Abstract
Background
A variety of markers have been proposed to identify breast cancer stem cells. Here, we used immunohistostaining and flow cytometry to analyze their interrelationships and to sort cells for tumorigenicity studies.
Methods
Cytokeratin, CD44, and CD90 were localized to primary breast cancer and normal breast (NB) tissue by immunohistostaining and related to CD117 and CD133 expression by flow cytometry. Immunodeficient NOD.CB17-Prkdcscid/J and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice were used to test tumorigenicity of sorted CD90+ low-light scatter, CD90+ high-light scatter, and CD90neg tumor cells.
Results
NB basal cells coexpressed CD44 and CD90. As cells transited luminally, CD44 was retained and downmodulated, and CD90 was lost and cytokeratin increased. In breast tumors, basal-like CD44+/CD90+ cells were localized to the tumor periphery, adjacent to CD90+ stroma. Like normal luminal cells, interior tumor cells were CD44+/CD90−. Immunophenotyping (CD44/CD90/CD117/CD133) of cytokeratin+ cells revealed no significant difference in expression between tumors and tumor-free breast. In both, CD133 was distributed approximately equally among CD44/CD90 subsets, whereas CD117 expression was highest in the basal-associated CD44+/CD90+ subset. Sorted CD90+ pleural effusion cells with lymphoid light scatter, 49% of which were CD44+, were uniquely tumorigenic in immunodeficient mice (100 cells/injection).
Conclusions
Our data demonstrate that all tumors contain a small population of CD44+/CD90+ cells, mimicking the phenotype of ductal-basal cells. These are localized to the tumor periphery, adjacent to CD90+ stroma. Among the nonhematopoietic, nonmesothelial cells found in metastatic pleural effusions, low-light scatter CD90+ cells are most potently tumorigenic, compared to high-scatter CD90+ cells and CD90− cells.
Keywords: cancer stem cells, breast cancer, flow cytometry, immunohistochemistry, immunofluorescence, cytokeratin, CD44, CD90, CD133, CD117
The cancer stem-cell hypothesis seeks to explain tumor self-renewal, therapy resistance, metastasis, and relapse in relation to the properties of normal tissue stem cells (1–3). Putative cancer stem and progenitor cells have been detected in a variety of epithelial cancers using markers that are associated with normal tissue stem cells but highly contextual (1,4– 12). These include the mesenchymal stem-cell markers CD44 (7) and CD90 (8,13), stem/progenitor markers CD133 (4,14), CD117 (15,16), ABC transporters (17,18), and aldehyde dehydrogenase (5,19). The relationships between these markers have not been well studied in any epithelial cancer, nor has expression on tumor tissues been compared with expression in normal tissues from which the tumors arise. In this report, we examine the expression of the stem/progenitor-associated markers CD44, CD90, CD117, and CD133 on cytokeratin+ hematopoietic (Heme) lineage negative populations identified in primary ductal breast cancer, adjacent tumor-free tissue, normal breast (NB) tissue removed during mammoplasty, and metastatic pleural effusions. The tissue locations of populations identified by multiparameter flow cytometry performed on disaggregated tissue were inferred in comparison with immunohistostaining performed on tissue sections. We determined that resting cytokeratin+/CD90+ cells (half of which express CD44) are localized to the periphery of tumor nests, mimicking the basal layer of normal ductal tissue.
In a subset of samples, we also examined the fidelity of marker expression following short-term in vitro selection under conditions designed for the propagation of human embryonic stem (ES) cells.
MATERIALS AND METHODS
Patient Samples
Specimens were collected under protocols exempted by the University of Pittsburgh Internal Review Board (UPCI 04-162, 05-06140), because they use only waste materials and deidentified clinical data. Malignant pleural effusions (MPEs, n = 3) were all positively identified as metastatic adenocarcinoma of the breast. Tissues used for flow cytometry included breast tumors (n = 24), adjacent tumor-free tissue (n = 24), and normal ductal tissue from mammoplasty (n = 3), all of which were processed within 2 h of surgery.
Sample Preparation
Single-cell suspensions were prepared from all solid tissues and pleural fluid. Tissues were minced with paired scalpels. Minced tissues and MPE were digested with type I collagenase (0.4% in RPMI 1640 medium, Cat. No. C-0130, Sigma Chemicals, St. Louis, MO) and DNase (350 KU/ml, Sigma Chemicals, Cat. No. D-5025) and disaggregated through 100 mesh stainless steel screens (17). Viable cells were concentrated and separated from erythrocytes and debris on a Ficoll–Hypaque gradient (Histopaque 1077, Sigma Chemicals). Erythrocytes were lysed using an ammonium chloride lysing solution (Beckman-Coulter, Fullerton, CA, Cat No. IM3630d).
Staining and Flow Cytometry
To minimize nonspecific binding of fluorochrome-conjugated antibodies, pelleted cell suspensions were prein-cubated for 5 min with neat decomplemented (56°C, 30 min) mouse serum (5 µl) (10). Before intracellular cytokeratin staining, cells were stained for surface markers [2 µl each added to the cell pellet and 15–30 min on ice; CD44-PE (Beckman-Coulter, Cat No. A32537), CD90-biotin (BD, Cat. No. 555594), streptavidin-energy-coupled dye (ECD; Beckman-Coulter, Cat. No. IM3326), Heme lineage cocktail: CD14-PECy5 (Beckman-Coulter, Cat. No. IM2640U), CD33-PECy5 (Beckman-Coulter, Cat. No. IM2647U), glycophorin A-PECy5 (BD Biosciences, Cat. No. 559944), CD133-allophycocyanin (APC; Miltenyi Biotech Cat. No. 130-090-854), CD117-phycoerythrin cyanine 7 (PC7; Beckman-Coulter, Cat. No. IM3698), CD45-allophycocyanin cyanine 7 (APCC7; BD, Cat. No. 557833)] and fixed with 2% methanol-free formaldehyde (Polysciences, Warrington, PA). Cells were then permeabilized with 0.1% saponin (Beckman-Coulter) in phosphate-buffered saline (PBS) with 0.5% human serum albumin (10 min at room temperature). Cell pellets were incubated with 5 µl of neat mouse serum for 5 min, centrifuged, and decanted. The cell pellet was disaggregated and incubated with 2 µl of antipan cytokeratin-fluorescein isothiocyanate (FITC; Beckman-Coulter, Cat. No. IM2356) for 30 min. Cell pellets were diluted to a cell concentration of 10 million cells/400 µl of staining buffer. DAPI (Sigma Chemicals, Cat. D1306) was added to a final concentration of 5 µg/ml before sample acquisition (10).
Nine-color analysis was performed using the three-laser, nine-color CyAn ADP cytometer (Beckman-Coulter, Miami, FL). An effort was made to acquire a total of 10 million cells per sample at rates not exceeding 10,000 events/s. DAPI was acquired in two fluorescence channels (FL6 and FL7), with PMT gain optimized for linear (cell cycle), and log (elimination of hypodiploid events) acquisition, respectively. The cytometer was calibrated to predetermined photomultiplier target channels before each use using eight-peak Rainbow Calibration Particles (Spherotech, Libertyville, IL, Cat. No. RCP-30-5A).
Offline compensation and analyses were performed using the VenturiOne analysis package using scalable parallel processing and designed specifically for multiparameter rare event problems (Applied Cytometry, Dinnington, Sheffield, UK). Data analysis was performed using methods that we have previously described in detail (20). Flow cytometric histograms of side scatter and fluorescence parameters are displayed on a four-decade logarithmic scale. Forward scatter and DAPI fluorescence for cell cycle are displayed on linear scales. Spectral compensation matrices were calculated for each staining combination within each experiment using single-stained mouse IgG capture beads (Becton Dickinson, Cat. No. 552843) for each tandem antibody and BD Calibrite beads for FITC, PE, and APC controls.
Flow-Cytometric Cell Sorting
Staining for surface markers was performed as described earlier using the following antibodies: CD326-FITC [epithelial cell adhesion molecule (EpCAM), human epithelial antigen, TACSTD1, Miltenyi Biotech Cat. No. 130-080-301]; CD44-PE, CD90-biotin, streptavidin-ECD, heme lineage cocktail-PECy5, and CD31-APC (R&D Systems, Cat. No.FAB 3567A); CD117-PC7 and CD45-APCC7 (BD, Cat. No. 557833). Stained cells were resuspended to 10 × 106 /ml in PBS with 2 mM EDTA, 0.5% BSA, and filtered through a 70-µm cap filter. DAPI was added immediately before sorting. Sorting was performed on a three-laser eight-colored Beckman Coulter legacy MoFlo equipped with a temperature-controlled SmartStation sample handler and a 100-µm nozzle. The cytometer was calibrated before each use using SpectrAlign beads and eight-peak Rainbow Calibration Particles. The sample was maintained at 4°C during sorting and sorted into ice-cold 100% newborn calf serum.
Tumor Xenografts
Female NOD.CB17-Prkdcscid/J (NOD/SCID, Cat. No. 001303) and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG, Cat. No. 005557) mice 6–8 weeks of age were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed five to a cage in a specific pathogen-free environment. Before injection of tumor cells, mice were anesthetized by methoxyflurane inhalation. For each injection, 100 sorted cells were admixed with 10,000 irradiated unsorted tumor (100 Gy from a 137Ce source) suspended in 25-µl ice-cold clarified MPE fluid plus 25 µl Matrigel (Becton Dickinson). Fifty microliters of ice-cold cell suspension were injected subcutaneously into the mammary fat pads (four injections per animal). Animals were examined twice weekly for behavioral changes and evidence of tumor and sacrificed ~6 months after injection.
Immunohistostaining
Paraffin sections (4–6-µm thickness) were prepared from embedded tissues. Both individual paraffin-embedded tissue specimens and specimens on tissue microarrays (TMAs) were evaluated. TMAs were constructed in-house using a manual tissue arrayer (Beecher Instruments, Sun Prairie, WI). The tumor TMA was constructed from 64 randomly selected well-characterized invasive breast carcinomas. Of these 64 cases, 52 were ductal, 9 lobular, 1 metaplastic, and 1 mixed ductal and lobular carcinoma. Three 0.6-mm tissue cores were obtained from one to two tissue blocks on each case. The benign breast TMA was constructed from 58 reduction mammoplasty specimens from 30 patients. Each case was represented with threefold redundancy similar to the tumor TMA. All interpretable samples were included in the analysis.
