Abstract
The integrated human immunodeficiency virus type 1 (HIV-1) genome is transcribed in a single pre-mRNA that is alternatively spliced into more than 40 mRNAs. We characterized a novel bidirectional exonic splicing enhancer (ESE) that regulates the expression of the HIV-1 env, vpu, rev, and nef mRNAs. The ESE is localized downstream of the vpu-, env-, and nef-specific 3′ splice site no. 5. SF2/ASF and SRp40 activate the ESE and are required for efficient 3′ splice site usage and binding of the U1 snRNP to the downstream 5′ splice site no. 4. U1 snRNP binding to the 5′ splice site no. 4 is required for splicing of the rev and nef mRNAs and to increase expression of the partially spliced env mRNA. Finally, our results indicate that this ESE is necessary for the recruitment of the U1 snRNP to the 5′ splice site no. 4, even when the 5′ splice site and the U1 snRNA have been mutated to obtain a perfect complementary match. The ESE characterized here is highly conserved in most viral subtypes.
Recent data on the human, mouse, and rat genomes indicate that at least 60% of the genes in these organisms are alternatively spliced. Posttranscriptional regulation of gene expression by alternative splicing generates greater protein diversity. Retroviral genomes are often extremely complex. Structural and regulatory viral proteins are encoded by mRNAs derived from a single pre-mRNA transcribed from the integrated proviral genome. Since viral genes often overlap, definition of any sequence as exonic or intronic is gene dependent. For instance, most of the coding sequences of the human immunodeficiency virus type 1 (HIV-1) gag and pol genes are located in a region that defines an intron for the tat, rev, env, nef, vpu, vpr, and vif mRNAs.
In HIV-1, the combination of several alternative 5′ and 3′ splice sites (ss) yield more than 40 mRNAs (11, 31). Nevertheless, approximately 50% of the viral pre-mRNAs leave the nucleus without being spliced. Several gene products are translated from unspliced (gag and pol) or partially spliced (env, vpu, vif, and vpr) viral mRNAs. Furthermore, the unspliced viral RNA is packaged into new virions as a viral genome. Unspliced or partially spliced cellular mRNAs are usually degraded in the nucleus, but in HIV-1, the viral regulatory protein Rev promotes the export of incompletely spliced viral RNAs to the cytoplasm (9, 16, 27, 30). Mechanisms regulating the synthesis of both unspliced and spliced viral messages and their export to the cytoplasm involve interactions between viral RNA sequences and the host cell. Indeed, interactions between the U1 snRNP and viral sequences are required for the stability and export of unspliced and partially spliced viral mRNAs (17, 25). In vitro studies have shown that the interaction between the U1 snRNA and an inactive 5′ ss could increase splicing at upstream 3′ ss in the viral mRNA (2). Alteration of the delicate balance between spliced and unspliced RNAs or disruption of the unspliced viral RNA export pathway can dramatically affect HIV-1 infectivity and pathogenesis (2, 12, 31, 42).
In metazoans, the 5′ and 3′ ss are precisely recognized and joined together by the splicing machinery. Nevertheless, their sequences can vary greatly, and often the splicing machinery does not recognize sequences that match consensus ss motifs. Thus, it was envisioned early that recognition of the appropriate exonic and intronic sequences are based on additional sequences and the relative position and strength of the ss. Several lines of evidence suggested that 5′ and 3′ ss are coupled across either the exons or the introns that they define. An exon and an intron definition model for ss pairing have both been proposed (3, 32, 43). These models suggest that ss are functionally coupled and the splicing machinery recognizes pairs of ss that are flanking, and thus defining, an exon or the ones flanking an intron. A family of arginine-serine-rich RNA binding proteins named SR proteins mediates the coupling between ss pairs. SR proteins have either one or two RNA binding domains at the N terminus and an SR-rich tail of varying length at the C terminus (13, 33). This SR-rich domain has been shown to be required in the interactions between SR proteins and essential splicing factors such as the U1 70,000-molecular-weight component of the U1 snRNP (18) and the U2AF65/35 heterodimer (43). In the early stages of the splicing reaction, the U1 snRNP binds the 5′ ss while the heterodimer U2AF65/35 binds the 3′ ss and the upstream polypyrimidine tract. The binding of these factors to the splicing substrate is thought to be the key step in splicing regulation.
To date, more than 12 human SR proteins have been identified, and all of them have been shown to complement splicing-deficient HeLa S100 cytosolic extracts in an in vitro splicing assay (13, 45). SR proteins have been shown to activate splicing in several systems by binding exonic splicing enhancers (ESE). ESEs have been defined as short exonic sequences, distinct from the ss, whose presence increases the splicing of a given exon (41). To identify functional ESE motifs, individual SR proteins have been analyzed by functional SELEX in vitro (6, 24, 38). A binding sequence score matrix has been derived to allow the detection of motifs recognized by SF2/ASF, SC35, SRp40, and SRp55 (5a, 24). Although most ESEs have been shown to promote either upstream 3′ ss or downstream 5′ ss recognition, in a few cases, it has been shown that a single ESE can simultaneously promote the recognition of both upstream and downstream ss, thus working as a bidirectional splicing enhancer (4).
In this work we will elucidate the complex regulation of the alternative HIV-1 exon 5, bordered by the alternative 3′ ss no. 4c, 4a, 4b, and 5 and the 5′ ss no. 4. Mutations of SR binding sites contained in a newly identified bidirectional ESE within exon 5 affect usage of the upstream alternative 3′ ss and thus expression of the rev, nef, and env mRNAs. Furthermore, according to previous results with this and other systems (7, 17, 25), we will show that stabilization of the U1 snRNP binding to the 5′ ss no. 4 by the ESE is also required for the expression of unspliced and partially spliced viral mRNAs. This indicates a role in the stabilization of the unspliced mRNA or possibly RNA export for the SR protein-dependent ESE.
MATERIALS AND METHODS
Oligonucleotides.
