Abstract
Transcriptional targeting is a desirable property for many gene transfer applications. Because endothelial cells line most blood vessels, they are attractive candidates for the introduction of therapeutic gene products. As a proof-of-concept study, we attempted to identify a synthetic, endothelial cell-specific promoter by use of a high-throughput screen involving self-inactivating (SIN) human immunodeficiency virus type 1 (HIV-1)-based vectors. Select duplex oligodeoxynucleotides recognized by transcription factors and located 5′ of endothelial cell-specific mRNA transcripts were randomly ligated and cloned upstream of a minimal ICAM-2 promoter driving enhanced green fluorescent protein (eGFP) in a SIN HIV-1-based vector. Vesicular stomatitis virus G protein-pseudotyped particles were prepared from a library of >106 vector recombinants and used to transduce an endothelial cell line. The highest eGFP expressers were repeatedly sorted, and the synthetic promoters were recovered and retested by a luciferase reporter. Several promoters were active and specific to endothelial cells of varied species, with high selectivity indexes and inducibility under hypoxia-mimetic conditions. One in particular was then introduced back into a SIN HIV-1-based vector to confirm its endothelial cell activity and specificity. This study suggests that SIN vectors may be used in a high-throughput manner to identify tissue-specific promoters of high activity, with potential applications for both transcriptional targeting and gene transfer.
A major goal of gene therapy is the introduction of genes of interest into desired cell types. Because elevated levels of protein expression are often favored, typically ubiquitously and highly active viral or cellular promoters and enhancers drive the transgene in viral and nonviral vectors for both ex vivo and in vivo gene delivery. Such nonregulated promoters can cause difficulties, highlighted by the recent experience in the SCID-X1 patients who received ex vivo retrovirally transduced hematopoietic stem cells and later developed acute lymphoblastic leukemia (11), presumably due to oncogenic activation of the LMO-2 gene by the intact murine leukemia virus long terminal repeat (LTR) of the retroviral vector (15).
Transcriptional targeting is one way of limiting mRNA and protein expression to a specific cell type, even if the gene transfer vector was introduced into multiple cell types. This can be accomplished by using a tissue-specific promoter with or without an enhancer. Naturally occurring promoters, such as the albumin (18, 35) or β-globin promoter (22, 27) (the latter typically includes the locus control and other regulatory regions), may be problematic due to their relatively low activity, large size, or leakiness (i.e., variable expression in nontarget cell types). Methods have now been developed to identify more active and cell-specific promoters (8, 20, 32).
For example, Li and colleagues used a luciferase reporter plasmid coupled to a minimal promoter to isolate novel skeletal muscle (SM) promoters (20). A few duplex oligonucleotides representing the binding sites of SM-specific and nonspecific transcription factors were randomly ligated and cloned upstream of a minimal SM promoter driving firefly luciferase. Approximately 1,000 plasmid clones were then individually tested by transient transfection into SM cells and readout in 96-well format by luciferase luminometry. By this method, several active and SM-specific promoters were identified. Of note, these promoters had no common structure or arrangement of the DNA binding elements (20).
Although skeletal myocytes constitute an attractive target, especially for intramuscular gene delivery, endothelial cells are also worthy of attention. Endothelial cells line essentially all major blood vessels (arteries and veins) and thus have direct access to the circulatory system (36). Potential gene products to be delivered comprise hormones, polypeptides, and other protein factors found in plasma such as insulin, growth hormone, or factor VIII. Others include angiogenic growth factors or angiostatic molecules for the treatment of ischemic or neovascular conditions, respectively. The promoter regions upstream of several endothelial cell-specific genes have been characterized, and most have binding sites for both specific and nonspecific transcription factors. Visual inspection of several of these promoters suggests that there is no common structure or arrangement of the transcription binding DNA elements (1, 3, 5, 10, 12, 14, 19, 21, 23, 30, 33, 34). As one example, the core active promoter for human ICAM-2 is only ∼340 bp in length (6) and has binding sites for the transcription factors NF-κB, SP1, GATA, and Ets (Fig. 1B).
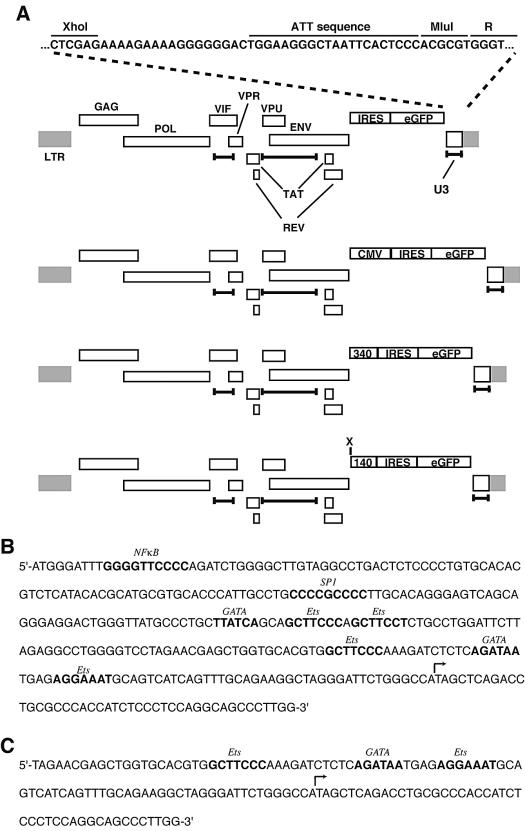
FIG. 1.
Structures of SIN HIV-based vectors and sequences of ICAM-2 promoters. (A) Schematic of SIN vectors. At top is the base vector, with U3 sequence and gene products as indicated. The bottom three are SIN vectors used in these studies; provirus deletions are indicated by the delimited bars. CMV and 340 denote the CMV IE enhancer and the ∼340-bp ICAM-2 promoters, respectively; X denotes the unique XbaI site just 5′ of the minimal 140 ICAM-2 promoter (140). (B) DNA sequence of ICAM-2 340 promoter. Sequence of ∼340-bp ICAM-2 promoter, beginning at −292 and ending at +44 (relative to major transcription start site), with elements bolded and identified in italic. (C) Minimal 140-bp ICAM-2 promoter, beginning at −95 and ending at +44; note absence of TATA box. DNA elements are identified, as are transcription start sites (bent arrows).
Because of their ability to efficiently transduce nondividing and terminally differentiated cells, human immunodeficiency virus (HIV)-based vectors have tremendous potential for gene therapeutic purposes (29). The most advanced of these vectors are self-inactivating (SIN) in that they have a large deletion in the 3′ LTR, which is duplicated to the 5′ end during transduction, thus inactivating both LTRs and necessitating an internal promoter to drive the transgene of interest (Fig. 1A) (24, 37). Typically, this internal promoter is highly active and nonspecific (e.g., the cytomegalovirus [CMV] immediate-early [IE] enhancer-promoter), but occasionally, cell-specific promoters are used (17, 22, 25, 27). Pseudotyping this class of vector with vesicular stomatitis virus glycoprotein G (VSV G) or amphotropic murine leukemia virus envelope increases vector stability and titer, broadens its host range, and allows the vector to transduce most mammalian cell types (29).