Tissue sections were heated (60°C, 20 min), deparaffinized (three washes in xylenes), rehydrated by successive washes in absolute ethanol, 90% ethanol, 75% ethanol, and deionized water and rinsed twice in Dako wash buffer (Dako).
For immunofluorescent staining, antigen retrieval was performed at 125°C for 20 min in pH 9.0-EDTA buffer (Dako). After two washes in DAKO wash buffer, the tissue sections were incubated for 60 min in a blocking solution (PBS, 5% goat serum, and 0.05% Tween 20) to reduce nonspecific antibody binding. All immunofluorescent staining was performed using primary antibodies CD44 [1:25 (15.2 µg/ml final concentration), Dako, Cat. No. M7082, clone DF1485] and CD90 [1:10 (6.2 µg/ml), BD Biosciences Cat. No. 550402, clone 5E10]. Universal Negative Control (Ready to use, Dako cat. No. N1698) was substituted for primary antibody. All primary antibodies and controls were incubated overnight at 4°C Tissue sections were washed twice using DAKO wash buffer before applying biotinylated secondary goat anti-mouse antibody [1:500 (1.58 µg/ml), Dako Cat. No. E0433] for 1 h at room temperature. Tissue sections were washed twice with Dako wash buffer and incubated with streptavidin-Cy3 [1:500 (2 µg/ml), Sigma Cat. No. 6402] for 30 min at room temperature. Slides were washed again, and tissue sections were incubated with pan-cytokeratin FITC-conjugated antibody [1:50 (28 µg/ml), clone C-11, Abcam Cat. No. ab11212] for 1 h at room temperature. Stained tissues sections were washed again twice in DAKO wash buffer, and nuclear staining was attained through 5-min incubation with DAPI (7.15 µM Invitrogen, Cat. No. D1306). Slides were washed twice in PBS-A and mounted in Prolong Gold antifade reagent (Invitrogen, Cat. No. P36934). Immunofluorescent staining was observed and photographed using an epifluorescence microscope (Nikon Eclipse TE 2000-U).
For immunohistochemical staining (mouse xenografts and supplement), antigen retrieval was performed at 125°C for 20 min in either pH 6.0 or pH 9.0-EDTA buffers (Dako) depending on the antigen. Immunohistochemistry on treated paraffin sections was performed by using the EnVisionTM+ Dual Link Kit (Dako). Endogenous Enzyme Block (Dako) was applied for 10 min to quench any endogenous peroxidase. After two washes, the tissue sections were incubated for 60 min in a blocking solution (PBS, 5% goat serum, and 0.05% Tween 20) to reduce nonspecific antibody binding. All immunohistochemistry experiments were performed using primary antibodies cytokeratin (CTK; 1:100 CTK in blocking solution clone MNF116, Dako, Cat. No. M0821), CD44 [1:25, Dako, Cat. No. M7082, clone DF1485, reported to bind to all CD44 isoforms (21)], estrogen receptor a (Ready to use, Dako, Cat. No. N1575), Ki67 (Ready to use, Dako, cat. No. N1633, clone, MIB-1), sonic hedgehog (1:100, Abcam, cat. No. AB53281), and CD90 (1:25, BD Biosciences Cat. No. 550402, clone 5E10). Universal Negative Control for n-series mouse primary antibodies (Ready to use, Dako Cat. No. N1698) was substituted for primary antibody as a negative control. All primary antibodies and controls were incubated for 1 h at room temperature. Tissue sections were washed before applying horseradish peroxidase-labeled polymer (Dako Cat. No. K4061) for 30 min at room temperature. Staining was completed by incubating the specimens with 3,3′-diaminobenzidine+ substrate-chromogen for 5–10 min (as determined by microscopic observation). Finally, tissue sections were washed twice in Dako wash buffer and cell nuclei stained for 3 min with hematoxylin (Dako Cat. No. S3302). Sections were washed in deionized water and sequentially immerged for 1 min in 75% ethanol, 90% ethanol, absolute ethanol, and thrice in xylene. Slides were mounted in nonaqueous medium (Cyto-sealTM280, Richard-Allan Scientific). Immunohistochemistry slides were photographed under brightfield microscopy using a digital camera and software (Spot Insight 2 Meg FW Color Mosaic model 18.2, Diagnostic Instruments) interfaced to a Nikon Labophot microscope.
In Vitro Culture Conditions for the Proliferation and Differentiation of Tumor-derived Cells
Single-cell suspensions were cultured in modified human ES cell medium at a density of 10,000–100,000 cells/96 well in 10-replicate wells and scored for growth and differentiation at 1, 2, 3, and 4 weeks. Human ES medium consisted of knock-out DMEM medium (Invitrogen Cat. No. 10829-018), 50 µM 2ME, 20% knock-out serum replacement (Invitrogen Cat. No. 10828028), 10 mM nonessential amino acids (Invitrogen Cat. No. 11140-050), and basic fibroblast growth factor 4 ng/ml (Invitrogen Cat. No. 13256-029). Individual culture wells were photographed and scored for the presence of colonies as well as cellular and colony morphologies. Confluent wells were trypsinized, split 1:3, and expanded in six well plates or harvested for flow cytometry.
RESULTS
Detection of CD44 and CD90 on Cytokeratin Positive Serial Sections from Tumor and Adjacent Nontumor Tissue
Flow cytometry permits analysis of multiple markers on single cells at the expense of histological context and selection artifact. To correlate our multiparameter flow cytometric data in context, we examined expression of key markers by immunohistostaining. Figure 1 shows immunofluorescent staining of an infiltrating ductal carcinoma and adjacent tumor-free breast ductal tissue. Serial sections were stained with antibodies directed against pan-cytokeratins, the adhesion molecule CD44, and the stem/progenitor marker CD90. Our analysis focused on CD44 (7,22) and CD90 (10,22), which have been proposed as key markers identifying breast cancer stem cells.
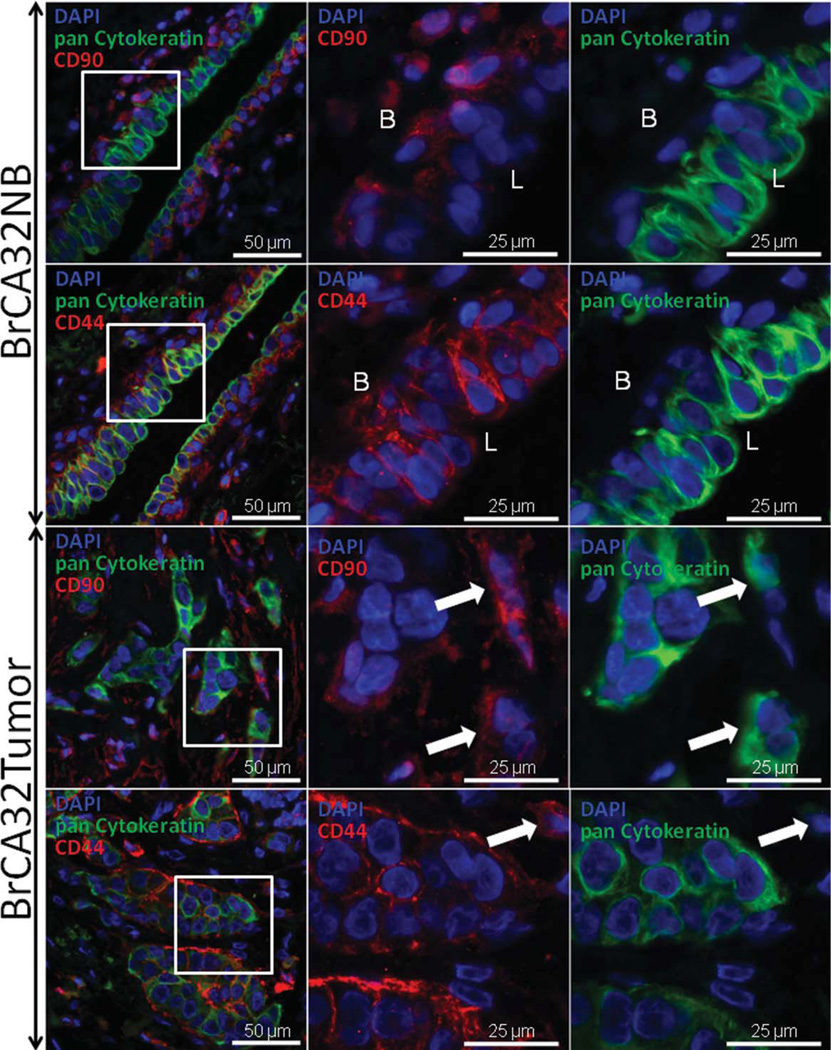
Fig. 1.
Expression of cytokeratin, CD44 and CD90, in NB ducts and invasive ductal carcinoma. Three-color immunofluorescent staining was performed on serial paraffin sections from adjacent tumor-free breast and a breast tumor from the same patient. The first column of photomicrographs represents a composite of DAPI, FITC, and Cy3 staining and was taken using a 10× objective. The white square indicates the field acquired in the second (DAPI plus Cy3) and third (DAPI plus FITC) columns using a 40× objective. In NB ducts, CD90 expression was limited to basal cells and scattered stromal/vascular cells. CD44 expression was present on basal (B) and luminal (L) ductal cells, whereas cytokeratin was not expressed on basal cells but increased during the basal to luminal transition. Scattered CD44bright/cytokeratin− cells (lymphocytes) were also seen in the stroma. Ductal adventitial cells were uniformly alpha smooth muscle actin (ASMA) positive (Supporting Information Fig. 2). CD90 was largely absent in tumor cells central to tumor nests, but was expressed on both cytokeratin+ tumor cells (arrows) and cytokeratin− stromal cells. CD44 was expressed in central and peripheral cytokeratin+ tumor cells, and less commonly in scattered cytokeratin− stromal cells (arrow). Arrows, cells coexpressing cytokeratin and CD90; cell outside the tumor nest expressing CD44 but not cytokeratin.