Oligonucleotides were synthesized and purified as previously described (34). The oligonucleotides are as follows: 183 (SV-env), 5′-ATCCTGCAGGGCTTTAGGCTTTGATCCC; 197 (5′ PCR primer), 5′-TAATACGACTCACTATAGGG; 1095 (SV-env), 5′-GGGCCTAGGAATTCTCTCTTGAGCTCGCAGTAAGTAGCTTAAGCTCTCCGAAGACAGTGGCAATGAGAGT; 1098 (env-GAR), 5′-AATTCAGGAAGAAGCGGAGACAGCGACGAAGAGCT; 1099 (env-GAR), 5′-GTCCTTCTTCGCCTCTGTCGCTGCTTC; 1373 (env-HIV no. 18), 5′-GAGCGAATTCTGGGCAAGTTTGTG; 1374 (env-HIV no. 18), 5′-TGATGAGCTCAGCCTCCTACTATC; 1424 (SV-env cs11), 5′-TTAAGATACTTACCTGGAGCT; 1425 (SV-env cs11), 5′-CCAGGTAAGTATC; 1457 (SV-env/E42 cs11), 5′-GAGCTTAAGATACTTACCTGTTTGATAGAGA; 1460 (dsx-GAR), 5′-CGAAAGGAAGAAGCGGAGACAGCGACGAAGAGTT; 1461 (dsx-GAR), 5′-CGAACTCTTCGTCGCTGTCTCCGCTTCTTCCTTT; 1544 (SV-1-env SF2− SRp40−), 5′-CTTGAAAGCGAAAGTAAAGC; 1559 (cs14 U1), 5′-CGAAGATCTCAAGATACTTACCTGGCAGGGGAGAT; 1583 (dsx-HIV no. 18), 5′-CGAAAAAATTATTCATAATGATAGTAGGAGGCTT; 1584 (dsx-HIV no. 18), 5′-CGAAGCCTCCTACTATCATTATGAATAATTTTTT; 1598 (env-SRp40−), 5′-AATTCAGGAAGAAGCGGATTTAGCGACGAAGAGCT; 1599 (env-SRp40−), 5′-CTTCGTCGCTAAATCCGCTTCTTCCTG; 1600 (env-SF2− SRp40−), 5′-AATTCAGGAATTAGCGGCGACTTCGCTTAAGAGCT; 1603 (env-SF2− SRp40−), 5′-CTTAGGCGAAGTCGCCGCTAATTCCTG; 1604 (dsx-SRp40−), 5′-CGAAAGGAAGAAGCGGATTTAGCGACGAAGAGTT; 1605 (dsx-SRp40−) 5′-CGAACTCTTCGTCGCTAAATCCGCTTCTTCCTTT; 1606 (dsx-SF2−), 5′-CGAAAGGAATTAGCGGAGACAGCGCTTAAGAGTT; 1607 (dsx-SF2−), 5′-CGAACTCTTAAGCGCTGTCTCCGCTAATTCCTTT; 1608 (dsx-SF2− SRp40−), 5′-CGAAAGGAATTAGCGGCGACTTCGCTTAAGAGTT; 1609 (dsx-SF2− SRp40−), 5′-CGAACTCTTAAGCGAAGTCGCCGCTAATTCCTTT; 1613 (env-SF2−), 5′-AATTCAGGAATTAGCGGAGACAGCGCTTAAGAGCT; 1614 (env-SF2−), 5′-CTTAAGCGCTGTCTCCGCTAATTCCTG; 1690 (SV-1-env SF2− SRp40−), 5′-ATCGAGCTCTTAAGCGAAGTCGCCGCTAATTCCTGCCGTAGGAG; 1698 (NLS MS2), 5′-GATCCCGGGATGGGCCGCAAAAAACGCCGCCAACGCCGGGCCCCACGGGGTTCTCATCAT; 1710 (MS2 binding site), 5′-AATTATTGACCGTACACCATCAGGGTACGCGAATTCATTAATGAGCT; 1711 (MS2 binding site), 5′-CATTAATGAATTCGCGTACCCTGATGGTGTACGGTCAAT; 1712 (cs23 U1), 5′-CGAAGATCTCCTTAAGATACTTACCTGGAGCTCGCAGGGGAGAT; 1768 (MS2-ΔSR), 5′-TCTAGACTCGAGTCATCCACCACCACCACCGTA; 1791 (MS2-SRp40), 5′-GGTGGATCCAGCAAAAGGCAC; 1792 (MS2-SRp40), 5′-AGACTCGAGTTAATTGCCACT; 1795 (MS2-SF2), 5′-GGTGGATCCAGAAGTCCAAGTTATGGAAGATCTC; 1796 (MS2-SF2), 5′-AGACTCGAGTTATGTACGAGAGCGAGATCTGC; 1916 (GAR), 5′-TGTACTACTTACTGCGAGCTCTTCGTCGCTGTCTCCGCTTCTTCCTGCCTATAGTGAGTCGTATT; 1917 (HIV no. 18), 5′-TGTACTACTTACTGCGAGGCCTCCTACTATCATTATGAATAATTTTGCCTATAGTGAGTCGTATTA; 1918 (SRp40−), 5′-TGTACTACTTACTGCGAGCTCTTCGTCGCTAAATCCGCTTCTTCCTGCCTATAGTGAGTCGTATTA; 1919 (SF2−), 5′-TGTACTACTTACTGCGAGCTCTTAAGCGCTGTCTCCGCTAATTCCTGCCTATAGTGAGTCGTATTA; 1920 (SF2− SRp40−), 5′-TGTACTACTTACTGCGAGCTCTTAAGCGAAAGTCGCCGCTAATTCCTGCCTATAGTGAGTCGTATTA; 1921 (3U), 5′-TGTACTACTAACTGCGAGCTCTTCGTCGCTGTCTCCGCTTCTTCCTGCCTATAGTGAGTCGTATTA.
Recombinant plasmids.
The GAR ESE and its mutations were inserted into the BstBI site of the dsx-ΔE plasmid (40) by using pairs of kinase-treated cDNA oligonucleotides.
For the construction of the parent HIV-1 glycoprotein expression plasmid SV-env, the subgenomic HIV-1 EcoRI-KpnI fragment (nucleotides [nt] 5742 to 6348, the nucleotides are numbered according to the parent sequence HIV NL43 in the HIV databank of Myers et al. [29]) of pNLA1 (37), which is a cDNA derivative of pNL4-3 (1), was substituted for a PCR-amplified linker fragment with primers 1095-183 and the BamHI-AvrII fragment (nt 8470 to 5206) was substituted for a BamHI-XbaI fragment of SV E/X tat− rev− (17), an XbaI-ClaI fragment of pASVT7pA Δint, and a ClaI-AvrII fragment of SVrev (17). Subsequently, the SacI-AflII fragment was substituted for a linker (1424-1425) containing the 5′ ss cs11 mutation (SV-env cs11). The GAR ESE and its mutations were constructed by insertion of pairs of cDNA oligonucleotides into the SacI-EcoRI vector fragment of SV-env (primers 1098-1099 [/GAR], 1598-1599 [/SRp40−], 1600-1603 [/SF2− SRp40−], and 1613-1614 [/SF2−]). For the construction of SV-env/HIV no. 18, the EcoRI-SacI fragment of SV-env was substituted for a PCR-amplified fragment of SV E/X tat− rev− (1373-1374). Plasmids SV-env/E42 and SV-env/E42 cs11 were constructed by substituting the SacI-AflII fragment of SV-env for a PCR-amplified fragment (primers 197-1153 [/E42] and 197-1457 [/E42 cs11]) of SV E/X tat− rev−. For the construction of SV-env/ex5, the SacI-KpnI fragment of SV-env/GAR was substituted for the SacI-KpnI fragment of SV-env/E42.
SVcrev was constructed by recloning the EcoRI-XhoI fragment from pUHcrev (35) into pSVT7.
For the construction of plasmid pUCBΔU1, the BglII-PstI fragment (nt 440 to 726) of the parent plasmid pUCBU1 (kindly provided by M.L. Hammarskjold) was substituted for a linker (1132-1133) containing unique BglII, PstI, and XhoI sites. For the mutations, the BglII-XhoI fragment (nt 440 to 464) of pUCBΔU1 was substituted for a PCR-amplified fragment with a 5′ PCR primer carrying the BglII site and the desired mutation, for example, 1712 (U1 cs23), and the 3′ PCR primer 1131.