We reasoned that it may be possible to combine the synthetic promoter strategy described above with a promoter-less SIN HIV-based vector to conduct a high-throughput screen to identify a promoter that is both highly active and cell type specific. Because of our interest in ischemic cardiovascular disease and angiogenic gene delivery, we focused on isolating a synthetic promoter for endothelial cells. Here we describe such a genetic screen and report on the identification and characterization of several endothelial cell-specific promoters, one of which is quite active and specific. This general method is applicable to other cell types and could be easily modified to identify transcriptionally active DNA fragments (promoter and enhancer elements) in a genome-wide screen.
MATERIALS AND METHODS
Plasmids.
The base SIN vector pHIV-IRES-eGFP-X-M was constructed by deleting ∼400 bp from the 3′ LTR of pHIV-APΔVifΔVprΔEnv (31) by PCR (leaving 21 bp of the ATT sequence and the R sequence intact) and replacing AP with a 1.4-kb internal ribosome entry site (IRES)-enhanced green fluorescent protein (eGFP) cassette (Fig. 1A). pHIV-CMV-IRES-eGFP-X-M was constructed by inserting a 1.0-kb fragment containing the CMV IE enhancer-promoter from pCI (Promega Biotec) just upstream of the IRES. pHIV-ICAM140-IRES-eGFP-X-M and pHIV-ICAM340-IRES-eGFP-X-M were similarly constructed by inserting 140- and ∼340-bp fragments of the ICAM-2 promoter, respectively, generated by PCR from human genomic DNA (Fig. 1B and C). The luciferase reporter pGL3-Basic was obtained from Promega Biotec, and PGL3-ICAM140 and PGL3-ICAM340 were constructed by inserting the identical fragments of the ICAM-2 promoter just upstream of the luciferase coding region.
Cells and vector supernatants.
All cells were grown at 37°C in a water-jacketed 5% CO2 incubator and passaged at 1:3 to 1:5 once confluence was attained (typically once or twice weekly). 293T and COS7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (Life Technologies or Gemini), penicillin, and streptomycin (complete DMEM). Bovine aortic endothelial cells (BAECs) and early passage human foreskin fibroblasts (HFFs) were gifts from K. Hirschi and L. Donehower (both of Baylor College of Medicine), respectively, and similarly maintained. Human aortic endothelial cells and human umbilical vein endothelial cells (HUVECs) were purchased from Clonetics and grown according to the supplier's instructions. Rhesus macaque choroidal endothelial cells (CRL-1780) were originally obtained from the American Type Culture Collection and maintained in Ham's F-12 medium with similar supplements. Rabbit endothelial venous cells (REVCs) (9) were a gift from Y. Gillespie (University of Alabama, Birmingham) and were passaged in complete DMEM.
HIV vector supernatants were produced as described previously by cotransfection of 293T cells with the appropriate plasmids by the calcium-phosphate method. After 72 h, supernatant was harvested and titers were determined on the relevant cell type. After another 3 to 5 days, the percentage or number of eGFP-positive cells was quantitated by either inverted epifluorescence microscopy or flow cytometry, with eGFP fluorescence measured in the FL1 channel (FACSCalibur; Becton-Dickinson).
Library construction and analysis.
Duplex oligodeoxynucleotides, each with XbaI-compatible ends (Table 1) and representing the binding sites of specific and general transcription factors, were treated with kinase at the 5′ ends and ligated in equimolar ratios. Products were size fractionated by horizontal agarose gel electrophoresis, and those 150 to 500 bp in size were gel extracted, ligated into XbaI-cleaved pHIV-ICAM140-IRES-eGFP-X-M, and transformed into electrocompetent Escherichia coli strain DH10B (Stratagene). Approximately 1.2 × 106 bacterial colonies from 40 large petri plates were scraped and pooled together, and purified plasmid DNA was prepared by the alkaline lysis method and CsCl ultracentrifugation (yield, >5.0 mg).
TABLE 1.
Nonspecific and specific duplex oligonculeotides used
| Elementa | Length (bp) | Sequenceb |
|---|---|---|
| Sp1 | 11 | 5′-CATGGGCGGGT-3′ |
| 3′-CCGCCCAGTAC-5′ | ||
| 5′-CATGAGGCGGG-3′ | ||
| 3′-TCCGCCCGTAC-5′ | ||
| HIF-1 plus enhancer | 39 | 5′-CATGCCACAGTGCATACGTGGGCTCCAACAGGTCCTCTT-3′ |
| 3′-GGTGTCACGTATGCACCCGAGGTTGTCCAGGAGAAGTAC-5′ | ||
| ETE box | 49 | 5′-CATGGTACTTCATACTTTTCATTCCAATGGGGTGACTTTGCTTCTGGAG-3′ |
| 3′-CATGAAGTATGAAAAGTAAGGTTACCCCACTGAAACGAAGACCTCGTAC-5′ | ||
| NF-κB | 14 | 5′-CATGGGGACTTTCC-3′ |
| CCCTGAAAGGGTAC-5′ | ||
| Shear stress | 16 | 5′-CATGGGTCTCGGTCTC-3′ |
| 3′-CCAGAGCCAGAGGTAC-5′ |
HIF, hypoxia inducible factor; ETE, endothelin-1 element.
Core elements are underlined.
As described above, plasmid vector DNA (100 μg) was cotransfected with an equivalent amount of VSV G expression plasmid into a single 15-cm-diameter plate of 293T cells at 50 to 60% confluence, and vector supernatant was harvested. The equivalent of 2 × 106 IU was used to transduce 2 × 107 choroid endothelial cells (estimated transduction efficiency of ∼10%), and after 5 days, the highest eGFP expressers were sorted on a FACSCalibur. Sorted cells were expanded, resorted, and expanded again. Genomic DNA was prepared by using the QIAGEN PCR template preparation kit, and synthetic promoters were recovered by PCR with DNA primers 5′-ACGTGGTACCGGATTTTGCTAAGATGGGTGGCGC-3′ and 5′-ATATGGATCCAAGGGCTGCCTGGAGGGAGATGGT-3′ and thermocycling conditions of 94°C for 30 s, 65°C for 60 s, and 72°C for 30 s (35 cycles). PCR products were cleaved with KpnI and BamHI and directionally cloned in bulk into KpnI and BglII-cleaved pGL3-Basic (Promega Biotec). Recombinant plasmids were purified by the QIAGEN method and transfected into different cell types with Lipofectamine 2000 reagent (Invitrogen). Luciferase activity was measured by luminometry. The synthetic promoters of greatest interest were sequenced by the automated dideoxy chain termination method.
To transfer these promoters back into the SIN vector, PCR was performed with purified luciferase plasmid DNA as the template and primers 5′-GACTGCGGCCGCGGTACCGGATTTTGCTAAG-3′ and 5′-GACTCCCGGGGCTTACTTAGATCGCAGATCC-3′ under thermocycling conditions of 94°C for 30 s, 61°C for 60 s, and 72°C for 30 s (35 cycles). DNA products were cloned into pCR2.1 (Invitrogen) and sequenced by using flanking oligonucleotide primers. Plasmid was cleaved with NotI-SmaI, and the synthetic promoter fragments were cloned directionally into NotI-SmaI-cleaved pHIV-IRES-eGFP-X-M.
