Abstract
In one of the earliest events in human cytomegalovirus (HCMV)-infected cells, the major immediate-early (IE) protein IE1 initially targets to and then disrupts the nuclear structures known as PML oncogenic domains (PODs) or nuclear domain 10. Recent studies have suggested that modification of PML by SUMO is essential to form PODs and that IE1 both binds to PML and may disrupt PODs by preventing or removing SUMO adducts on PML. In this study, we showed that in contrast to herpes simplex virus type 1 (HSV-1) IE110 (ICP0), the loss of sumoylated forms of PML by cotransfected IE1 was resistant to the proteasome inhibitor MG132 and that IE1 did not reduce the level of unmodified PML. Reduced sumoylation of PML was also observed in U373 cells after infection with wild-type HCMV and proved to require IE1 protein expression. Mutational analysis revealed that the central hydrophobic domain of IE1, including Leu174, is required for both PML binding and loss of PML sumoylation and confirmed that all IE1 mutants tested that were deficient in these functions also failed both to target to PODs and to disrupt PODs. These same mutants were also inactive in several reporter gene transactivation assays and in inhibition of PML-mediated repression. Importantly, a viral DNA genome containing an IE1 gene with a deletion [IE1(Δ290-320)] that was defective in these activities was not infectious when transfected into permissive fibroblast cells, but the mutant IE1(K450R), which is defective in IE1 sumoylation, remained infectious. Our mutational analysis strengthens the idea that interference by IE1 with both the sumoylation of PML and its repressor activity requires a physical interaction with PML that also leads to disruption of PODs. These activities of IE1 also correlate with several unusual transcriptional transactivation functions of IE1 and may be requirements for efficient initiation of the lytic cycle in vivo.
Human cytomegalovirus (HCMV), a member of the betaherpesvirus subfamily, typically causes nearly ubiquitous asymptomatic latent or persistent infections. However, primary infections of newborns and reactivation from latent infection in immunocompromised individuals, including recipients of organ transplantation and patients with AIDS, can lead to life-threatening problems with overt systematic and chronic disease (56, 65). During lytic cycle infection, HCMV gene expression occurs in a three-step sequential fashion with immediate-early (IE or α), delayed-early (DE or β), and late (L or γ) kinetics. Among the IE proteins, two nuclear regulatory phosphoproteins, IE1 (or IE72) and IE2 (or IE86), are the first and most abundantly expressed proteins and are synthesized by differential splicing from the same complex overlapping transcription unit within the major IE (MIE) locus (56).
The 72-kDa IE1 protein (491 amino acids) is encoded by exons 1, 2, 3, and 4 (UL123), whereas the 86-kDa IE2 protein (579 amino acids) is encoded by exons 1, 2, 3, and 5 (UL122), and the two proteins share 85 amino acids at the N terminus. IE2 is a specific DNA-binding protein that is essential for lytic cycle progression and directs transcription of downstream DE and L gene expression (52), and it may have multiple roles in the process of preparing the cell for progeny virus production. In contrast, studies using a mutant HCMV that lacks the entire exon 4 of IE1 revealed that although IE1 is important for progression of productive infection in permissive primary human fibroblast (HF) cells at low multiplicities of infection (MOI), it is not essential in cell culture at high MOI (24, 57). This defect of IE1 mutant virus infection in HF cells at low MOI has been attributed to a failure or delay in accumulation of viral DE gene products, and although the level of accumulation of IE2 protein was comparable to that for the wild-type virus, IE2 displayed an altered intranuclear localization pattern (3, 24, 57). Similar defects of the mutant virus were also observed after infection of U373 cells or of a U373-derived cell line overexpressing PML even at high MOI (3). A more recent study showed that the absence of IE1 during low-MOI infection leads to a reduction of accumulation of a broad range of DE mRNAs, suggesting that IE1 plays an important role in transactivation of DE genes at low MOI (23). One or both of the MIE proteins may also be involved in regulating the switch between latent and lytic infection (56).
Whereas IE2 has all of the characteristics of a typical hydrophilic nuclear transcriptional transactivator protein, in contrast, the earlier and more abundantly expressed IE1 protein is largely hydrophobic except for a highly acidic C-terminal domain but is also exclusively nuclear in intracellular localization and associates with metaphase chromosomes (42, 79). Under some conditions IE1 may positively autoregulate the MIE promoter through upstream NF-κB sites (17, 71), and a recent study showed that IE1 selectively induces nuclear RelB and p50 among NF-κB/Rel factors in vascular smooth muscle cells (34). Another aspect of the influence of IE1 on viral gene expression is its ability to synergistically augment the IE2-mediated transactivation of several viral DE genes in transient-cotransfection assays (14, 36, 40, 50, 51, 75). IE1 also activates some cellular promoters relatively weakly in reporter gene assays, including DNA polymerase α (Pol α), dihydrofolate reductase, and prointerleukin1β, and the human immunodeficiency virus long terminal repeat (LTR) and mouse mammary tumor virus (MMTV) LTR (27, 29, 38, 53, 78). Activation of the dihydrofolate reductase gene by IE1 may involve direct interaction with E2Fs and pocket proteins and their phosphorylation by an associated kinase activity of IE1 (53, 63, 69). IE1 was also suggested to activate AP-1 activity through a pathway involving MEKK1 (39). Direct protein interactions of IE1 with cellular transcription factors such as the hTAFII130 component of TFIID, SP-1, and CTF-1 have also been reported (27, 49, 50, 81).
A separate group of studies have demonstrated that IE1 transiently targets to the PML-associated nuclear bodies known as PML oncogenic domains (PODs), PML nuclear bodies, or nuclear domain 10 within the first 2 h after infection, and subsequently PML, Sp100, SUMO, and IE1 are all displaced from the PODs into the nucleoplasm within 3 to 4 h after wild-type virus infection (2, 4, 41, 79). PODs have been proposed to be involved in multiple cellular functions, including cell proliferation, transcriptional regulation, apoptosis, interferon responses, and maintenance of genome stability (12, 58, 70, 77, 83). Both PML and Sp100 are posttranslationally modified by covalent conjugation (sumoylation) with the small ubiquitin (Ub)-like modifier-1 protein (SUMO-1). Recent studies demonstrated that sumoylation of PML is required for its own punctate nuclear localization pattern and to form the mature PODs where other cellular proteins such as Sp100, Daxx, and CBP are recruited (31, 44, 48, 82). Furthermore, both IE1 and IE2 proteins are also both partially conjugated to SUMO-1 and possibly SUMO-2 in both infected and transfected cells (6, 28, 74, 80).
The integrity of PODs also appears to be affected by infection by most other DNA viruses, including adenovirus (13, 19, 43), herpes simplex virus type 1 (HSV-1) (22, 54, 55), and Epstein-Barr virus (1, 9), leading to disruption or reorganization of POD proteins at early times after lytic infection. The exact role of POD disruption by the regulatory proteins of DNA viruses is not clear as yet. However, the importance of POD disruption at very early times after virus infection is suggested by the findings that input simian virus 40, HSV-1, and HCMV viral genomes are deposited at the periphery of the PODs in infected cells; that HCMV IE transcripts are detected near the PODs (30, 32); and that initiation of formation of HCMV DNA replication compartments also occurs in IE2-positive domains adjacent to the PODs (5). Furthermore, in the absence of IE1 and in the presence of overexpressed PML, a large proportion of the HCMV IE2 protein, which normally transits rapidly through the PODs, either accumulates in or becomes trapped in the PODs for up to 36 h after infection (3). We also showed that in mammalian GAL4 fusion gene reporter assays with cotransfected cells, IE1 completely inhibits the powerful transcriptional repression function of PML (80).
Muller and Dejean originally showed that HCMV IE1 and HSV-1 IE110 can both cause the loss of sumoylation of cotransfected PML in DNA transfection assays with HeLa cells, suggesting that POD disruption in both virus infections may correlate with the ability of these two viral proteins to block the addition of or to remove SUMO from PML (60). The RING finger containing HSV-1 IE110 protein is now also known to have possibly two distinct intrinsic Ub E3 ligase activities, which are postulated to lead to proteasome-mediated degradation of PML, CENP-C, CENP-A, and the catalytic subunit of DNA protein kinase (11, 26, 76). PML (but not HCMV IE1) is also a RING finger protein, although no Ub E3 ligase activity has yet been ascribed to it. PML has several differentially spliced isoforms, and we have previously worked almost exclusively with the 560-amino-acid form, PML VI. However, the 633-amino-acid form, PML IV, contains an additional C-terminal domain that binds to p53 and certain histone deacetylases, and therefore we chose to include it in our current studies.
In the present study we set out to characterize the ability of HCMV IE1 to block or remove SUMO adducts from PML in comparison with this property of IE110. We also mapped domains of IE1 required for binding both to PML and for causing the loss of sumoylation of PML to investigate whether there is any correlation between IE1-PML interactions with POD and SUMO disruption. We also asked whether there might be a correlation between those activities and the various transactivation and derepression functions ascribed to IE1 and whether those activities are required for infectivity of the genome when the DNA alone is transfected into permissive cells.
MATERIALS AND METHODS
Cell culture and virus infection.
Vero cells, HeLa cells, 293T cells, and U373-MG cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. To generate stable U373 cells expressing hemagglutinin (HA)-tagged PML IV, a 633-amino-acid version of PML (33), monolayer cultures of the parental U373-MG cell line in six-well plates were transfected with 0.02 μg of plasmid pSV2-Neo plus 0.4 μg of plasmid pUS109 by use of Lipofectamine 2000 reagents (Invitrogen). After 3 days the cells were replated into 100-mm-diameter dishes and grown in the presence of 1 mg of G418 per ml. About 100 neomycin-resistant colonies were pooled and used for the experiments.