SV-1-env 3U was constructed by substituting the 5′ ss no. 4 containing SacI-NheI fragment of SV-1-env with the respective SacI-NheI fragment from the SV E/X tat− rev− 3U construct (17). For the construction of SV-1-env SF2− SRp40−, the EcoRI-SacI fragment of SV-1-env was substituted with a PCR-amplified fragment (primers 1544-1690) with the 3′ PCR primer carrying the SF2− SRp40− mutation.
For the construction of SV-env/MS2, the EcoRI-SacI fragment of SV-env was substituted for a pair of cDNA oligonucleotides (1710-1711) containing a mutated EcoRI site and a newly inserted EcoRI site to allow repetitive insertion of MS2 sites (SV-env/2MS2).
The XmaI-XhoI fragment of SV SD4/SA7 ds pA was substituted for a PCR-amplified fragment (primers 1698-1081 [SV NLS-MS2-9G8] and 1698-1768 [SV NLS-MS2-ΔSR] of pHISBIT-MS2-RS9G8 [kindly provided by B. R. Graveley]). Subsequently, the BamHI-XhoI fragment SV NLS-MS2-9G8 was substituted for a PCR-amplified fragment (primers 1791-1792 [SV-NLS-MS2-SRp40] and 1795-1796 [SV NLS-MS2-SF2]) of SV NLS-MS2-9G8.
In vitro pre-mRNA splicing assays and preparation of SR proteins.
Capped 32P-labeled runoff transcripts were synthesized by in vitro transcription with T7 RNA polymerase. HeLa cell and S100 extracts were prepared as described previously (28). Splicing reactions were performed in a total volume of 25 μl containing 15 μl of HeLa cell nuclear extract or S100 as described previously (28). The reaction mixtures were incubated at 30°C for 2 h. RNAs recovered from the splicing reaction mixtures were separated on an 8 M urea-6% polyacrylamide gel and visualized with an InstantImager (Packard). SR proteins were prepared from HeLa cells and calf thymus as previously described (44) and added to the splicing reaction mixtures as indicated.
Immobilization of RNA on agarose beads and RNA affinity assays.
RNAs were covalently linked to adipic acid dihydrazide agarose beads by modification of a published procedure (23) as previously described (5). 500 pmol of RNA was placed in a 400-μl reaction mixture containing 100 mM sodium acetate (pH 5.0) and 5 mM sodium m-periodate (Sigma). Reaction mixtures were incubated for 1 h in the dark at room temperature. The RNA was then ethanol precipitated and resuspended in 500 μl of 0.1 M sodium acetate (pH 5.0). Four hundred microliters of a 50% slurry of adipic acid dihydrazide agarose beads (Sigma) was washed four times in 10 ml of 0.1 M sodium acetate (pH 5.0) and pelleted after each wash at 300 rpm for 3 min in a clinical centrifuge. After the final wash, 500 μl of 0.1 M sodium acetate (pH 5.0) was added to the beads and the slurry was then mixed with the periodate-treated RNA and rotated for 12 h at 4°C. The beads with the bound RNA were then pelleted and washed three times in 1 ml of 2 M NaCl and three times in 1 ml of buffer D (20 mM HEPES-KOH [pH 7.6], 5% [vol/vol] glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol). The binding efficiency of RNA to the beads was between 70 and 80%, as determined with 5′ 32P-end-labeled RNA.
The beads containing immobilized RNA were incubated in a reaction mixture containing 250 μl of HeLa cell nuclear extract and 400 μl of buffer D for 20 min at 30°C. Beads were then pelleted by centrifugation at 106 × g for 3 min and washed four times with 1 ml of buffer D. After the final centrifugation, the proteins bound to the immobilized RNA were eluted by addition of 60 μl of protein sample buffer.
Cell culture and transfection.
HeLa-T4+ cells (26) were propagated and transfected as previously described (17) with FuGENE 6 (Roche Molecular Biochemicals), and the transfection efficiency was monitored by cotransfection of pGL3-control (Promega). The medium was changed after 24 h, and cells were harvested and prepared 48 h after transfection. For reverse transcription (RT)-PCR, cells were transfected with 1 μg of the respective SV-1-env plasmid and 1 μg of pXGH5 (36) as a control for transfection efficiency, and total RNA was isolated after 30 h.
Western blot analysis.
Cells were scraped from six-well plates into the medium, sedimented at 12,000 × g for 14 s, washed twice in phosphate-buffered saline and suspended in 200 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer (21). An aliquot from the phosphate-bufferd saline washing step was analyzed for luciferase activity (luciferase assay system; Promega). The protein concentration was measured by a Bradford protein assay (Bio-Rad) and adjusted in each sample to equal amounts of luciferase activity and protein amount by adding extracts of mock-transfected cells. Samples were subjected to electrophoresis on SDS-7% polyacrylamide gels and transferred to a polyvinylidene difluoride membrane (0.45-μm pore size, Immobilon P; Millipore). Protein detection was performed with a monoclonal mouse anti-gp120 antibody (1:500, NEA-9305 [DuPont] or 1:5,000, 87-133/026 [kindly provided by Dade Behring]) and visualized by a chemiluminescence detection system (ECL system and ECL hyperfilm [Amersham], Super Signal Ultra [Pierce]).
RT-PCR assay.
Isolation of total RNA was performed by using a modified guanidinium isothiocyanate protocol (8). Prior to RT, 4-μl RNA samples were subjected to DNase I digestion with 10 U of DNase I (Roche Molecular Biochemicals). After DNase I inactivation at 95°C for 10 min, 4.5 μl of the DNase I-digested RNA samples were reverse transcribed with 200 U of SuperScript III RNase H reverse transcriptase (Invitrogen) with 0.375 mM oligo(dT)15 (Roche Molecular Biochemicals) as a primer. As a negative control for the remaining plasmid DNA contamination of each sample, a second assay was performed as described above except that reverse transcriptase was replaced with double-distilled H2O. PCR was carried out with 1.25 U of AmpliTaq (Applied Biosystems) according to the manufacturer's protocol. Double-spliced and skipped RNA was detected with primer pair 1544-1542, and human growth hormone (hGH) mRNA was detected with primer pair 1225-1224. To determine the linear PCR amplification range allowing a semiquantitative estimation of the relative abundance of SV-1-env and hGH mRNA, a preliminary PCR test series was carried out with the same cDNA sample but varying the number of PCR cycles between 15 and 30. According to the obtained results, PCR analysis was performed with 26 cycles for SV-1-env and hGH PCR amplification.
PCR products were separated on 6% nondenaturing polyacrylamide gels, stained with ethidium bromide, and visualized with the Lumi-Imager F1 (Roche Molecular Biochemicals).
RESULTS
SRp40 and SF2/ASF activate the GAR ESE in vitro.