For reverse transcription (RT)-PCR, RNA was prepared at 72 h by using the Trizol reagent (Life Technologies) from Lipofectamine-transfected cells. An oligo(dT)18 primer was used for first-strand synthesis. For luciferase mRNA, PCR primers 5′-GAAGAGATACGCCCTGGTTCC-3′ and 5′-GTACATCGACTGAAATCCCCTG-3′ were used under thermocycling conditions of 94°C for 10 s, 52°C for 30 s, and 68°C for 30 s (25 cycles) and gave rise to a 400-bp product. For β-actin mRNA, PCR primers 5′-CCAGAGAGGACATTGTTGGC-3′ and 5′-TGGAGAAGAGCTATGAGCTGC-3′ were used under identical thermocycling conditions and gave rise to a 196-bp DNA fragment. DNA products were separated by horizontal agarose gel electrophoresis, stained with ethidium bromide, and quantitated by using National Institutes of Health ImageQuant software.
RESULTS
Construction of a complex synthetic promoter library.
We wished to improve the synthetic promoter strategy of Li and colleagues (20) by employing SIN HIV-based vectors and flow cytometry to achieve higher throughput. The overall strategy is illustrated schematically in Fig. 2. Because of our interest in endothelial cell gene delivery, we decided to focus our attention on endothelial cell-specific promoters. A number of endothelial cell-specific genes have been molecularly characterized, and some of the upstream transcriptional DNA elements have been identified and studied. Because a reasonably achievable library complexity and screening was limited to ∼2 × 106 clones, we chose five different duplex oligonucleotides, representing the binding sites of some of these nonspecific and specific transcription factors (Table 1). Because the arithmetic mean length of the chosen oligonucleotides was 25 bp and we were attempting to identify synthetic promoters of approximately 250 bp in size, we estimated that we could cover most of the sequence and permutation space in a single plasmid library.
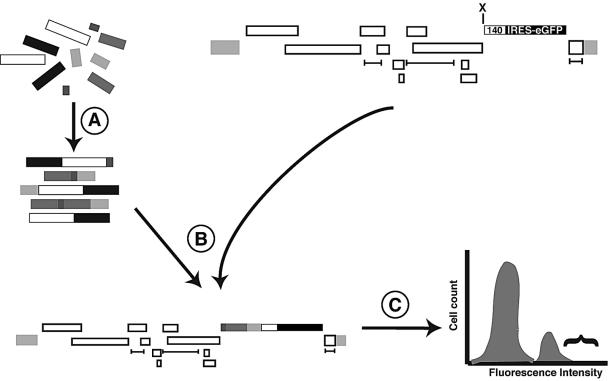
FIG. 2.
Strategy for library construction and screening. Step A is the ligation of the duplex oligonucleotides, step B is the cloning of those products in the 140 SIN vector, and step C is the production of VSV G-pseudotyped particles followed by endothelial cell transduction and flow cytometry (shown as a histogram for simplicity). Bracket indicates highest eGFP expressers to be positively sorted. Note that the positive cells represent a small fraction (<10%) of the overall population.
All five duplex oligonucleotides were ligated in equimolar ratios by their XbaI-compatible ends, size fractionated, and cloned upstream of an IRES-eGFP reporter in the SIN vector XbaI-cleaved pHIV-ICAM140-IRES-eGFP-X-M, which has 140 bp of the ICAM-2 promoter (Fig. 1A). Approximately 1.2 × 106 plasmid recombinants were scraped and pooled, and purified DNA was prepared. Two dozen of these were isolated individually and analyzed by restriction digestion by native vertical polyacrylamide gel electrophoresis. More than 90% had an insert, with an average size of 175 bp. In parallel with pHIV-CMV-IRES-eGFP-X-M and the other SIN vectors (Fig. 1A), pooled DNA was used to produce VSV G-pseudotyped replication-defective HIV particles by transient cotransfection of 293T cells.
Screening the synthetic promoter library.
The rhesus monkey choroid endothelial cell line was used to screen the synthetic promoter library. Because we wished to avoid double integrants that could confound later analyses, we chose a relatively low multiplicity of infection (MOI), such that <10% of the cells were transduced. The use of greater amounts of vector supernatant resulted in nonlinear increases in transduction efficiency, as has been reported for HIV-based vectors in rhesus cells (26) (thought to be related to a postentry block in replication) (Fig. 3). Because the 140 promoter has minimal activity and most of the synthetic promoters would be expected to be inactive, it was not surprising that only a few of the transduced cells had high eGFP activity (Fig. 4). Close inspection of library of transduced cells compared with HIV-ICAM140-IRES-eGFP-X-M (VSV G) transduced choroid cells suggested only a very subtle increase in eGFP expression in the case of the library.
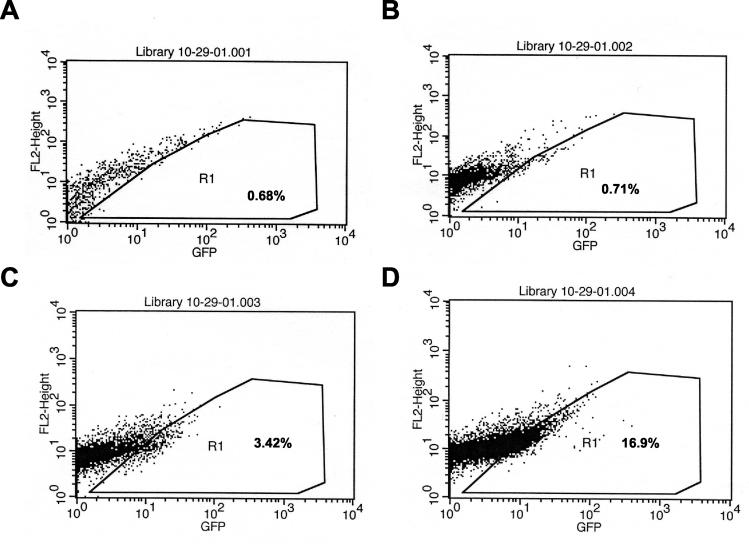
FIG. 3.
Transduction of choroid endothelial cells. Cells were transduced in plate (10-cm diameter) format (∼2 × 107 cells/plate) with increasing amounts of 140 vector supernatant, and 72 h later, cells were analyzed by flow cytometry. (A) Mock transduction; (B) 0.30 ml of vector supernatant; (C) 0.70 ml of vector supernatant; (D) 1.0 ml of vector supernatant. Percentages of cells falling into R1 are as indicated.
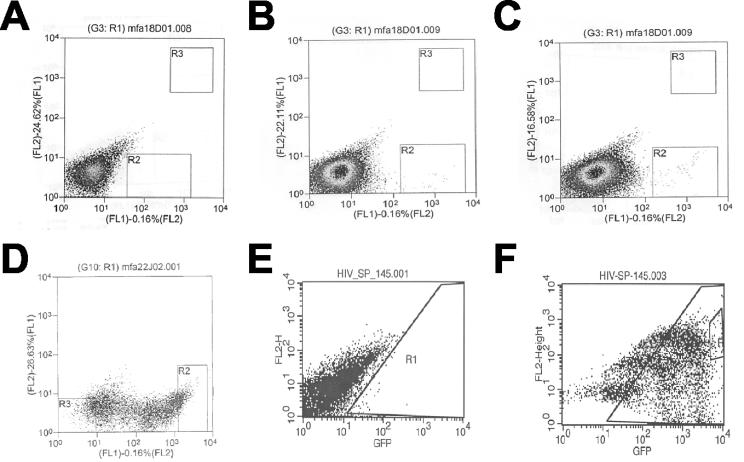
FIG. 4.