Virus stocks for wild-type HCMV(Towne) and IE1 deletion mutant HCMV(CR208) (24) were prepared as previously described (2). For experiments with immunoblot analysis after virus infection, U373 cells were seeded into six-well plates at 2 × 105 cells per well. On the next day, the cells were mock infected or infected with HCMV(Towne) or HCMV(CR208) at an MOI of 2.
Plasmid construction.
Expression plasmids pCMX-PML, encoding PML VI (a 560-amino-acid version of PML), and pSG5-PML IV, encoding a 633-amino-acid version of PML (33), were provided by Ronald M. Evans (The Salk Institute, San Diego, Calif.) and Kun-Sang Chang (The University of Texas M.D. Anderson Cancer Center, Houston), respectively. Expression plasmids for 5′ HA-tagged PML VI (pUS112) or 5′ HA-tagged PML IV (pUS109) were generated on a pSG5 (25) background by using the Gateway technology (Invitrogen).
Mammalian expression plasmids pJHA303 encoding wild-type IE1(1-491), pJHA304 for IE1(1-346), pJHA307 for IE1(1-231), and pJHA308 for IE1(Δ132-274) were generated by moving the cDNA fragments into a pSG5 background from yeast GAL4-A fusion plasmids pJHA239, pJHA300, pJHA255, and pJHA251 (2). pJHA346, encoding IE1(Δ290-320), was generated by deleting the SpeI fragment from pJHA303. pJHA423, encoding IE1(1-420), was generated by deleting the C-terminal region by PCR mutagenesis in a pJHA303 background. pJHA305, encoding IE1(Δ1-85), was generated by placing the PCR fragment containing the entire exon 4 into pSG5. The IE1(L174P) gene in pYX145 and the IE1(K450R) in pYX118, encoding point mutant IE1 proteins, were described previously (80). Expression plasmid pGH92, encoding IE110(ICP0) of HSV-1, and plasmid pGR169, encoding IE68(ICP22) of HSV-1, were described previously (59, 62). pJHA312, expressing Flag-tagged SUMO-1 in the pSG5 background, was described previously (6).
Yeast plasmids expressing the GAL4-DB/PML VI fusion (pJHA238) or GAL4-A/IE1(wt) (pJHA239) were previously described (2). Plasmids expressing GAL4-A/IE1(Δ290-320) (pJHA404) and GAL4-A/IE1(1-420) (pHR50) were generated by using the Gateway technology. Point mutations GAL4-A/IE1(L174P) in pYX146 and GAL4-A/IE1(K450R) in pYX144 were generated from pJHA239 with the Stratagene QuickChange site-directed mutagenesis protocol.
Reporter plasmid pLA12, containing the HCMV UL54(Pol)-luciferase reporter gene, and plasmid containing (GAL4)5/TK-LUC were previously described (6). Plasmid pMMTV-LUC, containing MMTV LTR-luciferase, and plasmid pDPALΔ5′, containing the human DNA Pol α-luciferase reporter gene, were gifts from Jae-Kyun Shin (Sungkyunkwan University, Suwon, Korea) and from Teresa Wang (Stanford University, San Francisco, Calif.), respectively.
BAC mutagenesis.
The reagents for conjugal transfer of sequences to bacterial artificial chromosome (BAC) DNA in Escherichia coli and the Towne HCMV-BAC (T-BAC) clone (52) were provided by H. Zhu (University of Medicine and Dentistry of New Jersey). Plasmid pRL45 contained a 6.6-kb SalI-EcoRI genomic DNA fragment harboring the complete MIE region (68) and was used as the template in IE1 mutagenesis. To create transfer vectors to mutate UL123 from HCMV, a 4.1-kb PvuII-SalI restriction fragment containing the IE1(K450R) allele or a 2-kb SacI-BamHI restriction fragment containing the IE1(Δ290-320) allele was cloned into pGS284, a derivative of the positive suicide selection vector pCV442 (52). The equivalent restriction fragments containing the wild-type IE1 allele were also cloned into pGS284 and used to rescue the UL123-mutated BACs. The procedure for conjugative transfer has been described elsewhere (18, 73). Briefly, to transfer DNA sequences in pGS284 to T-BAC, E. coli S17-λpir containing the GS284 donor plasmid was conjugated with a RecA+ derivative of E. coli DH10B (73) harboring the T-BAC DNA. Exoconjugates were selected sequentially with antibiotics and sucrose. The resultant mutant T-BAC DNAs were examined by DNA sequencing and restriction enzyme digestion.
Electroporation.
T-BAC DNAs were introduced into the permissive HF cells by using electroporation. For each transfection, 5 × 106 HF cells were suspended in 250 μl of medium plus 10% serum and mixed with 4 μg of BAC DNA, 1 μg of plasmid pCMV71 encoding pp71, and 1 μg of plasmid pEGFP-N1 (Clontech) in a 0.4-cm cuvette. Following electroporation at 250 V and 960 μF, the cells were plated in a 10-cm-diameter tissue culture plate. When the surviving cells became confluent, the cells were split 1:3 into new dishes. The cells were cultured at 37°C, and green fluorescent protein (GFP) spreading was monitored.
Yeast two-hybrid interaction assays.
The yeast strain Y190 was the host for rapid assays for lacZ expression by using an X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) filter assay or for quantitation of interaction by using a β-galactosidase assay. The X-Gal filter assay and the β-galactosidase assay were described previously (2).
Transient DNA transfection.
For immunoblot analysis, 293T cells were seeded into six-well plates, and DNA mixtures were introduced into subconfluent cells by using the N,N-bis-(2-hydroxyethyl)-2-aminoethanesulfonic acid-buffered saline version of the calcium phosphate procedure as described previously (66). For indirect immunofluorescence assay (IFA), Vero cells were seeded into two-well slide chambers and DNA mixtures were introduced into cells with FuGene 6 reagents (Roche). For luciferase reporter assay, U373 and HeLa cells were seeded into 12-well plates, and DNA transfection was carried out with Lipofectamine 2000 reagents (Invitrogen).
Antibodies.
Mouse monoclonal antibody (MAb) 6E1 against HCMV IE1 (exon 4) was purchased from Vancouver Biotech (Vancouver, British Columbia, Canada). Mouse MAb 8131, which detects epitopes present in both IE1 and IE2 (exons 2 and 3), was purchased from Chemicon (Temecula, Calif.). Rat anti-HA MAbs (3F10) either conjugated with peroxidase or labeled with fluorescein and anti-myc mouse MAb 9E10 were purchased from Roche. The rabbit antipeptide polyclonal antibody referred to as PML(C), directed against amino acids 484 to 498 of PML, was described previously (3).
Immunoblot analysis.
DNA-transfected or virus-infected cells were washed with phosphate-buffered saline, and total extracts were prepared by direct boiling of the cell pellets in sodium dodecyl sulfate (SDS) loading buffer. Clarified cell extracts from the equivalent of 5 × 104 cells were separated on an SDS-8% or -4 to 20% gradient polyacrylamide gel, followed by the standard procedure with the enhanced chemiluminescence system (Amersham).
IFA.
Cells were fixed in methanol or with 1% paraformaldehyde and permeabilized with 0.2% Triton X-100. All subsequent procedures were described previously (4). Slides were examined and photographed on a Zeiss Axiopgoto2 microscope.
Luciferase reporter assay.
For luciferase reporter assays, transfected U373-MG cells were lysed directly in 12-well plates by three freeze-thaw steps in 200 μl of 0.25 M Tris-HCl (pH 7.9) plus 1 mM dithiothreitol. The subsequent procedures were described previously (3). A TD-20/20 luminometer (Turner Designs) was used for a 10-s assay of the photons produced (measured in relative light units).
RESULTS
IE1 interferes with sumoylation of PML in a proteasome-independent manner.
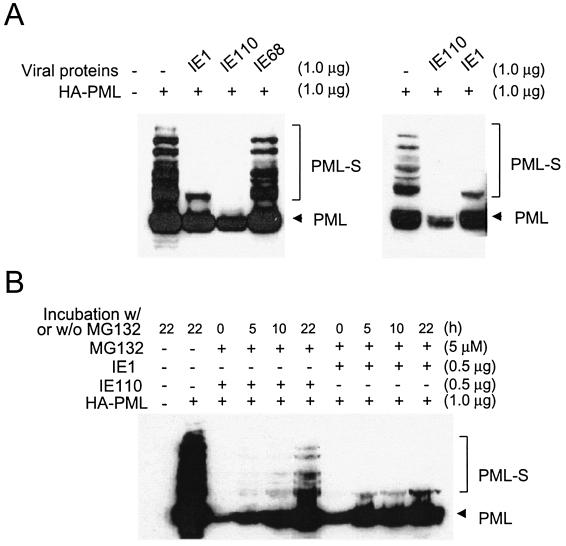
Expression of both HCMV IE1 and HSV-1 IE110(or ICP0) leads to the redistribution of PML from PODs in both virus-infected and DNA-transfected cells. However, the PML IFA signals totally disappear in HSV-1-infected cells (22, 54, 55), whereas these signals are merely displaced into the nucleoplasm in HCMV-infected cells (4, 41, 79, 80), which implies that a fundamentally different process may be involved. In addition, both proteins were also reported to cause loss of sumoylation of cotransfected PML in transient assays in HeLa cells, suggesting that a lack of sumoylation of PML may lead to disruption of PODs (60). For biochemical studies of multiple sumoylated forms of PML without the complications of there being seven different-size endogenous isoforms of unsumoylated PML (33), we compared the desumoylation activities of the IE1 and IE110 viral proteins in cotransfection assays with just the single HA-tagged PML VI isoform. We first needed to confirm that the exogenous HA-PML VI protein was indeed targeted to the PODs normally and whether it was also displaced from the PODs by IE1 as expected. When HA-PML VI alone was transfected into Vero or 293T cells, the HA signal exactly colocalized with the IFA signal obtained with anti-PML antibody, which also detects all of the other endogenous PML isoforms. When cotransfected with IE1, all of the detectable HA-PML was displaced from punctate bodies into the nucleoplasm together with IE1, demonstrating that the added HA-PML protein behaved normally (data not shown). Essentially these same properties in response to IE1 have also been observed with endogenous PML, as well as with both cotransfected HA-tagged PML-IV (633 amino acids, 98 kDa) and PML VI (560 amino acids, 68 kDa) in U373, Vero, HeLa, and 293T cells (data not shown).