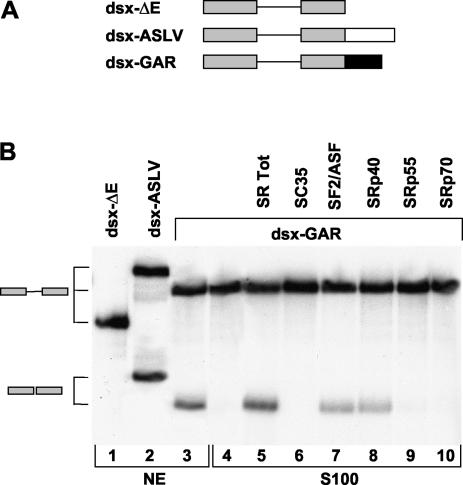
Previous results suggested that a purine-rich nucleotide motif, which we have termed GAR, within the HIV-1 exon 5 enhances expression of the env gene mRNA (Fig. 1) (10, 17). To characterize the putative splicing enhancer present in the GAR fragment, we inserted the 36-nt GAR sequence into the downstream exon of an enhancer-dependent splicing reporter substrate derived from the Drosophila melanogaster dsx gene (Fig. 2A). The intron in the dsx system is efficiently spliced only when an SR-dependent splicing enhancer is present in the downstream exon (15, 39, 40). Splicing of the parental dsx substrate (dsx-ΔE) is extremely weak (Fig. 2B, lane 1) because of a nonconsensus 3′ ss. Insertion of an SR-dependent splicing enhancer activates splicing, since SR proteins recruited by the ESE can stabilize the interaction of splicing factors with the weak 3′ ss (14, 40). As a control, an SR-dependent splicing enhancer derived from the avian sarcoma-leukosis virus (ASLV) was inserted into the downstream exon of the dsx substrate, generating the substrate dsx-ASLV, which was spliced efficiently (Fig. 2B, lane 2). Insertion of the GAR fragment to generate the dsx-GAR substrate increased splicing efficiency to a level similar to that obtained with the ASLV enhancer (Fig. 2B, lane 3), showing that the GAR sequence acts as a strong ESE.
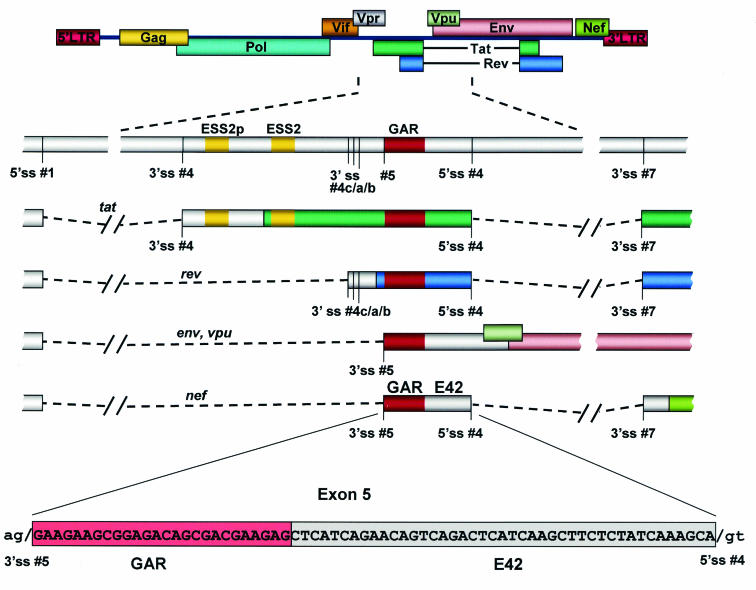
FIG. 1.
Schematic drawing of the HIV-1 genome. The region carrying tat and rev, coding exon 2, is enlarged (middle). The exonic splicing silencers regulating usage of 3′ ss no. 4, ESS2p and ESS2, are shown in yellow. The bidirectional exonic enhancer (red box) regulates usage of the upstream 3′ ss no. 4c/a/b and 5 as well as the downstream 5′ ss no. 4. The bidirectional ESE was named GAR for its rich purine content and is located within exon 5.
FIG. 2.
(A) The putative splicing enhancer present in the GAR fragment (black) was substituted for the ASLV enhancer (white) in the downstream exon of the enhancer-dependent splicing reporter substrate dsx-ΔE. (B) In vitro splicing reaction of the dsx substrates in HeLa cell nuclear extract (NE) (lanes 1 to 3) or splicing-deficient HeLa S100 cytoplasmic extract (lanes 4 to 10). S100 extract was complemented with total the SR protein preparation (lane 5) or with single SR proteins (lanes 6 to 10). All of the single SR proteins activated splicing of a β-globin splicing substrate with equal efficiency (data not shown).
To identify putative SR binding sites we analyzed the GAR region with the ESEfinder web interface (5a). Two SF2/ASF sites, one SRp55 site, and one SRp40 site were predicted to be present within the fragment. To determine whether these or other individual SR proteins promote splicing of the dsx-GAR transcripts, we complemented splicing-deficient HeLa S100 cytoplasmic extracts with single SR proteins. SR proteins are absent from the S100 extracts, and this leads to loss of splicing activity. The addition of SR proteins can complement this deficiency, recovering splicing activity (19). Only the total SR protein preparation, purified SF2/ASF, and SRp40 were able to efficiently complement the S100 extracts to activate splicing of the dsx-GAR substrate (Fig. 2B, lanes 5, 7, and 8). SRp70, SRp55, and SC35 were unable to stimulate splicing of the dsx-GAR RNA substrate (Fig. 2B, lanes 6, 9, and 10). As a control, all of the purified SR proteins activated splicing of a β-globin splicing substrate with equal efficiency (data not shown). These results indicate that the GAR fragment contains an SF2/ASF- and SRp40-dependent splicing enhancer.
To demonstrate that SF2/ASF and SRp40 interact with the GAR sequence on the binding sites predicted by the ESEfinder (5a), we mutated the putative SRp40 site, the two SF2/ASF sites, and all three sites as shown in Fig. 3A. As a control, we tested another fragment derived from the HIV-1 mRNA of identical length (Fig. 3A) (HIV no. 18). The HIV no. 18 fragment could not enhance splicing (Fig. 3B, lane 2). Mutation of the SRp40 binding site decreased the splicing efficiency about fourfold (Fig. 3B, lane 3) while mutation of the two SF2/ASF binding sites generated a sevenfold decrease in splicing efficiency (Fig. 3B, lane 4). Mutation of all three predicted SR protein binding sites produced a further decrease in splicing efficiency (Fig. 3B, lane 5). These results establish that the GAR sequence contains a complex SF2/ASF- and SRp40-dependent ESE that activates splicing at a weak upstream 3′ ss.
FIG. 3.
(A) Mutations of the SRp40 and SF2/ASF binding sites. A fragment derived from the HIV-1 env mRNA of identical length which was not predicted to contain an ESE (HIV no. 18) was used as a control fragment. Shaded boxes correspond to SF2/ASF binding sites, and nonshaded boxes correspond to SRp40 binding sites. A G-to-T mutation flanking the more 5′ predicted SF2/ASF binding site had been introduced to prevent generation of a new binding site in the cause of inactivating this site. (B) In vitro splicing assay of the dsx-GAR constructs in HeLa cells nuclear extracts.