In NB ducts, cytoplasmic cytokeratin staining was limited to luminal ductal cells with markedly bright staining of the apical surface. CD44 was strongly membrane-associated and was present in both basal and luminal cells of most samples (92%, Table 1), but stained with a gradient of decreasing intensity from basal to luminal. CD90 staining was both membrane-associated and cytoplasmic and followed a pattern similar to CD44, being strongly expressed in basal cells, but was low or absent on luminal cells in 80% of the samples tested.
Table 1.
Expression of CD44, CD90, and Cytokeratin in Benign Breast Paraffin Sections
| Basal |
Luminal |
|||||||
|---|---|---|---|---|---|---|---|---|
| Exp | Sample | Method | CD44 | CD90 | CTK | CD44 | CD90 | CTK |
| NB003R | Reduction | IHC/IF | + | + | − | Low | − | + |
| NB003L | Reduction | IHC/IF | + | + | − | Low | − | + |
| 32NB | Benign | IHC/IF | + | + | − | + | − | + |
| TMA1 | Benign | IF | + | + | − | Low | + | + |
| TMA2 | Benign | IF | + | + | − | + | − | + |
| TMA3 | Benign | IF | + | + | − | Low | − | + |
| TMA4 | Benign | IF | + | + | − | Low | Low | + |
| TMA5 | Benign | IF | + | + | − | Low | + | + |
| TMA6 | Benign | IF | + | + | − | Low | Low | + |
| TMA7 | Benign | IF | + | NA | − | Low | NA | + |
| TMA8 | Benign | IF | + | + | − | Low | Low | + |
| TMA9 | Benign | IF | + | + | − | Low | − | + |
| TMA11 | Benign | IF | + | + | − | + | − | + |
| TMA12 | Benign | IF | + | + | − | Low | − | + |
| TMA14 | Benign | IF | + | + | − | Low | Low | + |
| TMA15 | Benign | IF | + | + | − | Low | Low | + |
| TMA16 | Benign | IF | + | + | − | + | + | + |
| TMA17 | Benign | IF | + | + | − | Low | + | + |
| TMA18 | Benign | IF | + | + | − | Low | − | + |
| TMA20 | Benign | IF | + | + | − | − | − | + |
| TMA22 | Benign | IF | + | + | − | Low | Low | + |
| TMA23 | Benign | IF | + | + | − | + | + | + |
| TMA24 | Benign | IF | + | + | − | Low | − | + |
| TMA25 | Benign | IF | + | + | − | Low | + | + |
| TMA27 | Benign | IF | + | + | − | Low | − | + |
| TMA30 | Benign | IF | + | + | − | − | − | + |
| No. positive (%) | 26 (100%) | 25 (100%) | 0 (0%) | 5 + 19L (92%) | 6 + 6L (48%) | 26 (100%) | ||
Low, low-fluorescence intensity but positive compared to background and negative control samples; NA, not available (specimen was not interpretable). Individual TMA samples were prepared in replicate (three to six sections) and one to three photomicrographs (40× objective) were taken per section.
Morphologically, the majority of tumors demonstrated nests and trabeculae of malignant cells infiltrating the stroma (Fig. 1). The stroma consisted of fibroblastic connective tissue and small vessels. Almost all tumors demonstrated some degree of intratumoral lymphocytic infiltrate. Some samples showed the presence of normal unremarkable and hyperplastic ducts intermixed with the tumor.
The great majority of morphologically identifiable epithelioid tumor cells had clear cytoplasmic staining for cytokeratin. As in NB, CD44 staining was chiefly membrane-associated with highly variable intensity. Most samples (79%) expressed CD44 on smaller tumor cells located on the periphery of tumor nests. Eighty-eight percent had CD44+ large epithelioid cells within the central portions of the tumor. A proportion of infiltrating lymphocytes was also CD44+. CD44 was less commonly expressed on fibroblastic cells surrounding tumor nests or in the intratumoral stroma.
CD90 was both membrane-associated and cytoplasmic and detected on CD44-negative intratumoral stromal cells and on small vessels, where it marks mesenchymal stem cells (23). Within tumor nests and trabeculae, CD90 staining was most pronounced on the layer of cytokeratin negative fibroblastic cells surrounding the tumor capsules (CD44− in 19 of 24 samples). On the periphery of the tumor, CD90+ cells tended to form a double layer with uninterrupted fibroblastic cells on the outside and solitary or small clusters of CD90+ small epitheliod tumor cells on the inside. CD90 expression overlapped with CD44 in this peripheral region (peripheral tumor cells adjacent to fibroblastic cells) and, occasionally, in large anaplastic tumor cells outside well-defined tumor nests.
Flow-Cytometry Analytical Strategy
Mechanical and enzymatic digestion, required for flow cytometry of solid tissues, has the potential to introduce significant artifact. Dead cells, cellular fragments, and apoptotic cells can form a significant proportion of events in disaggregated tissues. Nonspecific antibody binding and autofluorescence of such events can interfere with interpretation. In addition to tumorigenic cells expressing CD44 and CD90, tumor samples contain stromal cells, reactive cells, and immune cells. The latter is concentrated in disaggregated tissue, because they are recovered with high efficiency relative to parenchymal and stromal cells. A further source of potential artifact is the tendency for malignant cells to cluster, even after disaggregation and enzymatic digestion. The presence of clusters causes an artifact in which markers expressed on discrete populations appear to be coexpressed. Figure 2, a disaggregated breast tumor sample, illustrates the analysis performed on all samples in this study and highlights the gating strategy used to circumvent these problems. Cell clusters were eliminated in comparison with forward scatter pulse height and width (20). Debris and hypodiploid (necrotic and apoptotic) cells were identified and eliminated on the basis of DNA staining on log DAPI fluorescence intensity. Finally, cells were eliminated, which express the pan Heme lineage marker CD45, CD14 [present on monocytes, macrophages, and mesothelial (Meso; 24) cells], or the myeloid and erythroid markers CD33 and glycophorin A, respectively. The denominator for all subsequent determinations in this study was non-Heme, non-Meso, singlet cells with at least 2N DNA, or subsets of this fraction. Lymphocytes (CD45bright/glycophorin A-/CD33-/CD14-) were used as an internal reference population to define the light-scatter properties of small resting cells and to locate the DAPI fluorescence intensity of cells having 2 N DNA content.
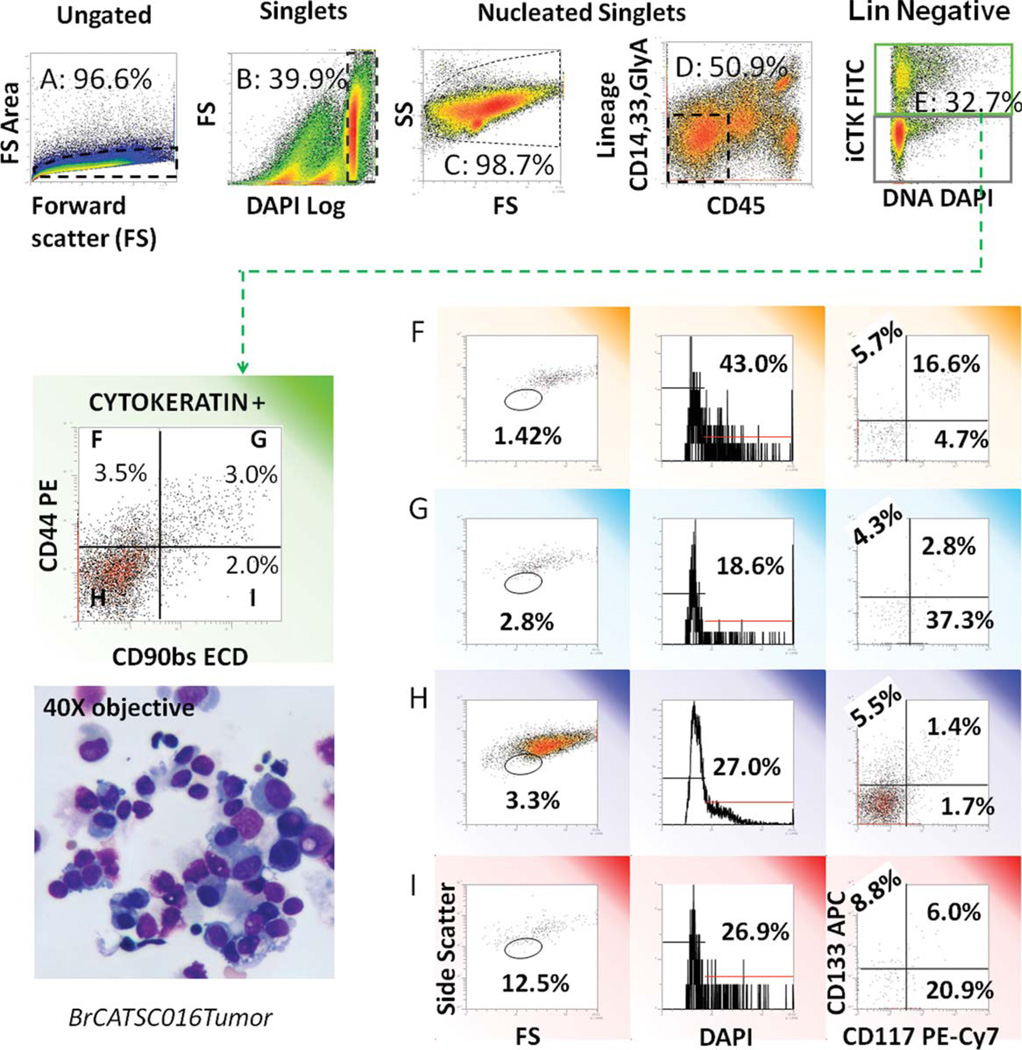
Fig. 2.