To obtain high-level transfection efficiency for the PML sumoylation assays, 293T cells were either transfected with HA-PML alone or cotransfected together with either IE1 or IE110. Total extracts were prepared by directly boiling the cell pellets in SDS loading buffer, followed by separation by SDS-polyacrylamide gel electrophoresis and immunoblot analysis with antibody against HA (Fig. 1A). Consistent with previous reports (35), highly modified forms of PML containing multiple SUMO adducts were detected in cells transfected with HA-PML alone. However, in both of the two separate experiments shown, most of the sumoylated forms (especially those with more than one attached SUMO moiety) did not accumulate in cells in which PML was cotransfected together with either IE1 or IE110. Nevertheless, the observed effects of IE1 and IE110 were very different, because the level of the unmodified form of PML was also greatly reduced by IE110, as previously reported (20), but it was not significantly affected by IE1. Moreover, in cells cotransfected with IE1 (but not with IE110), the single-SUMO-conjugated (monosumoylated) PML forms were more resistant to IE1 than the highly modified forms. Cotransfection with the IE68 (ICP22) nuclear protein of HSV-1, which did not affect the sumoylation of PML, was used as a specificity control.
FIG. 1.
Characterization of the desumoylation activity of IE1 in transient-DNA-transfection assays. (A) Comparison of the activities of HCMV IE1 and HSV-1 IE110 in abolishing sumoylation of PML. 293T cells were transfected with 1 μg of plasmid encoding HA-PML VI alone or cotransfected with 1 μg of plasmid encoding HCMV IE1, HSV-1 IE110, or HSV-1 IE68. At 48 h after transfection, extracts were prepared as described in Materials and Methods. Equal amounts of cell extracts were separated on an SDS-8% polyacrylamide gel, and immunoblot analysis was performed with a rat anti-HA MAb. The results of two separate experiments are shown to ensure that the levels of unmodified forms of PML are significantly reduced in cells that received IE110, but not IE1. (B) Effects of proteasome inhibitor MG132 on interference with sumoylation of PML by IE1 or IE110. 293T cells were transfected with 1 μg of plasmid encoding HA-PML VI alone or cotransfected with 0.5 μg of plasmid encoding either IE1 or IE110. At 17 h after infection, culture media were replaced with fresh media and cells were either further incubated for 22 h or incubated for 0, 5, 10, and 22 h with media containing 5 μM proteasome inhibitor MG132 before harvesting, extract preparation, and immunoblotting as described above. The unmodified form of PML is indicated by arrowheads, and sumoylated forms of PML are also indicated as PML-S.
The disruption of PODs by HSV-1 IE110, which itself encodes an intrinsic Ub E3 ligase activity (22, 54, 55), has been shown to correlate with the rapid proteasome-dependent degradation of both SUMO-modified and unmodified forms of PML. Furthermore, proteasome inhibitors such as MG132 proved to stabilize PML against IE110-induced degradation and prevent disruption of PODs in HSV-1-infected cells (16, 20, 64). In contrast, we have previously shown by IFA experiments that displacement of PML from PODs by IE1 is independent of proteasome activity, because it was not prevented by MG132, whereas the IE110-mediated effect was inhibited by MG132 in parallel transfected-cell controls (80). To also address this question at the protein level, we carried out a PML sumoylation assay similar to that described above in the presence of MG132 (Fig. 1B). Cotransfected 293T cells receiving plasmids encoding HA-PML and IE1 were compared to those receiving HA-PML and IE110 after 0, 5, 10, and 22 h of incubation with MG132. In cells cotransfected with IE110, the levels of high-molecular-weight SUMO-modified PML forms proved to partially recover when the cells were incubated in the presence of MG132. The incomplete effect of MG132 on the activity of IE110 in this experiment probably results from the relatively high expression level of IE110 in our transfected cells. In contrast, in cells cotransfected with IE1 there was no recovery at all in the levels of high-molecular-weight SUMO-modified forms of PML. This result further supports the concept that IE1 does not cause proteasome-mediated degradation of PML or require proteasome activity to prevent or remove accumulated SUMO adducts on PML. Therefore, HCMV IE1 uses a somewhat different mechanism to reduce the levels of sumoylated PML than does the HSV-1 IE110 protein.
IE1-mediated loss of SUMO-conjugated forms of PML in HCMV-infected cells.
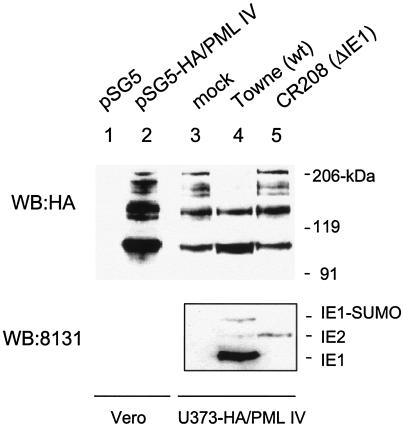
To investigate whether the IE1-mediated loss of sumoylated forms of PML also occurs in HCMV-infected cells, we needed to have an appropriately tagged form of PML in a stable permissive cell line, because the endogenous sumoylated forms of PML cannot easily be distinguished on the basis of size alone from the several different isoforms of unmodified PML. We have previously generated and used a stable HCMV-permissive U373 cell line that constitutively expressed high levels of PML VI and have shown that POD disruption and PML dispersal after HCMV infection still occurs (albeit more slowly than normal) (3). In preliminary experiments, when Vero cells were cotransfected with IE1 and HA-PML IV, a similar pattern of heavily reduced sumoylation of PML was observed as demonstrated above with HA-PML VI (data not shown). Therefore, to be able to unambiguously discriminate between endogenous PML bands on immunoblots that are sumoylated and those representing the differentially spliced unsumoylated isoforms, we also generated U373 cells that constitutively express HA-tagged PML IV. U373 cells were cotransfected with a plasmid encoding a neomycin-resistant marker and a plasmid encoding the HA-PML IV. Neomycin-resistant cells were pooled and used for virus infection. Stable constitutive expression of HA-PML in the U373 pooled cell line was analyzed by IFA with anti-HA antibody. The result showed that about 70% of the cells were positive for HA-PML (data not shown). When an extract prepared from the U373 pooled cell line was subjected to immunoblot analysis with anti-HA antibody, a typical ladder of sumoylated forms of HA-PML was observed (Fig. 2, lane 3), and the pattern was identical to that produced in Vero cells transiently transfected with HA-PML IV (Fig. 2, lane 2).
FIG. 2.
Effect of HCMV infection on sumoylation in the U373 cell line overexpressing PML. Lanes 1 and 2, control Vero cells were transfected with either vector (pSG5) or plasmid encoding HA-PML IV. At 48 h after transfection, the cells were harvested and total extracts were prepared as described for Fig. 1. Lanes 3 to 5, the U373 pooled cell line overexpressing HA-PML IV was either mock infected or infected with wild-type (wt) HCMV(Towne) or IE1 deletion HCMV(CR208) virus at an MOI of 2. Cells were harvested at 6 h after infection, and total extracts were prepared as described for Fig. 1. Equal amounts of cell extracts were separated on an SDS-8% polyacrylamide gel, and immunoblot analysis (WB) was performed with rat anti-HA MAb conjugated with peroxidase (top panel). The same filter membrane for lanes 3, 4, and 5 was stripped, and a second immunoblot analysis was carried out with MAb 8131, which detects both IE1 and IE2 (bottom panel).
The U373/PML IV cells were then mock infected or infected with either wild-type HCMV(Towne) or IE1 deletion mutant virus (CR208) at an MOI of 2.0. At 6 h after infection, cell extracts were prepared and the sumoylation pattern of HA-PML was analyzed (Fig. 2, lanes 3 to 5). Similar to the case for IE1-transfected cells, the levels of high-molecular-weight sumoylated forms of HA-PML IV disappeared in cells infected with wild-type HCMV(Towne), although again the monosumoylated form was unaffected. In contrast, the sumoylation pattern of PML was not affected at all in cells infected with IE1 deletion mutant virus (CR208). In a control experiment, both the lack of IE1 expression in CR208-infected cells and the expression levels of IE2 in Towne- and CR208-infected cells were confirmed by immunoblot analysis of the same filter membrane with MAb 8131, which detects both IE1 and IE2 (Fig. 2, bottom panel). The result showed that in CR208-infected cells, IE1 was not expressed at all and the level of IE2 was even higher than that in Towne-infected cells, suggesting that the lack of effect on PML sumoylation by CR208 virus was not related to a lower input virus concentration than for the Towne sample. This result demonstrates that the loss of the sumoylated forms of PML also occurs in HCMV-infected cells in an IE1-dependent manner.
Domains of IE1 required for desumoylation of PML.