The GAR ESE promotes binding of the U1 snRNP to the downstream 5′ ss no. 4.
We have previously shown that binding of the U1 snRNP to the 5′ ss no. 4 leads to efficient env expression, possibly by increasing the stability of the unspliced env mRNA (10, 17). To test the effect that the ESE contained in the GAR fragment has on the binding of the U1 snRNP to the 5′ ss no. 4, we introduced the mutations, summarized in Fig. 3A, in the SV-env/GAR expression vector (Fig. 4A). We transiently transfected HeLa-T4+ cells with the wild-type and mutated constructs. Since the env mRNA is efficiently exported to the cytoplasm only in the presence of Rev, the Rev expression vector SVcrev was cotransfected. Cells were harvested 48 h after transfection, and the env level was detected by Western blotting.
FIG. 4.
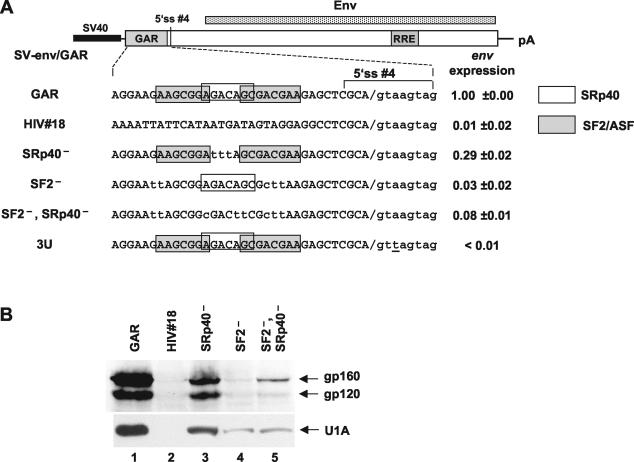
(A) Schematic drawing of the SV-env/GAR construct carrying the env open reading frame and the upstream GAR element. SF2 and SRp40 sites are indicated by boxes. SV40, SV40 early promoter; p(A), SV40 polyadenylation signal. Immunoblot analyses of env expression were quantified (right). (B, upper blot) Immunoblot analysis of glycoproteins expressed in HeLa-T4+ cells transfected with SVcrev, pGL3-control, and env expression vectors carrying different upstream sequences. (B, lower blot) RNA substrates were covalently linked to agarose beads and incubated in HeLa nuclear extracts. Proteins bound to the substrates were eluted, separated by SDS-polyacrylamide gel electrophoresis, and immunoblotted with U1A-specific antibody.
The wild-type GAR sequence led to efficient env expression (Fig. 4B, lane 1) while the control sequence HIV no. 18 substituted for the GAR sequence completely abolished it (Fig. 4B, lane 2). Inactivation of the predicted SRp40 binding site led to a threefold decrease in env expression (Fig. 4B, lane 3). The effect of mutations of the SF2/ASF and the combined SF2/ASF and SRp40 binding sites on env expression were more pronounced, reducing its level 30- and 12-fold, respectively (Fig. 4B, lanes 4 and 5). env expression correlated with U1 snRNP binding to the 5′ ss no. 4, as was shown by RNA affinity chromatography utilizing antibodies directed against the U1A component of the U1 snRNP (Fig. 4B, lower panel). Therefore, this result indicates that the same mutations affecting the GAR enhancer in the dsx-GAR in vitro splicing model are also affecting U1 snRNP binding to the 5′ ss no. 4 in vivo, suggesting that the SF2/ASF- and SRp40-dependent ESE is bidirectional. Indeed, it regulated splicing at the upstream 3′ ss in the dsx splicing reporter substrate and recruited U1 snRNP to the downstream 5′ ss no. 4 in vivo.
An MS2-SR/MS2 binding site system can functionally substitute for the GAR sequence.
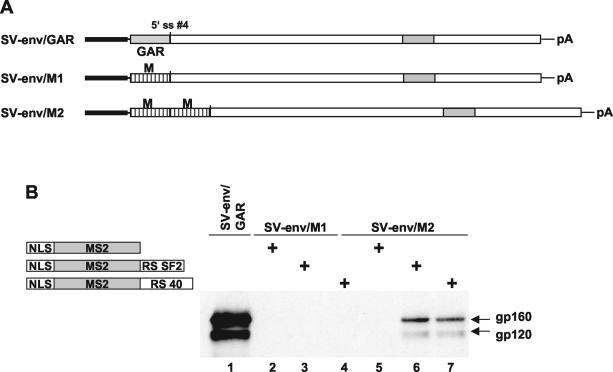
Analysis of env expression and U1 snRNP binding in Fig. 4 shows that mutation of both the SF2 and SRp40 binding sites induced a level of env expression slightly higher than that in the SF2 mutant alone (lanes 4 and 5). It is possible that the new mutations we inserted for the GAR region in the SF2− SRp40− mutant could be functionally active in increasing env expression by creating a suboptimal SR protein binding site, by increasing mRNA stability, or by some other mechanism. To confirm that SR protein binding upstream of the 5′ ss no. 4 is required for efficient env expression, the GAR sequence was substituted for either one or two binding sites for the MS2 bacteriophage coat protein in the viral substrate SV-env/GAR. Constructs expressing hybrid proteins, containing the MS2 protein and the SF2 or SRp40 RS domains, were cotransfected with the mutated viral substrates. This hybrid system has been previously shown to efficiently substitute for functional ESE-SR protein interactions (15). Surprisingly, substitution of the GAR sequence for one MS2 binding site did not induce env expression when the MS2-SF2 or MS2-SRp40 expression vectors were cotransfected (Fig. 5B, lanes 3 and 4). On the contrary, substitution of the GAR for two MS2 binding sites restored env expression after cotransfection with either the MS2-SF2 or the MS2-SRp40 expression vector (Fig. 5B, lanes 6 and 7). These results suggest that two distinct binding sites upstream of 5′ ss no. 4 are required for efficient U1 snRNP binding and env expression.
FIG. 5.
(A) Schematic drawing of the env expression constructs carrying the env open reading frame with the GAR region (SV-env/GAR) or the GAR region substituted for one (SV-env/M1) or two (SV-env/M2) MS2 binding sites (M). (B) Immunoblot analysis of glycoproteins expressed in HeLa-T4+ cells transfected with SV-env/GAR, SV-env/M1, or SV-env/M2. Viral expression vectors were cotransfected with expression vectors expressing the MS2 bacteriophage coat protein, the MS2 protein fused with the SF/ASF RS domain, or the MS2 protein fused with the SRp40 RS domain. NLS, nuclear localization signal.
Extended base pairing between 5′ ss no. 4 and the U1 snRNA is not sufficient for efficient env expression.
