Abstract
The severe acute respiratory syndrome-associated coronavirus (SARS-CoV) causes severe pneumonia with a fatal outcome in approximately 10% of patients. SARS-CoV is not closely related to other coronaviruses but shares a similar genome organization. Entry of coronaviruses into target cells is mediated by the viral S protein. We functionally analyzed SARS-CoV S using pseudotyped lentiviral particles (pseudotypes). The SARS-CoV S protein was found to be expressed at the cell surface upon transient transfection. Coexpression of SARS-CoV S with human immunodeficiency virus-based reporter constructs yielded viruses that were infectious for a range of cell lines. Most notably, viral pseudotypes harboring SARS-CoV S infected hepatoma cell lines but not T- and B-cell lines. Infection of the hepatoma cell line Huh-7 was also observed with replication-competent SARS-CoV, indicating that hepatocytes might be targeted by SARS-CoV in vivo. Inhibition of vacuolar acidification impaired infection by SARS-CoV S-bearing pseudotypes, indicating that S-mediated entry requires low pH. Finally, infection by SARS-CoV S pseudotypes but not by vesicular stomatitis virus G pseudotypes was efficiently inhibited by a rabbit serum raised against SARS-CoV particles and by sera from SARS patients, demonstrating that SARS-CoV S is a target for neutralizing antibodies and that such antibodies are generated in SARS-CoV-infected patients. Our results show that viral pseudotyping can be employed for the analysis of SARS-CoV S function. Moreover, we provide evidence that SARS-CoV infection might not be limited to lung tissue and can be inhibited by the humoral immune response in infected patients.
In the fourth quarter of 2002, a novel severe, acute pneumonia emerged in Guangdong province, China. The disease, termed severe acute respiratory syndrome (SARS) by the World Health Organization, experienced a global spread, with Asian countries being most severely affected. In May 2003, a novel coronavirus (CoV) was discovered in SARS patients (16, 31, 53) and shown to be the cause of the disease (19, 32). Due to coordinated containment measures like quarantine of patients and travel restrictions (22, 73), the spread of SARS was stopped by July 2003. By then, over 8,000 patients had developed SARS and more than 700 had died from the disease (72). It has been speculated that SARS might be a seasonal disease (49), and evidence has been presented that the SARS-associated CoV (SARS-CoV) might replicate in an animal reservoir (23). It is therefore conceivable that SARS-CoV might be reintroduced into the human population (49), a possibility that calls for the development of antiviral agents and vaccines in order to prevent future epidemics.
Based on antigenic cross-reactivity CoVs were initially subdivided into three groups. These results were largely confirmed upon subsequent phylogenetic analysis (35, 58); however, some deviations between the two classifications were observed. Although SARS-CoV exhibits a similar genome structure, it is only distantly related to known CoVs and constitutes the first member of either a new phylogenetic group (41, 57) or a new subgroup of group 2 CoVs (61). Thus, the approximately 29,750 nucleotides comprising the positive-strand RNA SARS-CoV genome encode the structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N) (41, 57), which are highly conserved among CoVs. The S, M, and E gene products are inserted into the viral envelope, and coexpression of M and E has been shown to be sufficient to drive budding of virus-like particles (67). However, further studies suggest that either M or E might be sufficient to mediate release of progeny virions (34, 40). CoVs usually bud into an intermediate compartment of the endoplasmic reticulum and the Golgi complex, followed by the release of virus particles from the infected cell by exocytosis (30, 66).
Generally, CoV infection of target cells is mediated by the S proteins, which exhibit characteristics of class I fusion proteins (3). They are trimeric type I integral membrane proteins, organized into subdomains necessary for receptor engagement (S1) and membrane fusion (S2) (20). S has been shown to be incorporated into virions by interactions with M (45, 51, 52). In infected cells and in S-transfected cells, S can be transported to the cell surface, where it mediates fusion with adjoining cells, resulting in the formation of syncytia (2, 11, 39). CoV S proteins facilitate entry into target cells by interacting with specific receptors on the cell surface. Thus, mouse hepatitis viruses engage carcinoembryonic antigen-cell adhesion molecules, human and feline CoVs bind to aminopeptidase N, and bovine CoVs recognize 9-O-acetylated sialic acids (20). The S protein was shown to be the sole determinant of the host range of CoVs, since the exchange of the ectodomain of S is sufficient to transfer target cell specificity (33).
Based on sequence comparison, the SARS-CoV S domain organization resembles that of other CoV S proteins, and SARS-CoV S is also predicted to be a class I fusion protein (41, 57). The angiotensin-converting enzyme 2 (ACE2) has been reported to function as a receptor for SARS-CoV (38). Indeed, ACE2 mRNA expression has been detected in lung and kidney tissues (24), and SARS-CoV S engagement of ACE2 might account for the infection of lung tissue in SARS patients (31, 32, 46) and the successful isolation of SARS-CoV S on macaque kidney cells (16, 31, 53); however, no detailed information on SARS-CoV tropism and its correlation with ACE2 expression is available so far.
The SARS-CoV S protein harbors 23 potential N-linked glycosylation sites (41, 57) which might limit recognition by the humoral immune response. SARS-CoV-infected individuals usually develop an antibody response against the virus (16, 31, 53), but it is unclear whether S-specific neutralizing antibodies are generated and whether the humoral immune response plays a key role in clearing SARS-CoV infection. The analysis of SARS-CoV S function should therefore provide valuable insights into SARS pathogenesis and might open possibilities for therapeutic intervention and vaccine development.
We used pseudotyped lentiviral particles (pseudotypes) bearing the SARS-CoV S protein to study viral tropism and determinants of S-mediated infection. Pseudotypes bearing the SARS-CoV S protein were able to enter cell lines of kidney and liver origin, and the latter were highly permissive to infection by replication-competent SARS-CoV, suggesting that SARS-CoV might target liver tissue in vivo. Pseudotype infection of cells was pH dependent, indicating that SARS-CoV particles might be endocytosed upon attachment to the surface of target cells and fuse with the membrane of endocytic vesicles. SARS-CoV S-mediated infection could be inhibited by rabbit sera from animals immunized with purified SARS-CoV and importantly by sera from SARS patients, demonstrating that S is a target for neutralizing antibodies.
MATERIALS AND METHODS
Plasmid construction and in vitro mutagenesis.
