Abstract
Murine gammaherpesvirus 68 (MHV-68) is genetically related to the human gammaherpesviruses, Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) and Epstein-Barr virus (EBV). It has been proposed as a model for gammaherpesvirus infection and pathogenesis. Open reading frame 31 (ORF31) is conserved among the Beta- and Gammaherpesvirinae subfamily, and there is no known mammalian homologue of this protein. The function of MHV-68 ORF31 and its viral homologues has not yet been determined. We described here a primary characterization of this protein and its requirement for lytic replication. The native MHV-68 ORF31 was detected at peak levels by 24 h postinfection, and the FLAG-tagged and green fluorescent protein fusion ORF31 were localized in the cytoplasm and nucleus in a diffuse pattern. Two independent experimental approaches were then utilized to demonstrate that ORF31 was required for lytic replication. First, small interfering RNA generated against ORF31 expression blocked protein expression and virus production in transfected cells. Then, two-independent bacterial artificial chromosome-derived ORF31-null MHV-68 mutants (31STOP) were generated and found to be defective in virus production in fibroblast cells. This defect can be rescued in trans by MHV-68 ORF31 and importantly by its KSHV homologue. A repair virus of 31STOP was also generated by homologous recombination in fibroblast cells. Finally, we showed that the defect in ORF31 blocked late lytic protein expression. Our results demonstrate that MHV-68 ORF31 is required for viral lytic replication, and its function is conserved in its KSHV homologue.
Members of the Gammaherpesvirinae subfamily are known for their ability to establish latent infection in lymphocytes and to cause acute infection in epithelial and fibroblast cells. This subfamily includes Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8), Epstein-Barr virus (EBV), and murine gammaherpesvirus 68 (MHV-68). KSHV and EBV are associated with several types of human diseases. KSHV is linked to Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma (7, 23, 33). EBV is associated with nasopharyngeal carcinoma, Burkitt's lymphoma, Hodgkin's disease, posttransplant lymphoproliferative disorder-like lymphoma, and certain types of T-cell lymphomas (29).
MHV-68, also referred to as γHV68, is a natural pathogen of wild rodents (4, 22, 28). The complete nucleotide sequence of MHV-68 has been determined and is similar to EBV but is more closely related to KSHV. The genome of MHV-68 encodes genes that are common to other members of the gammaherpesvirus family, as well as cellular gene homologues and genes unique to MHV-68 (19, 37). Unlike KSHV and EBV, MHV-68 establishes productive infections in a variety of fibroblast and epithelial cell lines and is capable of infecting laboratory mice, simplifying both in vitro and in vivo study of this gammaherpesvirus (10, 35). The study of conserved genes in MHV-68 will significantly contribute to the understanding of their roles in viral replication and in the pathogenesis of human gammaherpesviruses.
MHV-68 open reading frame 31 (ORF31) homologues have been found in gammaherpesviruses, EBV (BDLF4), KSHV (ORF31), and herpesvirus saimiri (ORF31), as well as in betaherpesviruses, human cytomegalovirus (UL92), and human herpesvirus 6 (U63). However, no homologue of ORF31 has been found in alphaherpesviruses or in mammalian genomes. The putative gene products of the ORF31 homologues are similar in size (200 to 225 amino acids), although their function is unknown (2, 3, 8, 16, 30, 37). The N terminus of these proteins contains six conserved cysteine and histidine residues that might form a zinc-binding domain, which is the only conserved domain apparent from sequence analysis. This conservation of ORF31 between two subfamilies of herpesviruses strongly suggests it plays an important role in the herpesvirus life cycle.
To investigate the role of ORF31 in viral replication, we decided to generate an ORF31-null MHV-68 mutant. In an initial effort to generate the ORF31-null mutant, we were unable to purify the mutant lacking ORF31 with the classical homologous recombination method that we have used to recover several other MHV-68 mutants (38; unpublished data). The inability to purify the ORF31-null mutant suggested that mutants lacking ORF31 are impaired for growth. To determine the role of ORF31 in MHV-68 replication, we chose to create a recombinant bacterial artificial chromosome (BAC) of MHV-68 from which an ORF31-null genome could be derived without the need for purification from wild-type (wt) MHV-68 in mammalian cells.
Recently, the cloning of several herpesviruses, including human cytomegalovirus, herpes simplex virus, pseudorabies virus, EBV, and KSHV, as a BAC has been described (5, 9, 13, 20, 21, 31, 32, 36, 40). BAC clones have also been reported for MHV-68, and several MHV-68 virus mutants have been generated successfully using the MHV-68 BAC system (1, 25, 27). Here we report the analysis of MHV-68 ORF31 protein expression and the generation and characterization of an ORF31-null MHV-68(BAC). Our results suggest that ORF31 is required for lytic viral replication.
MATERIALS AND METHODS
Viruses, cells, and plaque assays.
wt MHV-68 was originally obtained from the American Type Culture Collection (ATCC; VR1465). The working wt MHV-68 virus stock was generated by infecting BHK-21 cell (a baby hamster kidney fibroblast cell line) (ATCC CCL-10) at a multiplicity of infection (MOI) of 0.05 PFU per cell. The T-Rex 293 cell line (Invitrogen) is a human embryonic epithelial cell line that stably expresses the Tet repressor. BHK-21 cells were cultured in complete Dulbecco modification of Eagle medium (DMEM) containing 10% fetal bovine serum, supplemented with penicillin and streptomycin. T-Rex 293 cells were maintained in complete DMEM containing blasticidin (5 μg/ml). Plaque assay was performed in monolayers of BHK-21 cells overlaid with 1% methylcellulose as described previously (39).
DNA extraction and Southern blot analysis.
T-Rex-31 cells were seeded onto 24-well plates. Protein expression was induced by adding doxycycline (1 μg/ml) for 12 to 15 h prior to infection. The induced cells were then incubated with various viruses at an MOI of 4 for 1 h. Doxycycline was added and maintained in the culture postinfection (p.i.) until harvesting DNA. Total cellular DNA was isolated at 24 h p.i. Briefly, the infected cells were harvested and washed once with phosphate-buffered saline (PBS). The pellet was resuspended in Tris-EDTA buffer (10 mM Tris-HCl-1 mM EDTA [pH 8.0]). An equal volume of 2× lysis buffer (0.5% sodium dodecyl sulfate [SDS], 40 mM Tris-HCl, 20 mM EDTA, 200 mM NaCl) containing proteinase K (0.5 mg/ml) was added, and the suspension incubated at 56°C for at least 6 h. The sample was extracted twice with phenol-chloroform and once with chloroform-isoamyl alcohol (24:1). The resulting DNA was precipitated and dissolved in Tris-EDTA buffer. BAC plasmid DNA was isolated from DH10B Escherichia coli cultures by using a Qiagen Plasmid Midi kit (Qiagen, Inc., Valencia, Calif.) according to the manufacturer's modified protocol. For Southern blot analysis, DNAs were subjected to either BamHI or BglII digestion overnight and separated on 0.8% agarose gels. Gels were subjected to depurination, denaturation, and neutralization. DNAs on treated gels were transferred to charged nylon membranes (Amersham Pharmacia Biotech). The membranes were UV cross-linked and prehybridized at 65°C in the buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10× Denhardt solution, 0.5% SDS, and denatured salmon sperm DNA (50 μg/ml). Probes were generated by random priming method with [α-32P]dCTP and genomic viral DNA fragment as templates. For the probe used for Fig. 4C, the genomic viral DNA fragment was a PCR product (nucleotides [nt] 46264 to 50036) amplified with primers 31-11 and 31-12 (Table 1). For the probe used for Fig. 6B, an EcoRV-XbaI fragment (nt 46420 to 49951) of the MHV-68 genomic DNA was used as a template. The membranes were washed at 65°C in 2× SSC with 0.1% SDS, followed by 0.1× SSC with 0.1% SDS. Radioactivity was detected and quantitated by using a Storm imaging system (Molecular Dynamics, Sunnyvale, Calif.).