Analytical strategy for evaluation of light scatter, DNA content, and stem/progenitor markers in cytokeratin+ nonhematopoietic (non-Heme) tumor cells. A freshly excised tumor was disaggregated and prepared for flow cytometry as described. Five-hundred thousand events were acquired. Top panels: the gating strategy used to remove cell clusters, debris, red, and white blood cells is shown. From left to right: forward light scatter pulse analysis is used to eliminate cell clusters and retain singlet cells (A); the DNA stain DAPI is used to define nucleated cells with ≥2N DNA and eliminate subcellular debris (B); forward versus side light scatter is shown for total nucleated cells (C); CD45 versus a cocktail of CD14, CD33, and glycophorin A is used to gate out Heme cells (D); CD14+ is also expressed on mesothelial cells, which are present in pleural effusions. Cytokeratin positive epithelial cells are subsequently identified (E). This gating strategy was used for all subsequent analyses. Cytokeratin positive cells were sub-setted on the basis of CD90 and CD44 expression. Cell morphology (as determined by light scatter), DNA content (DAPI), and expression of the stem-cell markers CD117 and CD133 were measured on each CD44/CD90 subset. The inset in the lower left shows a cytocentrifuge preparation of the single-cell suspension prepared for flow cytometry. Apoptotic smudge cells and debris are removed from the flow cytometric analysis by the gating strategy. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Several markers have been proposed to identify stem and progenitor cells in solid tissues and epithelial tumors. Among these are CD44, CD90, CD133, and CD117. To reconcile these observations and establish the relationship between tissue stem/progenitor markers, and markers indicative of differentiation along the epithelial lineage, we evaluated their coexpression on cytokeratin positive epithelial cells. Cytokeratin+ cells were subsetted on the basis of CD44/CD90 expression, and these subpopulations, in turn, were assessed for the presence of low-light-scatter cells, DNA content, and expression of the stem/progenitor markers CD117 and CD133. The use of multiparameter flow cytometry permitted simultaneous determination of all markers.
Heterogeneity of CD44 and CD90 Expression on Cytokeratin+ Cells
Figure 3 summarizes cytokeratin expression, DNA content, and CD44/CD90 expression on Heme lineage negative cells from pleural effusions, disaggregated tumor, NB, and low-passage cultured tumor and NB. The histograms on the left illustrate the heterogeneity observed within and between-sample types. In these samples, the proportion of cytokeratin+ cells among Heme lineage negative cells ranged from >97% in the well-established breast tumor cell line BT474 to <1% in pleural effusions. Low-passage tumor and NB cultures also favored the outgrowth of cytokeratin negative cells. Among cytokeratin+ cells, solid breast tumors were characterized by their high proportion of cells with >2 N DNA content (cycling and/or aneuploid). Cytokeratin+ tumor cells had the highest proportion of cells with >2 N DNA content (41.7%) when compared with NB (9.4%, P = 0.08, two-tailed t-test). Grossly normal excised breast tissue showed a bimodal distribution of cells with high-DNA content, perhaps indicative of ductal hyperplasia in some samples. In this series, NB consisted of seven samples of adjacent histologically tumor-free tissue from mastectomy and two samples from breast reduction mammoplasty. Neither of the mammoplasty samples had a high proportion of cells with >2 N DNA content.
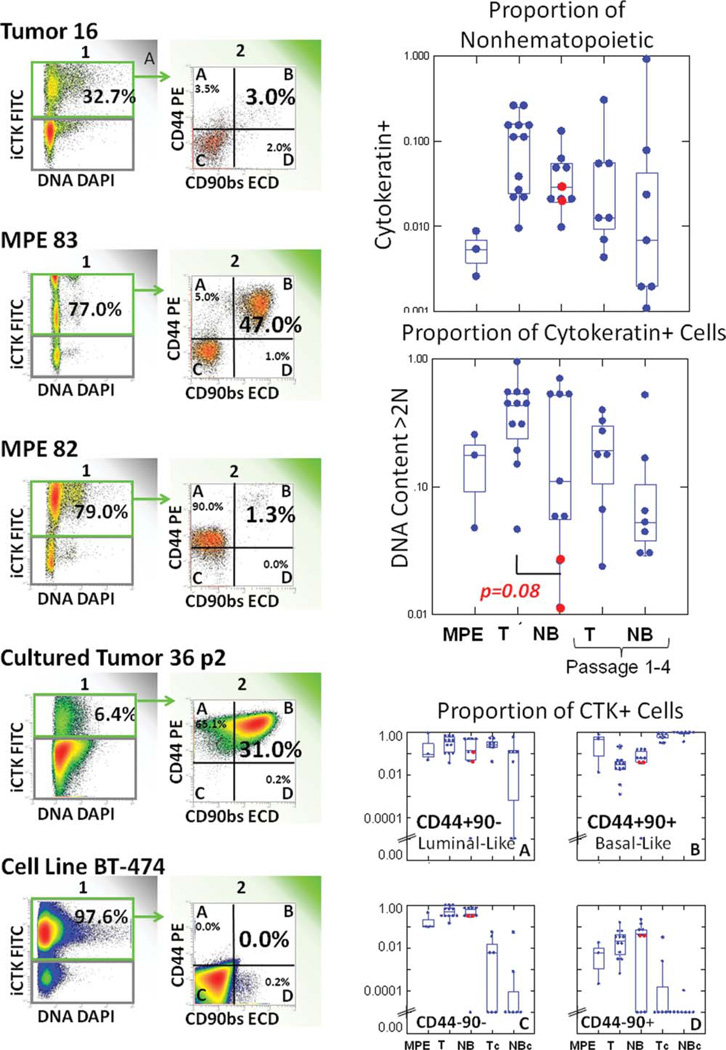
Fig. 3.
Stem/progenitor marker expression on cytokeratin positive non-Heme cells in primary breast cancer, two examples of MPE, short term cultured primary tumor, and a well-established epithelial breast cancer cell line. All events are gated on non-Heme cells as shown in Figure 2. The leftmost panel divides cells between cytokeratin negative (gray) and cytokeratin positive (green arrow) populations. The next histogram subdivides cytokeratin positive (green border) cells on the basis of CD44 and CD90 expression. The box plots on the right (from top to bottom) show the distribution of cytokeratin+ cells among non-Heme cells, DNA content, and CD44/CD90 subsets among cytokeratin+ cells. The data are plotted as dot box plots, where the Y-axis represents the proportion of positive cells on a logarithmic scale, and the dots represent individual observations. The waist indicates the group median, and the hinges (upper and lower boundaries of the box) indicate interquartile distances. The whiskers (bars) give the ranges, exclusive of outliers. Outliers (more than 1.5 times the hingespread from the median) are beyond the whiskers. Red dots indicate normal breast from reduction mammoplasty. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
In both disaggregated tumor and adjacent tumor-free tissue, a significant proportion of cytokeratin+ cells expressed CD44 without coexpression of CD90 (34.6% and 24.7%, respectively). On the basis of immunofluorescence (Fig. 1 and Tables 1 and 2), these populations can be identified as interior tumor cells and luminal ductal cells, respectively. Cells coexpressing CD44 and CD90, representing tumor cells at the invasive front and normal basal ductal cells, were less numerous (5.4 and 8.1%, respectively). CD44 and CD90 expression in cytokeratin+ metastatic pleural effusion cells varied from sample to sample. In two of three PE, the majority of cytokeratin+ cells coexpressed CD44 and CD90, reminiscent of basal ductal cells, and cells detected at the invasive front of tumor nests. In the third (MPE 82), the majority were CD44+/CD90−, resembling luminal ductal cells and cells in the tumor interior. Among short-term cultured cells (tumor and NB), virtually all cytokeratin+ cells were CD44+/CD90+, suggesting a selective advantage for in vitro propagation. CD44 (25) and CD90 (26) are also brightly and homogeneously expressed in the absence of cytokeratin in cultured mesenchymal stem cells. Neither CD44 nor CD90 was expressed in the cytokeratin+ cancer cell line BT474. The salient features are that (1) cytokeratin+ cells from tumor and adjacent tumor-free tissue, although morphologically disparate (Fig. 1), have similar expression profiles of CD44 and CD90; (2) these expression profiles can be correlated with immunofluorescence to identify the histological location of cells within tumors and normal tissue; (3) short-term culture favors the outgrowth of cytokeratin+/CD44+/CD90+ cells; (4) the well-established highly tumorigenic breast tumor cell line BT-474 requires neither CD44 nor CD90.
Table 2.
