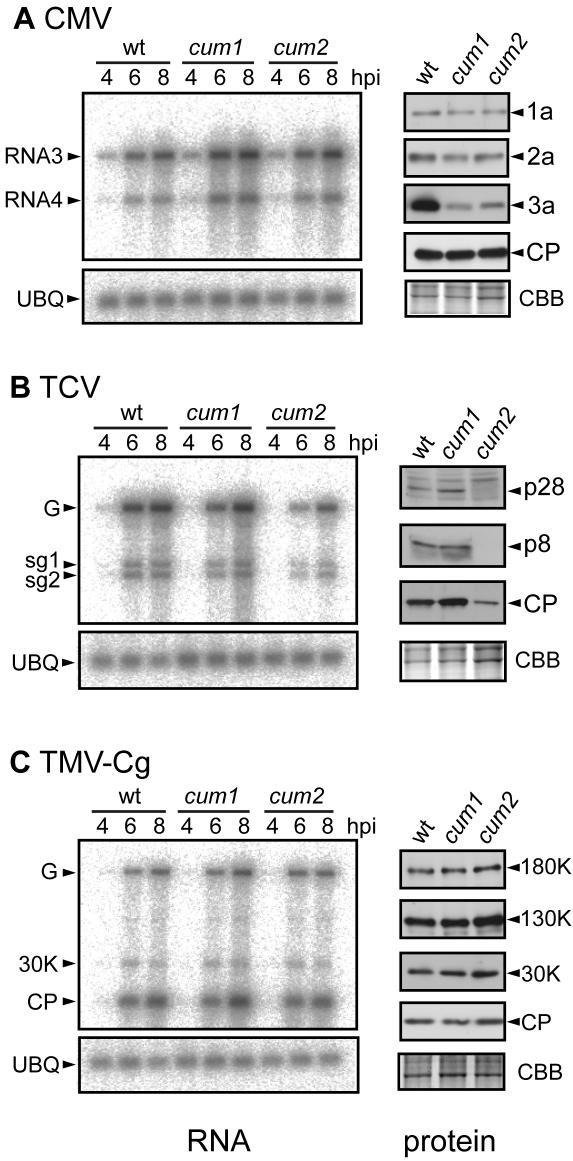
CMV encodes five proteins. Proteins 1a and 2a are involved in viral RNA replication and are synthesized by the translation of CMV RNA1 and RNA2, respectively. Protein 3a is involved in viral cell-to-cell movement and is synthesized by the translation of RNA3. Protein 2b, which suppresses gene silencing, and the CP are synthesized by the translation of the RNA2-derived subgenomic RNA4A and the RNA3-derived RNA4, respectively (Fig. 3A) (3, 8, 24). CMV RNA multiplication from 4 to 8 h postinoculation (hpi) and CP accumulation in cum1 and cum2 mutant protoplasts were similar to those in wild-type protoplasts (Fig. 4A), confirming published results (39, 40). As with the CMV CP, the accumulation of the 1a and 2a replication proteins in cum1 and cum2 mutant protoplasts was similar to that in wild-type protoplasts (Fig. 4A). In contrast, in cum1 and cum2 mutant protoplasts, 3a accumulation was only 20% (ranging from 6 to 64% of wild-type levels in seven independent experiments) and 30% (ranging from 16 to 67% of wild-type levels in six independent experiments) of that in wild-type protoplasts, respectively, at 8 hpi (Fig. 4A and data not shown). These results demonstrate that the cum1 and cum2 mutations affect protein 3a accumulation but not the accumulation of CMV RNA3, suggesting that the mutations affect the translation of CMV RNA3 or the stability of the 3a protein.
FIG. 3.
Structures of the CMV and TCV genomes. ORFs indicated by white boxes are translated, and those indicated by gray boxes are not translated. Note that CMV RNAs are 5′ capped, whereas TCV RNAs are not, and that neither viral RNA is polyadenylated.
FIG. 4.
Accumulation of CMV-, TCV-, and TMV-encoded proteins and RNA in cum1 and cum2 mutant and wild-type protoplasts. Protoplasts were prepared from liquid-cultured calli derived from seedlings of Col-0 wild-type (wt) and the cum1 and cum2 mutants. Protoplasts (4 × 106) were electroporated with either 7 μg of CMV-Y RNA, 5 μg of TCV RNA, or 20 μg of TMV-Cg RNA. Inoculated protoplasts were cultured for 4, 6, or 8 h for RNA analysis or for 8 h for protein analysis. The accumulation of CMV-Y RNA3, CMV-Y RNA4, and the TMV-Cg- and TCV-M-related RNAs was analyzed by Northern blot hybridization as described by Yoshii et al. (39). RNA bands were visualized with a Bio Imaging analyzer (BAS 1000; Fuji Photo Film, Tokyo, Japan). The accumulation of the virus-encoded proteins was analyzed by immunoblotting as described previously (39). Mock-inoculated wild-type protoplasts were simultaneously analyzed to identify plant-derived RNA and protein bands (data not shown). To confirm that similar amounts of RNA and protein were loaded in each lane, detection of ubiquitin mRNA (UBQ) and Coomassie brilliant blue staining of sodium dodecyl sulfate-polyacrylamide gels (CBB) were performed. (A) Accumulation of CMV-Y-related molecules. Positions of RNA3, RNA4, 1a, 2a, 3a, and the CP are indicated. (B) Accumulation of TCV-B-related molecules. Positions of the genomic RNA (G), subgenomic RNA1 (sg1), subgenomic RNA2 (sg2), and the proteins p28, p8, and the CP are indicated. (C) Accumulation of TMV-Cg-related molecules. Positions of the genomic RNA (G), the 30K and CP subgenomic RNAs, and the 180K, 130K, and 30K proteins and the CP are indicated.