Flow cytometry of transduced choroid cells. (A) Mock transduction; (B) cells transduced with 140 SIN vector; (C) cells transduced with library; (D) analysis post-first sort; (E) mock transduction for second sort; (F) analysis post-second sort. R2 in panel C was the initial sorting gate; R2 in panel D was the second sorting gate. Note that in both panels D and F there appear to be three distinct populations of unknown significance.
Transduced choroid cells were sorted for high eGFP expression, and the sorted cells were expanded and resorted for eGFP expression (Fig. 4). During the resort, some of the cells had low or no eGFP expression, which may be due to sorting error, silencing of the eGFP transgene in the transduced cells, or selective expansion of the eGFP-negative cell population due to cytotoxicity associated with the vector. After the resort, visual examination of the cells by epifluorescence microscopy revealed >95% of the cells to be eGFP positive, with stable transgene expression over a several month period, and genomic DNA was prepared from this population of cells.
Analysis of synthetic promoters.
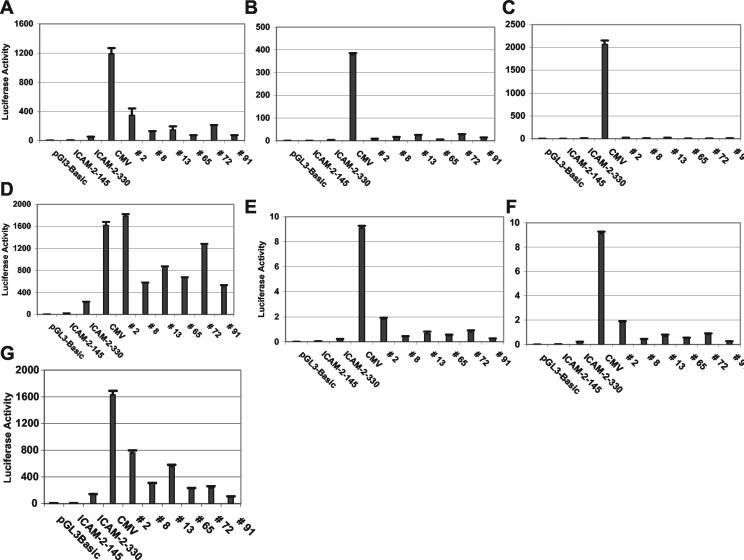
Because of potential confounders, including epigenetic phenomena, multiple vector integrants, or chromatin read-through transcription effects due to the genome integration site, we wished to recover and retest the synthetic promoters. Using flanking vector PCR primers and purified genomic DNA, the synthetic promoters were amplified in bulk and directionally cloned into a luciferase reporter plasmid. QIAGEN-purified plasmid DNA was individually prepared from ∼90 clones and transfected into a variety of cell types and lines, and luciferase activity was measured by luminometry. Approximately one-third of the clones had activity within 5-fold and most (74 of 86) had activity within 20-fold of the 140 promoter and were not further pursued (Fig. 5). A dozen clones, however, had markedly increased activity compared to 140 (20- to 115-fold greater) and comparable or superior activity to that of 340 (ICAM-2 promoter with full activity) (Fig. 5 and 6). Of the 11 that were further analyzed, 5 were duplicates, leaving 6 unique synthetic promoters. These clones were active in all endothelial cell types tested (not just the choroid cells) and had minimal activity in other cell types such as HFFs and COS7 and 293T cells (Fig. 6 and data not shown). As expected, the CMV IE enhancer-promoter had the greatest activity but was robust in all tested cell types, consistent with its known nonspecificity. Clone 2 was the most active, especially in the REVCs, and in other endothelial cells, it was one-fifth to one-third as active as the CMV IE enhancer-promoter (Fig. 6). The activity of clone 2 was typically 5- to 10-fold greater than that of 340 in endothelial cells.
FIG. 5.
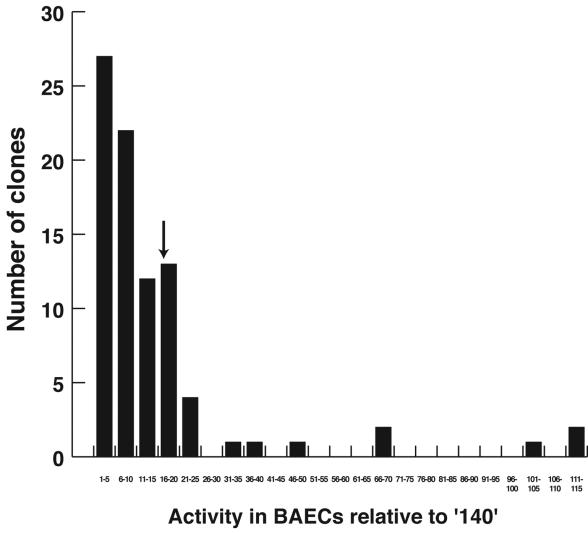
Binning of luciferase activity of tested clones. BAECs were transfected individually in six-well format with both the test clone and a Renilla luciferase normalization control; both luciferase activities were measured 72 h later and also normalized to the 140 minimal promoter. Note that the majority of clones have activity that is <20-fold greater than that of the minimal promoter, whereas some have activity more than 100-fold greater. The down arrow indicates the relative activity of the 340 ICAM-2 promoter. The CMV IE enhancer-promoter had activity that was ∼330-fold greater (i.e., off this scale).
FIG. 6.
Activity of clones in different cells. Cells of varied type and species were transfected in at least triplicate with the different clones, and luciferase activity was measured 72 h later and normalized with respect to a cotransfected Renilla luciferase control reporter plasmid. (A) BAECs; (B) REVCs; (C) COS-7 cells; (D) HUVECs; (E) human aortic endothelial cells; (F) HFFs; (G) choroid cells. For each panel, the different constructs used are shown at the bottom, and values represent mean relative light units ± standard error.
A selectivity index for each of the six synthetic promoters was calculated by dividing the activity of the clone in an endothelial cell by its activity in HFFs normalized by doing the same for the CMV IE enhancer-promoter. As shown in Table 2, the selectivity index for clone 2 ranged from ∼40 to ∼150 (depending upon the cell type), whereas the least selective was clone 91 (values ranged from ∼4.5 to ∼29). Even the latter compared favorably to 140 and 340, whose values ranged from ∼1.5 to ∼5.0 (contrasted to 1.0 to 1.3 for the CMV promoter).
TABLE 2.
Selectivity index for select clones
| Clone no. | Selectivity indexa for:
|
||
|---|---|---|---|
| BAECs | REVCs | Choroid cells | |
| 2 | 41 | 146.74 | 62.2 |
| 8 | 6.74 | 21.29 | 11.61 |
| 13 | 6.54 | 22.98 | 14.72 |
| 65 | 12 | 77.05 | 26.39 |
| 72 | 6.27 | 26.77 | 4.96 |
| 91 | 4.56 | 29.28 | 4.61 |
| ICAM-140 | 2.0 | 5.38 | 2.49 |
| ICAM-330 | 1.45 | 4.16 | 3.41 |
| CMV IE promoter | 1.00 | 1.33 | 1.34 |
The selectivity index is calculated as (the activity in the indicated cell type divided by the activity in HFFs), the result of which is divided by the CMV IE enhancer-promoter activity in BAECs divided by the CMV enhancer-promoter activity in HFFs. All activities are relative light units, based upon transient transfection.