Expression of CD44 and CD90 in Breast Tumor Paraffin Sections
| Tumor peripheral |
Tumor central (epithelial) |
Tumor peripheral (fibroblast) |
Instratumor (stromal) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exp | Histology/ER-PR status | Method | CD44 | CD90 | CD44 | CD90 | CD44 | CD90 | CD44 | CD90 |
| 16 | Invasive ductal Ca/ER+ PR− | IHC | − | + | + | + | − | + | − | + |
| 21 | Invasive ductal Ca/ER+ PR− | IHC | + | + | − | − | − | + | − | + |
| 22 | Invasive ductal Ca/ER+ PR weak | IHC | + | + | + | − | − | + | − | + |
| 26 | Invasive ductal Ca (w/papilloma)/ER+ PR+ | IHC | + | + | + | − | − | + | − | + |
| 27 | Invasive ductal Ca/ER+ PR+ | IHC | − | + | + | − | + | + | − | + |
| 31 | Invasive ductal Ca/ER+ PR+ | IHC | + | + | + | − | − | + | − | + |
| 32 | Invasive ductal Ca/ER+ PR+ | IHC/IF | + | + | + | − | − | + | − | + |
| 36 | Pleomorphic invasive lobular Ca/ER+ PR+ | IHC | − | + | + | − | − | + | + | + |
| 41 | Invasive ductal Ca/ER+ PR+ | IHC | + | + | + | − | + | + | + | + |
| 42 | Invasive ductal Ca/ER− PR− | IHC | + | + | + | − | − | + | − | + |
| TMA1 | Invasive ductal CA/ER+ PR+ | IF | + | − | + | − | − | + | − | + |
| TMA8 | Ductal/tubular/papillomatous CA/ER+ PR+ | IF | + | − | + | − | + | + | − | + |
| TMA12 | Invasive ductal CA/ER+ PR+ | IF | − | + | + | − | − | + | − | + |
| TMA16 | Ductal/tubular/papillomatous CA/ER+ PR+ | IF | + | − | + | − | − | + | − | + |
| TMA20 | Invasive ductal CA/ER+ PR+ | IF | + | − | + | − | − | + | + | + |
| TMA22 | Invasive ductal CA/ER+ PR+ | IF | + | + | − | − | − | + | + | + |
| TMA25 | Mixed ductal/lobular/ER+ PR+ | IF | + | + | + | + | − | − | − | + |
| TMA26 | Ductal/micropapillomatous CA/ER+ PR+ | IF | + | + | + | − | + | + | + | + |
| TMA27 | Invasive ductal CA/ER+ PR+ | IF | + | + | + | + | − | + | − | + |
| TMA31 | Invasive ductal CA/ER− PR− | IF | + | + | + | − | + | + | − | + |
| TMA36 | Invasive ductal CA/ER+ PR+ | IF | − | − | − | − | − | + | + | + |
| TMA43 | Invasive ductal CA/ER+ PR+ | IF | + | + | + | − | − | + | − | + |
| TMA46 | Ductal/tubular CA/ER+ PR | IF | + | + | + | + | − | + | + | + |
| TMA56 | Ductal/tubular CA/ER+ PR+ | IF | + | + | + | + | − | + | − | |
| No. positive (%) | 19(79%) | 19(79%) | 21(88%) | 4(17%) | 5(21%) | 23(96%) | 7(29%) | 23(96%) | ||
ER-PR, estrogen receptor progesterone receptor status. Tumor peripheral: nonfibroblastic cells immediately peripheral to morphologically identifiable tumor nests or trabeculae. Tumor central (epithelial): nests and trabeculae of epithelioid tumor cells. Tumor peripheral (fibroblast): fibroblastic cells in the immediate periphery of the tumor. Intratumor (stromal): fibroconnective tissue separating tumor nests. All these stromal cells were cytokeratin negative.
Light-Scatter Properties of CD44/CD90 Subsets of Cytokeratin+ Cells
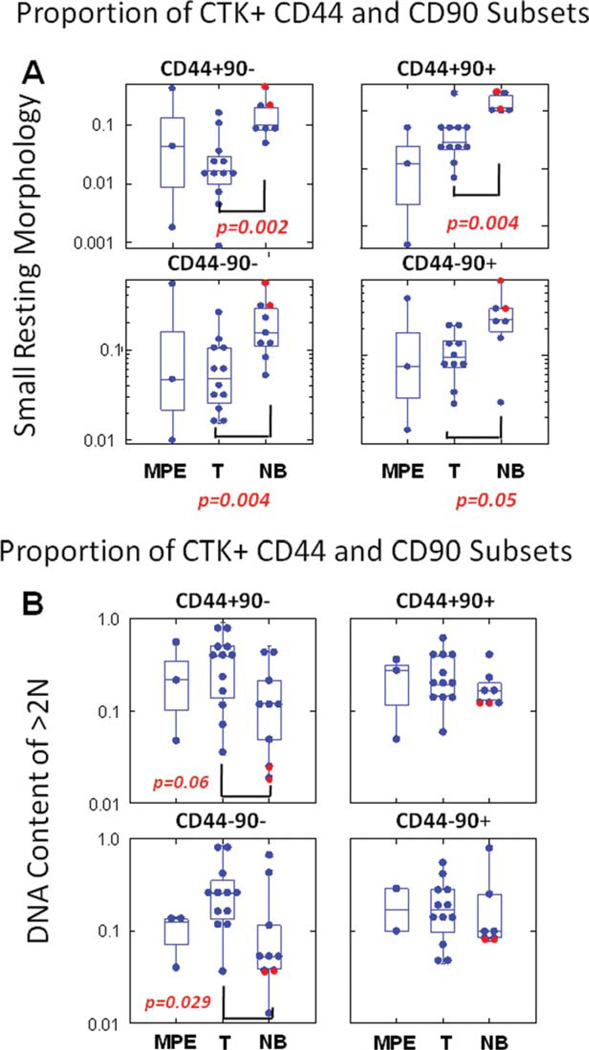
Low-angle (forward) and 90° (side) light scatter are physical properties that correlate with cell size and internal complexity, respectively. Among cytokeratin+ cells from disaggregated tissues, low-light-scatter cells are characterized by their relatively small size and high nucleus to cytoplasm ratio. NB tissue (mammoplasty and tumor free-resected breast) had the highest proportion of resting low-light-scatter cells (P = 0.002, compared to tumor; Fig. 4A, top left), in all CD44/90 subsets of cytokeratin+ cells, but low-light-scatter cells were most prominent in the basal-like CD44+/CD90+ subset. The two mammoplasty samples (red circles) were among those with the highest proportion of small resting cells.
Fig. 4.
Analysis of morphology and DNA content in CD44/CD90 subsets. Cytokeratin positive cells were defined as described in Figure 2. The top panels (A) represent the proportion of cells with small resting morphology (lymphoid light scatter) among cytokeratin positive CD44/CD90 subsets. The bottom panels (B) display the proportion of cells with greater than 2N DNA content (cycling and aneuploid cells). Passenger lymphocytes (CD45 bright, CD14/CD33/GlyA negative) were used as an internal standard for both low-light scatter cells and 2N DNA content. Determinations on ductal tissue from normal breast (mammoplasty) are shown as red circles. P values were calculated on log-transformed values using Student’s two-tailed t-test. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DNA Content of CD44/CD90 Subsets of Cytokeratin+ Cells
DAPI staining in gently permeabilized cells provides an estimate of DNA content and permits the simultaneous detection of surface markers. Cells with >2 N DNA content are either cycling or aneuploid. In all cytokeratin+ CD44/90 subsets, the proportion of cells with DNA content >2 N was greatest in tumor cells (Fig. 4B). The largest difference was in the CD44−/CD90− fraction where 30.7% (mean) of tumor cells was cycling/aneuploid, compared to 16.2% of adjacent tumor-free breast tissue cells (P = 0.029). Mammoplasty samples (red circles), which also had the highest proportion of low-light-scatter cells (Fig. 4A), had the lowest proportion of cells with >2 N DNA content.
Expression of the Stem/Progenitor Markers CD117 and CD133 on CD44/CD90 Subsets of Cytokeratin+ Cells
CD133 and CD117 expression in each CD44/CD90 subset of cytokeratin+ cells and the respective comparisons of each subpopulation between tumor and NB tissue or pleural effusions and NB tissue are shown in Table 3. The dataset consisted of 12 tumor samples, 3 MPE, and 9 tumor free or NB samples. P values were determined using Student’s two-tailed t-test on log-transformed proportions of each subpopulation. Both CD133 and CD117 were expressed on minor subpopulations of cytokeratin+ cells. Tumor and adjacent tumor-free ductal tissue had indistinguishable distributions of each CD133/CD117 subset. In contrast, metastatic pleural effusions lacked CD117+ cells, with or without coexpression of CD133 (Table 3, bolded P values). CD133+ cells constituted 10–19% of cytokeratin+ cells, but were distributed approximately equally between CD44/CD90 subsets. Most significantly, in both NB and tumor samples, the highest proportion of CD117+/CD133− cells was found among the CD44+/CD90+ fraction associated with basal ductal cells and the tumor invasive front (P < 0.0002, ANOVA, all contrasts significant).
Table 3.
CD133 and CD117 Expression on CD44/CD90 Subsets
| Tumor |
MPE |
NB |
T versus NB |
MPE versus NB |
||
|---|---|---|---|---|---|---|
| Subset | Denominator | Mean (LCI95, UCI 95) |
Mean (LCI95, UCI 95) |
Mean (LCI95, UCI 95) |
P value | P value |
| 133+ 117− | Cytok+ 44+ 90− | 6.9% (2.8, 17.0) | 14.4% (2.8, 73.6) | 7.2% (4.3, 12.3) | 0.931 | 0.312 |
| 133+ 117+ | Cytok+ 44+ 90− | 5.0% (1.9, 12.8) | 0.1% (0.0, 0.1) | 29.0% (18.3, 46.0) | 0.011 | 0.000 |
| 133− 117− | Cytok+ 44+ 90− | 38.9% (17.0, 88.9) | 71.8% (47.2, 109.2) | 39.1% (24.3, 62.8) | 0.994 | 0.201 |
| 133− 117+ | Cytok+ 44+ 90− | 8.2% (4.6, 14.6) | 0.1% (0.0, 1.8) | 10.2% (4.8, 21.8) | 0.658 | 0.003 |
| 133+ 117− | Cytok+ 44+ 90+ | 7.5% (4.2, 13.4) | 3.7% (0.2, 59.6) | 11.6% (6.5, 20.5) | 0.335 | 0.272 |
| 133+ 117+ | Cytok+ 44+ 90+ | 5.2% (2.9, 9.5) | 0.1% (0.1, 0.1) | 11.4% (4.9, 26.8) | 0.147 | 0.006 |
| 133− 117− | Cytok+ 44+ 90+ | 52.7% (43.6, 63.7) | 85.5% (67.9, 107.6) | 42.4% (30.2, 59.4) | 0.254 | 0.050 |
| 133−117+ | Cytok+ 44+ 90+ | 27.4% (18.6, 40.3) | 0.4% (0.1, 1.4) | 25.6% (15.8, 41.6) | 0.839 | 0.000 |
| 133+ 117− | Cytok+ 44− 90− | 6.5% (2.6, 16.1) | 44.0% (26.4, 73.2) | 11.0% (6.4, 18.7) | 0.383 | 0.021 |
| 133+ 117+ | Cytok+ 44− 90− | 3.2% (1.2, 8.2) | 0.1% (0.1, 0.2) | 13.7% (5.6, 33.1) | 0.052 | 0.001 |
| 133− 117− | Cytok+ 44− 90− | 0.2% (0.1, 0.3) | 48.9% (27.4, 87.1) | 48.9% (36.3, 65.8) | 0.790 | 0.996 |
| 133− 117+ | Cytok+ 44− 90− | 5.6% (2.8, 11.5) | 0.1% (0.1, 0.1) | 10.3% (5.9, 17.9) | 0.245 | 0.000 |
| 133+ 117− | Cytok+ 44− 90+ | 11.1% (6.5, 19.2) | 5.9% (0.9, 41.5) | 17.3% (9.4, 31.9) | 0.340 | 0.218 |
| 133+ 117+ | Cytok+ 44− 90+ | 8.0% (4.7, 13.5) | 0% (0, 0) | 15.7% (4.7, 52.4) | 0.276 | 0.000 |
| 133− 117− | Cytok+ 44− 90+ | 54.0% (43.8, 66.4) | 88.7% (79.3, 99.3) | 51.3% (36.9, 71.3) | 0.796 | 0.088 |
| 133− 117+ | Cytok+ 44− 90+ | 4.2% (0.6, 32.7) | 0% (0, 0) | 9.6% (3.4, 26.7) | 0.218 | 0.000 |
Geometric mean and 95% lower and upper confidence intervals are shown for the proportion of positive cells (subsets) among the specified denominator population. P values, determined by Student’s t-test (two tailed) on log transformed values are shown for comparisons of tumor versus normal breast (T_NB) and malignant pleural effusion versus normal breast (MPE_NB). Populations and P values are bolded where P< 0.002, the adjusted minimal value for significance, given 32 comparisons.