The cum1 and cum2 mutations affect the expression of proteins encoded by RNA carrying noncoding sequences of CMV RNA3.
Since eIF4E and eIF4G are involved in translation initiation, less efficient RNA3 translation may explain the strong reduction in 3a accumulation in the cum1 and cum2 mutant protoplasts. To test this possibility, capped mRNAs of FLUC containing either the 5′ UTR, 3′ UTR, or both UTRs of either CMV RNA3 or RNA4 were constructed because the UTRs of many viral mRNAs contain RNA sequence elements controlling translation efficiency (12). These chimeric FLUC RNAs were synthesized by in vitro transcription and electroporated into cum1, cum2, and wild-type protoplasts. The internal control, a capped RLUC RNA that carried 5′ and 3′ UTRs derived from an E. coli plasmid vector and a 3′ poly(A)30, was mixed with the FLUC RNA on electroporation and was used to correct for differences in general translation activity, protoplast quality, and electroporation efficiency. Six hours after electroporation, the cells were harvested and disrupted, and FLUC and RLUC activities were measured.
When capped FLUC RNAs carrying the 3′ UTRs of CMV RNA3 (construct V-3) and RNA4 (construct V-4) were electroporated into the wild-type protoplasts, the relative FLUC activities (the FLUC/RLUC ratios) were 50- and 100-fold higher, respectively, than the relative FLUC activity from FLUC RNA that did not contain any CMV RNA sequences (construct V+) (Table 1). This suggests that the 3′ UTRs of CMV RNA3 and RNA4 harbor elements that enhance the expression of encoded proteins. The relative FLUC activities expressed from V+ RNA in cum1 and cum2 protoplasts were not significantly different from that in wild-type protoplasts. In the cum1 and cum2 mutant protoplasts, however, the relative FLUC activities from V-3 RNA were significantly lower than that in the wild-type protoplasts (56 and 39% of the wild-type level, respectively). Similarly, the relative FLUC activities from V-4 RNA were significantly lower in the cum1 and cum2 mutants than in the wild-type protoplasts (42 and 32% of the wild-type level, respectively) (Table 1). Therefore, the cum1 and cum2 mutations each affected the function of the CMV 3′UTRs, which enhanced the expression of encoded proteins.
TABLE 1.
FLUC expression from RNAs harboring UTRs of CMV or TCV RNA, which were electroporated into cum1, cum2, and wild-type protoplasts
| Construct
|
Protoplast type | Relative FLUC activity (FLUC/RLUC ratio)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ cap | 5′ UTR | 3′ UTR | RNA name (template) | Expt 1 | Expt 2 | Expt 3 | Expt 4 | Expt 5 | Expt 6 | Expt 7 | Avg (% of wt) | |
| + | Vector | CMV RNA3 | V-3 (pMP-3′-/EcoRI) | wta | 2.9 × 10−1 | 1.6 × 10−1 | 1.9 × 10−1 | 2.1 × 10−1 | ||||
| cum1 | 1.5 × 10−1 | 9.7 × 10−2 | 1.1 × 10−1 | 1.2 × 10−1b (56) | ||||||||
| cum2 | 1.0 × 10−1 | 8.8 × 10−2 | 6.3 × 10−2 | 8.4 × 10−2b (39) | ||||||||