A eukaryotic expression vector for the SARS-CoV S envelope protein was constructed by reverse transcription-PCR of the SARS-CoV RNA genome (strain Frankfurt) with use of the SuperScript One step RT-PCR kit (Invitrogen Corp., San Diego, Calif.) and oligonucleotides SARS-S-3stop (TTATGTGTAATGTAATTTGACACCCC) and SARS-S-5 (CACCATGTTTATTTTCTTATTATTCTT). The resulting fragment was subcloned into pcDNA3.1/HisTopo (Invitrogen Corp.); the SARS-CoV leader sequence was thereafter introduced by PCR mutagenesis. In parallel the SARS-CoV S cDNA including the leader sequence was inserted into the pCAGGS vector (24). Expression plasmids encoding the envelope proteins of murine leukemia virus (MLV) and vesicular stomatitis virus (VSV G) have been described previously (60). The DNA sequence of all PCR-amplified sequences was confirmed by automated sequence analysis (ABI, Weiterstadt, Germany).
Cell culture, transfection, infection, and reporter assays.
The lymphatic cell lines C8166 and BL41 were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum. 293T, Vero, HEp-2, and HepG2 cells were maintained in Dulbecco's minimal essential medium (Gibco/BRL, Eggenstein, Germany) supplemented with 10% fetal calf serum; medium for Huh-7 cells was additionally supplemented with a 1% amino acid cocktail (Invitrogen Corp.).
Human immunodeficiency virus (HIV) pseudotypes were generated by cotransfection of 293T cells with the indicated envelope expression plasmids in combination with the pNL4-3-Luc-R−E− (9, 25) or the pNL4-3-GFP (56) plasmid as described previously (60). The culture supernatants were harvested 48 h after transfection, passed through 0.4-μm-pore-size filters, aliquoted, and stored at −80°C. For infection with pseudotypes carrying luciferase or green fluorescent protein (GFP) as a reporter gene, cells were seeded onto 96-well plates at densities ranging from 8 × 103 up to 1.2 × 104 and incubated with equal volumes of viral supernatants normalized for HIV capsid (p24) content by enzyme-linked immunosorbent assay (ELISA) (Murex; Abbott Diagnostics, Abbott Park, Ill.). For the determination of cell tropism, virus particles were concentrated on the cells by centrifugation for 2 h at 800 × g (50). Generally, the medium was replaced after 12 h, and luciferase activity was determined 72 h after transduction with a commercially available kit as recommended by the manufacturer (Promega, Madison, Wis.). For inhibition of lysosomal acidification, 293T or Huh-7 cells were preincubated for 30 min at 37°C with ammonium chloride or bafilomycin A at final concentrations of 1 to 15 mM and 20 to 100 nM, respectively. Thereafter, the indicated pseudotyped viruses, normalized for luciferase activity, were added for 6 h followed by complete renewal of the culture medium. Finally, for neutralization of SARS-CoV S-bearing pseudotypes by immune sera, equal volumes of viral supernatants, normalized for comparable luciferase activity upon infection in the absence of serum, were preincubated with serial dilutions of serum for 30 min and incubated with target cells for 7 h. Thereafter, the infection medium was removed, the cells were maintained in fresh medium for 72 h, and luciferase activity was determined as described above.
Antibodies, Western blotting, and fluorescence-activated cell sorting (FACS) analysis.
For production of SARS-CoV antigen, the supernatant of Vero cells infected with SARS-CoV (strain Frankfurt) was harvested 24 h after infection and cleared of cell debris by centrifugation. The SARS-CoV particles were further purified from the supernatant by ultracentrifugation for 4 h through a sucrose cushion. The pellet was resuspended, completely inactivated by heat treatment (1 h at 56°C) and gamma irradiation (30 kGy), and used to immunize rabbits to generate a polyclonal SARS-CoV antiserum. Sera from patients infected with SARS-CoV were purified from whole-blood samples. Anti-human and anti-rabbit horseradish peroxidase- or fluorescein-conjugated secondary antibodies were obtained from Dianova (Hamburg, Germany).
For Western blotting, transfected 293T cells were lysed in RIPA lysis buffer (0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 1% deoxycholate, 0.05 M Tris [pH 7.3], 0.15 M NaCl, 0.15 M phenylmethylsulfonyl fluoride), diluted in SDS Laemmli buffer, and boiled at 94°C for 10 min. Samples were electrophoresed in SDS-10% polyacrylamide gels, and the proteins were transferred onto nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany). Western blotting and chemiluminescence detection were performed according to the manufacturer's protocol (ECL Western detection kit; Amersham Pharmacia Biotech Europe, Freiburg, Germany).
For FACS analysis, the transfected 293T cells were incubated with anti-SARS-CoV antisera of human or rabbit origin in a total volume of 100 μl of FACS buffer. The reaction mixtures were incubated at 4°C for 30 min, followed by a washing step; thereafter, bound antibody was detected by adding 0.5 μg of an appropriate fluorescein-coupled secondary antibody (Dianova). Antibody binding was allowed to proceed for 30 min at 4°C, and then the cells were washed and SARS-S surface expression was quantitated by FACS analysis.
Indirect immunofluorescence analysis.
For infection with pseudotypes carrying GFP as a reporter gene, Huh-7 cells were plated onto 96-well plates at 104 cells per well and incubated with pseudotyped virus particles normalized for p24 content by ELISA (Murex; Abbott Diagnostics). After 72 h, cells were fixed with 4% paraformaldehyde for 15 min at room temperature, washed with phosphate-buffered saline, and monitored for GFP expression with a Zeiss Axiovert-200 microscope (Zeiss, Jena, Germany). All images were recorded with a Nikon COOLPIX-995 digital camera (Nikon GmbH, Düsseldorf, Germany) with identical aperture, exposure time, and white balance and processed by using the Adobe Photoshop package (Universal Imaging Corp., Brandywine, Pa.; Adobe Systems Inc.). Each experiment was performed in quadruplicate, repeated at least three times, and evaluated by two investigators. Alternatively, Huh-7 and Vero cells cultured on 15-mm-diameter sterile glass slides in 12-well dishes were infected with 25 μl of supernatant of SARS-CoV (Hong Kong and Frankfurt strains). Mock-infected cells were used as a negative control. After 24 h, cells were fixed and subjected to indirect immunofluorescence with a human anti-SARS antiserum followed by incubation with an anti-human fluorescein isothiocyanate-labeled antibody. Staining was analyzed by using a Zeiss LSM 510 confocal laser scanning microscope.
RESULTS
We functionally analyzed the S protein of SARS-CoV using viral pseudotyping, a well-established technology which allows the characterization of viral glycoproteins (GPs) upon incorporation into heterologous viral particles. Lentiviral particles pseudotyped with SARS-CoV S were generated by cotransfection of S expression plasmids with HIV reporter genomes harboring a defective env gene. Cell tropism, determinants of infection, and neutralization of the SARS-CoV S-harboring pseudotypes was investigated.