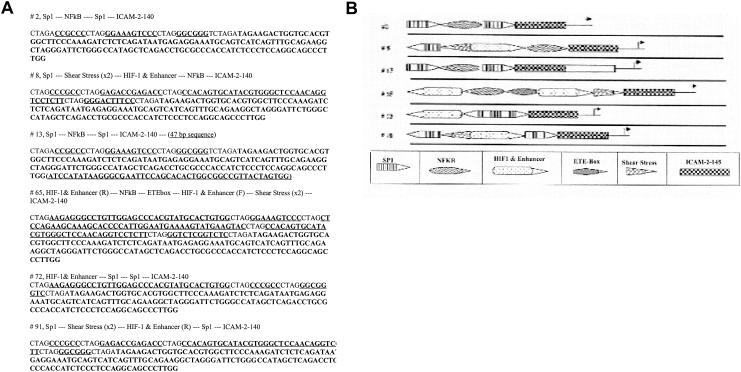
The synthetic promoters of these clones were DNA sequenced (Fig. 7A). A schematic of their organization indicates no common arrangement or structure, with the exception that clones 2 and 13 are clearly related (Fig. 7B). The extraneous sequence of clone 13 is likely derived from a contaminating plasmid, since it contains the sequence of multiple six cutter restriction endonucleases. Its precise origin is undefined, since a BLAST search reveals ∼80 DNA sequences with P values of <4 × 10−9. All of the sequenced clones appear to have a random assortment of the duplex oligonucleotides, with both specific and nonspecific DNA elements represented in both orientations and none clearly over- or underrepresented. To demonstrate that these promoters were acting at the level of mRNA accumulation, RT-PCR was performed, with clone 2 as a test case. As shown in Fig. 8, the greatest amount of RT-PCR product was obtained after transfection of clone 2, consistent with an increase in mRNA encoding luciferase. Although we cannot exclude effects on transcript elongation, stability, or half-life, it is most likely that the promoters act at the level of mRNA initiation.
FIG. 7.
Sequence and structure of select clones. (A) DNA sequences of indicated synthetic promoters. For clone 13, the sequence of uncertain origin is in parentheses. (B) Schematic of the structures of the indicated synthetic promoters.
FIG. 8.
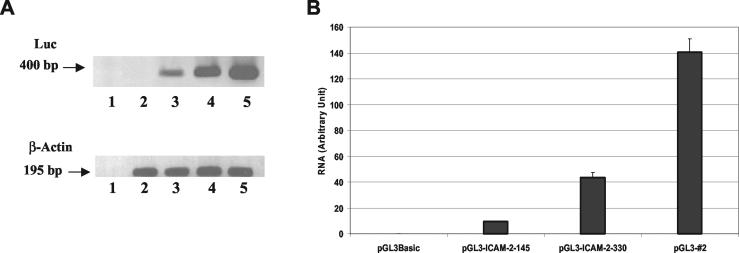
Quantitation of mRNA levels. (A) BAECs were transfected with different constructs, and 72 h later, RNA was prepared and RT-PCR for both luciferase (Luc) and β-actin was performed as described in the text. Lanes: 1, water control; 2, pGL3-Basic; 3, 140 minimal promoter; 4, 340 ICAM-2 promoter; 5, clone 2 synthetic promoter. (B) Quantitation of the results shown in panel A.
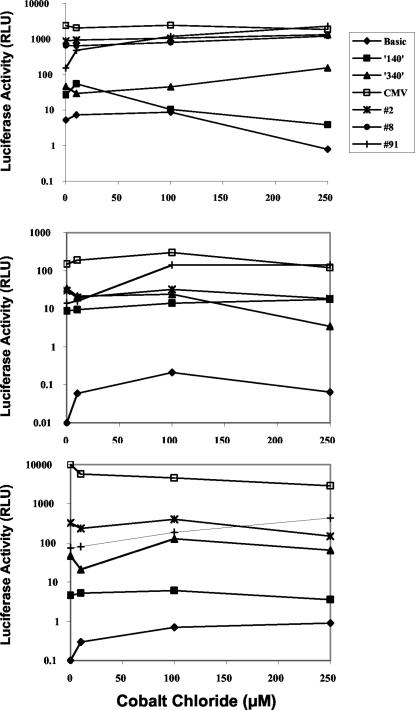
Several of the synthetic promoters have a hypoxia inducible factor 1 (HIF-1)-positive enhancer element, which is considered responsive to low oxygen tension. To demonstrate promoter activation in the presence of hypoxia, cells were transfected and then treated with increasing amounts of the hypoxia mimetic CoCl2 (2, 28). Several of the promoters were up-regulated two- to fivefold in this setting, specifically in endothelial cells, whereas the CMV promoter had uniformly high activity, irrespective of cell type and CoCl2 concentration (data not shown and Fig. 9). Clone 91 was the most responsive, with inductions of 15- to 20-fold in BAECs and less than 5- to 10-fold in other cell types (Fig. 9). Similar induction levels for clone 91 were observed in choroidal cells. Clone 2 was unresponsive to CoCl2 in all cell types.
FIG. 9.
The effect of hypoxia on synthetic promoter activity. Different cell types were transfected with the indicated luciferase constructs and exposed to increasing amounts of CoCl2 48 h later. Twenty-four hours later, luciferase activity was measured. Top panel, BAECs; middle panel, HFFs; bottom panel, 293T cells. Note the log scale for relative light units (RLU).
Several of the promoters (notably 340 and clones 2 and 8) have an NF-κB site, which should induce expression upon activation of that pathway. Both BAECs and HFFs were transiently transfected with these and control plasmids and then exposed to increasing amounts of tumor necrosis factor alpha (TNF-α; R & D Systems). Only clone 8 reproducibly had higher luciferase activity in the presence of TNF-α, which occurred in both cell types, although induction levels were only slight (data not shown). These results suggest that these synthetic promoters can respond to physiologic stressors, similar to endogenous endothelial cell-specific promoters, although the presence of a specific DNA element does not ensure responsiveness (it has to be empirically determined).