| + | Vector | CMV RNA4 | V-4 (pCP-3′/EcoRI) | wt | 5.9 × 10−1 | 2.5 × 10−1 | 5.4 × 10−1 | 4.6 × 10−1 | ||||
| cum1 | 2.9 × 10−1 | 1.1 × 10−1 | 1.8 × 10−1 | 1.9 × 10−1b (42) | ||||||||
| cum2 | 2.3 × 10−1 | 7.9 × 10−2 | 1.3 × 10−1 | 1.5 × 10−1b (32) | ||||||||
| + | CMV RNA3 | CMV RNA3 | 3-3 (pMP-5′+3′/EcoRI) | wt | 1.9 × 10−1 | 1.1 × 10−1 | 1.7 × 10−1 | 1.5 × 10−1 | ||||
| cum1 | 3.2 × 10−2 | 2.6 × 10−2 | 3.4 × 10−2 | 3.0 × 10−2b (20) | ||||||||
| cum2 | 1.4 × 10−2 | 1.6 × 10−2 | 1.2 × 10−2 | 1.4 × 10−2b (9) | ||||||||
| + | CMV RNA3 | CMV RNA4 | 3-4 (pMP-5′+CP-3′/EcoRI) | wt | 4.3 × 10−1 | 2.4 × 10−1 | 3.4 × 10−1 | 3.4 × 10−1 | ||||
| cum1 | 6.6 × 10−2 | 5.2 × 10−2 | 7.0 × 10−2 | 6.3 × 10−2b (19) | ||||||||
| cum2 | 9.0 × 10−2 | 3.8 × 10−2 | 4.9 × 10−2 | 5.9 × 10−2b (17) | ||||||||
| + | CMV RNA4 | CMV RNA4 | 4-4 (pCP-5′ + 3′/EcoRI) | wt | 7.0 × 10−1 | 4.0 × 10−1 | 6.7 × 10−1 | 5.9 × 10−1 | ||||
| cum1 | 3.9 × 10−1 | 2.6 × 10−1 | 3.9 × 10−1 | 3.5 × 10−1b (58) | ||||||||
| cum2 | 3.5 × 10−1 | 2.2 × 10−1 | 2.0 × 10−1 | 2.6 × 10−1b (43) | ||||||||
| + | Vector | Vector | V+ (pV-luc+/EcoICRI) | wt | 2.4 × 10−3 | 1.3 × 10−3 | 9.9 × 10−3 | 4.5 × 10−3 | ||||
| cum1 | 2.8 × 10−3 | 1.8 × 10−3 | 4.9 × 10−3 | 3.2 × 10−3c | ||||||||
| cum2 | 5.0 × 10−3 | 1.6 × 10−3 | 4.2 × 10−3 | 3.6 × 10−3c | ||||||||
| − | Vector | TCV G | V-G (p28/88-3′/SmaI) | wt | 8.9 × 10−3 | 3.5 × 10−3 | 1.6 × 10−2 | 8.6 × 10−3 | 9.3 × 10−3 | |||
| cum1 | 1.2 × 10−2 | 9.5 × 10−3 | 2.0 × 10−2 | 9.3 × 10−3 | 1.3 × 10−2d (138) | |||||||
| cum2 | 3.8 × 10−3 | 2.4 × 10−3 | 2.9 × 10−3 | 4.1 × 10−3 | 3.3 × 10−3e (35) | |||||||
| − | Vector | TCV Sg2 | V-Sg2 (p38-3′/SmaI) | wt | 1.3 × 10−2 | 5.3 × 10−3 | 2.3 × 10−2 | 1.7 × 10−2 | 1.4 × 10−2 | |||
| cum1 | 1.6 × 10−2 | 8.0 × 10−3 | 2.6 × 10−2 | 1.4 × 10−2 | 1.6 × 10−2c | |||||||
| cum2 | 4.8 × 10−3 | 3.0 × 10−3 | 4.4 × 10−3 | 6.0 × 10−3 | 4.5 × 10−3b (32) | |||||||
| − | TCV G | TCV G | G-G (p28/88-5′ + 3′/SmaI) | wt | 2.2 × 10−2 | 8.2 × 10−3 | 4.4 × 10−2 | 1.9 × 10−2 | 2.3 × 10−2 | |||
| cum1 | 4.4 × 10−2 | 3.8 × 10−2 | 7.3 × 10−2 | 3.5 × 10−2 | 4.8 × 10−2d (206) | |||||||
| cum2 | 1.3 × 10−2 | 7.3 × 10−3 | 1.0 × 10−2 | 8.3 × 10−3 | 9.6 × 10−3e (42) | |||||||
| − | TCV Sg2 | TCV Sg2 | Sg2-Sg2 (p38-5′ + 3′/SmaI) | wt | 4.5 × 10−2 | 1.9 × 10−2 | 8.6 × 10−2 | 4.1 × 10−2 | 4.8 × 10−2 | |||
| cum1 | 1.2 × 10−1 | 1.0 × 10−1 | 2.2 × 10−1 | 1.0 × 10−1 | 1.4 × 10−1d (283) | |||||||
| cum2 | 1.6 × 10−2 | 1.1 × 10−2 | 2.1 × 10−2 | 1.7 × 10−2 | 1.7 × 10−2b (35) | |||||||
| − | Vector | Vector | V−(pV-luc+/EcoICRI) | wt | 2.4 × 10−5 | 3.6 × 10−5 | 3.4 × 10−5 | 3.1 × 10−5 | ||||
| cum1 | 2.8 × 10−5 | 4.2 × 10−5 | 2.6 × 10−5 | 3.2 × 10−5c | ||||||||
| cum2 | 5.6 × 10−5 | 3.9 × 10−5 | 2.0 × 10−5 | 3.8 × 10−5c | ||||||||
wt, wild type.
Statistically significant (P < 0.05 by Student's paired t test, one-sided test) decrease in the FLUC/RLUC ratio compared with wild-type protoplasts.