Tumorigenicity of CD90+ Low-Light-Scatter Cells
Our previous experiments demonstrate that CD90 marks two populations in breast tumor (basal-like cytokeratin+ tumor cells and cytokeratin negative stromal/vascular cells). Because stromal/vascular cells (CD90+/cytokeratin negative) are largely absent in pleural effusions, CD90+ tumor cells can be sorted without recourse to cytokeratin, which is intracellular. Two independent experiments were performed using MPE from two patients with metastatic adenocarcinoma of the breast. Both samples had been cryopreserved and were separated on a Ficoll/Hypaque gradient before staining. All sort populations were gated on DAPI and forward-scattered to eliminate DAPI permeant dead cells, forward scatter pulse height and width to eliminate cell clusters, and CD45 versus heme lineage to eliminate blood and Meso cells. CD31 was used to eliminate vascular endothelial (Endo) cells. Three populations meeting the gating criteria mentioned earlier were sorted for tumorigenicity studies: (1) CD90+ cells with low (lymphocyte-like) light scatter; (2) CD90+ cells with highlight scatter; (3) the remaining CD31 negative non-Heme cells not included in populations 1 or 2. In this way, all viable non-Heme, non-Meso, and non-Endo cells were included among the sort populations. CD44, CD117, and EpCAM expression were measured as “outcome variables” (20) on sorted populations. Only 21% of viable pleural effusion cells met the criteria of non-Heme, non-Meso, and non-Endo (Table 3). Of these, 0.18% was CD90+ cells with low-light scatter (sort population 1) and 34% was CD90+ cells with high-light scatter (sort population 2), the remainder being CD90 negative (sort population 3). CD44 expression was highest on CD90+ high-side-scatter cells, whereas expression of the epithelial marker EpCAM was highest on CD90 negative cells. For each experiment, mice (five per cell population) were injected at four sites each with 100 sorted cells plus 10,000 irradiated unsorted MPE feeder cells, providing 40 opportunities for tumor growth of each population tested. Experiment 1 used NOD/SCID mice, whereas experiment 2 used NSG mice. In these experiments, where tumor cell dose was limited to 100 sorted cells, only CD90+ low-light-scatter cells (49% of which were CD44+) were tumorigenic (25% of sites in experiment 1 and 5% of sites in experiment 2). Pooling the results of the two experiments, enhanced tumorigenicity of the CD90+ low-light-scatter population was statistically significant (P = 0.02, Fisher’s two-tailed exact test).
DISCUSSION
The Cancer Stem Cell
Seven years after the initial report, suggesting the existence of epithelial cancer stem cells (7), the tumor stem-cell paradigm remains both intriguing and controversial. Markers such as CD44, CD133, CD90, MDR (10,14,27–29), [reviewed in (3)] and ALDH1 (5) have been proposed in a variety of epithelial tumors to identify a unique subset of cancer cells that is clonogenic (self-renewing) and, in some cases, therapy resistant. Identification of cancer stem cells relies in part on showing enriched tumorigenicity. This has been accomplished by sorting subpopulations from disaggregated tumor and injecting the sorted populations into immunodeficient mice. Demonstrating that a particular human tumor cell subset has enhanced tumorigenicity in a xenograft model is problematic. Factors influencing measured tumorigenicity include patient-to-patient variability (if freshly isolated cells are used), extreme in vitro selection (if cell lines are used), purity of the isolated population, injection site, vehicle and feeder cells, strain of immunodeficient mouse (30), and duration of observation. Additionally, enhanced tumorigenicity of a particular subset must be shown in relationship to some other population. If that population is an “unsorted” patient isolate, sorting on any marker that increases the proportion of epithelial cells (and depletes immune, vascular, and stromal cells) will demonstrate increased tumorigenicity. Because MPEs have a high proportion of immune cells, tumor disaggregation results in selection bias in favor of CD45+ cells (Fig. 2), resulting in a substantial selection effect. This selection bias could explain in part the concentration of tumorigenic cells in Heme lineage-negative CD133+-selected cells, compared to unselected disaggregated tumor, which is rich in reactive cells (31). In this study, we overcame this obstacle by comparing tumorigenicity within mutually exclusive populations that, in aggregate, represent all non-Heme, non-Meso, and non-Endo breast cancer pleural effusion cells (CD90+ low-light scatter, CD90+ high-light scatter, and CD90−).
Choice of Markers
Cytokeratin was among the first antigens associated with epithelial cancers (32) and was used here as a first cut to ensure the epithelial origin of the candidate populations. Likewise, stem/progenitor markers were chosen on the basis of published results in the cancer and normal adult tissue stem-cell literature. The hyaluronic acid receptor CD44, a ubiquitous adhesion molecule, was early implicated in tumor metastasis (33) and is the principal marker proposed by Clarke and his colleagues to identify tumorigenic breast cancer cells (7). CD44 is prominent on normal mesenchymal stem cells (34) and has been proposed to play a role in their migration and homing (35). We have proposed CD90 as a principal cancer stem/progenitor cell marker in a variety of epithelial cancers including lung and breast (10,22). CD45− CD90+ cells have recently been detected in liver tumors (13) and in the circulation of liver cancer patients (36). CD90 also appears to be a key lineage-independent adult tissue stem-cell marker. In this context, CD90 was first described on the most primitive murine (37,38) and human (39,40) Heme stem cells. It is also expressed on oval cells of the liver (41) and on perivascular stem cells (23,42), which are closely related to mesenchymal stem cells (43). In human breast, it is one of the key markers used to sort cells, which give rise to bipotent (basal and luminal) colonies in vitro (8). Mutations in CD117, variously known as c-kit or stem-cell factor receptor, are implicated in gastrointestinal stromal tumors (16) and myeloid leukemia (44). It has also been identified as a key marker for breast luminal progenitor cells and was highly expressed in BRCA1-associated preneoplastic tissue and tumors (45). Furthermore, we have documented the persistence of small (low-light scatter) CD44+ CD117+ tumor cells in the MPE of a patient treated with capecitabine (46). In normal biology, CD117 is an important cytokine receptor and signaling molecule, playing a role in survival, proliferation, and differentiation of Heme stem cells and a variety of other cell types. CD117 is also prominent in tissue mast cells (47), which are often present as tumor infiltrating cells. Thus, it is necessary to demonstrate the absence of CD45 expression before this can be interpreted as an epithelial stem/progenitor marker. In the bone marrow, CD133 marks Heme stem/progenitor cells (48). It has also been implicated on putative cancer stem cells in a variety of tumors (4,49,50) and is present on human prostate epithelial basal cells (12). From this brief survey, it is evident that the proteins generally considered as stem-cell markers are present across a wide variety of cell types and require anatomical and pathological contexts for their interpretation.
Tumor Heterogeneity
Immunohistostaining studies of freshly excised tumor illustrate that tumors are heterogeneous and consist of tumor cells, phenotypically and genetically heterogeneous in themselves (14,51,52), plus reactive cells including vessels, stroma, and infiltrating immune cells. Some samples also include areas of more normal appearing ductal structures, benign lesions, or carcinoma in situ. All these elements are represented in disaggregated tumor used for flow cytometry and, ultimately, tumorigenicity studies. It is therefore important to localize populations detected in disaggregated tissue by a method that preserves histologic relationships. Immunohistostaining can be used to validate and localize markers and search for combinations of markers that map to unique locations. In this study, we determined that small cytokeratin+/CD90+ cells (often coexpressing CD44) are found uniquely at the periphery of tumor nests and in normal basal ductal cells. Furthermore, we showed that proliferating (Ki67+) cells are distributed throughout tumor nests, but are rarely if ever on the periphery (see Supporting Information Fig. 1). Additionally, we learned that cytokeratin−/CD90+ cells map to the stroma, whereas CD44, a marker coexpressed with CD90 on bone marrow mesenchymal stem cells (25), is only occasionally associated with tumor stroma (Fig. 1, Table 2).