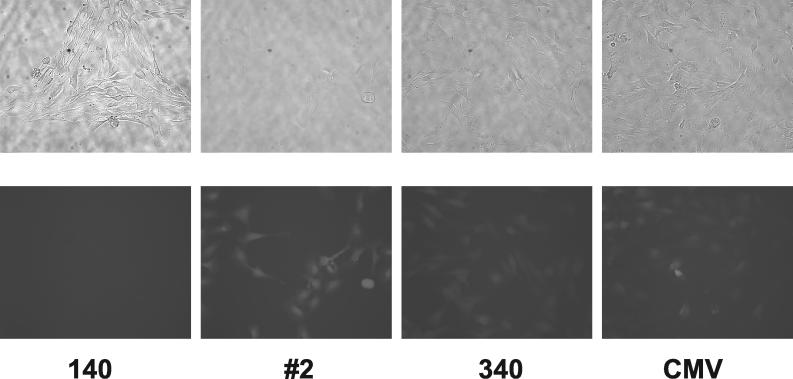
Finally, we wished to demonstrate that the synthetic promoters were still active after placement back into the SIN HIV-based vector. For this, clone 2 was PCR amplified and cloned upstream of IRES-eGFP in the base vector pHIV-IRES-eGFP-X-M. VSV G-pseudotyped vector particles were produced and used to transduce both endothelial and nonendothelial cell lines. As shown in Fig. 10, choroidal cells transduced with pHIV-#2-IRES-eGFP-X-M (VSV G) were brighter than cells transduced with either pHIV-ICAM140-IRES-eGFP-X-M (VSV G) or pHIV-ICAM340-IRES-eGFP-X-M (VSV G) and had similar brightness to cells transduced with pHIV-CMV-IRES-eGFP-X-M (VSV G). Nonendothelial cells that were transduced with similar amounts of vector showed little eGFP fluorescence, consistent with the previously observed specificity of the synthetic promoters. Both HFFs and HUVECs were transduced at a low MOI (to avoid multiple integrants) with the same four SIN vectors and then analyzed by flow cytometry. As shown in Table 3, neither clone 2 or 340 was active in HFFs, whereas clone 2 was clearly active in the endothelial cells. Note the compressed dynamic range in that the mean fluorescence intensity (MFI) ratio for cells transduced with the CMV vector was only 6.7. These results suggest that, as expected, the synthetic promoters maintain both their activity and specificity in the setting of a SIN HIV-based vector.
FIG. 10.
Transduction of choroid cells with SIN vectors. The images in the top row are phase-contrast images, and those in the bottom row are corresponding fluorescence images with a GFP filter. 140, cells transduced with HIV-ICAM140-IRES-eGFP-X-M (VSV G); #2, cells transduced with pHIV-#2-IRES-eGFP-X-M (VSV G); 340, cells transduced with HIV-ICAM340-IRES-eGFP-X-M (VSV G); CMV, cells transduced with HIV-CMV-IRES-eGFP-X-M (VSV G). Transduction rates for the latter three were approximately 50%. No fluorescent cells were visible after mock transduction (data not shown).
TABLE 3.
Transduction of primary cells by SIN vectors
| Cell type | Vectora | Amt used (ml)b | % Trans- ductionc | MFI(+)d | MFI(−)e | Ratiof |
|---|---|---|---|---|---|---|
| HFF | 140 | 0.25 | 0.57 | 22.2 | 8.3 | 2.67 |
| 330 | 0.25 | 2.69 | 26.34 | 9.88 | 2.67 | |
| Clone 2 | 0.25 | 0.69 | 24.02 | 8.06 | 2.98 | |
| CMV | 0.1 | 10.46 | 58.18 | 12.82 | 4.53 | |
| HUVEC | 140 | 0.01 | 2.35 | 16.31 | 6.15 | 2.65 |
| 330 | 0.01 | 5.8 | 17.98 | 6.77 | 2.66 | |
| Clone 2 | 0.025 | 6.85 | 25.54 | 6.7 | 3.81 | |
| CMV | 0.01 | 7.47 | 42.26 | 6.3 | 6.7 |
All SIN vectors were pseudotyped with VSV G.
Transductions were performed overnight in a six-well format with the indicated amount of vector supernatant (2-ml total volume).
Percentage of transduction as determined by flow cytometry 72 h later.
MFI of the positive cells.
MFI of the negative cells.
MFI(+) divided by MFI(−).
DISCUSSION
Targeting of gene transfer vectors may be advantageous for different therapeutic applications. For retroviral and lentiviral vectors, it is problematic to achieve targeting at the level of viral or cellular entry, and efficient targeting of a specific genomic integration site is not yet technically feasible. Use of tissue-specific promoters has achieved a certain level of transcriptional targeting, especially in the setting of SIN vectors. Often, however, these promoters have modest levels of activity (especially when compared to a very active viral promoter) or the sheer size of the promoter-enhancer makes high-titer vector production difficult. For these reasons, we sought to identify a novel, cell-specific promoter of short length and superior activity.
We chose to concentrate our efforts on endothelial cells because of their proximity to the bloodstream and their involvement in angiogenesis and vasculogenesis, not to mention other disease processes (4). The method we developed, however, can be generalized to any cell type that is culturable and for which there is rudimentary knowledge of positively acting transcriptional control elements. A number of endothelial cell-specific genes are known and characterized, and we were able to choose from a number of DNA binding elements located upstream of these genes.
We also sought to increase the sampled sequence space of synthetic promoters, especially when compared to previously developed methods where only approximately 1,000 promoters were tested. To accomplish this aim, we used a SIN HIV-based vector such that bulk construction and testing of a library of 106 recombinants was quite manageable. We realized that the law of diminishing returns would become operational at some point during the screen in that we expected to recover synthetic promoters that had activity comparable to that of the CMV IE, but we doubted that we would unearth promoters that had fivefold-greater activity. We fully anticipated, however, finding promoters that had activity similar if not greater than that of a highly active endothelial cell-specific promoter, namely that of ICAM-2.
One might argue why conduct this experimental exercise if only promoters of strength similar to that of ICAM-2 will be identified. We would argue that (i) there is reasonable probability of recovering much more active and specific promoters (higher selectivity indexes), (ii) the synthetic promoters might be regulated differently and thus serve diverse therapeutic purposes, (iii) cataloging such a collection might allow a determination of what makes a promoter endothelial cell specific, especially when they are all very compact, and (iv) it shows as proof-of-concept that such a screen can be performed and it may be quite useful in cell types in which much less is known regarding transcriptional regulatory mechanisms (e.g., dendritic cells and macrophages).
The success of this type of approach most certainly depends upon the initial choice of vector and transcription elements. Although the SIN vector used here is not the most advanced, it has a very extensive U3 deletion such that only rarely are positive cells observed after transduction with the promoter-less variant, and these may be due to read-through transcription or other integration site effects (e.g., integration near a fortuitous promoter). Furthermore, the amount of p24 (capsid) found in the supernatant of cells transduced with this SIN vector is essentially background. In addition, the titer of the CMV-driven vector is >106 IU/ml, making large-scale screens facile. The more difficult decision was the choice of the DNA binding elements and basal promoter. We realized that beginning with a promoter with too much activity might be self-defeating, as would one that had no activity at all. Thus, we compromised with a 140-bp minimal promoter, recognizing that others might prove superior, an approach similarly taken by Li and colleagues (20). This promoter clearly had some activity (Fig. 3 and 4) but not an overwhelming amount, making the identification of more active promoters relatively easy. This promoter does not have a canonical TATA element, and it is certainly possible that the outcome of the screen would have been different had the promoter included that element.
The outcome of the library screen obviously also depended upon the choice of DNA binding elements, of which there was a very wide selection, even when limited to only those located upstream of endothelial cell-specific genes. Because we wished to cover a certain sequence/permutation space, we restricted the number to five, used in an equistoichiometric ratio, and we also focused on relatively short DNA fragments. In addition, we decided to include two nonspecific and three specific elements, since (i) all examined endothelial promoters have a mixture of both, (ii) a similar scheme was successful in the case of skeletal myocytes, and (iii) we thought this combination might optimize both activity and specificity. The fact that we had a modicum of success with this approach suggests that it did work, although we cannot exclude the possibility that other not dissimilar methods would have yielded better results.