Not statistically significant (P > 0.5 by Student's paired t-test) difference from the value for wild-type protoplasts.
Statistically significant (P < 0.05 using Student's paired t-test, one-sided test) increase in the FLUC/RLUC ratio, as compared with wild-type protoplasts.
Borderline statistically significant (P < 0.08 using Student's paired t-test, one-sided test) decrease in the FLUC/RLUC ratio, as compared with wild-type protoplasts.
When capped FLUC RNAs carrying the 5′ UTR of RNA3 and 3′ UTR of RNA3 or RNA4 (construct 3-3 or 3-4) were used for electroporation, the relative FLUC activities expressed in the cum1 and cum2 protoplasts were significantly lower than the activity in the wild-type protoplasts (for 3-3, 20 and 9% of the wild-type level, respectively; for 3-4, 19 and 17% of the wild-type level, respectively) (Table 1). The reductions in relative FLUC activity by the cum1 and cum2 mutations were greater for constructs 3-3 and 3-4 than for V-3 and V-4. Conversely, when capped FLUC RNA carrying both the 5′ and 3′ UTRs of CMV RNA4 (construct 4-4 in Table 1) was electroporated, the relative FLUC activities expressed in the cum1 and cum2 protoplasts were significantly lower than that in the wild-type protoplasts (58 and 43% of the wild-type level in cum1 and cum2 protoplasts, respectively). However, the reductions in the relative FLUC activity by the cum1 and cum2 mutations were similar to, or even smaller than, that for the construct V-4 (Table 1). These results suggest that the 5′ UTR sequence of CMV RNA3, but not that of RNA4, reduces the relative FLUC activity more drastically in the cum1 and cum2 protoplasts than in the wild-type protoplasts. In summary, the cum1 and cum2 mutations affected the expression of protein products from RNA carrying the noncoding sequences of CMV RNA3 more strongly than that from RNA carrying the noncoding sequences of CMV RNA4. This explains, at least partly, why CMV 3a protein accumulation, but not CP accumulation, is significantly reduced in CMV-infected cum1 and cum2 protoplasts. Furthermore, because the levels of CMV RNA3 accumulation in CMV-inoculated cum1 and cum2 protoplasts were similar to those in CMV-inoculated wild-type protoplasts, the stability of CMV RNA3 is unlikely to be affected by the cum1 and cum2 mutations. Therefore, the cum1 and cum2 mutations probably affect the translation of CMV RNA3.
Reduced accumulation of TCV-related RNA and proteins in cum2 protoplasts.
To understand how the cum2 mutation inhibits TCV multiplication in plants, we examined the accumulation levels of TCV-related RNA and TCV-encoded proteins in cum2 protoplasts. TCV encodes five proteins: p28 and p88 are involved in viral RNA replication and are synthesized by the translation of the genomic RNA, p88 is synthesized by the readthrough of the termination codon of p28, p8 and p9 are involved in viral cell-to-cell movement and are synthesized by the translation of subgenomic RNA1, and p38 is the CP and is synthesized by the translation of subgenomic RNA2 (Fig. 3B) (5, 15). As Fig. 4B shows, the accumulation of TCV-encoded p28, p8, and p38 (CP) at 8 hpi and the increase in TCV-related RNA accumulation from 4 to 8 hpi were reduced in cum2 protoplasts (lower than 40% of the respective wild-type levels in five to eight independent experiments), but not in cum1 protoplasts, compared to wild-type protoplasts (Fig. 4B and data not shown). This is inconsistent with the results reported by Yoshii et al. (40). To find a plausible explanation for these conflicting data, we checked the genotypes of the protoplasts by using the CAPS analysis (for details, see Materials and Methods) and found that the cum2 protoplasts used by Yoshii et al. (40) were heavily contaminated with wild-type or cum2 heterozygous protoplasts (reference 40 and data not shown). We confirmed that the wild-type, cum1, and cum2 seeds used in this study each had correct genotypes of the cum1 and cum2 loci by using the CAPS analysis (for details, see Materials and Methods). Therefore, the accumulation of TCV-related RNA and TCV-encoded proteins is reduced in cum2 protoplasts. For CMV and TMV, the accumulation of viral RNA and CP in noncontaminated cum2 protoplasts was similar to that in wild-type protoplasts (Fig. 4A and C).
The cum2 mutation affects the expression of protein products encoded by RNAs carrying noncoding sequences of TCV RNA.
In order to obtain insight into the inhibition mechanisms of TCV multiplication by the cum2 mutation, we examined the effect of the cum1 and cum2 mutations on the expression of protein products encoded by FLUC RNA mimicking the TCV RNA. Since the 5′ terminus of the TCV RNA is naturally uncapped, uncapped FLUC RNAs containing UTRs from TCV genomic RNA or subgenomic RNA2 were prepared. These RNAs were electroporated into the wild-type, cum1, and cum2 protoplasts with the capped RLUC internal control RNA described above, and the expressed FLUC and RLUC activities were determined 6 h after electroporation.