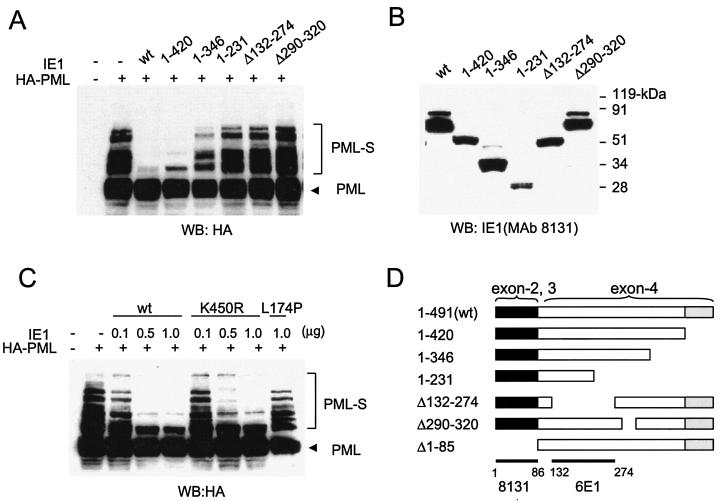
To investigate the domain requirements within IE1 to desumoylate PML, cotransfection assays were carried out with a set of variant IE1 proteins containing truncation or deletion mutations (Fig. 3A). The different deletion mutant IE1 proteins used in this experiment are summarized in the diagram in Fig. 3D. Similar to the case for wild-type IE1 (which in this experiment also removed the monosumoylated form), a truncation mutant, IE1(1-420), that lacks the acidic C-terminal region still caused a nearly complete loss of the sumoylated forms of PML. However, the C-terminal truncation mutant IE1(1-346) showed an intermediate effect, and IE1(1-231) failed to desumoylate PML. In addition, two internally deleted IE1 proteins that lack either the region from amino acid position 132 to 274 or that from position 290 to 320 also failed to abolish sumoylation of PML. As expected, an N-terminal truncation mutant, IE1(Δ1-85), that lacks both one of the activation domains (67) and a nuclear localization signal encompassing exons 2 and 3 and therefore accumulates in the cytoplasm (79) had little effect on sumoylation levels of PML (data not shown). These results indicate that the acidic C-terminal region from position 421 to 491 is dispensable but that most of the central hydrophobic domain of IE1 from codon 132 to 346 needs to be intact for the activity to desumoylate. A role for the N terminus could not be determined because of the complication of the loss of the nuclear localization signal motif.
FIG. 3.
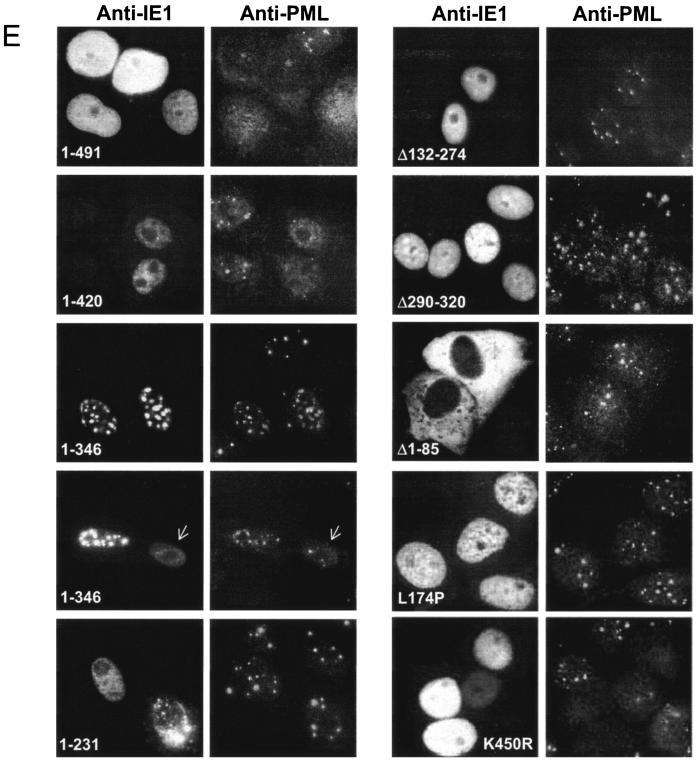
Domains of IE1 required for desumoylation of PML and disruption of PODs. (A) PML sumoylation assays with the deletion mutant IE1 proteins. 293T cells were transfected with 1 μg of plasmid (pUS112) encoding HA-PML alone or cotransfected with 1 μg of plasmid encoding wild-type (wt) IE1, IE1(1-420), IE1(1-346), IE1(1-231), IE1(Δ132-274), or IE1(Δ290-320). At 48 h after transfection, a PML sumoylation assay was carried out as described for Fig. 1. (B) Expression levels of the mutant IE1 proteins. The same cell extracts described for panel A were separated on an SDS-4 to 20% gradient polyacrylamide gel, and immunoblot analysis (WB) was carried out with mouse MAb 8131 against IE1. (C) 293T cells were transfected with a plasmid encoding HA-PML or cotransfected with various amounts of plasmid encoding wild-type IE1 (0.1, 0.5, or 1.0 μg), IE1(K450R) (0.1, 0.5, or 1.0 μg), or IE1(L174P) (1.0 μg) protein. At 48 h after transfection, cell extracts were prepared and immunoblot analysis was carried out with anti-HA MAb as described for panel A. Unmodified and sumoylated (PML-S) forms of PML are indicated. (D) Structures of the IE1 proteins used in domain mapping experiments. 1-491 (wild-type IE1 in pJHA303), 1-420 (pJHA423), 1-346 (pJHA304), 1-231 (pJHA307), Δ132-274 (pJHA308), Δ290-320 (pJHA346), and Δ1-85 (pJHA305) are illustrated. Coding regions corresponding to each exon of IE1 are shown at the top. Closed bars represent exons 2 and 3, and gray bars represent the C-terminal acidic domains. The estimated map locations for epitopes recognized by MAbs 8131 and 6E1 are shown at the bottom. (E) Localization patterns of the mutant IE1 proteins and their effects on distribution of the endogenous PML proteins. Vero cells were transfected with plasmids encoding wild-type IE1, IE1(1-420), IE1(1-346), IE1(1-231), IE1(Δ132-274), IE1(Δ290-320), IE1(Δ1-85), IE1(L174P), or IE1(K450R) and fixed in methanol at 48 h after transfection, followed by double-label IFA for IE1 and PML. IE1 was detected with mouse MAb 8131 or 6E1 and fluorescein isothiocyanate-labeled anti-mouse immunoglobulin G. PML was detected with rabbit polyclonal antibody PML(C) and rhodamine-coupled anti-rabbit immunoglobulin G.
IE1 itself is also modified by SUMO-1, and the major sumoylation site has been mapped to a lysine residue at position 450 (74, 80). To investigate the expression levels of sumoylated and unmodified forms of mutant IE1 proteins, the same extracts used for immunoblot analysis of PML in Fig. 3A were subjected to immunoblotting with mouse MAb 8131, which recognizes exons 2 and 3 of IE1 (Fig. 3B). As expected, the C-terminal truncation mutants IE1(1-420) and IE1(1-231), which lack the acidic domain containing the lysine residue at position 450, were not modified by SUMO; however, a weak band that migrated more slowly than the unmodified form was detected in cell extracts transfected with IE1(1-346). The slowly migrating band in cells transfected with IE1(1-346) appeared not to be a sumoylated form, because unlike with the wild-type IE1, this form was not increased at all when IE1(1-346) was coexpressed with SUMO-1 (data not shown). Interestingly, IE1(Δ132-274) gave no sumoylated band, suggesting that the deleted domain is also needed for sumoylation of the lysine residue at position 450. However, IE1(Δ290-320) did produce a sumoylated band in an abundance similar to that for the wild-type IE1 protein. Given that IE1(Δ290-320) failed to desumoylate PML, this result is consistent with the previous findings that sumoylation of IE1 is not essential for POD disruption (74, 80).
Two important point mutant IE1 proteins have been previously described. A mutant with a substitution of Leu to Pro at position 174 (L174P) is defective for POD disruption as well as for POD targeting and for loss of sumoylation of PML (60). However, a mutant with a substitution of Lys to Arg at position 450 (K450R), which was not sumoylated at all, still retained the ability to disrupt PODs (and presumably therefore to target them) when observed by IFA (74, 80). Neither of these mutants has been tested previously in transactivation assays. In our PML loss-of-sumoylation assays, IE1(K450R) retained the ability to efficiently desumoylate PML in a dose-dependent manner, whereas IE1(L174P) failed to do so (Fig. 3C). These data are consistent with the results of previous but less extensive studies that suggest a correlation between the absence of sumoylation on PML and disruption of PODs (60).
Correlation between IE1-PML protein-protein interactions and interference with sumoylation as well as with targeting to PODs, but not necessarily with disruption of PODs.
We next characterized the abilities of all eight mutant IE1 proteins used in the PML sumoylation assay to disrupt PODs. Vero cells were transfected with plasmids encoding wild-type or mutant IE1 proteins, and double-label IFA for IE1 and PML was carried out (Fig. 3E). Consistent with previous observations (2, 79), both wild-type IE1 and IE1(1-420) disrupted PODs, confirming that the acidic C-terminal region is dispensable for both desumoylation of PML and disruption of PODs. Just as in the PML sumoylation assay, the effect of IE1(1-346) was intermediate. In most transfected cells, IE1(1-346) distributed as a nuclear punctate form that virtually totally colocalized with PML in large PODs without disrupting them, whereas in a smaller subset of the cells, which instead showed a greatly reduced number of PODs (arrows), it localized as a diffuse form. These two different localization patterns were previously reported as apparently inconsistent results (2, 79), but our studies here show that IE1(1-346) actually displays an intermediate ability to desumoylate PML and to disrupt PODs. Nevertheless, IE1(1-346) dramatically retained the ability to still target to PODs in those cells that did not show disruption of their PODs. We also found that IE1(Δ290-320), which did not desumoylate PML, failed to disrupt PODs, but unlike IE1(1-346), it did not target to PODs (although some of it concentrates in nuclear patches, these do not colocalize with PODs). In addition, IE1(K450R) disrupted PODs, whereas IE1(1-231), IE1(Δ132-274), IE1(Δ1-85), and IE1(L174P) all failed to do so and also did not colocalize with PODs. Although it is possible that the effects of wild-type or mutant IE1 proteins on the integrity of PODs may be affected by the expression levels of the proteins, our results with an extensive set of deletion or point mutant IE1 proteins show that the failure of mutant IE1 proteins to target to and disrupt PODs (Fig. 3E) largely correlates with the absence of negative effects on the sumoylation of PML (Fig. 3A and C). These results are also in agreement with the previous finding that IE1 abolishes sumoylation of PML, leading to POD disruption, and that mutant IE1(L174P) both had no effect on sumoylation of PML and failed to produce POD disruption (60).