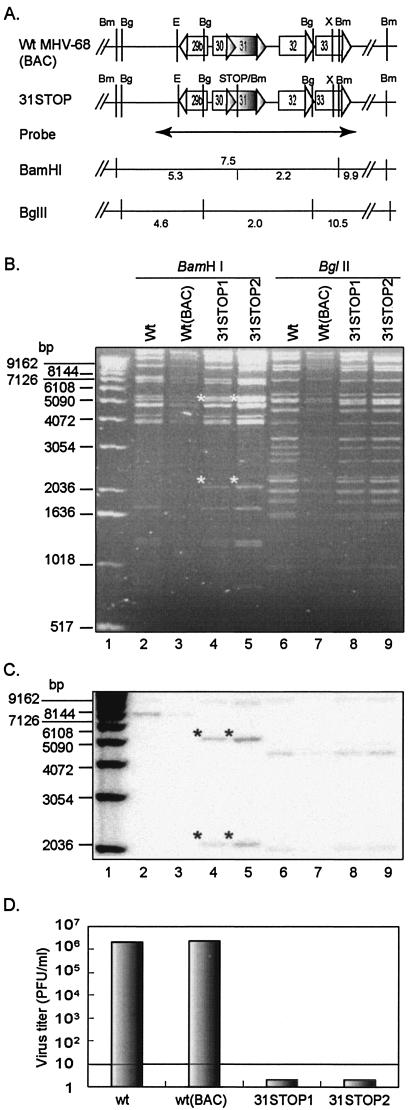
FIG.4.
Analysis of recombinant MHV-68(BAC) DNAs. (A) Schematic representation of the structure of ORF31 and its flanking ORFs, the predicted restriction enzyme sites of BamHI (Bm), BglII (Bg), EcoRV (E), and XbaI (X), position of a probe (nt 46264 to 50036) and the predicted restriction fragments detected by the probe in Southern blotting analysis. Predicted fragments of BamHI-digests from wt MHV-68(BAC) are indicated above the line, and those from 31STOP are below the line. (B) Restriction enzyme digestion of the recombinant MHV-68(BAC) DNA. wt MHV-68 viral DNA (wt) was isolated from extracellular virions, and wt MHV-68(BAC) and 31STOP DNAs were isolated from E. coli strain DH10B transformed with BAC plasmids. Two clones of the ORF31-null MHV-68(BAC) (31STOP1 and 31STOP2) were included. DNAs were digested with BamHI or BglII, as indicated at the top of the panel, and then visualized by electrophoresis and ethidium bromide staining. The expected bands of 2.1 and 5.3 kb derived from BamHI digestion of 31STOP plasmid are indicated by asterisks at the upper left corners of the bands. (C) The DNAs shown in panel B were blotted and hybridized with the [α32P]dCTP-labeled probe shown in panel A. The expected bands derived from 31STOP BamHI digests are indicated by asterisks at the upper left corners of the bands. (D) Deficiency of infectious viral production by ORF31-null MHV-68(BAC). BHK-21 cells were transfected with wt MHV-68 virion DNA or wt MHV-68(BAC), 31STOP1, or 31STOP2 plasmid DNA. Supernatant was collected at 5 days [wt and wt(BAC)] or 7 days (31STOP1 and 31STOP2) p.i. Virus titer was determined by plaque assay in BHK-21 cells in duplicate. The detection level by plaque assay was 10 PFU/ml.
TABLE 1.
Nucleotide sequences of primers used to generate recombinant plasmids
| Primer | Sequence (5′ to 3′)a | Genomic positionb (nt) |
|---|---|---|
| 31-01 | GGCGGATCCGTATAAAAGATGCAAGCG | 47713 to 47729 |
| 31-02 | GGAGGAGCGGCCGCTTCTAAATACATCCATTTC | 48309 to 48291 |
| 31-03 | TTGGGTACCGTATAAAAGATGCAAGCG | 47713 to 47729 |
| 31-04 | AAGGCAGGATCCTTATTCTAAATACATCC | 48312 to 48296 |
| 31-05 | GGGCGGATCCACCATGGACTACAAAGACGATGACGACAAGATGTATAAAAGATGCAAGc | 47710 to 47727 |
| 31-06 | GAAGGAGCGGCCGCTTATTCTAAATACATCC | 48312 to 48296 |
| 31-07 | TTGGGTACCATGTATAAAAGATGCAAGCG | 47710 to 47729 |
| 31-08 | GCAGGATCCTTATTCTAAATACATCC | 48312 to 48296 |
| 31-09 | CCCTCCGGAATGTCACAAAACAGAAAG | 50763 to 50780 (K) |
| 31-10 | GAGGATCCCTACGTATCTTTCGTTG | 51437 to 51421 (K) |
| 31-11 | GTATGGAGCATATAAGTGC | 46264 to 46282 |
| 31-12 | GAACCAACAACTGGCAGGTC | 50036 to 50017 |
| 31-13 | CCGATGCATGTGATTGCGCTCTGTTTG | 47272 to 47289 |
| 31-14 | TTTGGATCCTCAATTAATCAAGCACGTTATAGGCCTCTGd | 47791 to 47774 |
| 31-15 | GTGCTTGATTAATTGAGGATCCAAAATTTGTACCAGTGTCd | 47792 to 47809 |
| 31-16 | GGTGTGCATGCGTTGGCATAGATTGAC | 48213 to 48198 |
Restriction enzyme sites are in italics; sequences that are present in MHV-68/KSHV genome are underlined.
Genomic position refers to the position in the published sequence of the MHV-68 (U97553) or KSHV genome (U75698) (30, 37). K, position in the KSHV genome.
The FLAG tag coding sequence is indicated in bold face.
The sequence of the stop codon is indicated in bold face.
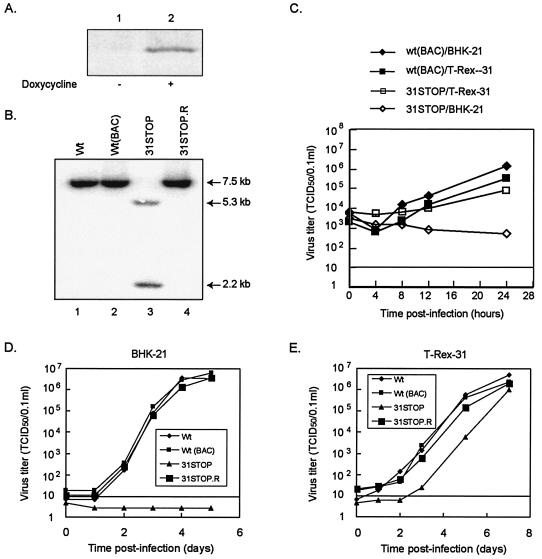
FIG. 6.