Uncapped FLUC RNA containing the 5′ and 3′ UTR sequences derived from a plasmid vector (construct V−) expressed only low levels of relative FLUC activity. The relative FLUC activities in the cum1 and cum2 mutant protoplasts were not significantly different from that in the wild-type protoplasts (Table 1). Replacement of the vector-derived 3′ UTR with the 3′ UTRs of TCV genomic RNA (construct V-G) and subgenomic RNA2 (construct V-Sg2) drastically increased the relative FLUC activity in wild-type protoplasts (Table 1). In the constructs carrying the 3′ UTRs of TCV RNA, replacement of the 5′ UTRs from the vector-derived sequence with that from TCV genomic and subgenomic RNAs (constructs G-G and Sg2-Sg2) further increased the FLUC expression in the wild-type protoplasts (Table 1). These results are consistent with the conclusions of Qu and Morris (25) that the 3′ UTR of TCV RNA contains a major TE and that the 5′ and 3′ UTRs of TCV genomic or subgenomic RNA synergistically enhance translation.
For the constructs containing the TCV 3′ UTRs (V-G, V-Sg2, G-G, and Sg2-Sg2), the relative FLUC expression in cum2 mutant protoplasts was moderately, but significantly, reduced compared to that in the wild-type protoplasts (32 to 42% of the respective wild-type levels) (Table 1). These results suggest that the cum2 mutation affects the expression of TCV-encoded proteins. Interestingly, in the cum1 mutant protoplasts, the relative FLUC expression from constructs V-G, G-G, and Sg2-Sg2 was significantly higher than that in the wild-type protoplasts (Table 1).
DISCUSSION
In this study, we demonstrated that CUM1 and CUM2 encode eIF4E and eIF4G, respectively. Inoculation of CMV RNA into the cum1 and cum2 mutant protoplasts revealed that the mutations affected 3a protein accumulation but not the accumulation of CMV RNA and the 1a, 2a, and coat proteins (Fig. 4A). Moreover, the expression of protein products from the capped chimeric FLUC RNA carrying the 5′ and 3′ UTRs of CMV RNA3 (construct 3-3 in Table 1) was severely reduced, but the expression from the capped chimeric FLUC RNA carrying the 5′ and 3′ UTRs of CMV RNA4 (construct 4-4 in Table 1) was only mildly affected by the cum1 and cum2 mutations. For TCV, the accumulations of viral RNA and proteins were all reduced in TCV-infected cum2 mutant protoplasts as far as we examined (Fig. 4B). Electroporation experiments with uncapped chimeric FLUC RNA carrying the 3′ UTR of TCV RNA demonstrated that the cum2 mutation affected the expression of protein products from this RNA (Table 1).
eIF4E- and eIF4G-related genes in A. thaliana.
The A. thaliana genome contains five genes that encode eIF4E-related proteins: eIF4E (CUM1), eIFiso4E (26), a novel cap-binding protein (28), and two eIF4E-like proteins (At1g29550 and At1g29590). At1g29550 and At1g29590 are closely related to eIF4E and are expressed at the mRNA level (Arabidopsis Information Resource [http://www.arabidopsis.org/] and M. Yoshii, R. Kozuka, S. Naito, and M. Ishikawa, unpublished results) but remain to be analyzed further. The presence of these homologs may explain why the loss of eIF4E (CUM1) due to a nonsense mutation is not lethal to A. thaliana. Mutations in the eIF4E and eIFiso4E genes confer resistance against potyviruses in several plant species (9, 21, 23, 27). One of these mutations, lsp1 in A. thaliana, inhibits the multiplication of turnip mosaic virus at the single-cell level, suggesting that eIFiso4E is required for turnip mosaic virus gene expression or replication (21). However, it is not known whether lsp1 inhibits the translation of turnip mosaic virus RNA.
The A. thaliana genome contains at least three genes that encode eIF4G-related proteins: eIF4G (CUM2) and two eIFiso4G proteins. The two A. thaliana eIFiso4G genes (At2g24050 and At5g57870) are expressed at the mRNA level (Arabidopsis Information Resource). In wheat, eIF4E and eIF4G form the eIF4F complex, and eIFiso4E and eIFiso4G form the eIFiso4F complex (4). Since the cum1 and cum2 mutations have similar effects on the expression of CMV 3a protein, eIF4F probably has a specialized function in the process, which eIFiso4F or the other eIF4F-like complexes perform less efficiently. The eIF4G protein not only binds to eIF4E but also binds to eIF4A helicase and eIF3, and the cum2-1 mutation (1,327th Pro to Ser) resides in a region that corresponds to the domain identified to be important for the binding to eIF4A and eIF3 in human eIF4GI (17). Therefore, the cum2 mutation may affect the binding of eIF4G to eIF4A or eIF3.
Effects of the cum1 and cum2 mutations on protein expression from CMV RNA.