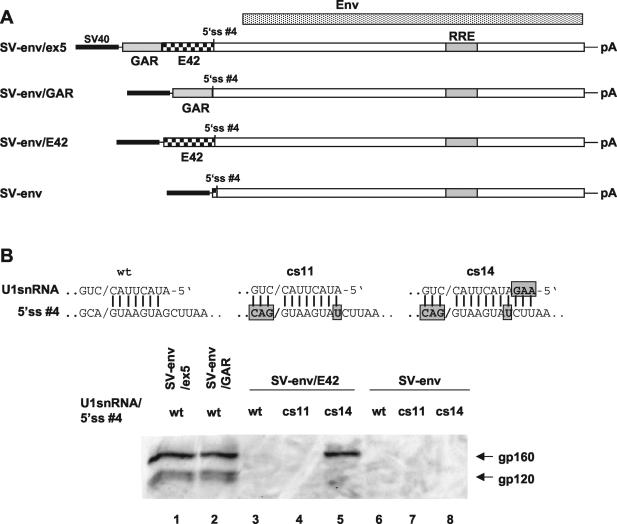
To further investigate the role of the GAR ESE in env expression and U1 snRNP recruitment, we analyzed the constructs summarized in Fig. 6A. As expected, the presence of the full exon 5 or the GAR fragment alone is required for the efficient expression of env (Fig. 6B, lanes 1 and 2). On the contrary, in the construct carrying the E42 fragment (SV-env/E42) or in the absence of exon 5 sequences (SV-env), env is not expressed (Fig. 6B, lanes 3 and 6).
FIG. 6.
(A) Schematic drawing of the env expression constructs carrying the env open reading frame with either the complete exon 5 sequence (SV-env/ex5), only the GAR region (SV-env/GAR), or the 3′ half of exon 5 (SV-env/E42) or without the exon 5 sequence (SV-env) upstream of 5′ ss no. 4. SV40, SV40 early promoter; pA, SV40 polyadenylation signal. (B) Immunoblot analysis of glycoproteins expressed in HeLa-T4+ cells transfected with SVcrev, pGL3-control, or env expression vectors and an expression plasmid coding for wild-type U1 snRNA (wt) or U1 snRNAs with additional matching nucleotides (cs14). A perfect match with wild-type U1 snRNA was achieved by mutating the 5′ ss (cs11). Additional base pairs are indicated by boxes.
U1 snRNP binding to the 5′ ss no. 4 is stabilized by a 7-nt base pairing. Since SR proteins interacting with ESEs are known to stabilize the interaction between the U1 snRNP and a weak 5′ ss, we wanted to test the possibility that an increased base pairing between the 5′ ss no. 4 and the U1 snRNA will compensate for the absence of the GAR ESE. In cs11 we increased the base pairing between the U1 snRNA and the 5′ ss no. 4 to 11 nt, mutating the 5′ ss no. 4 (Fig. 6B). In cs14, the base pairing was increased to 14 nt by cotransfecting a suppressor U1 snRNA, as indicated in Fig. 6B. Increasing the base pairing to 11 nt in both SV-env/E42 and SV-env did not recover env expression (Fig. 6B, lanes 4 and 7). Surprisingly, extending the base pairing to 14 nt in the SV-env/E42 did recover env expression (Fig. 6B, lane 5) while no effect on env levels was detected in the SV-env construct when the base pairing was increased to 14 nt (Fig. 6B, lane 8).
These results indicate that a stable interaction between the 5′ ss no. 4 and the U1 snRNP leading to env expression is dependent on the presence of the upstream GAR ESE even when the base pairing between the 5′ ss and the U1 snRNA is extended. Furthermore, it appears that the E42 fragment contains a suboptimal ESE, since it can promote env expression when the base pairing between the U1 snRNA and the 5′ ss is increased to 14 nt.
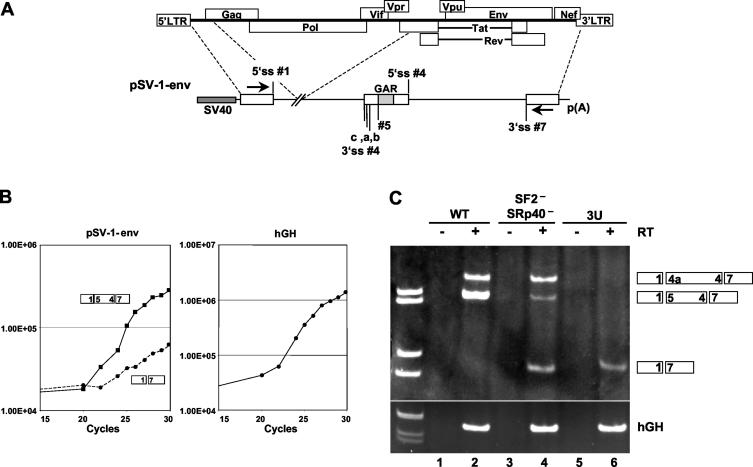
The GAR ESE is required for efficient exon 5 and rev exon 2 inclusion.
Next we analyzed the ability of the GAR ESE to regulate inclusion of exon 5 and rev exon 2 in a 3 exon-2 intron minigene. Exon 5 is generated by splicing at 3′ ss no. 5 in combination with 5′ ss no. 4 to produce nef-specific mRNAs. rev exon 2, also named rev coding exon 1, is generated by splicing at either 3′ ss no. 4c, 4a, or 4b in combination with 5′ ss no. 4. We transfected HeLa-T4+ cells with the subgenomic HIV construct SV-1-env shown in Fig. 7A. The SV-1-env construct contains HIV-1 exon 1, a deletion in the gag-pol region, and the full rev-env-nef coding region. The splicing pattern of this construct was analyzed by RT-PCR analysis, carried out within the linear range of amplification for the viral template and with hGH substrate as a control (Fig. 7B). Identification of the splice junctions were obtained by sequencing of the gel-isolated PCR fragments. The wild-type substrate SV-1-env efficiently expressed rev exon 2 and exon 5 (Fig. 7C, lane 2), utilizing the 3′ ss no. 4a and 5 in combination with the 5′ ss no. 4. Mutation of the SRp40 and SF2/ASF binding sites in the SV-1-env constructs down-regulated inclusion of rev exon 2 and exon 5 (Fig. 7C, lane 4). Furthermore, a new splicing product was originated by the joining of 5′ ss no. 1 with 3′ ss no. 7, thus skipping the intervening exon. Interestingly, mutation of the 5′ ss no. 4 completely inhibited splicing of both exon 5 and rev exon 2 (Fig. 7C, lane 6). This result shows that both the GAR ESE and a strong 5′ ss are required for efficient splicing of the viral substrate and correct definition of exon 5 and rev exon 2. Interestingly, under these experimental conditions, usage of 3′ ss no. 4b and 4c has not been detected in significant amounts. This could be due to the fact that these ss are rarely utilized by the splicing machinery in vivo.
FIG. 7.
(A) Schematic diagram of the SV-1-env construct. Start codons for the expression of gag, tat, and rev were mutated. SV-1-env 3U contains a mutation in position +3 of 5′ ss no. 4. The positions of the primers used for RT-PCR are indicated by arrows. (B) Determination of the linear amplification range for SV-1-env and hGH RT-PCR assays. (C) RT-PCR assay. HeLa-T4+ cells were transfected with SV-1-env (lanes 1 and 2), SV-1-env SF2− SRp40− (lanes 3 and 4), or SV-1-env 3U (lanes 5 and 6) and cotransfected with pXGH5 expressing the hGH mRNA as an internal control for transfection efficiency. WT, wild type; +, present, −, absent.