Propagation of MHV-68 mutant viruses in an ORF31 complementing cell line. (A) A complementing cell line (T-Rex-31) was established by using a Tet-on inducible mammalian expression system. ORF31 expression was induced by adding doxycycline (1 μg/ml) for 24 h. Lysate from noninduced (−) or induced (+) cells was analyzed by Western blot with anti-FLAG monoclonal antibody. (B) Southern blotting analysis of MHV-68 recombinant viral DNA. The doxycycline-induced T-Rex-31 cells were infected with wt MHV-68, wt MHV-68(BAC), ORF31-null (31STOP), or revertant of ORF31-null (31STOP.R) viruses at an MOI of 4. Total cellular DNA was isolated at 24 h p.i. and subjected to Southern blotting with an [α-32P]dCTP-labeled probe to an EcoRV-XbaI fragment (nt 46420 to 49951) of the MHV-68 genomic sequence. Hybridizing bands are indicated by arrows. (C) Single-step growth curves of MHV-68 mutants. BHK-21 (diamonds) or T-Rex-31 (squares) cells were infected with wt MHV-68(BAC) (solid) or 31STOP (open) at MOIs of 4. The input virus was quantitated by serial dilution in T-Rex-31 cells for virus titer and by plaque assay in BHK-21 cells to determine the presence of 31STOP revertant; none was detected. The whole culture (including cells and supernatant) was harvested at the indicated times and then quantitated by serial dilution in T-Rex-31. TCID50 was scored at 7 days p.i. (D and E) Multiple-step growth curves of MHV-68 mutants. BHK-21 (D) or T-Rex-31 cells (E) were infected with viruses as indicated in the figure with MOIs of 0.01. The culture was harvested, and virus titers were determined as described for panel C. For both single- and multiple-step growth curves, data were the average of two independent experiments, and virus titers were determined in duplicate.
Molecular cloning.
The MHV-68 ORF31 was cloned into pET-30b(+) (Novagen, Madison, Wis.) to construct the pET-30b/ORF31 for expression of the His-tagged ORF31 in E. coli strain BL21. ORF31 coding sequence was amplified by using primers 31-01 and 31-02 (Table 1), excised by using BamHI and NotI, and cloned into the BamHI-NotI sites of the pET-30b(+).
Plasmid pFLAG-mORF31 was constructed by inserting the full-length MHV-68 ORF31 into the BamHI-KpnI sites of the vector pFLAG-CMV2 (Kodak). The inserted ORF31 sequence was amplified by PCR with primers 31-03 and 31-04 (Table 1). The plasmid pcDNA5/TO/F-ORF31 was constructed by using a similar strategy, in which the full-length ORF31 fused to the FLAG coding sequence of pFLAG-CMV2 was inserted into the BamHI-NotI sites of pcDNA5/TO (Invitrogen). The inserted ORF31 sequence was amplified by PCR with primers 31-05 and 31-06 (Table 1).
Plasmid pEGFP-m31 was constructed by inserting the full-length MHV-68 ORF31 into the KpnI-BamHI sites of pEGFP-C1 vector that is used to express a protein of interest fused to the C terminus of green fluorescent protein (GFP; Clontech). The full-length ORF31 was amplified by using primers 31-07 and 31-08 (Table 1). Plasmid pEGFP-k31 was constructed by using a similar strategy, by which the full-length KSHV ORF31 was inserted into the BspEI-BamHI sites of the pEGFP-C1 vector. The KSHV ORF31 was PCR amplified by using total DNA isolated from BC-1 cells (latently infected with KSHV) as a template and primers 31-9 and 31-10 (Table 1).
The suicide shuttle vector used for BAC mutagenesis, pGS284, was kindly provided by G. Smith and L. Enquist (Princeton University) (31). To construct the shuttle plasmid for the generation of ORF31-null MHV-68(BAC), a 1.0-kb fragment was prepared by a two-step PCR and inserted into the NsiI-SphI sites of pGS284. The 1.0-kb PCR fragment contained an insertion of triple stop codons and a BamHI site located between nt 47791 and 47792 of the viral genome and ca. 500-bp nucleotides on each side of the insertion homologous to the viral genome. Briefly, primers 31-13 and 31-14 were used for amplification of the PCR fragment 31STOPa and primers 31-15 and 31-16 for 31STOPb (Table 1). These two PCR fragments were then used as templates to generate the 31STOPc fragment, with primers 31-13 and 31-16 (Table 1). The 31STOPc was digested with NsiI and SphI, ligated with the NsiI-SphI fragment of pGS284 and then electroporated into S17λpir E. coli cells (GS111). The resulting plasmid, pGS284/31STOP, was screened by colony PCR and BamHI digestion of the PCR product to verify the insertion in the shuttle plasmid. The inserted fragment was sequenced to verify that the stop codon-BamHI site insertion was in the right position.
To construct plasmid pSVL-3500, the MHV-68 genomic sequence (nt 46264 to 50036) was amplified by PCR with the primers 31-11 and 31-12 (Table 1) and then digested with EcoRV and XbaI. The EcoRV-XbaI fragment (nt 46420 to 49951) was cloned into pSVL vector to obtain the pSVL-3500, which was used for homologous recombination to rescue the ORF31-null mutant.
The inserted DNA fragment in each construct was sequenced to confirm that there were no additional mutations, deletions, or insertions in the MHV-68 coding sequences.
Cell transfection.
Plasmid DNA was prepared with standard method by using the Qiagen Plasmid Midi kit according to the manufacturer's recommendations (Qiagen, Inc., Valencia, Calif.). For BHK-21 or NIH/3T3 cell transfection, ca. 2 × 105 cells were transferred to 24-well culture plates 1 day prior to the experiment. Plasmid DNA or BAC DNA (0.4 μg per transfection) was transfected into cells by using Lipofectamine Plus reagent (Gibco). Transfection of T-Rex 293, T-Rex-31, or 293T cells was carried out with Lipofectamine 2000 according to the manufacturer's recommendations (Gibco). Cells were usually assayed at 2 days posttransfection (p.t.) unless otherwise indicated. Transfection of 293T cells with small interfering RNA (siRNA) was performed as described previously (15).
Generation of an inducible ORF31-expressing cell line.
The 293 Tet-on cell line (T-Rex 293) was purchased from Clontech and maintained in medium containing blasticidin (5 μg/ml). The Tet-on inducible mammalian expression system allowed for regulated expression of the gene of interest by adding or removing the tetracycline or doxycycline, such that the addition of the antibiotics results in expression of the gene of interest. T-Rex 293 cells grown on a 10-cm plate were transfected with plasmid pcDNA5/TO/F-ORF31 by using Lipofectamine 2000 reagent. Two days after transfection, cells were passaged and selected for stable transformants in 10% fetal bovine serum-DMEM containing hygromycin (200 μg/ml). Medium was changed every 4 days until stable cell colonies were observed. Cell colonies that were resistant to both blasticidin and hygromycin treatment were transferred to 24-well plates and expanded. Cells from each colony were tested for basal and induced expression of ORF31 by Western blot with anti-FLAG monoclonal antibody. Clone 29, which showed minimum background and high inducible expression of ORF31, was designated T-Rex-31 and chosen for future studies. T-Rex-31 was maintained in medium containing blasticidin (5 μg/ml) and hygromycin (100 μg/ml).
Construction of wt MHV-68(BAC) plasmid.