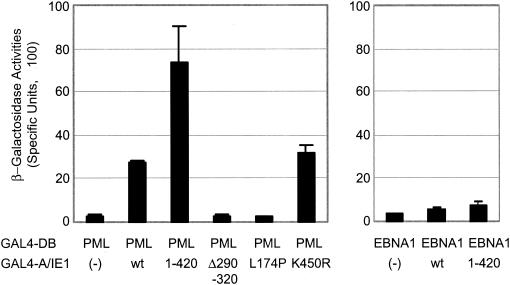
We previously showed that IE1(1-346) retained the ability to bind to PML in a yeast two-hybrid interaction assay, whereas neither IE1(1-231) nor IE1(Δ132-274) did so (2). The abilities of the other mutant IE1 proteins used in this study to bind to PML were tested in similar yeast two-hybrid interaction assays (Fig. 4). The results showed that IE1(Δ290-320) failed to bind to PML, whereas IE1(K450R) still bound to PML just as efficiently as did the wild-type protein. Most significantly, the IE1(L174P) point mutant had also lost the ability to bind to PML. Interestingly, IE1(1-420) gave stronger binding to PML than did wild-type IE1 by two to threefold, suggesting an inhibitory role of the C-terminal acidic domain of IE1 on PML binding in yeast. A similar inhibitory effect of the C-terminal acidic domain on PML binding was previously observed with IE1(1-346) (2). Our data show that the central hydrophobic domain of IE1 but not the acidic C-terminal domain is needed to bind to PML in yeast and that this property correlated fully with both the activities to desumoylate PML and to target to PODs. In addition, no mutants that failed to bind to PML resulted in POD disruption. However, binding to PML alone is not necessarily sufficient for POD disruption, as exemplified by IE1(1-346), which still targets to PODs in most cells but does not disrupt them (Fig. 3E).
FIG. 4.
Yeast two-hybrid interaction assays between PML and mutant IE1 proteins. The yeast Y190 cells were cotransformed with plasmids encoding either GAL4-DB/PML VI or GAL4-DB/EBNA-1 (2) and GAL4-A alone (pACTII), GAL4-A/wild-type IE1, GAL4-A/IE1(1-420), GAL4-A/IE1(Δ290-320), GAL4-A/IE1(L174P), or GAL4-A/IE1(K450R). Transformants were assayed for their β-galactosidase production. The average values and standard errors for β-galactosidase units in duplicated assays are shown. Assays of the interaction between Epstein-Barr virus and IE1 were used as negative interaction controls.
Desumoylation of PML also correlates with the transactivation and PML repression functions of IE1 in cotransfection assays.
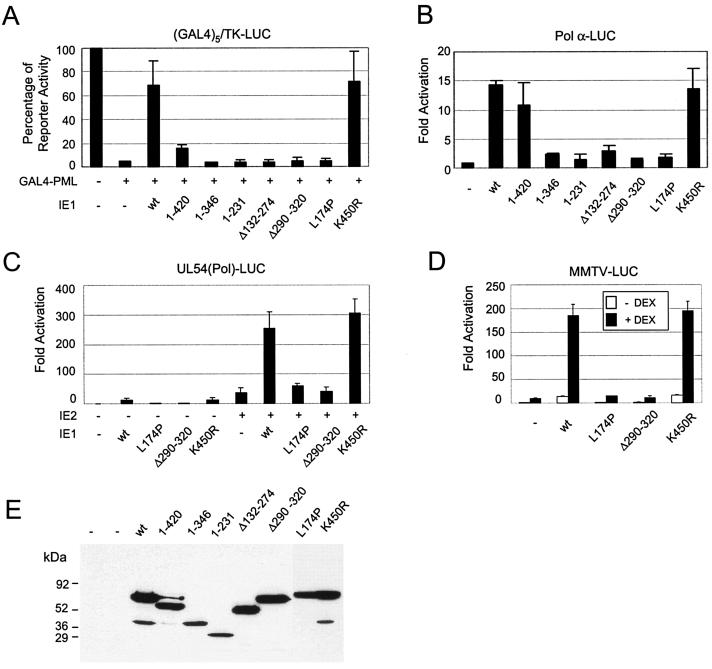
We previously demonstrated that IE1 counteracts the strong transcriptional repressor activity of PML in mammalian DNA-tethering assays with GAL4 fusion proteins. Data with two point mutant IE1 proteins (K450R and L174P) again suggested that this activity of IE1 also correlates with POD disruption (80). However, to extend the connection, we also tested the PML repression activities of all of the deletion mutant IE1 proteins used in the PML desumoylation assays described above (Fig. 5A). Again, both wild-type IE1 and IE1(K450R), but not IE1(L174P), efficiently inhibited the transcriptional repressor activity of GAL4-PML in cotransfected HeLa cells. The results with deletion mutant IE1 proteins showed that although IE1(1-420) partially inhibited the repressor activity of PML, all of the others failed to inhibit PML-mediated repression. The expression levels of the wild-type and mutant IE1 proteins in the cell extracts were confirmed by immunoblot analysis with MAb 8131, which detects an epitope mapping within exons 2 and 3 of IE1 (Fig. 5E). Note that the sumoylated forms of IE1 were not detected within cell extracts that were prepared by the freezing and thawing procedure used for luciferase assays. These results suggest that the activity of IE1 in both binding to and desumoylating PML correlates well with inhibition of PML repression, and again, the acidic C-terminal region of IE1, although not essential, proved to contribute to efficient inhibition.
FIG. 5.
Effects of PML desumoylation on transactivation and PML repression functions of IE1. (A) Effects of wild-type or mutant IE1 proteins on the transcriptional repressor activity of PML. HeLa cells were transfected with 0.5 μg of reporter plasmid containing a (GAL4)5/TK-LUC gene or cotransfected with 0.4 μg of plasmid encoding GAL4-PML alone or together with effector plasmid encoding wild-type (wt) IE1, IE1(1-420), IE1(1-346), IE1(1-231), IE1(Δ132-274), IE1(Δ290-320), IE1(L174P), or IE1(K450R). At 36 h after transfection, total cell extracts were prepared and assayed for luciferase activity. Shown are mean values with standard errors for the percentage of luciferase activity from three independent assays. (B) Effects of mutant IE1 proteins on transactivation of the cellular DNA Pol α promoter. U373-MG cells were transfected with 0.4 μg of reporter plasmid (pDPALΔ5′) containing a DNA Pol α-luciferase reporter gene and 1.0 μg of effector plasmid encoding wild-type or mutant IE1 proteins as described above. At 48 h after transfection, total cell extracts were prepared and assayed for luciferase activity. Luciferase activities are indicated as fold activation over the basal level of each reporter gene. The results shown are the mean values along with standard errors from three independent experiments. (C) Effects of mutant IE1 proteins on augmentation of transactivation of the HCMV Pol promoter (Pol-LUC) by IE2. U373-MG cells were transfected with 0.3 μg of target reporter plasmid (pLA12) encoding the HCMV UL54 (Pol) promoter-driven Pol-LUC gene and 0.3 μg of effector plasmid encoding wild-type IE1, IE1(L174P), IE1(Δ290-320), or IE1(K450R) either alone or together with 0.3 μg of effector plasmid encoding wild-type IE2 (pJHA124). At 48 h after transfection, total cell extracts were prepared and assayed for luciferase activity as described for panel B. (D) Effects of wild-type or mutant IE1 on transactivation of the MMTV promoter. U373-MG cells were transfected with 0.4 μg of reporter plasmid (pMMTV-LUC) containing MMTV LTR-luciferase reporter gene and 0.2 μg of plasmid encoding wild-type IE1, IE1(L174P), IE1(Δ290-320), or IE1(K450R). At 24 h after transfection, culture media were replaced with medium with or without 1 μM dexamethasone (DEX). After incubation for another 24 h, total cell extracts were prepared and assayed for luciferase activity. Luciferase activities are indicated as fold activation over the basal level of each reporter gene and are shown as averages from duplicated experiments. (E) Expression levels of the mutant IE1 proteins. The same cell extracts described for panel A were separated on an SDS-10% polyacrylamide gel, and immunoblot analysis was carried out with MAb 8131.
To investigate whether the ability of IE1 to desumoylate PML also matches with the several described positive transactivation activities of IE1, we carried out target reporter gene assays with the same set of wild-type and mutant IE1 proteins. U373-MG cells were cotransfected with a target reporter plasmid expressing luciferase under the control of the human DNA Pol α promoter (Pol α-LUC) together with effector plasmids expressing either wild-type or mutant IE1 proteins. The luciferase readouts showed that wild-type IE1 activated the Pol α promoter up to 15-fold and that only two of the mutant IE1 proteins, IE1(1-420) and IE1(K450R), retained some or all of the transactivation activity. In contrast, all of the other mutant IE1 proteins, including IE1(1-346), IE1(1-231), IE1(Δ132-274), IE1(Δ290-320), and, especially, the point mutant IE1(L174P), all of which had lost the ability to desumoylate PML as well as to disrupt PODs, also all failed to activate the Pol α promoter (Fig. 5B).
In two other variations of this assay, IE1 has been shown both to augment IE2-mediated transactivation of viral delayed early promoters and to directly transactivate the MMTV LTR by an unknown mechanism (78). Therefore, similar reporter assays were carried out with HCMV Pol(UL54)-LUC and MMTV LTR-LUC reporter genes. As expected, even wild-type IE1 had little activity on its own on the viral Pol promoter, but it boosted the effect of IE2 about sixfold. Similarly, in the presence of dexamethasone, wild-type IE1 alone transactivated the MMTV reporter gene 10-fold. However, again the results showed that both IE1(L174P) and IE1(Δ290-320) had lost both transactivation functions, whereas IE1(K450R) still retained levels of transactivation function similar to that of wild-type IE1 (Fig. 5C and D). Therefore, the results from all of these reporter assays imply that the activity of IE1 in regulation of transcription correlates directly with the ability to bind to PML as well as to desumoylate PML and to target or disrupt PODs.