DISCUSSION
In HIV-1, efficient viral replication is dependent on a highly controlled regulation of alternative splicing. A combination of weak 3′ ss, splicing enhancers, and splicing silencers is responsible for the correct splicing of more than 40 viral mRNAs. In this work, we analyzed a central region of the viral genome where a cluster of 4 alternative 3′ ss regulates the splicing of the rev, vpu, env, and nef genes. Regulation of these 3′ ss is determined by a downstream SR protein-dependent ESE we have termed GAR. The GAR element appears to act as a bidirectional splicing enhancer, since it also stabilizes the interaction of U1 snRNP with the downstream 5′ ss no. 4. This interaction is not only required for correct splicing of the mRNA species coding for tat, rev, and nef but also for proper expression of the partially spliced env mRNA. It is thought that the binding of the U1 snRNP to this 5′ ss can increase the stability of the viral mRNA (7, 17, 25); nevertheless, a synergistic effect on the Rev-dependent nuclear export of the unspliced mRNA cannot be ruled out. Utilizing a combination of computational (5a) and in vitro approaches (Fig. 2 and 3), we identified SR proteins SF2/ASF and SRp40 as the factors activating the GAR splicing enhancer.
Although several SR protein-dependent ESE have been demonstrated to enhance either 3′ or 5′ ss selection, examples of splicing enhancer sequences able to direct splicing factors to both upstream 3′ ss and downstream 5′ ss are scarce (4, 22). Having shown the activity of the GAR ESE on upstream 3′ ss in the dsx system, the bidirectional activity of the GAR splicing enhancer was confirmed by in vivo transient transfection assays. Mutation of the GAR sequence and its substitution of two MS2 binding sites in the MS2-RS system regulated the usage of the 5′ ss no. 4 and env expression (Fig. 4 and 5).
Further proof of the essential role of the GAR ESE in the expression of the partially spliced env mRNA is given by the experiments aimed at increasing the base pairing between the 5′ ss no. 4 and the U1 snRNA (Fig. 6). Surprisingly, in the absence of the GAR ESE, even an extended base pairing of 11 nt between the 5′ ss and the U1 snRNA was not sufficient to restore env expression. Furthermore, if the sequence we named E42 located downstream of the GAR ESE was also deleted, env expression could not be restored, even when the base pairing between the 5′ ss no. 4 and the U1 snRNA was increased to 14 nt. This may indicate that a suboptimal enhancer is present in the E42 fragment. Therefore, an SR-dependent ESE upstream of the 5′ ss no. 4 appears to be required for U1 snRNP binding, which results in env expression. Furthermore, when in the SV-env/E42 expression construct, the base pairing between the U1 snRNA and the 5′ss no. 4 was increased to 14 nt, env expression was recovered. This indicates that both upstream enhancer sequences and base pairing between the 5′ ss and the U1 snRNP are required for efficient env expression. Nevertheless, it is also possible that assembly of the 5′ cap structure at the 5′ end of the substrate could sterically hinder the assembly of the U1 snRNP at the 5′ ss in the SV-env substrates.
Analysis of the GAR ESE function in a subgenomic 3 exon-2 intron minigene showed that it is essential for the proper inclusion of the rev/nef exon 2. A mutation in the GAR ESE induced skipping of the rev/nef-specific central exon. It also appears that the choice of the 3′ ss no. 5, the more proximal with respect to the GAR ESE, is downregulated the most (Fig. 7). A similar result with complete inhibition of the central exon inclusion could be achieved by mutations of 5′ ss no. 4 (Fig. 7) (10). This result correlates well with previous observations suggesting that a complex set of interactions by the splicing machinery must occur, which spans the exon and its 3′ and 5′ ss (3). Furthermore, lack of binding of SR proteins to the GAR ESE and mutations inactivating the 5′ ss no. 4 both appear to downregulate the total amount of spliced mRNA. It is conceivable that although part of the precursor RNA is spliced so as to skip the central exon, the major part does not undergo splicing. Lack of the ESE will further reduce the stability and/or efficient export of the unspliced mRNA.
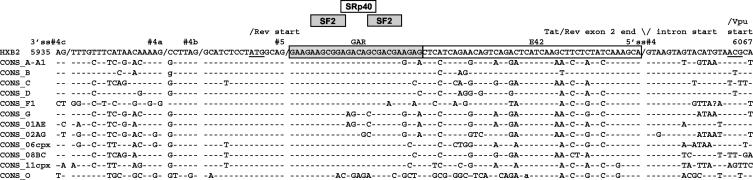
A proper understanding of the mechanisms and the cellular factors regulating the viral mRNA processing could reveal new targets for antiviral therapy. Due to the high mutation rate of the viral genome, virus isolates differ greatly and virus strains that share a higher degree of homology are grouped into subtypes. Sequence differences among the HIV-1 strains and subtypes suggest that regulation of splicing may differ in different strains. Surprisingly, analysis of the viral sequences of different viral subtypes have shown that, with the exception of the evolutionary distant O subtype, the GAR sequence is remarkably well conserved compared to the E42 fragment (Fig. 8). Since the GAR and the E42 fragment are both within the tat/rev coding region, the extended sequence conservation within the GAR fragments indicates an added selective pressure, maintaining an unchanged ESE sequence among different isolates. This is confirmed by mutation of the GAR ESE, which results in the skipping of the first coding exon of Tat and the use of the terminal 3′ ss no. 7. Given its key function in viral gene expression, this conserved viral sequence could be considered a potential target for antiviral therapy.
FIG. 8.
Consensus sequences for each HIV-1 subtype were aligned (20). The GAR ESE, the E42 fragment, and the predicted SF2/ASF and SRp40 binding sites are indicated. The 3′ and 5′ ss and Rev ATG translation initiation site are indicated. Since the GAR and the E42 fragment are both within the rev coding region, the extended sequence conservation within the GAR fragment indicates an added selective pressure to maintain the ESE sequence unchanged among different isolates.
Acknowledgments
We thank K. Strebel and M. Martin for providing plasmid pNLA1, M. Gething for pSVT7, B. R. Graveley for pHISBIT-MS2-RS9G8, R. Axel for HeLa-T4+ cells (through the MRC AIDS Directed Program Reagent Project), A. Zahler for SR proteins, A. Krainer for recombinant SF2, and Dade Behring for the anti-gp120 monoclonal antibody (87-133/026). We are also grateful to Imke Meyer and Maria Thieme for excellent technical assistance, Carolin Konermann for cloning MS2-SR expression vectors, and K. Köhrer and S. Scheuring for plasmid sequencing.