The BAC-containing wt MHV-68 plasmid was generated in our lab (T.-T. Wu et al., unpublished data). Briefly, pBeloBAC was engineered to be flanked by two loxP sites and contain a puromycin resistance gene-expressing cassette. The resulting clone, termed pBeloBAC(loxP+Puro), was linearized between two loxP sites, blunt ended, and inserted into a 4.2-kb MHV-68 genomic fragment derived from the left end of the viral genome, without disrupting any known ORFs. This plasmid was cotransfected into BHK-21 cells with MHV-68.GFP virion DNA, which contains a GFP expression cassette inserted at the same location as pBAC(loxP+Puro) (38). A recombinant virus containing the BAC(loxP+Puro) cassette was generated by homologous recombination in BHK-21 cells and screened for non-GFP-expressing virus. Circular replication intermediates of the viral genome, called pMHV-68(BAC), was isolated from the infected BHK-21 cells and transformed into E. coli cells by electroporation (12). Restriction enzyme digestion and electrophoresis analyses showed several pMHV-68(BAC) clones with similar digestion patterns as the wt MHV-68 genomic DNA. One of these clones (clone 7) was used for the reconstitution of wt MHV-68(BAC) in BHK-21 cells and for the homologous recombination in E. coli to construct the ORF31-null MHV-68(BAC).
Construction of ORF31-null MHV-68(BAC) plasmid.
The ORF31-null MHV-68(BAC) plasmid, 31STOP, was generated by allelic exchange in E. coli, as described by Smith and Enquist (31). The donor strain was GS111 E. coli cells carrying the shuttle plasmid, pGS284/31STOP, and the recipient strain was GS500 (recA+) harboring pMHV-68(BAC). The shuttle plasmid contains a positive selection marker of ampicillin resistance gene and a negative selection marker SacB that causes toxicity to the bacteria in the presence of sucrose. A chloramphenicol resistance marker is located within the BAC sequence residing in the MHV-68 genome. Conjugation was performed by cross-streaking the donor and the recipient strains. Cointegrates were selected by growth in the presence of chloramphenicol (34 μg/ml) and ampicillin (50 μg/ml) and allowed to resolve by overnight growth in the presence of chloramphenicol alone to ensure maintenance of the pMHV-68(BAC). After resolution, negative selection against the bacteria retaining the shuttle plasmid was performed by growing bacteria in the presence of 5% sucrose and chloramphenicol on Luria-Bertani plates lacking NaCl. The resulting colonies were streaked on Luria-Bertani plates containing either chloramphenicol or ampicillin. The recombinants that had lost the integrated shuttle plasmid were chloramphenicol resistant and ampicillin sensitive. The incorporation of the stop codon in ORF31 was determined by PCR and restriction enzyme digestion screening for the insertion of the BamHI site engineered next to the stop codon.
Generation of wt MHV-68(BAC) and ORF31-null MHV-68(BAC) viruses.
wt MHV-68(BAC) was reconstituted in BHK-21 cells by transfection of the cells with pMHV-68(BAC). The ORF31-null MHV-68(BAC) mutant virus was generated in an inducible cell line expressing ORF31 (T-Rex-31) by transfection of the cells with the 31STOP BAC plasmid. Cytopathic effect (CPE) appeared at 2 days p.t. in both of the wt MHV-68(BAC)-transfected BHK-21 and of the 31STOP-transfected T-Rex-31 cells. The whole culture (including supernatant and cells) was harvested at 6 days p.t. Viruses were prepared by three rounds of freezing and thawing and clearing of cell debris by low-speed centrifugation. Working virus stocks were generated by infecting BHK-21 and T-Rex-31 cells (MOI = 0.05) for wt MHV-68(BAC) and 31STOP, respectively. Viruses were quantitated by plaque assay and serial dilution in both BHK-21 and T-Rex-31 cells.
Generation of the 31STOP MHV-68(BAC) revertant.
To generate the revertant of 31STOP, 31STOP BAC plasmid was cotransfected with pSVL-3500 into BHK-21 cells. At 3 days p.t. when CPE appeared in 30% of the cells, the supernatant containing the recombinant viruses was harvested and used for screening for the revertant of 31STOP. The revertant, termed 31STOP.R, was confirmed by Southern blot analysis of the BamHI-digested viral DNA with a probe corresponding to the genomic region encoding ORF31 and its flanking sequences (nt 46420 to 49951). This virus was purified for three rounds by limiting dilution in BHK-21 cells to obtain a homogenous virus stock. The working virus stock was prepared in BHK-21 cells at an MOI of 0.05.
Viral protein expression and replication kinetics after infection with wt or ORF31-null viruses.
Noncomplementing (BHK-21) or complementing (T-Rex-31) cells were seeded onto 12-well plates 1 day prior to infection. Since no obvious cell toxicity was observed in T-Rex-31 cells upon drug induction, doxycycline was added to the medium when T-Rex-31 cells were seeded. Cells were mock infected or infected at MOIs of 4 for the single-step growth curve, 0.01 for the multiple-step growth curve, and 0.5 for Western blotting analysis. For infection, cells were incubated with viral inocula for 1 h at 37°C. After 1 h of incubation, the inoculum was removed, the cells washed three times with PBS, and fresh medium was added to the culture. The whole culture (including cells and supernatant) was harvested at various times p.i. Virus was prepared by three rounds of freezing and thawing and clearing of cell debris by low-speed centrifugation and then quantitated by serial dilution in T-Rex-31 cells. The time point at which the culture was harvested right after removal of the inocula, followed by a wash with PBS, was considered 0 h p.i. Infected-cell lysate was analyzed by Western blotting at 2 days p.i. with polyclonal rabbit antibodies to ORF26, ORF45, and ORF65.
Antibodies, Western blotting, and fluorescence and indirect immunofluorescence assays.
The BL21 strain (ATCC 47092) of E. coli was transformed with pET-30b/ORF31, and protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The His-tagged ORF31 was confirmed by Western blotting with anti-His antibody and then purified by nickel-nitrilotriacetic acid metal affinity chromatography (Qiagen, Valencia, Calif.). The purified ORF31 protein was injected into one rabbit for antibody production (Covance Research Products, Denver, Colo.). Polyclonal antibodies to MHV-68 ORF26, ORF45, or ORF65 were generated in our lab (15). The mouse monoclonal antibodies to FLAG or His epitopes and β-actin were purchased from Sigma (St. Louis, Mo.).
For Western blotting, cell extracts were analyzed with the following primary antibodies: rabbit polyclonal antibody to ORF31 (1:200), ORF26 (1:500), ORF45 (1:500), or ORF26 (1:500) or monoclonal antibody to FLAG tag, His tag, or actin. Anti-rabbit or anti-mouse immunoglobulin G (IgG) conjugated with horseradish peroxidase (Amersham Pharmacia Biotech) was used as secondary antibody. The proteins were detected by chemiluminescence detection (ECL Plus System; Amersham Pharmacia Biotech), and the signals were detected by using a Storm imaging system (Molecular Dynamics).