S is expressed at the surface of transiently transfected 293T cells.
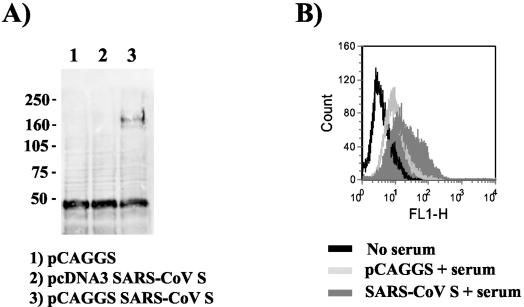
The generation of retroviral pseudotypes bearing SARS-CoV S requires that S is expressed on the cell surface to allow nonspecific incorporation into the membrane of budding viral particles. In order to examine whether the SARS-CoV S protein can be expressed on the cell surface, we transiently transfected 293T cells with pcDNA3 and pCAGGS vectors containing the SARS-CoV S gene. The pcDNA3 plasmid is a commonly used expression vector harboring a cytomegalovirus promoter. Expression of S in pCAGGS is driven by a combination of the cytomegalovirus enhancer and the chicken β-actin promoter; however, the S transcript produced by this construct contains an intron (48). In order to detect S expression, a rabbit serum generated by immunization with purified SARS-CoV particles was employed. In agreement with published reports (74) a specific band of approximately 170 to 180 kDa was detected by Western blotting in lysates from cells transfected with the pCAGGS-based S expression construct (Fig. 1A, lane 3). No specific signal was detected in control-transfected cells and in cells transfected with S inserted into pcDNA3 (lane 2), suggesting that the presence of an intron in the S transcript enhances expression. Additionally, the presence of SARS-CoV S on the surface of transfected cells was quantified by FACS. Expression of S was readily detected in S-pCAGGS-transfected cells but not in S-pcDNA3-transfected or control cells (Fig. 1B and data not shown), indicating that S can be transported to the cell surface after transient expression.
FIG. 1.
Transient expression of the SARS-CoV S protein. (A) Western blot analysis of SARS-CoV S expression. 293T cells were transfected either with the empty eukaryotic expression vector pCAGGS (lane 1) or the SARS-CoV S expression constructs pcDNA3-S (lane 2) or pCAGGS-S (lane 3). After lysis in RIPA buffer, the extracts were separated by SDS-10% polyacrylamide gel electrophoresis and detected in Western blot analyses with a polyclonal rabbit serum directed against SARS-CoV particles. Numbers at left are molecular masses in kilodaltons. (B) Surface expression of SARS-CoV S protein. 293T cells were transfected with the empty expression plasmid pCAGGS or the pCAGGS-S construct. Thereafter, cells were prepared for FACS analysis and incubated with the polyclonal rabbit serum in order to detect the S protein. Results of one experiment representative of three are shown.
The SARS-CoV S protein mediates entry into liver- and kidney-derived cell lines.
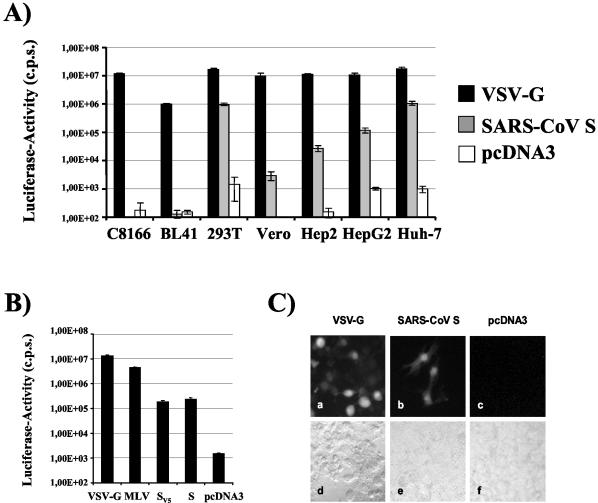
Culture supernatants containing pseudotyped particles bearing the SARS-CoV S protein or control particles lacking a GP were p24 normalized and used for infection of a panel of cell lines. Pseudotypes carrying VSV G served as a positive control. All cell lines were efficiently infected by VSV G pseudotypes (luciferase activity, ≥1.00E + 06), while infection by control pseudotypes, obtained after cotransfection of pcDNA3 or pCAGGS, was inefficient (luciferase activity, <2.00E + 03) (Fig. 2A and data not shown). Pseudotypes bearing the SARS-CoV S GP infected the hepatoma cell lines HepG2 and Huh-7 as well as the kidney-derived cell line 293T with high efficiency (luciferase activity, >1.00E + 05). Robust infection was also detected with the HeLa-derived cell line HEp-2. In contrast, SARS-CoV S-mediated infection of the lymphocytic cell lines C8166 and BL41 was equal to or below that observed with control particles bearing no GP (Fig. 2A), suggesting that these cell lines do express very low amounts of receptor(s) or no receptor(s) for SARS-CoV S. Entry of SARS-CoV S-bearing pseudotypes into Vero cells, which have been used for the initial isolation of SARS-CoV (16, 31), was repeatedly more efficient than entry of control viruses; however, infectious entry was about a hundred- to a thousandfold less efficient than entry into 293T or Huh-7 cells. Infection experiments with p24-normalized virus stocks and Huh-7 cells as targets confirmed that pseudotypes generated by cotransfection of S were at least 100-fold more infectious than were control particles and about 10- to 100-fold less infectious than particles bearing the amphotropic MLV and the pantropic VSV GP (Fig. 2B). Also, pseudotypes bearing the wild-type S protein or an S variant with a V5/His antigenic tag added to the C terminus of S exhibited comparable infectivity, indicating that the antigenic tag does not affect function (Fig. 2B). Finally we assessed the infectivity of the pseudotypes with an HIV reporter construct harboring GFP. Huh-7 cells were incubated with p24-normalized virus stocks, and infected cells were detected by fluorescence microscopy. In agreement with data obtained with luciferase reporter viruses, VSV G-mediated infection was efficient since a substantial portion of target cells expressed GFP (Fig. 2C, panels a and d). Cells expressing GFP were also readily detected upon SARS-CoV S-mediated infection (panels b and e); however, clearly fewer cells were GFP positive than were target cells transduced with VSV G-bearing pseudotypes. Infection with control particles did not result in significant transduction (panels c and f).
FIG. 2.