This work was supported by the NIH/NIAID grant 5R01AI052820-02 to M.C., by a Deutsche Forschungsgemeinschaft grant to H.S. (SCHA 909/1-1 and SCHA 909/2-1), and by a grant from the Heinz-Ansmann-Stiftung and a short-term fellowship of the Boehringer Ingelheim Fonds to M.F., who also had a research fellowship from the Duesseldorf Entrepreneurs Foundation. S.K. was supported by a Graduierten Fellowship from the University of Duesseldorf.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt, B. A., D. Hesslein, L. J. Chang, and C. M. Stoltzfus. 1994. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 14:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270:2411-2414. [DOI] [PubMed] [Google Scholar]
- 4.Bourgeois, C. F., M. Popielarz, G. Hildwein, and J. Stevenin. 1999. Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol. Cell. Biol. 19:7347-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Cartegni, L., J. Wang, Z. Zhu, M. Q. Zhang, and A. R. Krainer. 2003. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31:3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavaloc, Y., C. F. Bourgeois, L. Kister, and J. Stevenin. 1999. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5:468-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, D. D., and P. A. Sharp. 1989. Regulation by HIV Rev depends upon recognition of splice sites. Cell 59:789-795. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freund, M., C. Asang, S. Kammler, C. Konermann, J. Krummheuer, M. Hipp, I. Meyer, W. Gierling, S. Theiss, T. Preuss, D. Schindler, J. Kjems, and H. Schaal. 2003. A novel approach to describe a U1 snRNA binding site. Nucleic Acids Res. 31:6963-6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furtado, M. R., R. Balachandran, P. Gupta, and S. M. Wolinsky. 1991. Analysis of alternatively spliced human immunodeficiency virus type-1 mRNA species, one of which encodes a novel tat-env fusion protein. Virology 185:258-270. [DOI] [PubMed] [Google Scholar]
- 12.Gottlinger, H. G., T. Dorfman, E. A. Cohen, and W. A. Haseltine. 1992. The role of the tnv protein and tnv RNA splicing signals in replication of HIV-1 IIIB isolates. Virology 189:618-628. [DOI] [PubMed] [Google Scholar]
- 13.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graveley, B. R., K. J. Hertel, and T. Maniatis. 1998. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 17:6747-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graveley, B. R., and T. Maniatis. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1:765-771. [DOI] [PubMed] [Google Scholar]
- 16.Hope, T. J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365:186-191. [DOI] [PubMed] [Google Scholar]
- 17.Kammler, S., C. Leurs, M. Freund, J. Krummheuer, K. Seidel, T. O. Tange, M. K. Lund, J. Kjems, A. Scheid, and H. Schaal. 2001. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA 7:421-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohtz, J. D., S. F. Jamison, C. L. Will, P. Zuo, R. Luhrmann, M. A. Garcia-Blanco, and J. L. Manley. 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368:119-124. [DOI] [PubMed] [Google Scholar]
- 19.Krainer, A. R., A. Mayeda, D. Kozak, and G. Binns. 1991. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66:383-394. [DOI] [PubMed] [Google Scholar]
- 20.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, P. A. Marx, F. McCutchan, J. W. Mellors, and S. M. Wolinsky. 2001. HIV sequence compendium, LA-UR 02-2877. Theoretical Biology and Biophysics Group. Los Alamos National Laboratory, Los Alamos, N.M.
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lam, B. J., and K. J. Hertel. 2002. A general role for splicing enhancers in exon definition. RNA 8:1233-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langland, J. O., S. M. Pettiford, and B. L. Jacobs. 1995. Nucleic acid affinity chromatography: preparation and characterization of double-stranded RNA agarose. Protein Expr. Purif. 6:25-32. [DOI] [PubMed] [Google Scholar]
- 24.Liu, H. X., M. Zhang, and A. R. Krainer. 1998. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12:1998-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, X. B., J. Heimer, D. Rekosh, and M. L. Hammarskjold. 1990. U1 small nuclear RNA plays a direct role in the formation of a rev-regulated human immunodeficiency virus env mRNA that remains unspliced. Proc. Natl. Acad. Sci. USA 87:7598-7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47:333-348. [DOI] [PubMed] [Google Scholar]
- 27.Malim, M. H., S. Bohnlein, J. Hauber, and B. R. Cullen. 1989. Functional dissection of the HIV-1 Rev trans-activator-derivation of a trans-dominant repressor of Rev function. Cell 58:205-214. [DOI] [PubMed] [Google Scholar]
- 28.Mayeda, A., and A. R. Krainer. 1999. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 118:309-314. [DOI] [PubMed] [Google Scholar]
- 29.Myers, G., B. Korber, B. H. Hahn, K. T. Jeang, J. W. Mellors, F. E. McCutchan, L. E. Henderson, and G. N. Pavlakis. 1995. Human retroviruses and AIDS. A compilation and analysis of nucleic acid and amino acid sequences, LA-UR 02-2877. Theoretical Biology and Biophysics Group. Los Alamos National Laboratory, Los Alamos, N.M.
- 30.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 31.Purcell, D. F., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robberson, B. L., G. J. Cote, and S. M. Berget. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 10:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanford, J. R., D. Longman, and J. F. Caceres. 2003. Multiple roles of the SR protein family in splicing regulation. Prog. Mol. Subcell. Biol. 31:33-58. [DOI] [PubMed] [Google Scholar]
- 34.Schaal, H., M. Klein, P. Gehrmann, O. Adams, and A. Scheid. 1995. Requirement of N-terminal amino acid residues of gp41 for human immunodeficiency virus type 1-mediated cell fusion. J. Virol. 69:3308-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaal, H., P. Pfeiffer, M. Klein, P. Gehrmann, and A. Scheid. 1993. Use of DNA end joining activity of a Xenopus laevis egg extract for construction of deletions and expression vectors for HIV-1 Tat and Rev proteins. Gene 124:275-280. [DOI] [PubMed] [Google Scholar]
- 36.Selden, R. F., K. B. Howie, M. E. Rowe, H. M. Goodman, and D. D. Moore. 1986. Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol. Cell. Biol. 6:3173-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature 328:728-730. [DOI] [PubMed] [Google Scholar]
- 38.Tacke, R., and J. L. Manley. 1995. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 14:3540-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian, M., and T. Maniatis. 1992. Positive control of pre-mRNA splicing in vitro. Science 256:237-240. [DOI] [PubMed] [Google Scholar]
- 40.Tian, M., and T. Maniatis. 1994. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 8:1703-1712. [DOI] [PubMed] [Google Scholar]
- 41.Watakabe, A., K. Tanaka, and Y. Shimura. 1993. The role of exon sequences in splice site selection. Genes Dev. 7:407-418. [DOI] [PubMed] [Google Scholar]
- 42.Wentz, M. P., B. E. Moore, M. W. Cloyd, S. M. Berget, and L. A. Donehower. 1997. A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type 1 induces dominant aberrant splicing and arrests virus production. J. Virol. 71:8542-8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061-1070. [DOI] [PubMed] [Google Scholar]
- 44.Zahler, A. M. 1999. Purification of SR protein splicing factors. Methods Mol. Biol. 118:419-432. [DOI] [PubMed] [Google Scholar]
- 45.Zahler, A. M., W. S. Lane, J. A. Stolk, and M. B. Roth. 1992. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6:837-847. [DOI] [PubMed] [Google Scholar]