For fluorescence and indirect immunofluorescence assay, BHK-21 or NIH 3T3 cells grown on 24-well plates were transfected with plasmid DNA. To examine the subcellular localization of the GFP-ORF31 fusion protein, the transfected cells were transferred at 24 h p.t. to coverslips laying on 24-well plate and then incubated for 5 h, allowing cells to spread. The cells were then fixed in 2% paraformaldehyde for 10 min, washed twice with PBS, and mounted with Prolong Antifade kit (Molecular Probes, Eugene, Oreg.). To examine the cellular localization of FLAG-tagged ORF31, an indirect immunofluorescence assay was applied. Briefly, the fixed cells were permeabilized in 0.2% Triton X-100 for 5 min on ice, blocked with 10% normal goat serum for 30 min, and incubated with anti-FLAG monoclonal antibody for 1 h, followed by incubation with goat anti-mouse IgG conjugated with Cy3 (Jackson Immunoresearch Laboratory, West Grove, Pa.) for 30 min. Cells were washed twice with PBS for 10 min between each reaction. All of the reactions except permeabilization were performed at room temperature. The fluorescent signals were visualized with a confocal laser scanning microscope (Leica).
RESULTS
Expression of MHV-68 ORF31 in mammalian cells.
To examine the expression of ORF31 in transfected cells, we constructed a plasmid that expresses MHV-68 ORF31 fused to a FLAG epitope at the N terminus (pFLAG-m31) and a plasmid that expresses ORF31 fused to the C terminus of GFP (pEGFP-m31). These plasmids were transfected into 293T cells, and protein expression was examined by Western blotting with anti-FLAG and anti-GFP monoclonal antibodies sequentially. As shown in Fig. 1A, the FLAG-tagged MHV-68 ORF31 (FLAG-m31) was expressed as an ∼29-kDa protein (lane 4). The GFP protein shifts from ∼30- and ∼34-kDa doublets (lane 3) to ∼50 and ∼54-kDa doublets (GFP-m31, lane 5), showing a difference of ∼20 kDa. This difference is close to the size of MHV-68 ORF31 predicted from its coding nucleotide sequence (22 kDa).
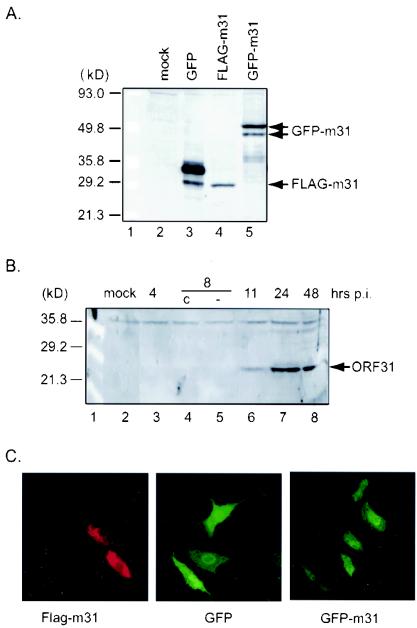
FIG. 1.
Expression and cellular localization of ORF31 in mammalian cells. (A) Western blotting analysis of ORF31 expressed from plasmid-transfected cells. 293T cells were mock transfected (lane 2) or transfected with pEGFP-C1 vector (lane 3), a plasmid expressing FLAG-tagged MHV-68 ORF31 (pFLAG-m31) (lane 4), or a plasmid expressing an MHV-68 ORF31 GFP fusion protein (pEGFP-m31) (lane 5). Cell extracts were analyzed by Western blotting with anti-GFP and sequentially with anti-FLAG antibody. Expression of FLAG-tagged ORF31 and GFP-m31 fusion proteins are indicated by arrows. (B) Western blotting analysis of the MHV-68 ORF31 protein in virus-infected cells. BHK-21 cells were infected with MHV-68 at an MOI of 5. Cell lysate was collected at the indicated times p.i. and subjected to Western blotting with rabbit polyclonal antibody against ORF31. In lanes 4 and 5, cells were treated with cycloheximide (c) or left untreated (−). The specific ORF31 signal is indicated by an arrow. (C) Cellular localization of ORF31 in plasmid-transfected cells. Cells were transfected with pFLAG-m31 (left), pEGFP-C1 (middle), or pEGFP-m31 (right) and then fixed at 24 h p.t. FLAG-tagged ORF31 was analyzed by indirect immunofluorescence assay with anti-FLAG monoclonal antibody, followed by incubation with goat anti-mouse IgG conjugated with Cy3. The fluorescence of GFP and GFP fusion protein was visualized after fixation. Images of FLAG-m31, GFP, and GFP-m31 protein were obtained with a confocal laser microscope (Leica).
To examine the expression of native ORF31 in virus-infected cells, polyclonal antibody against ORF31 was prepared in a rabbit and detected the 29-kDa protein expressed from pFLAG-m31 plasmid-transfected cells, which was similar to the FLAG-m31 detected by anti-FLAG antibody (data not shown). This polyclonal antibody was then used to examine the expression kinetics of the native ORF31 in MHV-68-infected BHK-21 cells (Fig. 1B). The antibody detected an ∼27-kDa band (∼2 kDa smaller than the FLAG-tagged ORF31), which first appeared at 11 h p.i. (lane 6) and peaked at 24 h p.i. (lane 7). The difference between the molecular masses detected by Western blotting and that predicted from its coding nucleotide sequence may be due to unknown modification(s). A background band of ∼37 kDa was also detected at all time points, indicating equal loading of the total protein.
Since there is no structural information available for ORF31, several algorithms were used to predict the secondary structure of the protein from the amino acid sequence. Both Tmpred and TMHMM algorithms predict that ORF31 contains one transmembrane helix (residues 150 to 168). According to the prediction, the N-terminal residues, amino acids 1 to 149, face the cytoplasm, and residues 150 to 168 (18 amino acids) represent the putative transmembrane helix. To address the subcellular localization of ORF31, indirect immunofluorescence analysis was performed with BHK-21 cells transiently expressing FLAG-m31. In the tagged protein, the FLAG epitope should remain in the cytoplasm as part of the intracellular domain. BHK-21 cells transfected with pFLAG-m31 were fixed and incubated with anti-FLAG antibody. After 1 h of incubation, the cells were incubated with the anti-mouse IgG-Cy3 conjugates. Detection of ORF31 with a confocal laser microscope indicated a pattern of even staining in the cytoplasm and nucleus (Fig. 1C, left panel). It should be pointed out that no intense staining was noted at the cell membrane. To verify the subcellular localization of ORF31, we examined the GFP fluorescence in NIH 3T3 cells transfected with pGFP-m31, which expresses MHV-68 ORF31 fused to the GFP protein at the N terminus. The GFP fluorescence from the GFP-m31 fusion protein was also evenly distributed between the cytoplasm and the nucleus (Fig. 1C, right panel), similar to the nonfused GFP expressed from vector (middle panel). This pattern was also observed in pEGFP-m31-transfected live cells (data not shown).
Downregulation of MHV-68 lytic replication by siRNA targeted to ORF31.