Tropism and infectivity of SARS-S-bearing lentiviral pseudotypes. (A) Cellular tropism of SARS-CoV S pseudotypes. SARS-CoV S-mediated infection of a panel of common cell lines was assessed. For infection, 3 ng of p24-normalized pseudotypes carrying either the pantropic VSV G as positive control, no GP (pcDNA3) as negative control, or the SARS-CoV S GP was added to the cells followed by a 2-h centrifugation at 800 × g in order to concentrate the infectious material on the cells. After 72 h, cells were lysed and luciferase activity was determined in the cell extracts. Results of a representative experiment performed in quadruplicate are shown. Comparable results were obtained in at least three different experiments with independent virus stocks. (B) Relative infectivity of pseudotypes bearing VSV G, MLV GP, and SARS-CoV S. Huh-7 cells were infected with culture supernatants containing 1 ng of viral antigen, and luciferase activity was measured 3 days after infection. Results of a representative experiment carried out in quadruplicate are shown; comparable results were obtained in an independent experiment. (C) Efficiency of transduction of Huh-7 cells with p24-normalized GFP reporter viruses. Huh-7 cells were spin infected for 2 h with 25 ng of pseudotypes bearing VSV G or SARS-CoV S or control pseudotypes lacking an envelope protein (pcDNA3). After 72 h, cells were fixed and analyzed by immunofluorescence. Results of a representative experiment carried out in quadruplicate are shown. The results were confirmed in two independent experiments.
The hepatoma cell line Huh-7 is susceptible to SARS-CoV infection.
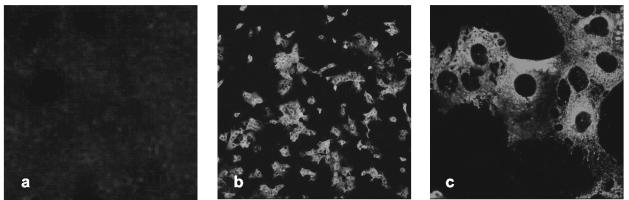
Next, we addressed the question whether efficient pseudotype infection of hepatoma cells correlates with SARS-CoV infection of these cells. For this, Huh-7 cells were incubated either with supernatants containing SARS-CoV of the Frankfurt strain or with control supernatant. After 24 h the cells were stained with human anti-SARS-CoV antiserum and analyzed by confocal microscopy. As shown in Fig. 3, staining of a substantial portion of Huh-7 cells infected with SARS-CoV was readily detected; however, no staining of cells infected with control supernatant was observed (Fig. 3 and data not shown). A similar result was obtained with SARS-CoV of the Hong Kong strain and with Vero cells, which are known to be susceptible to SARS-CoV (data not shown). Consequently, the hepatic cell tropism of SARS-CoV S-bearing lentiviral pseudotypes is reflected by live SARS-CoV particles, indicating that these cells could also be targeted in infected individuals.
FIG. 3.
Infection of hepatoma cells by replication-competent SARS-CoV. Huh-7 cells were mock infected (a) or infected with SARS-CoV (Frankfurt strain) (b and c) and stained with human anti SARS-CoV antiserum 24 h after infection. The cells were analyzed with a confocal laser scanning microscope (Zeiss LSM 510) at magnifications of ×10 (a and b) and ×63 (c).
Entry of pseudotypes bearing S into the kidney-derived cell line 293T and the hepatoma cell line Huh-7 is pH dependent.
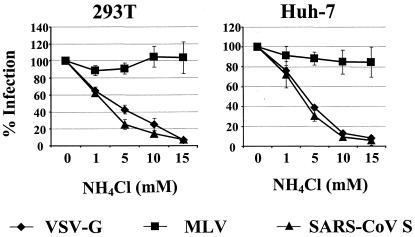
Enveloped viruses usually use two strategies to overcome the energy barrier associated with fusing the viral and the cellular membranes, an essential step in the infection process. First, the engagement of cell surface receptors by the viral GP can trigger conformational changes in the GP which lead to membrane fusion (13). Second, viruses can be endocytosed, and the low pH value in endocytic vesicles stimulates conformational changes in the GP required for fusion (26). Thus, entry mediated by the MLV and HIV GPs is known to be pH independent, while entry mediated by VSV G is pH dependent (42, 43). We investigated the impact of pH on SARS-CoV S-mediated infection by using ammonium chloride and bafilomycin A1, well-characterized inhibitors of vacuolar acidification (59). The indicated target cells were preincubated with rising concentrations of ammonium chloride or bafilomycin A1 and infected by pseudotypes harboring either VSV G, MLV GP, or SARS-CoV S (Fig. 4 and data not shown). The viral input was standardized for comparable luciferase production upon infection of target cells in the absence of inhibitor. Preincubation of 293T cells (left panel) with ammonium chloride did not reduce entry of MLV GP pseudotypes, while infection by pseudotypes harboring VSV G and S was reduced in a dose-dependent manner, reaching a maximum inhibition of at least 90%. Comparable results were obtained with Huh-7 cells (right panel) and when bafilomycin A was used as an inhibitor; however, bafilomycin A treatment also diminished MLV GP-mediated entry into Huh-7 cells, likely due to toxic side effects (Fig. 4 and data not shown). Thus, treatment with lysosomotropic agents inhibits S-mediated infection, arguing for a role of pH in S-driven membrane fusion.
FIG. 4.
Influence of inhibitors of acidification on SARS-CoV infection. Target cells were preincubated for 30 min with the indicated concentrations of ammonium chloride. Subsequently the cells were infected with pseudotypes carrying VSV G, MLV GP, or SARS-CoV S protein, which had been normalized for comparable luciferase activity upon infection of target cells in the absence of inhibitor. After 6 h, virus and inhibitor were removed from the cells, and fresh medium was added. Luciferase activity was determined in cell extracts after 72 h. The averages of three independent experiments performed in quadruplicate are shown. Error bars indicate standard errors of the means.
An antiserum raised against purified SARS-CoV neutralizes infection by SARS-CoV S-bearing pseudotypes.