Sources of Artifact
Performing flow cytometry on disaggregated tissues is subject to several significant sources of error, all of which have been ignored in the current cancer stem-cell literature: (1) comparison of intact and disaggregated tissue reveals a large selection toward infiltrating mononuclear cells, which survive mechanical disruption and enzymatic digestion far better than tightly associated epithelial and stromal cells (Fig. 2). This bias could also favor tumorigenic cells if they are smaller, less morphologically complex, or less tightly bound than nontumorigenic epithelial cells. Importantly, frequency estimates of tumorigenic cells are greatly influenced by selection bias imposed by tissue disaggregation; (3) tumor cells in ascites or malignant effusions naturally exist in small clusters. Even digestion and mechanical disaggregation do not result in a pure single-cell suspension. This problem is compounded by the tendency of disaggregated cells to recluster when suspended at high concentration. Although this problem can be addressed by doublet discrimination, the possibility exists that some of the most biologically important cells are eliminated from the analysis; (4) light-scatter analysis alone does a very poor job of discriminating living cells from cellular debris and dying cells. The use of DAPI as a DNA stain reveals that as much as 60% of singlet events (Fig. 2) are hypodiploid and therefore not analyzable; (5) nonspecific antibody binding can be eliminated by preincubation with mouse serum before staining and by negative gating on irrelevant lineage markers (in this case, those identifying Heme cells). In Figure 2, only 19.4% of raw acquired events pass these stringent criteria, and only 32.7% of these was cytokeratin+ and therefore epithelial tumor-cell candidates.
Comparison of Stem/Progenitor Marker Expression in Tumor and Adjacent Breast Tissue
In the present series, CD44 and CD90 staining were prominent by immunohistostaining on subsets of morphologically identifiable tumor cells and on normal ductal cells. In normal ductal tissue, the CD44+/CD90+ phenotype was distinctly basal (Table 1) and consistent with progenitor cells. As cells mature and move to the lumen, they tend to lose CD90 and gain cytokeratin. In agreement with Raouf’s analysis, which used an in vitro colony assay to measure clonogenic and differentiation potential, CD44 expression was not a discriminating feature between these states (8). In this study, expression of CD44 and CD90 in tumor cells was largely analogous to that observed in the normal duct. CD44 was detected on a variable proportion of morphologically identifiable tumor cells and present in both central and peripheral portions of most tumor nests. In contrast, CD90, which may be identified with basal ductal progenitor cells, was associated with a relatively small population of tumor cells at the periphery of nests as well as on adjacent cytokeratin negative fibroblastic cells and stromal/vascular cells. Under culture conditions, which favor retention of embryonic cells (see methods), freshly isolated breast tumor cells retained cytokeratin expression but rapidly assumed a CD44bright/CD90bright phenotype reminiscent of mesenchymal stem cells (Fig. 3).
One of the strengths of flow cytometry is the ability to measure the expression of multiple markers on single cells. CD133 did not distinguish between basal and luminal subsets or their presumptive tumor equivalents, raising concerns about its utility as a stem/progenitor marker in breast or breast tumor. In contrast, CD117 expression was prominent in the basal and basal-like CD44+/CD90+ subset of normal ductal and tumor cells, respectively. CD117+ cells were rare in aggressive MPE. Despite the fact that high-resolution immunophenotyping was able to resolve multiple discrete populations based on simultaneous measurement of CD44, CD90, CD117, and CD133 on cytokeratin+ cells, comparison of marker expression patterns in tumor and adjacent tumor-free tissue (or NB tissue) yielded no significant differences. The conservation of such striking similarities suggests that these markers may play similar functional roles in tumor and the normal tissues from which they are derived.
Size Matters
The association of low-light scatter, small size, and bland morphology with a multipotent resting state is best documented among normal Heme stem cells, where morphologic complexity and potentiality are inversely correlated (53). We previously showed that CD44+/CD90+ low-light-scatter breast cancer cells sorted from MPE were tumorigenic when ≤60 cells were injected (22). In this previous study, high-light-scatter CD44+/90+ cells were tested at a dose reflecting their relative prevalence (633–13,000 cells/injection) and were also tumorigenic. In this study, we standardized the number of cells injected (100 cells/site) to have an unbiased comparison of clonogenic frequency. In both studies, all populations were admixed with 10,000 heavily irradiated homologous tumor cells and suspended in a matrigel vehicle to prevent cell loss and potentially provide a niche effect. The present results demonstrate that only CD90+ low-light-scatter cells are tumorigenic when injected in limiting number. Notably, only 34% was EpCAM+ and 49% was CD44+. Furthermore, neither the more prevalent CD90+ high-light-scatter population (28% EpCAM+ and 73% CD44+) nor the most prevalent CD90 negative population (89% EpCAM+ but only 6% CD44+) were tumorigenic at 100 cells/site. CD44+ breast cancer cells have also been shown to be enriched for tumorigenic cells in a similar xenotransplant model (7,54,55). The finding that CD44 and CD90 expression coincides in relatively rare cells of small size located on the periphery of tumor nests and adjacent to CD90+ stroma, implies that cells at the invasive front, having a basal-like phenotype, are highly tumorigenic.
Working Hypothesis
We have used the data from our immunohistostaining, analytical flow cytometry, and tumorigenicity studies to develop a working hypothesis concerning tumor growth: tumor nests expand from the outside. Small cytokeratin+/CD44+/CD90+ cells on the periphery of tumor nests are basal cell equivalents, representing the best candidate for breast cancer stem/progenitor cells. As such, they give rise to CD44+/CD90− cells with high-proliferative capacity, which migrate “luminally” toward the center of tumor nests and proliferate. Under the mechanical stresses imposed by proliferation, the nest expands as a unit such that the small peripheral cytokeratin+/CD44+/CD90+ breast cancer stem cells in concert with the adjacent CD90+ fibroblastic cells surrounding the tumor form the invasive front. The fibroblastic cells provide paracrine signals (56) and a niche for adjacent cancer stem/progenitor cells. Armed with proteases, cells at the invasive front dissect their way though surrounding tissue (57), activating latent extracellular matrix-associated growth factors (58) in their path. Following therapy, dormant tumor, hypothesized to be small resting CD44+/CD90+/CD117+ cells (46), persists as solitary cells or in small clusters.
Supplementary Material
Table 4.
Characteristics and Tumorigenicity of Pleural Effusion Sort Populations
| Sort population |
Outcome parameters (pct of sort population) |
Tumorigenicity |
||||
|---|---|---|---|---|---|---|
| Population | Pct of viable cells |
Pct of non− (Heme, Endo, and Meso) |
CD44 | EpCAM | CD117 | Tumors/sites injected (Pct) |
| CD90+ LSLOW | 0.04% ± 0.01% | 0.18% ± 0.06% | 48.68% ± 18.85% | 34.06% ± 15.12% | 0.00% ±0.0 % | 6/40 (15%) |
| CD90+ LSHIGH | 7.13% ± 0.73% | 33.66% ± 3.39% | 72.92% ± 7.20% | 27.63% ±4.96 % | 0.00% ± 0.0% | 0/40 |
| CD90− | 14.01% ± 0.71% | 66.17% ± 3.45% | 5.55% ± 0.41% | 89.01% ± 0.71% | 0.03% ± 0.01% | 0/40 |
| Total | 21.18% ± 0.04% | 100.00% ± 0.0% | ||||
Immunophenotypic data are shown for experiment 1, in which three data files were collected during the beginning, midpoint, and end of the sort. Means and standard deviations from analyses of the three data files are shown. Tumorigenicity data are pooled results of two independent experiments in which mice were injected with 100 sorted tumor cells admixed with 10,000 irradiated (100 Gy) unsorted cells. In experiment 1 (NOD/SCID recipients), injection of CD90+ low-light-scatter cells resulted in tumors in 5 of 20 injection sites. Irradiated unsorted tumor (10,000 cells) injected into 20 sites in a total of five mice failed to cause tumors. In experiment 2 (NSG mice), the frequency of tumors was 1 in 20 sites. All xenograft tumors were confirmed human cytokeratin+ by immunohistochemistry with a noncrossreactive antibody (not shown).
ACKNOWLEDGMENTS
Vera Donnenberg is a Congressionally Directed Medical Research Program Era of Hope Scholar. The authors thank Ms. Melanie Pfeifer, Amber McCauslin, and Lisa Bailey for their expert technical assistance.
Grant sponsor: Department of Defense; Grant numbers: BC032981, BC044784; Grant sponsors: Hillman Foundation, Glimmer of Hope Foundation.
Footnotes
Additional supporting information may be found in the online version of this article.