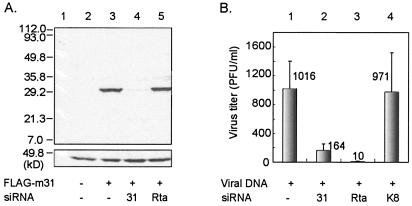
siRNAs have been widely used to inhibit expression of cellular, as well as viral genes in mammalian cells by inducing homology-dependent degradation of cognate mRNA (11, 14, 17, 26). Specific siRNAs have been used to demonstrate the essential role for viral proteins Rta and ORF45 in MHV-68 lytic replication (15). To examine whether reduction of ORF31 expression effects MHV-68 lytic replication, we designed a siRNA targeted against ORF31 (siRNA-ORF31). An siRNA targeting ORF50 that is critical for MHV-68 viral replication (18, 34, 38, 39) (siRNA-Rta) and an siRNA targeting KSHV K8 that has no homologue in MHV-68 (siRNA-K8) were used as controls. First, we examined the expression level of ORF31 in the presence of siRNA-ORF31 after cotransfection. The plasmid pFLAG-m31 was transfected into 293T cells with or without siRNA, and protein expression was analyzed by Western blotting with anti-FLAG antibody (Fig. 2A). Expression of ORF31 was significantly reduced by siRNA-ORF31 (lane 4) and, importantly, not affected by a nonspecific siRNA (siRNA-Rta, lane 5). Next, we tested MHV-68 viral replication in the presence of siRNA-ORF31. 293T cells were cotransfected with MHV-68 viral DNA and either siRNA-ORF31, siRNA-Rta, or siRNA-K8. Supernatants were collected at 48 h p.t., and virus titers were determined by plaque assay. As shown in Fig. 2B, MHV-68 viral progeny production was significantly reduced in the presence of either siRNA-ORF31 (column 2) or siRNA-Rta (column 3) but not in the presence of siRNA-K8 (column 4). Inhibition of viral replication by siRNA-Rta but not by siRNA-K8 indicates the specificity of RNA interference (RNAi), since Rta is known to play a crucial role during viral replication and KSHV K8 has no homologue in MHV-68. Reduction of virus production by siRNA-ORF31 suggests that ORF31 plays a critical role in MHV-68 replication.
FIG. 2.
Effect of siRNA targeted to ORF31 on viral protein expression and replication. (A) 293T cells were mock-transfected (lane 2) or transfected with plasmid pFLAG-m31 in the absence (lane 3) or presence of siRNA targeted to ORF31 (lane 4) or Rta (lane 5). At 48 h p.t., cell lysate was analyzed by Western blotting with anti-FLAG antibody (upper). The blot was reprobed with β-actin antibody to confirm equal loading (lower). (B) MHV-68 viral DNA was transfected into BHK-21 cells without (column 1) or with siRNA targeted against ORF31 (column 2), Rta (column 3), or nonspecific gene KSHV K8 (column 4). Supernatant was collected at 48 h p.t., and virus titers were determined by plaque assay on BHK-21 cells. The value is the average of three independent experiments. The detection sensitivity of the assay is 10 PFU/ml.
Disruption of ORF31 in an MHV-68 BAC vector.
To further study the essential role of ORF31 in MHV-68 lytic replication, we constructed an ORF31-null mutant by an allelic exchange method described previously (31). We have cloned the entire MHV-68 genome as a BAC, in which the full-length BAC sequence was inserted between nt 1838 and 1839 of the viral genome without disruption of any known gene (Fig. 3A). The insertion of the BAC sequence in this region does not affect viral growth in vitro compared to wt MHV-68 (as shown in Fig. 6D and E). Next, we sought to disrupt the MHV-68 ORF31 by inserting translation termination codons and a BamHI site between nt 47791 and 47792 of the viral genome (79 nt downstream of the translation start codon for ORF31) (Fig. 3B). The introduction of triple stop codons ensures that any translation would stop and not produce a functional gene product. The incorporation of the BamHI site next to the stop codon allowed us to screen the positive clones in bacteria and confirm the insertion. Recombinant MHV-68(BAC) clones were selected as described in Materials and Methods and analyzed by BamHI or BglII digestion, electrophoresis, and ethidium bromide staining. The predicted fragments of restriction enzyme digestion in the region of ORF31 and its flanking ORFs are shown in Fig. 4A. In BamHI digests, the fragment pattern of wt MHV-68(BAC) was similar to wt MHV-68 (Fig. 4B, lanes 2 and 3). The stop codon-BamHI site insertion in ORF31 yielded two bands of 5.3 and 2.2 kb, instead of one 7.5-kb band; these two bands were readily apparent in two clones of the 31STOP BamHI digests (lanes 4 and 5). In BglII digests compared to wt MHV-68 viral DNA fragments, a 2.9-kb band was replaced in wt MHV-68(BAC) by two additional predicted bands of 5.0 kb due to the BAC cassette insertion at the left end of viral genome (lanes 6 and 7). No other rearrangements were detected in BglII-digested 31STOP BAC DNA compared to wt MHV-68(BAC) (lanes 8 and 9). The DNA fragments around the insertion site were further analyzed by Southern blotting by using a 3.5-kb probe (nt 46264 to 50036) as shown in Fig. 4A. The result confirmed the expected pattern of hybridizing bands in the BamHI-digested wt MHV-68 viral DNA and wt MHV-68(BAC) (7.5- and 9.9-kb) versus 31STOP (2.2-, 5.3-, and 9.9-kb) (Fig. 4C, lanes 2 through 5). Consistent with the gel electrophoresis and ethidium bromide staining result, no additional or unexpected hybridizing bands were detected in BglII-digested 31STOP compared to wt MHV-68 viral DNA and MHV-68(BAC) DNA (lanes 6 through 9). We conclude that the stop codons and BamHI site have been successfully inserted into the MHV-68 ORF31.
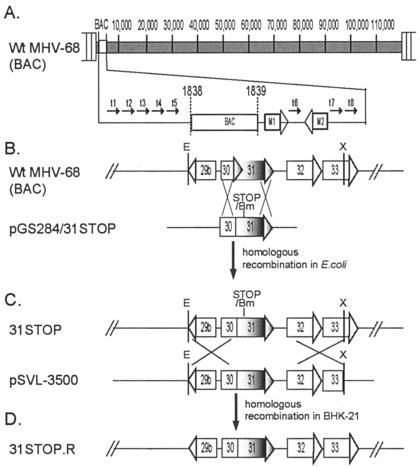
FIG. 3.
Construction of wt MHV-68(BAC), ORF31-null MHV-68(BAC), and the null revertant. (A) Representative diagram of the genomic sequence of wt MHV-68(BAC). The BAC sequence was inserted between nt 1838 and 1839 of the viral genome without disrupting any known genes. (B) ORF31 and adjacent ORFs are indicated in the wt MHV-68(BAC). Restriction enzyme sites of EcoRV (E) and XbaI (X) in the indicated DNA fragment are shown. The suicide shuttle plasmid pGS284/31STOP contains an insertion of stop codon-BamHI restriction site in the ORF31 coding sequence (between nt 47791 and 47792), and sequences flanking the insertion for homologous recombination. Homologous recombination between wt MHV-68(BAC) and pGS284/31STOP was carried out in E. coli, resulting in an ORF31-null MHV-68(BAC) plasmid, 31STOP. (C) Plasmid pSVL-3500 contains an EcoRV-XbaI fragment of the MHV-68 genomic sequence (nt 46420 to 49951) spanning the ORF31. Homologous recombination between 31STOP and pSVL-3500 was carried out in BHK-21 cells to obtain the revertant of the 31STOP mutant, 31STOP.R. (D) Schematic representation of the structure of 31STOP.R.
31STOP is deficient in viral replication.