Inclusion of the 3′ UTRs of CMV RNA3 or RNA4 in capped chimeric FLUC RNA enhanced the relative FLUC expression, and the enhancement conferred by the CMV 3′ UTRs was affected by the cum1 and cum2 mutations (Table 1). This suggests that eIF4E and eIF4G are involved in this enhancement and that eIFiso4E and eIFiso4G can carry out this process, albeit less efficiently.
In yeast, mutations in the eIF4E and eIF4G genes that strongly inhibit translation initiation also increase the rate of mRNA deadenylation and decapping, the key steps in a major mRNA decay pathway (31, 32). In addition, in plants, mRNA stability is often correlated with translation (33). Therefore, it is possible that the cum1 and cum2 mutations lead to the instability of mRNA carrying CMV UTRs. However, CMV RNA accumulated to wild-type levels in CMV-infected cum1 and cum2 mutant protoplasts (Fig. 4A). Therefore, the cum1 and cum2 mutations probably affect the translation of RNA carrying the 3′ noncoding sequence of CMV RNA, rather than the stability of this RNA.
Capped, polyadenylated mRNA can be circularized through linkage of the 3′ poly(A) tail, poly(A)-binding protein, eIF4G, eIF4E, and 5′ cap, and this circularization is thought to be important for efficient translation (36; reviewed in reference 29). Therefore, like poly(A) tails, the 3′ UTRs of CMV RNA may interact with the eIF4F complex directly or indirectly, contributing to efficient translation of CMV RNA through circularization of the mRNA.
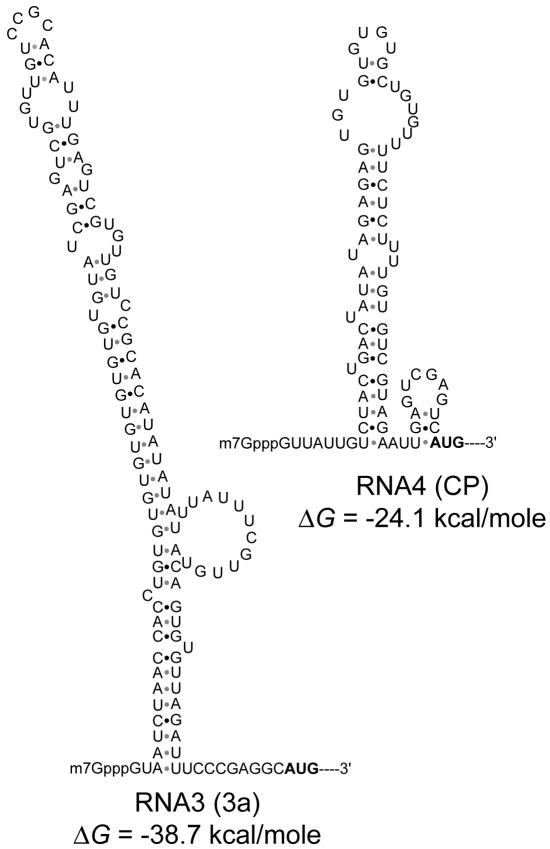
The presence of a stable secondary structure in the 5′ UTR inhibits translation initiation (18). The 5′ UTR of CMV RNA3 harbors more stable secondary structures than do the 5′ UTRs of CMV RNA1, RNA2, and RNA4 (Fig. 5 and data not shown). It has been reported that chimeric FLUC RNA that contains the CMV RNA3 5′ UTR is translated less efficiently in vitro than mRNA that contains the 5′ UTRs of other CMV RNA segments (19). Consistent with these observations, we showed that FLUC RNA containing the CMV RNA3 5′ UTR expressed less FLUC activity in vivo than that containing the 5′ UTR of CMV RNA4 (FLUC/RLUC ratios of 3.4 × 10−1 and 5.9 × 10−1 for constructs 3-4 and 4-4, respectively [Table 1]).
FIG. 5.
Predicted secondary structures of the 5′ UTRs of CMV RNA3 and RNA4. Secondary structures were predicted by using MFOLD version 2.3 (41). The free energies (ΔG) of the 5′ UTR structures were calculated by using the default settings and a folding temperature of 23°C.
We also demonstrated that the relative FLUC activity expressed from chimeric RNA carrying the 5′ UTR of RNA3 and the 3′ UTR of RNA3 or RNA4 (construct 3-3 or 3-4 in Table 1) was more severely affected by the cum1 and cum2 mutations than that from FLUC RNA carrying only the 3′ UTRs of CMV RNA (constructs V-3 or V-4 in Table 1). This suggests that eIF4F facilitates the in vivo translation of RNA containing the CMV RNA3 5′ UTR, which has a strong secondary structure, more efficiently than does eIFiso4F. In keeping with this possibility, Gallie and Browning (13) demonstrated that eIF4F promotes the in vitro translation of RNA with highly structured 5′ UTRs more efficiently than eIFiso4F does. Therefore, the observed inefficient expression of protein products from CMV RNA3 in the cum1 and cum2 mutants probably reflects an additive effect of the independent responses to the mutations, which the 5′ and 3′ UTRs of CMV RNA3 show on translation.
Effects of the cum2 mutation on translation of the TCV RNA.