Use of the SIN HIV-based vector coupled with flow cytometry of transduced cells allowed the construction and analysis of a plasmid library with >106 recombinants. The present library screen was 3 orders of magnitude greater than previous high-throughput methods in which individual clones were analyzed by luciferase luminometry (20). A library of 107 recombinants would be more difficult but not infeasible to construct and screen. Although it is probable that more active and specific promoters would be present in a larger library, it is also likely that the law of diminishing returns would apply here as well.
Could such a screen identify negative regulatory DNA elements, which are likely just as important as positive ones? One could certainly begin with an active promoter and insert DNA sequences between it and the reporter; the problem is that, for most cell types and genes, even fundamental knowledge regarding these sorts of sequences is lacking. Oligomers could be inserted (although construction of such a library could be difficult), but unfortunately, there is no ideal method for performing the screen. For example, at a low MOI, most cells will not be transduced and appear to be nonexpressers, and at a high MOI, multiple integrants will obscure the results. Even if oligomers with negative regulatory activity were to be identified, it may be difficult to discern their import and physiologic relevance.
It was not feasible, and we did not attempt, to individually clone and test every single PCR product obtained from genomic DNA prepared from the resorted eGFP-positive cells. Thus, we are not certain whether in the end we truly sampled the sequence space, and it is conceivable that more active and specific promoters were present but were not isolated. Because several thousand cells were initially positively sorted, we would have had to characterize thousands of PCR products, which was beyond the scope of this work. Of note, 5 of the 11 most-active promoters were duplicates, suggesting that the screen was partially saturating. In addition, two of the isolated clones (2 and 13) were quite similar, although we cannot exclude the possibility that those two clones are related artifactually (i.e., faulty cloning or PCR error).
Of the ∼90 PCR products that were recovered from the genomic DNA, 43% had activity 10-fold higher and 14% had activity 20-fold higher than that of the 140 clone. For the other 57%, it is uncertain why they survived the library screen, although possibilities include sorting error (likely accounts for a minority of the clones), read-through transcription or fortuitous promoter due to vector integration into an active transcriptional unit (especially since the reporter had a preceding IRES), or inadvertency due to the presence of more than a single vector integrant (i.e., one of the other vector integrants carried the active synthetic promoter). Although we used a relatively low MOI to reduce the probability of obtaining double and triple integrants, at some level this problem is unavoidable due to the Poisson distribution and the fact that many monkey cells show cooperative transduction kinetics (Fig. 3). The latter is presumably due to saturation of one or more factors present in nonhuman primate cells that eliminate a postentry step during HIV replication (7, 13, 16, 26), which was unappreciated when this screen was initiated. A lower MOI (e.g., 0.01) could have been used, which would have increased the probability of single integrants, but then a greater number of cells would have had to be analyzed to identify a similar number of high expressors. Thus, we decided to employ a higher MOI, realizing that a second test of activity would be required to screen out false positives. Much higher MOIs would presumably have led to even a higher false-positive rate.
Despite all the caveats outlined above, we were able to identify several active and endothelial cell-specific promoters. All of these had very high selectivity indexes, especially compared to the 340 and 140 clones. We confirmed for at least one of them (clone 2) that the promoter was acting at the level of mRNA accumulation (and presumably at the level of transcript initiation). This same promoter functioned as expected when placed back into the SIN vector, suggesting that its activity was not artifactual. Of greatest interest is that its simple structure and sequence belied its selectivity. The fact that both it and clone 13 (ignoring the attached sequence of uncertain origin) consist solely of nonspecific elements suggests that the specificity was imparted by the minimal 140 promoter, which has both nonspecific and specific elements. We have not performed mutagenesis studies of clone 2 to determine the functional regions, and it is conceivable but unlikely that somehow a site for an unknown specific transcription factor was created from hybrid sequences.
Casual inspection revealed no obvious structure or arrangement of the isolated active and specific synthetic promoters. All of the duplex oligonucleotides were represented, although admittedly this may not accurately reflect the tested sequence or permutation space, since it is a highly biased sampling. Some of the elements were in reverse orientation, but without further functional analyses, it is uncertain which elements or arrangements are critical for the activity or specificity of any individual clone. Several of the promoters had the HIF-1-positive enhancer element, and a subset of those was quite responsive to hypoxia-mimetic conditions, with activity inductions of 5- to 20-fold, with the highest induction levels in endothelial cells (presumably due to the presence of the transcription factor HIF-1), although the most inducible of these had increased activity in other cell types. Clone 8, which contains an NF-κB site, was induced less than fivefold in the presence of TNF-α, although other promoters with NF-κB were not induced. Thus, the presence of a DNA element did not guarantee functionality. This also suggests that, despite their artificial nature, some of the synthetic promoters are capable of responding to physiologic stressors and conditions to which endothelial cells may be subject. This may be useful in tissue ischemic or low-blood-flow conditions in which oxygen content is reduced.
In summary, we were able to use a SIN HIV-based vector to construct a complex library of short synthetic promoters and identified several that were both quite active and specific in endothelial cells of varied mammalian species, with high selectivity indexes. The simplicity of several of them was quite unexpected, and others responded to physiologic conditions to which endothelial cells are often exposed. Further characterization of these may provide insight into the mRNA expression control mechanisms of endothelial genes, and at present, some may prove useful for transcriptional targeting of gene therapy vectors now in use.
Acknowledgments
We thank K. Hirschi, Y. Gillespie, and L. Donehower for generous reagent gifts.
This work was supported by an American Heart Association Beginning Grant-in-Aid (9960097Y) and a Grant-in-Aid (0150793Y). C.D. was supported by a research agreement with AstraZeneca Pharmaceuticals and in part by T32 AI07456. R.E.S. is an Edward Mallinckrodt, Jr., Foundation Scholar.