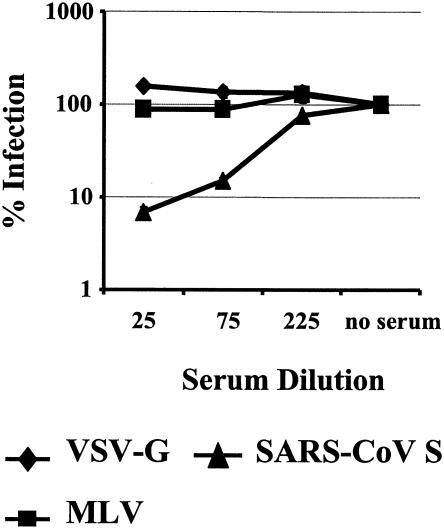
If SARS-CoV S is a target for neutralizing antibodies, vaccines based on native S might hold potential. Therefore, we tested the neutralization capacity of a rabbit serum raised against purified SARS-CoV particles. Since presumably intact virus particles were used as immunogen, one would expect the generation of conformation-dependent S-specific antibodies which might inhibit S function. Pseudotypes harboring SARS-CoV S or VSV G were normalized for comparable infection of target cells in the absence of serum, preincubated with different dilutions of either anti-SARS-CoV or a control rabbit serum, and used for infection of Huh-7 cells. Preincubation with anti-SARS-CoV serum but not with the control serum specifically reduced S-mediated infection by more than 90%, while infection driven by MLV GP or VSV G remained largely unaffected (Fig. 5 and data not shown). Thus, infection by SARS-CoV S-harboring pseudotypes can be inhibited by an anti-SARS-CoV immune serum, indicating that S can be targeted by the humoral immune response and confirming that infection by SARS-CoV S pseudotypes is driven by S.
FIG. 5.
Neutralization of SARS-CoV S-mediated infection by polyclonal rabbit serum raised against purified SARS-CoV. Pseudotypes bearing VSV G, MLV GP, or SARS-CoV S protein, which had been normalized to equal luciferase activity upon infection of target cells in the absence of immune serum, were incubated for 30 min with the indicated dilutions of the polyclonal rabbit serum. Thereafter, the virus-antibody mixture was added to Huh-7 cells for 7 h followed by complete removal of the culture medium. Luciferase activity was determined in cell extracts after 72 h. Results of a representative experiment carried out in triplicate are shown. Similar results were obtained in two independent experiments.
SARS patients generate S-specific neutralizing antibodies.
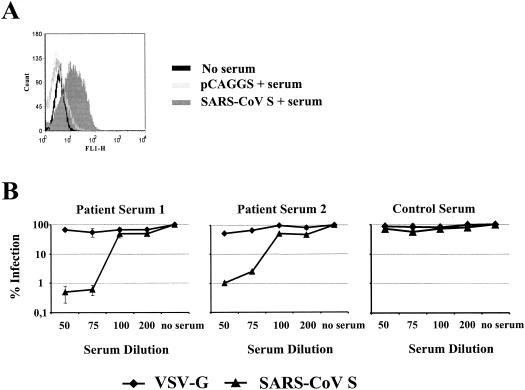
The S protein is known to be a major target of the humoral and cellular immune response in CoV infection (8, 63, 70), and a protective effect of S-specific neutralizing antibodies has been documented (5, 44). For that reason, we asked if sera from SARS patients recognize SARS-CoV S protein and neutralize infection by SARS-CoV S-bearing pseudotypes. The presence of S-specific antibodies in serum from a convalescent SARS patient was analyzed by FACS. Specific staining was observed with S-expressing cells, indicating that S-specific antibodies are generated in SARS patients (Fig. 6A). Next we examined if sera from SARS patients can neutralize S-driven infection. The neutralization assay was carried out as described above. Sera from two SARS patients potently inhibited SARS-CoV S-mediated infection at dilutions of 1:50 and 1:75, whereas no appreciable inhibition of VSV G-driven infection was observed (Fig. 6B). Moreover, neither SARS-CoV S- nor VSV G-mediated entry was efficiently inhibited by human sera from healthy donors, confirming a specific inhibition of SARS-CoV pseudotypes by SARS patient sera (Fig. 6B). Thus, SARS-CoV S is targeted by neutralizing antibodies generated by SARS patients, indicating that the humoral immune response might play a role in the clearance of the infection.
FIG. 6.
Inhibition of SARS-CoV S-mediated infection by sera from SARS patients. (A) Detection of cell-surface-expressed SARS-CoV S protein by SARS patient serum. The SARS-CoV S protein was transiently expressed in 293T cells as described in the legend to Fig. 1. The cells were stained with serum from a convalescent SARS patient, and staining was analyzed by FACS. Results of a representative experiment are shown; similar results were obtained in an independent experiment. (B) Neutralization of S-bearing pseudotypes by sera from SARS patients. Pseudotypes bearing the VSV G or SARS-CoV S protein, which had been normalized to equal luciferase activity upon infection of target cells in the absence of serum, were preincubated for 30 min with serial dilutions of sera from convalescent SARS patients or control serum from a healthy donor. The virus-antibody mixture was incubated with Huh-7 cells for 7 h followed by complete removal of the culture medium. After 72 h, luciferase activity was determined in cell extracts. Each experiment was performed in triplicate; results of one out of two experiments are shown.
DISCUSSION
A novel CoV has been identified as the cause of SARS (16, 31, 53). In order to understand SARS pathogenesis and to develop therapies and vaccines, it is necessary to analyze the function of SARS-CoV proteins important for viral replication. The S protein of CoVs is inserted into the viral envelope and mediates fusion of the viral and cellular membranes, a process essential for productive infection (20). Moreover, S is a determinant of viral pathogenesis (20, 55, 64). We investigated SARS-CoV S function employing lentiviral pseudotypes, i.e., replication-defective lentiviral particles competent for a single round of infection mediated by SARS-CoV S. Pseudotypes bearing SARS-CoV S infected kidney- and liver-derived cell lines with high efficiency, and the latter were also susceptible to infectious SARS-CoV, suggesting that these organs might be infected by SARS-CoV in vivo. Inhibition of vacuolar acidification strongly reduced infection by SARS-CoV S-bearing pseudotypes, indicating that low pH is required for S-driven infectious entry. Infection by SARS-CoV S-harboring pseudotypes was blocked by SARS patient sera, indicating that S is targeted by neutralizing antibodies during SARS-CoV infection. In summary, we describe a high-throughput system to study SARS-CoV S function. Our results indicate that SARS-CoV might have a broader tropism than initially appreciated and that the neutralizing antibody response could play an important role in clearing the infection.
The assembly and release of progeny HIV virions are believed to occur at the plasma membrane in most infected cells; however, budding into intracellular compartments has also been documented (1). Therefore, the successful generation of HIV-based pseudotypes requires that the GP of interest be expressed at the plasma membrane, where it can be nonspecifically incorporated into viral particles. Expression of SARS-CoV S with a standard plasmid (pcDNA3.1) proved unsuccessful; however, this problem was circumvented by employing the pCAGGS vector for S expression. Upon pCAGGS-driven expression an intron is incorporated upstream of the S transcript. Under these conditions, SARS-CoV S could be detected on the cell surface and in the lysates of transfected cells. Enhanced expression due to the presence of an intron has been reported for other human GPs (6, 28, 29). It has therefore been suggested that the association of certain RNAs with the cellular splicing machinery is necessary for efficient nuclear export or RNA stability or to prevent aberrant splicing events (28, 29).