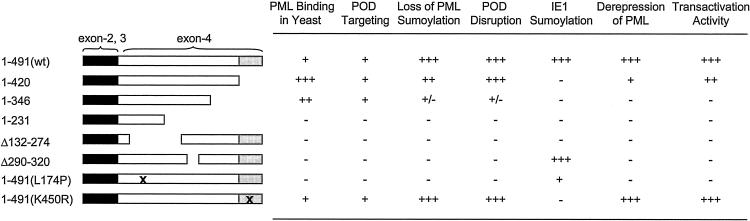
The relative levels of activities of wild-type or mutant IE1 proteins characterized in this study on PML binding in yeast, POD targeting, loss of PML sumoylation, POD disruption, sumoylation itself, derepression of PML, and transactivation functions are summarized in Fig. 6.
FIG. 6.
Summary of the activities of wild-type or mutant IE1 proteins. Several activities of the wild-type (wt) or mutant IE1 proteins characterized in this study are summarized at a semiquantitative level, including PML binding in yeast two-hybrid assays, POD targeting by IFA, loss of PML sumoylation in cotransfection assays, POD disruption in IFA, ability to function as a target for sumoylation, and derepression of PML or positive transactivation functions on other promoters in transient reporter gene cotransfection assays. The PML binding activities of mutants IE1(1-346), IE1(1-231), and IE1(Δ132-274) were shown previously (2). Sumoylation of IE1(L174P) was originally shown by Muller and Dejean (60), but we found that the sumoylation level of IE1(L174P) is reduced to only 30% that of the wild-type protein (data not shown). Domain mapping of IE1 for POD disruption was also reported by Wilkinson et al. (79).
Construction of recombinant T-BAC clones.
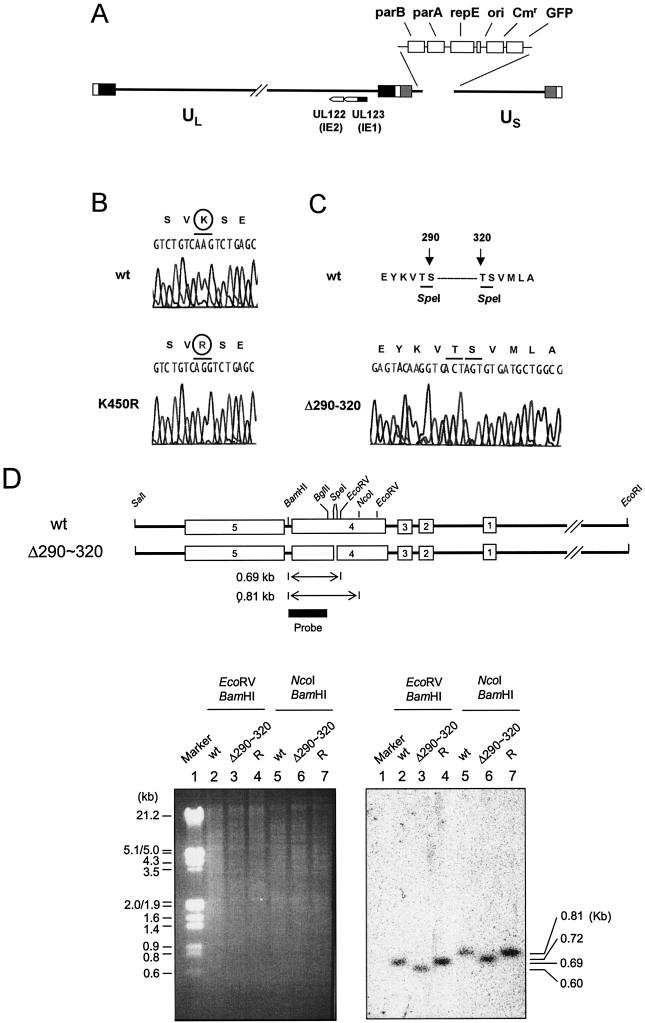
To study whether the abilities of IE1 to interfere with sumoylation of PML and to regulate transcription in target reporter gene assays may also be required for efficient viral growth, we generated mutant HCMV-BAC clones that encode mutant forms of IE1 that are deficient in PML-associated activities. We used a Towne HCMV-BAC (T-BAC) clone as a template for mutagenesis. This T-BAC clone has a 9-kb deletion from the dispensable portion of the US region (from US1 to US12) but instead contains a GFP expression cassette in the deleted region to facilitate the study of virus replication (Fig. 7A) (52).
FIG.7.
Construction of recombinant HCMV-BAC genomes encoding mutant IE1 proteins. (A) Genome structure of the parent Towne HCMV-BAC plasmid used in this study. The F plasmid sequences containing the replication origin (ori), replication and partitioning functions (repE, parA, and parB), chloramphenicol resistance (Cmr), and the GFP eukaryotic expression cassette, which have been substituted in place of the US1-to-US12 gene region, are indicated as boxes. The locations of UL122 (IE2) and UL123 (IE1) are also indicated. (B and C) Relevant DNA sequences of the wild-type (wt) and recovered mutant IE1 alleles of the HCMV-BAC plasmids. Genomic DNA containing either wild-type IE1(1-491) or the mutant IE1 alleles IE1(K450R) (B) and IE1(Δ290-320) (C) were PCR amplified from BAC DNAs and sequenced with specific primers. The two SpeI sites at amino acid positions 290 and 320 in wild-type IE1 are also indicated (C). (D) Genome structure of the Towne HCMV-BAC containing the deleted IE1(Δ290-320) allele. Top, the 6.6-kb EcoRI-SalI regions encompassing the MIE locus and the locations of the restriction enzyme sites used for deletion (SpeI) and mapping by Southern blot analysis (EcoRV, NcoI, and BamHI) are shown. Bottom, restriction fragment DNA patterns obtained following EcoRV-BamHI (lanes 2, 3, and 4) or NcoI-BamHI (lanes 5, 6, and 7) digestion of three HCMV-BAC DNAs (wild type [lanes 2 and 5], Δ290-320 [lanes 3 and 6], and the revertant [R] for Δ290-320 [lanes 4 and 7]) are shown (left). The 530-bp probe (BglII-BamHI fragment) used for Southern blot analysis (right) is shown in the top panel. The sizes of λ-HindIII/EcoRI are shown in the marker lane.
Both IE1(K450R) and IE1(Δ290-320) were successfully introduced into T-BAC clones in place of the wild-type IE1 gene. The mutated allele with flanking sequences for recombination was cloned into a transfer vector (pGS284) and conjugated into RecA+ E. coli harboring the T-BAC clone. Exoconjugates were selected and analyzed for correct generation of the mutant forms. To select and isolate recombinant T-BAC genomes containing the desired mutations, DNA fragments expected to contain the mutated allele were amplified by PCR and directly sequenced (Fig. 7B and C). In addition, the lack of any other apparent alterations in the viral genomes was checked by comparing the restriction fragment patterns of viral genomes in wild-type and mutant T-BACs. In the case of the IE1(Δ290-320) mutant, Southern blot analysis was carried out with restriction fragments of the viral genome (Fig. 7D). The appropriate DNA hybridization probe detected a 0.60-kb EcoRV-BamHI fragment and a 0.72-kb NcoI-BamHI fragment, as would be expected from a mutant HCMV-BAC clone containing the 90-bp deletion (Fig. 7D, lanes 3 and 6). A revertant T-BAC clone for IE1(Δ290-320) T-BAC was also generated by allelic exchange of the mutated allele with the wild-type DNA fragment cloned in pGS284. The correct conversion of the mutant allele back into the wild-type allele was confirmed by Southern blot analysis (Fig. 7D, lanes 4 and 7) and direct DNA sequencing (data not shown).
An HCMV-BAC clone containing the IE1(Δ290-320) mutation, but not IE1(K450R), is not infectious.
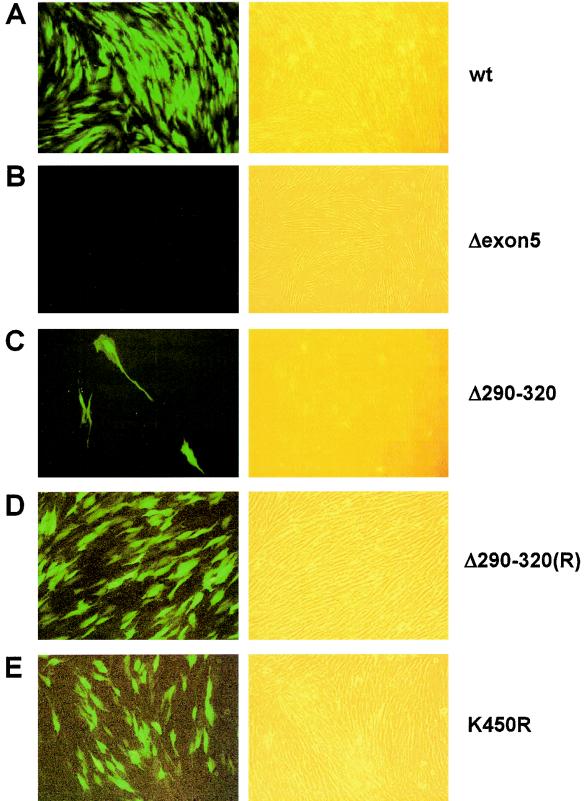
To investigate whether T-BAC genomes containing encoding IE1(K450R) or IE1(Δ290-320) are infectious, we electroporated purified T-BAC-wt, T-BAC-Δ290-320, or T-BAC-K450R into permissive HF cells and monitored the electroporated cultures for viral growth, which was judged by spreading of GFP signals and cytopathic effects on the cells. T-BAC-Δexon5, which has a deletion of exon 5 within IE2 and was shown previously to be noninfectious (52), was used as a negative control in these experiments. For each electroporation, expression plasmid DNA encoding either HCMV pp71 or GFP were both cotransfected together with the BAC plasmid DNA to enhance the efficiency of activation of the MIE promoter (8) and to monitor electroporation efficiency, respectively. The electroporation experiments were repeated at least five times for each mutant T-BAC, and the transfection efficiency, as judged by the initial transient GFP signals from the transfected cells, was measured to be 10 to 15% in all experiments. This transient GFP signal disappeared within the first week after electroporation.