REFERENCES
- 1.Arai, H., K. Nakao, K. Takaya, K. Hosoda, Y. Ogawa, S. Nakanishi, and H. Imura. 1993. The human endothelin-B receptor gene. Structural organization and chromosomal assignment. J. Biol. Chem. 268:3463-3470. [PubMed] [Google Scholar]
- 2.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 3.Bu, X., and T. Quertermous. 1997. Identification of an endothelial cell-specific regulatory region in the murine endothelin-1 gene. J. Biol. Chem. 272:32613-32622. [DOI] [PubMed] [Google Scholar]
- 4.Carmeliet, P., and R. K. Jain. 2000. Angiogenesis in cancer and other diseases. Nature 407:249-257. [DOI] [PubMed] [Google Scholar]
- 5.Collins, T., M. A. Read, A. S. Neish, M. Z. Whitley, D. Thanos, and T. Maniatis. 1995. Transcriptional regulation of endothelial cell adhesion molecules: NF-kappa B and cytokine-inducible enhancers. FASEB J. 9:899-909. [PubMed] [Google Scholar]
- 6.Cowan, P. J., D. Tsang, C. M. Pedic, L. R. Abbott, T. A. Shinkel, A. J. d'Apice, and M. J. Pearse. 1998. The human ICAM-2 promoter is endothelial cell-specific in vitro and in vivo and contains critical Sp1 and GATA binding sites. J. Biol. Chem. 273:11737-11744. [DOI] [PubMed] [Google Scholar]
- 7.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edelman, G. M., R. Meech, G. C. Owens, and F. S. Jones. 2000. Synthetic promoter elements obtained by nucleotide sequence variation and selection for activity. Proc. Natl. Acad. Sci. USA 97:3038-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman, C. K., and G. Y. Gillespie. 1995. Physiologic responses of REVC, a continuous rabbit endothelial vascular cell. J. Vasc. Res. 32:31-40. [DOI] [PubMed] [Google Scholar]
- 10.Gumina, R. J., N. E. Kirschbaum, K. Piotrowski, and P. J. Newman. 1997. Characterization of the human platelet/endothelial cell adhesion molecule-1 promoter: identification of a GATA-2 binding element required for optimal transcriptional activity. Blood 89:1260-1269. [PubMed] [Google Scholar]
- 11.Hacein-Bey-Abina, S., C. von Kalle, M. Schmidt, F. Le Deist, N. Wulffraat, E. McIntyre, I. Radford, J. L. Villeval, C. C. Fraser, M. Cavazzana-Calvo, and A. Fischer. 2003. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 348:255-256. [DOI] [PubMed] [Google Scholar]
- 12.Hata, Y., E. Duh, K. Zhang, G. S. Robinson, and L. P. Aiello. 1998. Transcription factors Sp1 and Sp3 alter vascular endothelial growth factor receptor expression through a novel recognition sequence. J. Biol. Chem. 273:19294-19303. [DOI] [PubMed] [Google Scholar]
- 13.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda, T., K. Wakiya, and M. Shibuya. 1996. Characterization of the promoter region for flt-1 tyrosine kinase gene, a receptor for vascular endothelial growth factor. Growth Factors 13:151-162. [DOI] [PubMed] [Google Scholar]
- 15.Kohn, D. B., M. Sadelain, and J. C. Glorioso. 2003. Occurrence of leukaemia following gene therapy of X-linked SCID. Nat. Rev. Cancer 3:477-488. [DOI] [PubMed] [Google Scholar]
- 16.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 100:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostic, C., F. Chiodini, P. Salmon, M. Wiznerowicz, N. Deglon, D. Hornfeld, D. Trono, P. Aebischer, D. F. Schorderet, F. L. Munier, and Y. Arsenijevic. 2003. Activity analysis of housekeeping promoters using self-inactivating lentiviral vector delivery into the mouse retina. Gene Ther. 10:818-821. [DOI] [PubMed] [Google Scholar]
- 18.Kuriyama, S., M. Yoshikawa, S. Ishizaka, T. Tsujii, K. Ikenaka, T. Kagawa, N. Morita, and K. Mikoshiba. 1991. A potential approach for gene therapy targeting hepatoma using a liver-specific promoter on a retroviral vector. Cell Struct. Funct. 16:503-510. [DOI] [PubMed] [Google Scholar]
- 19.Lee, M. E., M. S. Dhadly, D. H. Temizer, J. A. Clifford, M. Yoshizumi, and T. Quertermous. 1991. Regulation of endothelin-1 gene expression by Fos and Jun. J. Biol. Chem. 266:19034-19039. [PubMed] [Google Scholar]
- 20.Li, X., E. M. Eastman, R. J. Schwartz, and R. Draghia-Akli. 1999. Synthetic muscle promoters: activities exceeding naturally occurring regulatory sequences. Nat. Biotechnol. 17:241-245. [DOI] [PubMed] [Google Scholar]
- 21.Liu, Y., S. R. Cox, T. Morita, and S. Kourembanas. 1995. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells. Identification of a 5′ enhancer. Circ. Res. 77:638-643. [DOI] [PubMed] [Google Scholar]
- 22.May, C., S. Rivella, J. Callegari, G. Heller, K. M. Gaensler, L. Luzzatto, and M. Sadelain. 2000. Therapeutic haemoglobin synthesis in beta-thalassaemic mice expressing lentivirus-encoded human beta-globin. Nature 406:82-86. [DOI] [PubMed] [Google Scholar]
- 23.Meacock, S., R. Pescini-Gobert, J. F. DeLamarter, and R. Hooft van Huijsduijnen. 1994. Transcription factor-induced, phased bending of the E-selectin promoter. J. Biol. Chem. 269:31756-31762. [PubMed] [Google Scholar]
- 24.Miyoshi, H., U. Blomer, M. Takahashi, F. H. Gage, and I. M. Verma. 1998. Development of a self-inactivating lentivirus vector. J. Virol. 72:8150-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi, H., M. Takahashi, F. H. Gage, and I. M. Verma. 1997. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc. Natl. Acad. Sci. USA 94:10319-10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pawliuk, R., K. A. Westerman, M. E. Fabry, E. Payen, R. Tighe, E. E. Bouhassira, S. A. Acharya, J. Ellis, I. M. London, C. J. Eaves, R. K. Humphries, Y. Beuzard, R. L. Nagel, and P. Leboulch. 2001. Correction of sickle cell disease in transgenic mouse models by gene therapy. Science 294:2368-2371. [DOI] [PubMed] [Google Scholar]
- 28.Piret, J. P., D. Mottet, M. Raes, and C. Michiels. 2002. CoCl2, a chemical inducer of hypoxia-inducible factor-1, and hypoxia reduce apoptotic cell death in hepatoma cell line HepG2. Ann. N. Y. Acad. Sci. 973:443-447. [DOI] [PubMed] [Google Scholar]
- 29.Quinonez, R., and R. E. Sutton. 2002. Lentiviral vectors for gene delivery into cells. DNA Cell Biol. 21:937-951. [DOI] [PubMed] [Google Scholar]
- 30.Ronicke, V., W. Risau, and G. Breier. 1996. Characterization of the endothelium-specific murine vascular endothelial growth factor receptor-2 (Flk-1) promoter. Circ. Res. 79:277-285. [DOI] [PubMed] [Google Scholar]
- 31.Sutton, R. E., H. T. Wu, R. Rigg, E. Bohnlein, and P. O. Brown. 1998. Human immunodeficiency virus type 1 vectors efficiently transduce human hematopoietic stem cells. J. Virol. 72:5781-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tornoe, J., P. Kusk, T. E. Johansen, and P. R. Jensen. 2002. Generation of a synthetic mammalian promoter library by modification of sequences spacing transcription factor binding sites. Gene 297:21-32. [DOI] [PubMed] [Google Scholar]
- 33.Valdenaire, O., E. Rohrbacher, and M. G. Mattei. 1995. Organization of the gene encoding the human endothelin-converting enzyme (ECE-1). J. Biol. Chem. 270:29794-29798. [DOI] [PubMed] [Google Scholar]
- 34.Wilson, D. B., D. M. Dorfman, and S. H. Orkin. 1990. A nonerythroid GATA-binding protein is required for function of the human preproendothelin-1 promoter in endothelial cells. Mol. Cell. Biol. 10:4854-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, X., J. Holschen, S. C. Kennedy, and K. P. Ponder. 1996. Retroviral vector sequences may interact with some internal promoters and influence expression. Hum. Gene Ther. 7:159-171. [DOI] [PubMed] [Google Scholar]
- 36.Yancopoulos, G. D., S. Davis, N. W. Gale, J. S. Rudge, S. J. Wiegand, and J. Holash. 2000. Vascular-specific growth factors and blood vessel formation. Nature 407:242-248. [DOI] [PubMed] [Google Scholar]
- 37.Zufferey, R., T. Dull, R. J. Mandel, A. Bukovsky, D. Quiroz, L. Naldini, and D. Trono. 1998. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 72:9873-9880. [DOI] [PMC free article] [PubMed] [Google Scholar]