To generate 31STOP MHV-68(BAC) virus, we transfected BHK-21 cells with 31STOP BAC plasmid, since this cell line is permissive to MHV-68 replication and is routinely used to study MHV-68 in vitro. The transfected cells were observed daily for up to 7 days. Cells transfected with wt MHV-68 virion DNA or MHV-68(BAC) DNA showed complete CPE at 5 days p.t. No obvious CPE was detected in 31STOP-transfected cells at 7 days p.t. Supernatant from each transfection was collected for quantification of virus production. As shown in Fig. 4D, titers of virus produced from transfection with wt MHV-68 or wt MHV-68(BAC) reached ca. 2 × 106 PFU/ml. However, titers of virus produced from transfection with either of the two independent 31STOP BAC clones were below the detection level (<10 PFU/ml). A further two rounds of blinded passage of the supernatant harvested from the 31STOP-transfected cultures did not result in the development of CPE. These results suggested the dependence of 31STOP viral replication on the presence of ORF31 protein.
To examine whether MHV-68 ORF31 could rescue viral replication of the ORF31-null MHV-68(BAC) mutant, 31STOP was cotransfected with pEGFP-m31 into BHK-21 cells. At 5 days p.t., the supernatant of the transfectant was harvested for quantification of infectious virus production by using serial dilution in an inducible cell line expressing ORF31 (T-Rex-31), and the cells were lysed for Western blot to analyze viral protein expression. Expression of lytic viral proteins (ORF45, ORF26, and ORF65) was dramatically reduced in cells transfected with 31STOP plus GFP (Fig. 5B, lane 4). However, expression of lytic proteins was rescued by cotransfection with pEGFP-m31 (Fig. 5B, lane 5). Consistently, the titer of viruses produced from transfection with 31STOP plus GFP was below the level of detection, and that from cotransfection with 31STOP plus pEGFP-m31 was similar to wt MHV-68(BAC) (Fig. 5C, columns 1 through 3). The FLAG-tagged ORF31 was also tested for its ability to rescue the ORF31-null mutant virus and showed a similar result to the GFP-m31 fusion protein (data not shown). We concluded that MHV-68 ORF31 efficiently rescued the ORF31-null MHV-68(BAC) mutant in trans.
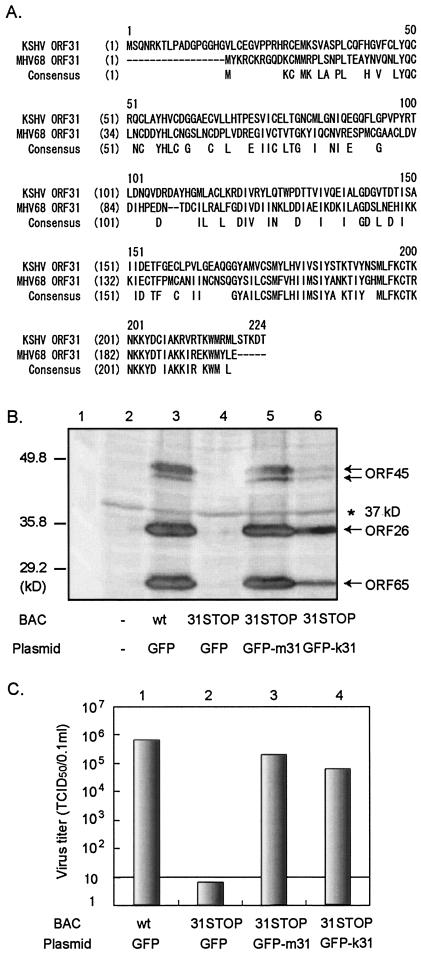
FIG. 5.
Rescue of ORF31-null MHV-68(BAC) mutant after cotransfection with MHV-68 ORF31 or it KSHV homologue. (A) Alignment between the predicted amino acid sequence of MHV-68 ORF31 and the KSHV homologue. The conserved sequence is shown under the aligned sequences. There is 31.3% identity between these two sequences. (B) Expression of lytic proteins from transfected fibroblast cells. BHK-21 cells were mock transfected (lane 2) or transfected with wt MHV-68(BAC) plus vector (GFP) (lane 3), 31STOP plus vector (lane 4), 31STOP plus pEGFP-m31 (lane 5), or 31STOP plus a plasmid expressing a KSHV ORF31 GFP fusion protein (pEGFP-k31; lane 6). At 3 days p.t., cell lysates were analyzed by Western blotting withpolyclonal antibodies to MHV-68 ORF65, ORF26, and ORF45. Specific bands detected by each antibody are indicated by arrows. A 37-kDa background band (*) indicates equal loading in each of the lanes. (C) Viral replication in transfected fibroblast cells. BHK-21 cells were transfected as described for panel B. The supernatant was collected at 3 days p.t., and virus titers were determined by serial dilution in T-Rex-31 cells. The detection level by the method used in this experiment was 10 TCID50/0.1 ml.
31STOP virus is rescued by providing KSHV ORF31.
Amino acid sequence alignment showed a 31% identity between MHV-68 ORF31 and the KSHV homologue (Fig. 5A). We therefore tested whether KSHV ORF31 was able to rescue the ORF31-null MHV-68(BAC) mutant. Full-length KSHV ORF31 was cloned into pEGFP-C1 vector to generate a KSHV ORF31 protein fused at the C terminus of GFP. This construct, pEGFP-k31, was cotransfected into BHK-21 cells with 31STOP. The fluorescence from GFP-k31 had a diffused pattern in the cytoplasms and nuclei of the transfected cells, similar to MHV-68 ORF31 (data not shown). Surprisingly, cotransfection of pEGFP-k31 restored the replication to the ORF31-null mutant, as indicated by expression of the lytic viral proteins, ORF26, ORF45, and ORF65 (Fig. 5B, lane 6) and the viral progeny production of MHV-68 (Fig. 5C, column 4). These results strongly indicate that ORF31 function is conserved between MHV-68 and KSHV.
Propagation of ORF31-null mutant virus in ORF31-inducible cells.
To propagate the ORF31-null MHV-68 mutant virus, we established a Tet-on inducible cell line (T-Rex-31) that expresses the FLAG-m31 upon induction with doxycyline. As shown in Fig. 6A, the FLAG-tagged ORF31 was expressed by T-Rex-31 in the presence (lane 2) but not in the absence (lane 1) of doxycycline. This cell line was then used to complement the ORF31-null MHV-68(BAC) mutant virus. The doxycycline-induced T-Rex-31 cells were transfected with the 31STOP plasmid. At 6 days p.t. the transfected cells developed severe CPE, indicating the replication of the mutant virus. However, there is a possibility that a revertant virus arose during the transfection with 31STOP. To test this possibility, the cell culture (including supernatant and cells) was harvested and used to infect fresh T-Rex-31, the parental T-Rex 293, or noncomplementing fibroblast BHK-21 cells. CPE was readily observed in the doxycycline-induced T-Rex-31 cells but was not observed in noninduced T-Rex-31, T-Rex 293, or BHK-21 cells, supporting the conclusion that the CPE was the result of replication of mutant virus and not of a wt revertant. To verify the genotype of the ORF31-null mutant, we performed a Southern blotting analysis on total cellular DNAs isolated from virus-infected T-Rex-31 cells by using a probe to an EcoRV-XbaI fragment (nt 46420 to 49951) of the viral genome. The expected hybridizing bands were apparent in BamHI-digested wt MHV-68(BAC) (7.5-kb) versus 31STOP (5.3 and 2.2 kb) (Fig. 6B, lanes 1 through 3), confirming the maintenance of the stop codon-BamHI insertion in the 31STOP viral genome. Therefore, the ORF31-null MHV-68(BAC) mutant was successfully propagated in the complementing cell line.