Qu and Morris (25) have shown that the genomic RNA of TCV lacks a 5′ cap structure and that the 5′ and 3′ UTRs of the genomic and subgenomic RNAs of TCV enhance their translation. Our results with wild-type protoplasts confirm their conclusions and suggest that the 3′ UTRs of TCV RNA contain a strong cap-independent TE and that the 5′ UTRs of TCV RNA contain a weak cap-independent TE.
The cum2 mutation affected protein expression from uncapped RNA containing the TCV 3′ UTR (Table 1). This suggests that enhanced expression of encoded proteins by the 3′ UTR of TCV RNA involves eIF4G. The cum1 mutation did not adversely affect, and in fact enhanced, the functioning of the 3′ UTR of TCV (Table 1). Therefore, eIF4E is dispensable for this mechanism or may even have an inhibitory effect on the functioning of the 3′ UTR of TCV. Since the enhanced relative FLUC expression conferred by the TCV 3′ UTR still occurs in the cum2 mutant (compare V-G or V-Sg2 with V− in Table 1), eIFiso4G may carry out this process more weakly, or the cum2 mutant allele may have residual activity. It has been shown that eIF4F participates in the cap-independent and 3′ TE-dependent translation of BYDV RNA in vitro (35). If a common mechanism underlies the translation of BYDV and TCV RNAs, it is plausible that the cum2 mutation affects the expression of TCV-encoded proteins via translation inhibition. However, our results alone do not eliminate the possibility that the cum2 mutation affects the stability of TCV RNA.
The results with CMV suggest that the cum1 and cum2 mutations affect the production of the 3a protein and that reduced accumulation of 3a causes delayed CMV multiplication in plants. Similarly, the results with TCV suggest that the cum2 mutation primarily affects the production of the p28 replication protein of TCV and that reduced accumulation of p28 inhibits the TCV RNA multiplication within a cell and in plants. However, future experiments should carefully test whether the observed phenotypes are in a direct cause-and-effect relationship. In addition, these mutations might alter the expression of many proteins, so that the observed phenotypes of the cum1 and cum2 mutants might be caused by alterations in the expression of other host proteins. In fact, eIF4E is overexpressed with the loss of eIFiso4E (9). Therefore, it is possible that eIFiso4E and eIFiso4G are overexpressed to compensate for the loss of eIF4E or eIF4G. The effect of the depletion of eIF4E or eIF4G may be examined more directly by using an in vitro translation system.
Acknowledgments
We thank D. Shibata, Y.-G. Liu, C. Masuta, T. J. Morris, A. E. Simon, and the Arabidopsis Biological Resource Center for materials and K. Komoda for help with antibody preparation. We also thank D. B. Goto and F. van Werven for careful readings of the manuscript and K. Fujiwara for general assistance.
This work was supported in part by grants from the Japan Society for the Promotion of Science to M.I.
REFERENCES
- 1.Allen, E., S. Wang, and W. A. Miller. 1999. Barley yellow dwarf virus RNA requires a cap-independent translation sequence because it lacks a 5′ cap. Virology 253:139-144. [DOI] [PubMed] [Google Scholar]
- 2.Bechtold, N., and G. Pelletier. 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82:259-266. [DOI] [PubMed] [Google Scholar]
- 3.Brigneti, G., O. Voinnet, W.-X. Li, L.-H. Ji, S.-W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Browning, K. S. 1996. The plant translational apparatus. Plant Mol. Biol. 32:107-144. [DOI] [PubMed] [Google Scholar]
- 5.Carrington, J. C., L. A. Heaton, D. Zuidema, B. I. Hillman, and T. J. Morris. 1989. The genome structure of turnip crinkle virus. Virology 170:219-226. [DOI] [PubMed] [Google Scholar]
- 6.Chiba, Y., R. Sakurai, M. Yoshino, K. Ominato, M. Ishikawa, H. Onouchi, and S. Naito. 2003. S-Adenosyl-l-methionine is an effector in the posttranscriptional autoregulation of the cystathionine gamma-synthase gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 100:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danthinne, X., J. Seurinck, F. Meulewaeter, M. van Montagu, and M. Cornelissen. 1993. The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol. Cell. Biol. 13:3340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding, S.-W., B. J. Anderson, H. R. Haase, and R. H. Symons. 1994. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198:593-601. [DOI] [PubMed] [Google Scholar]
- 9.Duprat, A., C. Caranta, F. Revers, B. Menand, K. S. Browning, and C. Robaglia. 2002. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32:927-934. [DOI] [PubMed] [Google Scholar]
- 10.Gallie, D. R., and V. Walbot. 1990. RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev. 4:1149-1157. [DOI] [PubMed] [Google Scholar]
- 11.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5:2108-2116. [DOI] [PubMed] [Google Scholar]
- 12.Gallie, D. R. 1996. Translational control of cellular and viral mRNAs. Plant Mol. Biol. 32:145-158. [DOI] [PubMed] [Google Scholar]
- 13.Gallie, D. R., and K. S. Browning. 2001. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 276:36951-36960. [DOI] [PubMed] [Google Scholar]