When HF cells received T-BAC-wt or T-BAC-K450R, the GFP signals (from the GFP cassette within the viral genome) began to reappear at 3 to 4 weeks after electroporation and spread into the surrounding cells (Fig. 8A and E). Consistent with the in vitro data showing that IE1(K450R) retained all activities of the wild-type protein (74, 80; this study), this result suggests that sumoylation of IE1 is not essential for reconstitution of virus from the input naked DNA genome in cultured HF cells. In contrast, when cells received T-BAC-Δ290-320, the GFP signals reappeared within only a very few cells and did not spread into the surrounding cells (Fig. 8C). However, this defect was completely rescued when cells received the revertant T-BAC-Δ290-320(R) (Fig. 8D). In the negative control, the T-BAC-Δexon5 culture failed to reexpress GFP in any cells (Fig. 8B). These results indicate that deletion between codons 290 and 320 in IE1 indeed interfered greatly with recovery of infectious virus from the input mutant genome, and they suggest that the correlated deficiencies in IE1(Δ290-320) activities in desumoylating PML and in regulating transcription may be required functions for efficient initiation of viral growth in cultured HF cells.
FIG. 8.
Infectivities of transfected HCMV-BAC DNAs. HF cells were electroporated with T-BAC-wt, T-BAC-Δexon5, T-BAC-Δ290-320, T-BAC-Δ290-320(R), or T-BAC-K450R and monitored for spreading of GFP signals (see text). GFP images (left panels) and their phase-contrast images (right panels) were photographed at 4 weeks after electroporation. Representative images from at least three independent experiments are shown. HF cells that received T-BAC-Δ290-320 usually gave no GFP-positive cells in most microscopic fields. The image shown in panel C was selected to include three GFP-positive cells, demonstrating that the few GFP-positive cells did not spread into the surrounding cells.
DISCUSSION
In the present study, we have demonstrated that IE1 efficiently inhibits the accumulation of sumoylated forms of PML in both DNA-cotransfected cells and wild-type HCMV-infected cells but not in cells infected with HCMV with an IE1 deletion. However, unlike IE110 cotransfection or HSV-1 infection, neither IE1 alone nor HCMV infection significantly reduced the levels of the unmodified form of PML, and the PML-desumoylating activity of IE1 was not significantly blocked by treatment with the proteasome inhibitor MG132 when measured by immunoblotting. Therefore, HCMV and HSV-1 appear to use different mechanisms to desumoylate.
By analysis of mutant forms of the IE1 protein, we found that, although the acidic C-terminal region is dispensable, most of the central hydrophobic region of IE1 needs to be intact for both binding to PML and desumoylation of PML to occur. We also found that mutant IE1 proteins that fail to bind to or desumoylate PML were all also unable to target to or disrupt PODs, strongly supporting the previous finding that disruption of PODs by herpesvirus proteins is linked to desumoylation of PML (60). Only a single mutant fails to show a complete correlation between these four activities: IE1(1-346), although it binds to PML and partially desumolates PML, usually still targets to PODs but fails to disrupt them (implying that another function, perhaps the acidic tail itself, is needed in addition to PML binding for efficient disruption). The PML-desumoylating and POD disruption activities of IE1 also appear to be tightly related to its functional activities both in inhibiting the transcriptional repressor activity of PML and in transactivating several cellular and viral promoters. These activities are also lost in IE1(1-346) and IE1(L174P). Most importantly, we found that a recombinant HCMV-BAC genome encoding a mutant, IE1(Δ290-320), that has lost all of these biochemical activities was not infectious in cultured fibroblast cells. Therefore, we suggest that both the activity of IE1 in disrupting PODs and its associated regulation of transcription are necessary for the efficient progress of infection. In contrast, an HCMV-BAC genome encoding the sumoylation-deficient IE1(K450R) was fully infectious under the same conditions, suggesting that, consistent with the previous findings from in vitro assays (74, 80), sumoylation of IE1 is not essential for the viral genome to be infectious.
The PODs appear to be involved in the regulation of cellular gene expression. The major POD component PML is known to act as a transcriptional cofactor or repressor (83). IE1 interferes with the powerful intrinsic transcriptional repressor activity of PML in mammalian GAL4 DNA-tethering assays (80). Our results here show that all mutant IE1 proteins that do not bind to or desumoylate PML also failed to inhibit the transcriptional repression activity of PML in the same assays. This effect also appears likely to explain or contribute to the several transcriptional up-regulation activities mediated by IE1 that we have investigated here. The transcriptional cofactor CBP can sometimes be found to be associated with PODs (45). Therefore, it is conceivable that disruption of PODs by IE1 may affect CBP-mediated gene regulation. We confirmed that HCMV IE1 activates the MMTV promoter containing glucocorticoid-responsive elements, which requires CBP as a cofactor for activation (78). It is plausible that CBP is released upon POD disruption, leading to activation of the MMTV promoter. Furthermore, augmentation of IE2-mediated transactivation of the HCMV UL54(Pol) promoter, as well as transactivation of both the MMTV promoter and the cellular DNA Pol α promoter, by IE1 all correlated with the ability of IE1 both to bind to PML and to disrupt PODs. These results suggest that displacement of PML by IE1 may regulate a broad range of gene expression involving POD components as cofactors.
Another impact of POD disruption on gene regulation could be to counteract gene silencing associated with heterochromatin. Members of the Sp100 family proteins that are associated with PODs have been shown to directly bind to heterochromatin protein 1 (HP1), suggesting that PODs are involved in regulating HP1-mediated gene silencing (46, 47, 72). Therefore, POD disruption by IE1 (which also releases Sp100 and Sp100-HMG from the PODs [data not shown]) may relieve such constraints on gene expression. Finally, Wang et al. (77) have reported that mouse PML−/− knockout cells both fail to mount interferon responses and are deficient in FAS-mediated and other apoptotic pathways. Whether disruption of PODs affects these events is unknown as yet.
It is not yet clear exactly how IE1 reduces the sumoylation levels of PML. One potential mechanism could be that IE1 prevents formation of sumoylated forms of PML, perhaps through its physical interaction with PML. Because IE1, unlike IE110, is also a substrate for SUMO modification, IE1 could potentially compete with PML for essential cellular components of the sumoylation system. Indeed, POD disruption by the ZTA protein in Epstein-Barr virus infection is also proteasome independent, and ZTA has been suggested to compete with PML for SUMO (1). However, a competition scenario appears unlikely for HCMV, based on the observations that, unlike PML (and IE2), IE1 does not bind to either SUMO or its conjugation enzyme Ubc9 in yeast two-hybrid interaction assays (80) and that, as shown here, IE1(Δ290-320) is still sumoylated to a level equivalent to that for the wild-type IE1 protein but without being able to affect the sumoylation level of PML (Fig. 3). Obviously, prevention of PML sumoylation could certainly occur in the cotransfection experiments, where newly synthesized HA-tagged PML may not ever enter the PODs in the presence of IE1, but it is difficult to imagine that endogenous PML is turned over sufficiently rapidly for this to account for the complete loss of PML from the PODs within just 3 h in wild-type HCMV-infected cells. A second explanation might simply be that the binding of IE1 to PML causes disaggregation and loss of PML protein complexes from PODs, which are subsequently subjected to SUMO proteases when not in the PODs. Removal of SUMO from newly expressed HA-PML by cotransfected SUMO proteases does occur efficiently even in the absence of IE1 (H.-R. Lee and J.-H. Ahn, unpublished data), but whether they can also remove SUMO from endogenous POD-associated PML is still unknown.
Two more likely potential mechanisms would be for IE1 to actively promote removal of SUMO from PML, either directly or indirectly. POD structure supposedly depends upon at least a core of sumoylated forms of PML, and other sumoylated proteins are actively recruited to PODs. IE1 may indirectly recruit an isopeptidase or a protease that specifically either desumoylates or degrades just the sumoylated forms of PML in vivo. Recently, IE110-induced recruitment of SENP1 (a SUMO protease) to PODs was reported to occur in cotransfection experiments, suggesting a possible role for a SUMO protease in desumoylation of PML during HSV-1 infection also, although there was no attempt to demonstrate a physical interaction between SENP1 and IE110 (7). Finally, IE1 may directly desumoylate PML. All known SUMO proteases contain the well conserved triad catalytic residues (His, Asp, and Cys) in their C-terminal regions (10, 37). Although there is no obvious matching motif within IE1, we cannot at present exclude the possibility that IE1 itself may act as a novel SUMO-specific protease.
Phosphorylation of PML also appears to be linked to the sumoylation status of PML. A phosphatase inhibitor, calyculin A, was shown to inhibit arsenic trioxide (As2O3)-triggered PML sumoylation (61), and PML is specifically phosphorylated in the M phase of the cell cycle, with a corresponding complete loss of sumoylation (21). With regard to this point, it is plausible that the reported associated kinase activity of IE1 (63) might be involved in the loss of sumoylation of PML. Those authors suggested that IE1(L174P) disrupts a proposed ATP binding site spanning amino acids 173 to 179. However, we confirmed here that IE1(L174P) itself fails to bind to PML in yeast two-hybrid interaction assays (60), and IE1(Δ290-320) is also deficient in binding to PML, suggesting an alternative explanation for the inactivity of this mutant. Conceivably, either IE1 binding to or IE1 phosphorylation of PML could block the sumoylation of PML without invoking a SUMO protease activity.