SARS is characterized by a severe pneumonia, and pathological changes in the lung are readily detectable in SARS patients. Thus, among other manifestations, hemorrhagic inflammation of pulmonary alveoli, proliferation of lung epithelial cells, and an influx of macrophages into alveoli have been documented (36, 46). Moreover, infected cells have been identified in lung tissue (31, 37, 46). SARS is therefore mainly recognized as a respiratory disease. However, it has been reported that SARS pathogenesis is not limited to alveolar damage. Notably, several reports indicate that one prominent pathological change associated with SARS is altered liver function (12, 17, 36, 53, 65). Hereby, the levels of liver-specific enzymes like alanine aminotransferase were found to be elevated in SARS patients, especially upon the manifestation of severe disease. But how does SARS-CoV infection impact liver function? Here, we present evidence that SARS-CoV S mediates efficient entry into hepatoma cell lines and that SARS-CoV can productively infect these cells, suggesting that infection of hepatocytes in vivo might contribute to alterations in liver function. Systematic analysis of liver tissue from SARS patients is therefore required to investigate if SARS-CoV targets hepatocytes in SARS patients and if infection correlates with dysregulation of liver metabolism. In situ analysis of infected cells in autopsy tissues from SARS patients indeed demonstrated the presence of viral RNA in hepatocytes near the central vein, further substantiating the speculation that SARS-CoV infects liver cells in vivo (75).
Similarly to the S-mediated entry into hepatoma cell lines, the efficient infection of kidney-derived cell lines by S-bearing pseudotypes suggests that pathological changes in the kidney, which can be associated with SARS (12, 16, 31), might at least in part be attributed to productive infection of kidney cells. Indeed, the replication of SARS-CoV in a macaque kidney cell line in vitro and the successful reisolation of SARS-CoV from kidney tissue from infected patients (16, 31) suggest that the virus replicates in these cells in vivo. In contrast, the inability of SARS-CoV S-bearing pseudotypes to target B- and T-cell lines indicates that these cells might be resistant to SARS-CoV infection in vivo, due to either the absence of a functional receptor or differential intracellular transport of virus particles, leading to nonproductive infection. In agreement with the first hypothesis, ACE2 expression in lymphoid tissues was found to be limited (24). Consequently, infection of T or B cells might not contribute to the pronounced lymphopenia observed in SARS patients (10, 37, 53, 71; D. A. Fisher, T. K. Lim, Y. T. Lim, K. S. Singh, and P. Tambyah, Letter, Lancet 361:1740, 2003; N. S. Panesar, Letter, Lancet 361:1985, 2003). Nevertheless, evidence for SARS-CoV replication in peripheral blood mononuclear cells has been reported, and the potential SARS-CoV infection of lymphoid cells in vivo requires further analysis. In summary, SARS-CoV S-bearing pseudotypes exhibit a differential tropism for human cell lines, and the efficient infection of liver- and kidney-derived cell lines may have implications for SARS pathogenesis.
Enveloped viruses employ two strategies to enter target cells. One involves GP-mediated engagement of receptors on the cell surface, which triggers fusion of the viral and cellular membranes (13). Alternatively, receptor engagement can trigger endocytosis followed by transport into an endocytic compartment, where the acid pH triggers the fusion activity in GP (59). Inhibitors of vacuolar acidification are well-established tools to assess if fusion by a viral GP is pH dependent (14, 15, 54). Treatment with ammonium chloride or bafilomycin A1, an inhibitor of vacuolar proton pumps (4, 69), demonstrated that SARS-CoV S-mediated infection of the kidney-derived cell line 293T and the hepatoma cell line Huh-7 depends on low pH. In contrast, low pH was not required for SARS-CoV S-mediated cell-cell fusion (38, 74); however, it has not been investigated if low pH can enhance fusion. The discrepancy between these findings could be attributed to the differences in the experimental systems used. Thus, overexpression of both S and the ACE2 receptor, which is required to obtain efficient cell-cell fusion in conjunction with the large interacting membrane surfaces during cell-cell fusion, might provide an optimized environment in which even inefficient membrane fusion events could be visualized. In contrast, endogenous expression of receptor on target cells and the limited interacting membrane surfaces can be important constraints of infectious entry of viral particles. Moreover, cell-type-specific differences might play a role. For example, herpes simplex virus GPs are well known to drive cell-to-cell fusion; nevertheless, entry into some cell lines has been shown to be pH dependent (47, 62). Assessment of the pH dependence of cellular entry of SARS-CoV particles is therefore required to define the trigger of S-mediated membrane fusion.
One of the major questions in SARS is how the immune system manages to control the infection in the majority of patients, while about 10% develop a severe form of the disease with a usually lethal outcome. The humoral and cellular immune response in CoV infections is mainly directed against the viral S protein (8, 63, 70). SARS patients usually also develop a readily detectable antibody response (16, 31, 53) which is at least in part directed against S, M, and N (7, 27, 68); however, its neutralizing ability is largely unknown. In order to gain insight into that question, we analyzed a rabbit serum raised against purified SARS-CoV particles. This serum specifically neutralized SARS-CoV S-mediated infection, demonstrating that the S protein of SARS-CoV can also be targeted by neutralizing antibodies. Furthermore, sera from convalescent SARS patients inhibited SARS-CoV S- but not VSV G-mediated infection with high efficiency, indicating the production of S-specific neutralizing antibodies. The humoral immune response might therefore play an important role in controlling and clearing SARS-CoV infection. Moreover, the S protein in its native conformation might be a suitable candidate for vaccine approaches. Indeed, immunization of macaques with adenoviruses coding for S, M, and N triggered the production of neutralizing antibodies, providing at least some indirect proof that S-based vaccines hold promise (21).