- 14.Gingras, A.-C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 15.Hacker, D. L., I. T. D. Pretty, N. Wei, and T. J. Morris. 1992. Turnip crinkle virus genes required for RNA replication and virus movement. Virology 186:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Hagiwara, Y., K. Komoda, T. Yamanaka, A. Tamai, T. Meshi, R. Funada, T. Tsuchiya, S. Naito, and M. Ishikawa. 2003. Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 22:344-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imataka, H., and N. Sonenberg. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17:6940-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak, M. 1986. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 83:2850-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon, C. S., and W.-I. Chung. 2000. Differential roles of the 5′ untranslated regions of cucumber mosaic virus RNAs 1, 2, 3 and 4 in translational competition. Virus Res. 66:175-185. [DOI] [PubMed] [Google Scholar]
- 20.Lahser, F. C., L. E. Marsh, and T. C. Hall. 1993. Contributions of the brome mosaic virus RNA-3 3′-nontranslated region to replication and translation. J. Virol. 67:3295-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lellis, A. D., K. D. Kasschau, S. A. Whitham, and J. C. Carrington. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12:1046-1051. [DOI] [PubMed] [Google Scholar]
- 22.Liu, Y. G., and R. F. Whittier. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674-681. [DOI] [PubMed] [Google Scholar]
- 23.Nicaise, V., S. German-Retana, R. Sanjuan, M.-P. Dubrana, M. Mazier, B. Maisonneuve, T. Candresse, C. Caranta, and O. LeGall. 2003. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol. 132:1272-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palukatis, P., M. J. Roossinck, R. G. Dietzgen, and R. I. B. Francki. 1992. Cucumber mosaic virus. Adv. Virus Res. 41:281-348. [DOI] [PubMed] [Google Scholar]
- 25.Qu, F., and T. J. Morris. 2000. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 74:1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez, C. M., M. A. Freire, C. Camilleri, and C. Robaglia. 1998. The Arabidopsis thaliana cDNAs coding for eIF4E and eIF(iso)4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J. 13:465-473. [DOI] [PubMed] [Google Scholar]
- 27.Ruffel, S., M.-H. Dussault, A. Palloix, B. Moury, A. Bendahmane, C. Robaglia, and C. Caranta. 2002. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32:1067-1075. [DOI] [PubMed] [Google Scholar]
- 28.Ruud, K. A., C. Kuhlow, D. J. Goss, and K. S. Browning. 1998. Identification and Characterization of a novel cap-binding protein from Arabidopsis thaliana. J. Biol. Chem. 273:10325-10330. [DOI] [PubMed] [Google Scholar]
- 29.Sachs, A. 2000. Physical and functional interactions between the mRNA cap structure and the poly(A) tail, p. 447-465. In N. Sonenberg, J. W. B. Hershey, and M. Mathew (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schaad, M. C., R. J. Anderberg, and J. C. Carrington. 2000. Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 273:300-306. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz, D. C., and R. Parker. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5247-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz, D. C., and R. Parker. 2000. Interaction of mRNA translation and mRNA degradation in Saccharomyces cerevisiae, p. 807-825. In N. Sonenberg, J. W. B. Hershey, and M. Mathew (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sullivan, M. L., and P. J. Green. 1993. Post-transcriptional regulation of nuclear-encoded genes in higher plants: the roles of mRNA stability and translation. Plant Mol. Biol. 23:1019-1104. [DOI] [PubMed] [Google Scholar]
- 34.Tsujimoto, Y., T. Numaga, K. Ohshima, M. Yano, R. Ohsawa, D. B. Goto, S. Naito, and M. Ishikawa. 2003. Arabidopsis TOBAMOVIRUS MULTIPLICATION (TOM) 2 locus encodes a transmembrane protein that interacts with TOM1. EMBO J. 22:335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S., K. S. Browning, and W. A. Miller. 1997. A viral sequence in the 3′-untranslated region mimics a 5′cap in facilitating translation of uncapped mRNA. EMBO J. 16:4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135-140. [DOI] [PubMed] [Google Scholar]
- 37.Wu, B., and K. W. White. 1999. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J. Virol. 73:8982-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamanaka, T., T. Ohta, M. Takahashi, T. Meshi, R. Schmidt, C. Dean, S. Naito, and M. Ishikawa. 2000. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 97:10107-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshii, M., N. Yoshioka, M. Ishikawa, and S. Naito. 1998. Isolation of an Arabidopsis thaliana mutant in which accumulation of cucumber mosaic virus is delayed. Plant J. 13:211-219. [DOI] [PubMed] [Google Scholar]
- 40.Yoshii, M., N. Yoshioka, M. Ishikawa, and S. Naito. 1998. Isolation of an Arabidopsis thaliana mutant in which the multiplication of both cucumber mosaic virus and turnip crinkle virus is affected. J. Virol. 72:8731-8737. (Author's correction, 77:8178, 2003.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]