The direct correlation between the activity of IE1 in desumoylating PML (and in disrupting PODs) and the various transactivation and derepression functions of IE1 strongly supports the idea that PODs and their components contribute to regulation of transcription, perhaps including the pathways needed for interferon to induce an antiviral state. Indeed, Chee et al. (15) have recently shown that PML mediates interferon-based antiviral effects against HSV-1. POD disruption by viral IE nuclear regulatory proteins expressed very early in infection almost certainly assists in the progression of productive infection by modulating aspects of both viral and cellular gene expression pathways and would obviously be advantageous if it also contributed to blocking of interferon-mediated pathways. The underlying detailed mechanism of IE1-dependent desumoylation of PML and identification of other potential target cellular (or viral) genes affected by POD disruption by IE1 remain worthy subjects for further investigation.
Acknowledgments
This research was supported by a Korea Research Foundation grant (KRF-2001-015-DP0451) and by a Molecular and Cellular BioDiscovery Research Program grant from the Ministry of Science and Technology, South Korea, to J.-H.A. and by grant RO1 AI24576 from the National Institutes of Health to G.S.H.
We thank Teresa Wang (Stanford University, San Francisco, Calif.) for providing a Pol α-LUC target reporter construct. We are grateful to Edward S. Mocarski (Stanford University, Stanford, Calif.) for samples of the IE1 deletion CR208 virus and its parent HCMV(Towne).
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J. H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol 18:4899-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. [DOI] [PubMed] [Google Scholar]
- 4.Ahn, J. H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn, J. H., W. J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn, J. H., Y. Xu, W. J. Jang, M. J. Matunis, and G. S. Hayward. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 75:3859-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey, D., and P. O'Hare. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951-2964. [DOI] [PubMed] [Google Scholar]
- 8.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell, P., P. M. Lieberman, and G. G. Maul. 2000. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J. Virol. 74:11800-11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. [DOI] [PubMed] [Google Scholar]
- 11.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbone, R., M. Pearson, S. Minucci, and P. G. Pelicci. 2002. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 21:1633-1640. [DOI] [PubMed] [Google Scholar]
- 13.Carvalho, T., J. S. Seeler, K. Ohman, P. Jordan, U. Pettersson, G. Akusjarvi, M. Carmo-Fonseca, and A. Dejean. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, C. P., C. L. Malone, and M. F. Stinski. 1989. A human cytomegalovirus early gene has three inducible promoters that are regulated differentially at various times after infection. J. Virol. 63:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 17.Cherrington, J. M., and E. S. Mocarski. 1989. Human cytomegalovirus IE1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 19.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 20.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett, R. D., P. Lomonte, T. Sternsdorf, R. van Driel, and A. Orr. 1999. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112:4581-4588. [DOI] [PubMed] [Google Scholar]
- 22.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gawn, J. M., and R. F. Greaves. 2002. Absence of IE1 p72 protein function during low-multiplicity infection by human cytomegalovirus results in a broad block to viral delayed-early gene expression. J. Virol. 76:4441-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus IE1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green, S., P. Isseman, and E. Sheer. 1988. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 16:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagglund, R., C. Van Sant, P. Lopez, and B. Roizman. 2002. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA 99:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayhurst, G. P., L. A. Bryant, R. C. Caswell, S. M. Walker, and J. H. Sinclair. 1995. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J. Virol. 69:182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunninghake, G. W., B. G. Monks, L. J. Geist, M. M. Monick, M. A. Monroy, M. F. Stinski, A. C. Webb, J. M. Dayer, P. E. Auron, and M. J. Fenton. 1992. The functional importance of a cap site-proximal region of the human prointerleukin 1 beta gene is defined by viral protein trans-activation. Mol. Cell. Biol. 12:3439-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, H. Y., C. Petrovas, and G. E. Sonenshein. 2002. RelB-p50 NF-κB complexes are selectively induced by cytomegalovirus immediate-early protein 1: differential regulation of Bcl-x(L) promoter activity by NF-κB family members. J. Virol. 76:5737-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamitani, T., K. Kito, H. P. Nguyen, H. Wada, T. Fukuda-Kamitani, and E. T. Yeh. 1998. Identification of three major sentrinization sites in PML. J. Biol. Chem. 273:26675-26682. [DOI] [PubMed] [Google Scholar]
- 36.Kerry, J. A., M. A. Priddy, and R. M. Stenberg. 1994. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J. Virol. 68:4167-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, K. I., S. H. Baek, and C. H. Chung. 2002. Versatile protein tag, SUMO: its enzymology and biological function. J. Cell Physiol. 191:257-268. [DOI] [PubMed] [Google Scholar]
- 38.Kim, S., S. S. Yu, and V. N. Kim. 1996. Essential role of NF-kappa B in transactivation of the human immunodeficiency virus long terminal repeat by the human cytomegalovirus 1E1 protein. J. Gen. Virol. 77:83-91. [DOI] [PubMed] [Google Scholar]
- 39.Kim, S., S. S. Yu, I. S. Lee, S. Ohno, J. Yim, and H. S. Kang. 1999. Human cytomegalovirus IE1 protein activates AP-1 through a cellular protein kinase(s). J. Gen. Virol. 80:961-969. [DOI] [PubMed] [Google Scholar]
- 40.Klucher, K. M., M. Sommer, J. T. Kadonaga, and D. H. Spector. 1993. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol. Cell. Biol. 13:1238-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 42.Lafemina, R. L., M. C. Pizzorno, J. D. Mosca, and G. S. Hayward. 1989. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology 172:584-600. [DOI] [PubMed] [Google Scholar]
- 43.Lai, H. K., and K. L. Borden. 2000. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene 19:1623-1634. [DOI] [PubMed] [Google Scholar]
- 44.Lallemand-Breitenbach, V., J. Zhu, F. Puvion, M. Koken, N. Honore, A. Doubeikovsky, E. Duprez, P. P. Pandolfi, E. Puvion, P. Freemont, and H. de The. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193:1361-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.LaMorte, V. J., J. A. Dyck, R. L. Ochs, and R. M. Evans. 1998. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA 95:4991-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lechner, M. S., G. E. Begg, D. W. Speicher, and F. J. Rauscher III. 2000. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol. Cell. Biol 20:6449-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehming, N., A. Le Saux, J. Schuller, and M. Ptashne. 1998. Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl. Acad. Sci. USA 95:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li, H., and J. D. Chen. 2000. PML and the oncogenic nuclear domains in regulating transcriptional repression. Curr. Opin. Cell Biol. 12:641-644. [DOI] [PubMed] [Google Scholar]
- 49.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukac, D. M., J. R. Manuppello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Margolis, M. J., S. Pajovic, E. L. Wong, M. Wade, R. Jupp, J. A. Nelson, and J. C. Azizkhan. 1995. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J. Virol. 69:7759-7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 55.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 56.Mocarski, E. S. 2001. Cytomegalovirus and their replication, vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 57.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mu, Z. M., K. V. Chin, J. H. Liu, G. Lozano, and K. S. Chang. 1994. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol. Cell. Biol. 14:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mullen, M. A., D. M. Ciufo, and G. S. Hayward. 1994. Mapping of intracellular localization domains and evidence for colocalization interactions between the IE110 and IE175 nuclear transactivator proteins of herpes simplex virus. J. Virol. 68:3250-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Hare, P., and G. S. Hayward. 1985. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J. Virol. 53:751-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pajovic, S., E. L. Wong, A. R. Black, and J. C. Azizkhan. 1997. Identification of a viral kinase that phosphorylates specific E2Fs and pocket proteins. Mol. Cell. Biol. 17:6459-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pass, R. F. 2001. Cytomegalovirus, vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 66.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pizzorno, M. C., M. A. Mullen, Y. N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pizzorno, M. C., P. O'Hare, L. Sha, R. L. LaFemina, and G. S. Hayward. 1988. trans-activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J. Virol. 62:1167-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poma, E. E., T. F. Kowalik, L. Zhu, J. H. Sinclair, and E. S. Huang. 1996. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J. Virol. 70:7867-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quignon, F., F. De Bels, M. Koken, J. Feunteun, J. C. Ameisen, and H. de The. 1998. PML induces a novel caspase-independent death process. Nat. Genet. 20:259-265. [DOI] [PubMed] [Google Scholar]
- 71.Sambucetti, L. C., J. M. Cherrington, G. W. Wilkinson, and E. S. Mocarski. 1989. NF-kappa B activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 8:4251-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seeler, J. S., A. Marchio, D. Sitterlin, C. Transy, and A. Dejean. 1998. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. USA 95:7316-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spengler, M. L., K. Kurapatwinski, A. R. Black, and J. Azizkhan-Clifford. 2002. SUMO-1 modification of human cytomegalovirus IE1/IE72. J. Virol. 76:2990-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, Z. G., D. Ruggero, S. Ronchetti, S. Zhong, M. Gaboli, R. Rivi, and P. P. Pandolfi. 1998. PML is essential for multiple apoptotic pathways. Nat. Genet. 20:266-272. [DOI] [PubMed] [Google Scholar]
- 78.Wienzek, S., and M. Dobbelstein. 2001. Viral and cellular factors that target the promyelocytic leukemia oncogenic domains strongly activate a glucocorticoid-responsive promoter. J. Virol. 75:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilkinson, G. W., C. Kelly, J. H. Sinclair, and C. Rickards. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79:1233-1245. [DOI] [PubMed] [Google Scholar]
- 80.Xu, Y., J. H. Ahn, M. Cheng, C. M. apRhys, C. J. Chiou, J. Zong, M. J. Matunis, and G. S. Hayward. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J. Virol. 75:10683-10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yurochko, A. D., M. W. Mayo, E. E. Poma, A. S. Baldwin, Jr., and E. S. Huang. 1997. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-κB promoters. J. Virol. 71:4638-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]
- 83.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]