In summary, our results provide novel insights into the range of target cells susceptible to SARS-CoV S-driven infection in vitro and argue for a role for low pH in triggering S-driven membrane fusion during infectious viral entry. Moreover, we show that SARS patients generate S-specific neutralizing antibodies, arguing for a role of the humoral immune response in clearing the infection. Pseudotyped viruses proved to be a suitable tool to study SARS-CoV S function and will be highly useful for many applications, e.g., high-throughput screening for inhibitors of SARS-CoV S-mediated entry under standard biosafety conditions. However, one needs to take into account that the efficiency of S incorporation into the viral envelope might differ between pseudotypes and SARS-CoV particles. Future studies are therefore required to determine how neutralization data with pseudotypes compare to those obtained with replication-competent SARS-CoV. Moreover, the SARS-CoV M and E proteins are likely incorporated into the viral envelope and might also be targeted by the humoral immune response. Indeed, antibodies directed against M have been shown to elicit neutralizing activity in the presence of complement (18). The development of a reporter system allowing the assessment of S, M, and E function will thus be an important and challenging task.
Acknowledgments
We thank B. Fleckenstein and J. Behrens for encouragement and support. We are grateful to H. W. Doerr, H. Rabenau, and W. Lim for providing SARS-CoV strains and viral RNA; N. Landau for pNL4-3-Luc-R−E−; H. Walter for the gift of serum samples from healthy individuals; N. Finze for p24 ELISA; T. C. Pierson and R. F. Siliciano for the gift of NL4-3-GFP; J. B. Bosch and P. Rottier for comments; and T. C. Pierson for critically reading the manuscript. We are especially indebted to G. Simmons for communicating unpublished results, for providing pCAGGS, and for not giving us TL.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB466) to H.H., A.M., T.G., M.G., and S.P. and by a grant from the BMBF (DLR: OIGC0101) to K.Ü.
REFERENCES
- 1.Amara, A., and D. R. Littman. 2003. After Hrs with HIV. J. Cell Biol. 162:371-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos, E. C., L. Heijnen, W. Luytjes, and W. J. Spaan. 1995. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology 214:453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch, B. J., R. Van Der Zee, C. A. De Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman, E. J., A. Siebers, and K. Altendorf. 1988. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA 85:7972-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchmeier, M. J., H. A. Lewicki, P. J. Talbot, and R. L. Knobler. 1984. Murine hepatitis virus-4 (strain JHM)-induced neurologic disease is modulated in vivo by monoclonal antibody. Virology 132:261-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman, B. S., R. M. Thayer, K. A. Vincent, and N. L. Haigwood. 1991. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 19:3979-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Che, X. Y., W. Hao, L. W. Qiu, Y. X. Pan, Z. Y. Liao, H. Xu, J. J. Chen, J. L. Hou, P. C. Woo, S. K. Lau, Y. Y. Kwok, and Z. Huang. 2003. Antibody response of patients with severe acute respiratory syndrome (SARS) to nucleocapsid antigen of SARS-associated coronavirus. Di Yi Jun Yi Da Xue Xue Bao 23:637-639. [PubMed] [Google Scholar]
- 8.Collins, A. R., R. L. Knobler, H. Powell, and M. J. Buchmeier. 1982. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology 119:358-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 10.Cui, W., Y. Fan, W. Wu, F. Zhang, J. Y. Wang, and A. P. Ni. 2003. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin. Infect. Dis. 37:857-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Groot, R. J., R. W. Van Leen, M. J. Dalderup, H. Vennema, M. C. Horzinek, and W. J. Spaan. 1989. Stably expressed FIPV peplomer protein induces cell fusion and elicits neutralizing antibodies in mice. Virology 171:493-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, Y., H. Wang, H. Shen, Z. Li, J. Geng, H. Han, J. Cai, X. Li, W. Kang, D. Weng, Y. Lu, D. Wu, L. He, and K. Yao. 2003. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 200:282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276:229-237. [DOI] [PubMed] [Google Scholar]
- 14.Drose, S., and K. Altendorf. 1997. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 200:1-8. [DOI] [PubMed] [Google Scholar]
- 15.Drose, S., K. U. Bindseil, E. J. Bowman, A. Siebers, A. Zeeck, and K. Altendorf. 1993. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32:3902-3906. [DOI] [PubMed] [Google Scholar]
- 16.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. M. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. M. E. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 17.Duan, Z. P., Y. Chen, J. Zhang, J. Zhao, Z. W. Lang, F. K. Meng, and X. L. Bao. 2003. Clinical characteristics and mechanism of liver injury in patients with severe acute respiratory syndrome. Zhonghua Gan Zang Bing Za Zhi 11:493-495. (In Chinese.) [PubMed] [Google Scholar]
- 18.Fleming, J. O., R. A. Shubin, M. A. Sussman, N. Casteel, and S. A. Stohlman. 1989. Monoclonal antibodies to the matrix (E1) glycoprotein of mouse hepatitis virus protect mice from encephalitis. Virology 168:162-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fouchier, R. A. M., T. Kuiken, M. Schutten, G. van Amerongen, J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. M. E. Osterhaus. 2003. Aetiology—Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, W., A. Tamin, A. Soloff, L. D'Aiuto, E. Nwanegbo, P. D. Robbins, W. J. Bellini, S. Barratt-Boyes, and A. Gambotto. 2003. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet 362:1895-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerberding, J. L. 2003. Faster… but fast enough? Responding to the epidemic of severe acute respiratory syndrome. N. Engl. J. Med. 348:2030-2031. [DOI] [PubMed] [Google Scholar]
- 23.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 24.Harmer, D., M. Gilbert, R. Borman, and K. L. Clark. 2002. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 532:107-110. [DOI] [PubMed] [Google Scholar]
- 25.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helenius, A., J. Kartenbeck, K. Simons, and E. Fries. 1980. On the entry of Semliki Forest virus into BHK-21 cells. J. Cell Biol. 84:404-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, T. Y., S. L. Wu, S. E. Cheng, Y. C. Wei, S. P. Huang, and C. Y. Hsiang. 2004. Antigenicity and receptor-binding ability of recombinant SARS coronavirus spike protein. Biochem. Biophys. Res. Commun. 313:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, M. T., and C. M. Gorman. 1990. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 18:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, M. T., and C. M. Gorman. 1990. The simian virus 40 small-t intron, present in many common expression vectors, leads to aberrant splicing. Mol. Cell. Biol. 10:1805-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. X. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. Leduc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 32.Kuiken, T., R. A. M. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. S. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. M. Peiris, and A. D. M. E. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviruses, p. 1163-1185. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 36.Lang, Z., L. Zhang, S. Zhang, X. Meng, J. Li, C. Song, L. Sun, and Y. Zhou. 2003. Pathological study on severe acute respiratory syndrome. Chin. Med. J. 116:976-980. [PubMed] [Google Scholar]