LITERATURE CITED
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 3.Donnenberg VS, Donnenberg AD. Multiple drug resistance in cancer revisited: The cancer stem cell hypothesis. J Clin Pharmacol. 2005;45:872–877. doi: 10.1177/0091270005276905. [DOI] [PubMed] [Google Scholar]
- 4.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 5.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, Houvenaeghel G, Extra JM, Bertucci F, Jacquemier J, Xerri L, Dontu G, Stassi G, Xiao Y, Barsky SH, Birnbaum D, Viens P, Wicha MS. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16:45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, Aparicio S, Marra M, Eaves C. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Liu AY, Roudier MP, True LD, Liu AY, Roudier MP, True LD. Heterogeneity in primary and metastatic prostate cancer as defined by cell surface CD profile. Am J Pathol. 2004;165:1543–1556. doi: 10.1016/S0002-9440(10)63412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg VS, Landreneau RJ, Donnenberg AD. Tumorigenic stem and progenitor cells: Implications for the therapeutic index of anti-cancer agents. J Control Release. 2007;122:385–391. doi: 10.1016/j.jconrel.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YN, Zhang J, He QH, Dai X, Shen L. Isolation and characterization of epithelial progenitor cells from human fetal liver. Hepatol Res. 2008;38:103–113. doi: 10.1111/j.1872-034X.2007.00163.x. [DOI] [PubMed] [Google Scholar]
- 12.Rizzo S, Attard G, Hudson DL. Prostate epithelial stem cells. Cell Prolif. 2005;38:363–374. doi: 10.1111/j.1365-2184.2005.00356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu W, Lam CT, Poon RT, Fan ST, Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PWK, Lam CT, Poon RTP, Fan ST. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Wright M, Calcagno A, Salcido C, Carlson M, Ambudkar S, Varticov-ski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Went PT, Dirnhofer S, Bundi M, Mirlacher M, Schraml P, Mangialaio S, Dimitrijevic S, Kononen J, Lugli A, Simon R, Sauter G. Prevalence of KIT expression in human tumors. J Clin Oncol. 2004;22:4514–4522. doi: 10.1200/JCO.2004.10.125. [DOI] [PubMed] [Google Scholar]
- 16.Sarlomo-Rikala M, Kovatich AJ, Barusevicius A, Miettinen M. CD117: A sensitive marker for gastrointestinal stromal tumors that is more specific than CD34. Mod Pathol. 1998;11:728–734. [PubMed] [Google Scholar]
- 17.Donnenberg VS, Meyer EM, Donnenberg AD. Measurement of multiple drug resistance transporter activity in putative cancer stem/progenitor cells. In: Yu J, editor. Methods in Molecular Biology. Springer, NY: Humana Press; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- 19.Visus C, Ito D, Amoscato A, Maciejewska-Franczak M, Abdelsalem A, Dhir R, Shin DM, Donnenberg VS, Whiteside TL, DeLeo AB. Identification of human aldehyde dehydrogenase 1 family member A1 as a novel CD8+ T-cell-defined tumor antigen in squamous cell carcinoma of the head and neck. Cancer Res. 2007;67:10538–10545. doi: 10.1158/0008-5472.CAN-07-1346. [DOI] [PubMed] [Google Scholar]
- 20.Donnenberg AD, Donnenberg VS. Rare-event analysis in flow cytometry. Clin Lab Med. 2007;27:627–652. viii. doi: 10.1016/j.cll.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 21.Horny HP, Menke DM, Kaiserling E. Neoplastic human tissue mast cells express the adhesion molecule CD44/HCAM. Virchows Arch. 1996;429:91–94. doi: 10.1007/BF00192430. [DOI] [PubMed] [Google Scholar]
- 22.Donnenberg VS, Luketich JD, Landreneau RJ, DeLoia JA, Basse P, Donnenberg AD. Tumorigenic epithelial stem cells and their normal counterparts. Ernst Schering Found Symp Proc. 2006;5:245–263. doi: 10.1007/2789_2007_054. [DOI] [PubMed] [Google Scholar]
- 23.Zimmerlin L, Donnenberg VS, Pfeifer ME, Meyer EM, Peault B, Rubin JP, Donnenberg AD. Stromal vascular progenitors in adult human adipose tissue. Cytometry A. 2010;77:22–30. doi: 10.1002/cyto.a.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross JA, Ansell I, Hjelle JT, Miller-Hjelle MA, Dobbie JW. Phenotypic and functional characterisation of human mesothelial cells. Tissue Antig. 2000;55(Suppl):65–66. [Google Scholar]
- 25.Dvorakova J, Hruba A, Velebny V, Kubala L. Isolation and characterization of mesenchymal stem cell population entrapped in bone marrow collection sets. Cell Biol Int. 2008;32:1116–1125. doi: 10.1016/j.cellbi.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 26.Jones EA, Kinsey SE, English A, Jones RA, Straszynski L, Meredith DM, Markham AF, Jack A, Emery P, McGonagle D. Isolation and characterization of bone marrow multipotential mesenchymal progenitor cells. Arthrit Rheum. 2002;46:3349–3360. doi: 10.1002/art.10696. [DOI] [PubMed] [Google Scholar]
- 27.Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, Dinulescu DM, Connolly D, Foster R, Dombkowski D, Preffer F, MacLaughlin DT, Donahoe PK. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci. 2006;103:11154–11159. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engelmann K, Shen H, Finn OJ. MCF7 side population cells with characteristics of cancer stem/progenitor cells express the tumor antigen MUC1. Cancer Res. 2008;68:2419–2426. doi: 10.1158/0008-5472.CAN-07-2249. [DOI] [PubMed] [Google Scholar]
- 29.Harris MA, Yang H, Low BE, Mukherje J, Guha A, Bronson RT, Shultz LD, Israel MA, Yun K. Cancer stem cells are enriched in the side population cells in a mouse model of glioma. Cancer Res. 2008;68:10051–10059. doi: 10.1158/0008-5472.CAN-08-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Altmannsberger M, Osborn M, Holscher A, Schauer A, Weber K. The distribution of keratin type intermediate filaments in human breast cancer. An immunohistological study. Virchows Archiv B Cell Pathol. 1981;37:277–284. doi: 10.1007/BF02892576. [DOI] [PubMed] [Google Scholar]
- 33.Haynes BF, Liao HX, Patton KL. The transmembrane hyaluronate receptor (CD44): Multiple functions, multiple forms. Cancer Cells. 1991;3:347–350. [PubMed] [Google Scholar]
- 34.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 35.Zhu H, Mitsuhashi N, Klein A, Barsky LW, Weinberg K, Barr ML, Demetriou A, Wu GD. The role of the hyaluronan receptor CD44 in mesenchymal stem cell migration in the extracellular matrix. Stem Cells. 2006;24:928–935. doi: 10.1634/stemcells.2005-0186. [DOI] [PubMed] [Google Scholar]
- 36.Yang ZF, Ngai P, Ho DW, Yu WC, Ng MNP, Lau CK, Li MLY, Tam KH, Lam CT, Poon RTP, Fan ST. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47:919–928. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- 37.Muller-Sieburg CE, Whitlock CA, Weissman IL. Isolation of two early B lymphocyte progenitors from mouse marrow: A committed pre-pre-B cell and a clonogenic Thy-1-lo hematopoietic stem cell. Cell. 1986;44:653–662. doi: 10.1016/0092-8674(86)90274-6. [DOI] [PubMed] [Google Scholar]
- 38.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 39.Craig W, Kay R, Cutler RL, Lansdorp PM. Expression of Thy-1 on human hematopoietic progenitor cells. J Exp Med. 1993;177:1331–1342. doi: 10.1084/jem.177.5.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baum CM, Weissman IL, Tsukamoto AS, Buckle AM, Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proc Natl Acad Sci USA. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrera MB, Bruno S, Buttiglieri S, Tetta C, Gatti S, Deregibus MC, Bussolati B, Camussi G. Isolation and characterization of a stem cell population from adult human liver. Stem Cells. 2006;24:2840–2850. doi: 10.1634/stemcells.2006-0114. [DOI] [PubMed] [Google Scholar]
- 42.Zannettino ACW, Paton S, Arthur A, Khor F, Itescu S, Gimble JM, Gronthos S. Multipotential human adipose-derived stromal stem cells exhibit a perivascular phenotype in vitro and in vivo. J Cell Physiol. 2008;214:413–421. doi: 10.1002/jcp.21210. [DOI] [PubMed] [Google Scholar]
- 43.Young HE, Steele TA, Bray RA, Hudson J, Floyd JA, Hawkins K, Thomas K, Austin T, Edwards C, Cuzzourt J, Duenzl M, Lucas PA, Black AC., Jr Human reserve pluripotent mesenchymal stem cells are present in the connective tissues of skeletal muscle and dermis derived from fetal, adult, and geriatric donors. Anat Rec. 2001;264:51–62. doi: 10.1002/ar.1128. [DOI] [PubMed] [Google Scholar]
- 44.Moore MAS. Converging pathways in leukemogenesis and stem cell self-renewal. Exp Hematol. 2005;33:719–737. doi: 10.1016/j.exphem.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat M-L, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ, Fox SB, Yan M, French JD, Brown MA, Smyth GK, Visvader JE, Lindeman GJ. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 46.Donnenberg VS, Donnenberg AD. Therapeutic index and the cancer stem cell paradigm. In: Bagley R, Teicher B, editors. Stem Cells and Cancer Series: Cancer Drug Discovery and Development. Springer, NY: Humana Press; 2009. [Google Scholar]
- 47.Edwards ST, Cruz AC, Donnelly S, Dazin PF, Schulman ES, Jones KD, Wolters PJ, Hoopes C, Dolganov GM, Fang KC. c-Kit immuno-phenotyping and metalloproteinase expression profiles of mast cells in interstitial lung diseases. J Pathol. 2005;206:279–290. doi: 10.1002/path.1780. [DOI] [PubMed] [Google Scholar]
- 48.Handgretinger R, Gordon PR, Leimig T, Chen X, Buhring HJ, Niethammer D, Kuci S. Biology and plasticity of CD133+ hematopoietic stem cells. Ann NY Acad Sci. 2003;996:141–151. doi: 10.1111/j.1749-6632.2003.tb03242.x. [DOI] [PubMed] [Google Scholar]
- 49.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkel-man RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 50.Maitland NJ, Collins AT. Prostate cancer stem cells: A new target for therapy. J Clin Oncol. 2008;26:2862–2870. doi: 10.1200/JCO.2007.15.1472. [DOI] [PubMed] [Google Scholar]
- 51.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, Parker LM, Anderson KS, Harris LN, Garber JE, Richardson AL, Schnitt SJ, Nikolsky Y, Gelman RS, Polyak K. Mol Definit Breast Tumor Heterog. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 52.Wicha M. Cancer stem cell heterogeneity in hereditary breast cancer. Breast Cancer Res. 2008;10:105. doi: 10.1186/bcr1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrews RG, Singer JW, Bernstein ID. Precursors of colony-forming cells in humans can be distinguished from colony-forming cells by expression of the CD33 and CD34 antigens and light scatter properties. J Exp Med. 1989;169:1721–1731. doi: 10.1084/jem.169.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 55.Ginestier C, Korkaya H, Dontu G, Birnbaum D, Wicha MS, Charafe-Jauffret E. [The cancer stem cell: The breast cancer driver] Med Sci (Paris) 2007;23:1133–1139. doi: 10.1051/medsci/200723121133. [DOI] [PubMed] [Google Scholar]
- 56.Elenbaas B, Weinberg RA. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp Cell Res. 2001;264:169–184. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 57.Westermarck J, Kahari V-M. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999;13:781–792. [PubMed] [Google Scholar]
- 58.Taipale J, Keski-Oja J. Growth factors in the extracellular matrix. FASEB J. 1997;11:51–59. doi: 10.1096/fasebj.11.1.9034166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.