- 37.Lee, N., D. Hui, A. Wu, P. Chan, P. Cameron, G. M. Joynt, A. Ahuja, M. Y. Yung, C. B. Leung, K. F. To, S. F. Lui, C. C. Szeto, S. Chung, and J. J. Sung. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986-1994. [DOI] [PubMed] [Google Scholar]
- 38.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo, Z., and S. R. Weiss. 1998. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology 244:483-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maeda, J., A. Maeda, and S. Makino. 1999. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffin, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. E. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 42.McClure, M. O., M. Marsh, and R. A. Weiss. 1988. Human immunodeficiency virus infection of CD4-bearing cells occurs by a pH-independent mechanism. EMBO J. 7:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClure, M. O., M. A. Sommerfelt, M. Marsh, and R. A. Weiss. 1990. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71:767-773. [DOI] [PubMed] [Google Scholar]
- 44.Nakanaga, K., K. Yamanouchi, and K. Fujiwara. 1986. Protective effect of monoclonal antibodies on lethal mouse hepatitis virus infection in mice. J. Virol. 59:168-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen, V. P., and B. G. Hogue. 1997. Protein interactions during coronavirus assembly. J. Virol. 71:9278-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicholls, J. M., L. L. M. Poon, K. C. Lee, W. F. Ng, S. T. Lai, C. Y. Leung, C. M. Chu, P. K. Hui, K. L. Mak, W. Lim, K. W. Yan, K. H. Chan, N. C. Tsang, Y. Guan, K. Y. Yuen, and J. S. M. Peiris. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361:1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicola, A. V., A. M. McEvoy, and S. E. Straus. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 49.Normile, D., and M. Enserink. 2003. SARS in China—tracking the roots of a killer. Science 301:297-299. [DOI] [PubMed] [Google Scholar]
- 50.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Opstelten, D. J., P. de Groote, M. C. Horzinek, H. Vennema, and P. J. Rottier. 1993. Disulfide bonds in folding and transport of mouse hepatitis coronavirus glycoproteins. J. Virol. 67:7394-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Opstelten, D. J., M. J. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez, L., and L. Carrasco. 1994. Involvement of the vacuolar H+-ATPase in animal virus entry. J. Gen. Virol. 75:2595-2606. [DOI] [PubMed] [Google Scholar]
- 55.Phillips, J. J., M. M. Chua, G. F. Rall, and S. R. Weiss. 2002. Murine coronavirus spike glycoprotein mediates degree of viral spread, inflammation, and virus-induced immunopathology in the central nervous system. Virology 301:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pierson, T. C., Y. Zhou, T. L. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. X. Tong, A. Tamin, L. Lowe, M. Frace, J. L. Derisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 58.Siddell, S. G. 1995. The Coronaviridae. Plenum Press, New York, N.Y.
- 59.Sieczkarski, S. B., and G. R. Whittaker. 2002. Dissecting virus entry via endocytosis. J. Gen. Virol. 83:1535-1545. [DOI] [PubMed] [Google Scholar]
- 60.Simmons, G., J. D. Reeves, C. C. Grogan, L. H. Vandenberghe, F. Baribaud, J. C. Whitbeck, E. Burke, M. J. Buchmeier, E. J. Soilleux, J. L. Riley, R. W. Doms, P. Bates, and S. Pohlmann. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115-123. [DOI] [PubMed] [Google Scholar]
- 61.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stohlman, S. A., C. C. Bergmann, R. C. van der Veen, and D. R. Hinton. 1995. Mouse hepatitis virus-specific cytotoxic T lymphocytes protect from lethal infection without eliminating virus from the central nervous system. J. Virol. 69:684-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taguchi, F., H. Kubo, H. Takahashi, and H. Suzuki. 1995. Localization of neurovirulence determinant for rats on the S1 subunit of murine coronavirus JHMV. Virology 208:67-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tong, Y. W., C. B. Yin, X. P. Tang, and W. D. Jia. 2003. Changes of liver function in patients with serious acute respiratory syndrome. Zhonghua Gan Zang Bing Za Zhi 11:418-420. (In Chinese.) [PubMed] [Google Scholar]
- 66.Tooze, J., S. Tooze, and G. Warren. 1984. Replication of coronavirus MHV-A59 in sac− cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 33:281-293. [PubMed] [Google Scholar]
- 67.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, J., J. Wen, J. Li, J. Yin, Q. Zhu, H. Wang, Y. Yang, E. Qin, B. You, W. Li, X. Li, S. Huang, R. Yang, X. Zhang, L. Yang, T. Zhang, Y. Yin, X. Cui, X. Tang, L. Wang, B. He, L. Ma, T. Lei, C. Zeng, J. Fang, J. Yu, J. Wang, H. Yang, M. B. West, A. Bhatnagar, Y. Lu, N. Xu, and S. Liu. 2003. Assessment of immunoreactive synthetic peptides from the structural proteins of severe acute respiratory syndrome coronavirus. Clin. Chem. 49:1989-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Werner, G., H. Hagenmaier, H. Drautz, A. Baumgartner, and H. Zahner. 1984. Metabolic products of microorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J. Antibiot. (Tokyo) 37:110-117. [DOI] [PubMed] [Google Scholar]
- 70.Williamson, J. S., and S. A. Stohlman. 1990. Effective clearance of mouse hepatitis virus from the central nervous system requires both CD4+ and CD8+ T cells. J. Virol. 64:4589-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong, R. S., A. Wu, K. F. To, N. Lee, C. W. Lam, C. K. Wong, P. K. Chan, M. H. Ng, L. M. Yu, D. S. Hui, J. S. Tam, G. Cheng, and J. J. Sung. 2003. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ 326:1358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.World Health Organization. 2003. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. www.who.int/csr/sars/country/table2003_09_23/en/print.html/html.
- 73.World Health Organization. 2003. WHO-recommended measures for persons undertaking international travel from areas affected by severe acute respiratory syndrome (SARS). Wkly. Epidemiol. Rec. 78:97-120. [PubMed] [Google Scholar]
- 74.Xiao, X., S. Chakraborti, A. S. Dimitrov, K. Gramatikoff, and D. S. Dimitrov. 2003. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 312:1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, Q. L., Y. Q. Ding, J. L. Hou, L. He, Z. X. Huang, H. J. Wang, J. J. Cai, J. H. Zhang, W. L. Zhang, J. Geng, X. Li, W. Kang, L. Yang, H. Shen, Z. G. Li, H. X. Han, and Y. D. Lu. 2003. Detection of severe acute respiratory syndrome (SARS)-associated coronavirus RNA in autopsy tissues with in situ hybridization. Di Yi Jun Yi Da Xue Xue Bao 23:1125-1127. (In Chinese.) [PubMed] [Google Scholar]