Generation of a 31STOP revertant in BHK-21 cells.
BHK-21 cells transfected with 31STOP did not develop CPE, indicating that the mutant virus with a stop codon insertion in ORF31 is unable to replicate. We thought to restore the function of ORF31 in the null virus by homologous recombination with wt ORF31. This was accomplished by cotransfection of BHK-21 cells with 31STOP BAC plasmid and pSVL-3500, which carries the MHV-68 sequence spanning the mutation (Fig. 3C). By cotransfection with plasmid pSVL-3500, the cells developed CPE, the rescue of 31STOP mutant was achieved, and a replication-competent viral revertant was obtained. The revertant virus, designated 31STOP.R, was purified for three rounds in BHK-21 cells by limiting dilution to make homogeneous virus. To confirm that the rescue of the 31STOP is due to the homologous recombination rather than to complementation in trans or insertion at a second locus, we performed a Southern blotting analysis of total DNAs isolated from T-Rex-31 cells infected with 31STOP.R by using a probe derived from one EcoRV-XbaI fragment of (nt 46420 to 49951) of the viral genome. An expected 7.5-kb band was detected by the probe, which suggested that the revertant appeared as a result of repair of the mutation by homologous recombination between the wt and mutant viral DNA (Fig. 6B, lane 4). These results further confirmed that the replication defect of 31STOP was due to the inserted mutation in ORF31.
Replication kinetics of the 31STOP virus.
To study the effect of ORF31 on viral infection, we infected noncomplementing (BHK-21) and complementing (T-Rex-31) cells with the wt MHV-68, wt MHV-68(BAC), the ORF31-null, and null revertant viruses. To determine the titers of the four input viruses, we performed plaque assay and serial dilution in T-Rex-31 and BHK-21 cells. Plaques were formed in BHK-21 cells infected with all viruses, except the ORF31-null mutant (31STOP). We also monitored the appearance of CPE in serial dilutions of the four input viruses. Consistent with the plaque assay result, no CPE was detected in BHK-21 cells infected with the 31STOP mutant. The titers of input wt MHV-68, wt MHV-68(BAC), and the null revertant (31STOP.R) were approximately two- to threefold higher in BHK-21 cells than in T-Rex-31 cells. We decided to use titers of the entire input viruses determined in T-Rex-31 cells in the infection experiment described below.
To examine whether an MHV-68 one-step growth curve is affected by the loss of function of ORF31, we infected both BHK-21 and doxycycline-induced T-Rex-31 cells with wt MHV-68(BAC) or 31STOP at an MOI of 4. At the indicated times p.i., the whole culture (including supernatant and cells) was harvested, and the virus titer was determined. As shown in Fig. 6C, there were no significant differences in virus titers at 0 h p.i. (P > 0.05). In both BHK-21 and T-Rex-31 cells, wt MHV-68(BAC) had entered productive phase by 8 h p.i. and reached peak by 24 h p.i. with titers of ca. 106 50% tissue culture infective dose(s) (TCID50)/0.1 ml and 3 × 105 TCID50/0.1 ml, respectively, showing no significant difference (P > 0.05). Although the 31STOP mutant had similar initial titers (0 h p.i.) in both BHK-21 and T-Rex-31 cells, it did not initiate production of infectious virus in BHK-21 cells by 24 h p.i., as indicated by decreasing of virus titers during the course of infection. However, in T-Rex-31 cells, ORF31-null virus had increasingly produced infectious viral progeny by 8 h p.i. and reached a titer of ca. 7 × 104 TCID50/0.1 ml at 24 h p.i., which was >100-fold higher than the titer of the mutant virus produced in BHK-21 cells (P < 0.05).
To examine the viral multiple-growth curve, we infected BHK-21 and T-Rex-31 cells with wt MHV-68, wt MHV-68(BAC), 31STOP, or 31STOP.R virus at an MOI of 0.01 and harvested the total virus (including supernatant and cells) at the indicated times p.i. Virus titer was determined by serial dilution in T-Rex-31 cells. In BHK-21 cells (Fig. 6D), replication of wt MHV-68, wt MHV-68(BAC), and 31STOP.R had entered production phase by 2 days p.i. and reached a plateau phase by about 4 to 5 days p.i. with a titer of approximately 3.7 to 6 × 106 TCID50/0.1 ml. On the other hand, titers of viruses produced from 31STOP-infected BHK-21 cells were below detection level (<10 TCID50) at every time point p.i. In contrast, in doxycycline-induced T-Rex-31 cells (Fig. 6E), progeny production from all virus infections, including 31STOP, had increased by 2 days p.i. and reached a peak by 7 days p.i. at titers of ca. 106 TCID50/0.1 ml. No significant differences were observed in virus titers between each virus (P > 0.05), although a delay in the replication kinetics was observed in 31STOP. This result indicated that infection in T-Rex-31 cells can overcome the ORF31 defect in 31STOP replication. The delay in viral replication kinetics of 31STOP in T-Rex-31 may be caused by the limited expression of ORF31 protein in the complementing cell line. The results shown in both multiple- and single-growth curves further confirmed that ORF31 is required for viral replication.
Deficiency of late viral protein expression in noncomplementing cells infected with 31STOP mutant.
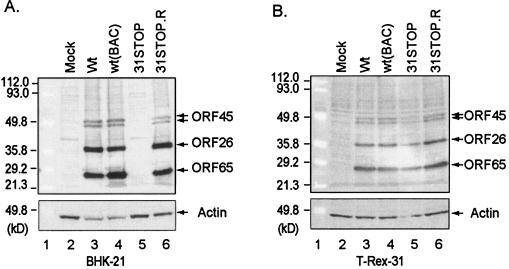
To further define the defect in the ORF31-null MHV-68 mutant, we assessed viral protein expression in noncomplementing (BHK-21) and complementing (T-Rex-31) cells infected with wt or mutant MHV-68 viruses. Prior to this experiment, we established that the 31STOP mutant is unable to replicate in the uninduced T-Rex-31 cells and its parental cell line (data not shown). BHK-21 and the induced T-Rex-31 cells were infected with wt MHV-68, wt MHV-68(BAC), 31STOP, or 31STOP.R viruses (MOI = 0.5). At 2 days p.i., expression of the lytic viral proteins was analyzed by Western blotting with specific polyclonal antibodies to capsid proteins ORF26 and ORF65 and tegument protein ORF45 (6). ORF26, ORF45, and ORF65 were abundantly expressed in BHK-21 cells infected with all viruses, except the 31STOP mutant (Fig. 7A, lanes 3 through 6). Importantly, these lytic viral proteins were expressed equally in T-Rex-31 cells (Fig. 7B, lanes 3 through 6). These results were consistent with the transient complementation assay (Fig. 5B).
FIG. 7.
Lytic viral protein expression in noncomplementing and complementing cells. BHK-21 (A) or T-Rex-31 (B) cells were infected with wt MHV-68, wt MHV-68(BAC), ORF31-null mutant (31STOP), or the null revertant (31STOP.R) (MOI = 0.5) as indicated at the top of each panel. At 2 days p.i., cell lysates were analyzed by Western blotting with polyclonal antibodies to MHV-68 ORF45, ORF26, and ORF65 that recognized specific bands as indicated on the right of the panels. Actin was probed to provide loading controls.
DISCUSSION
ORF31 is conserved between the Beta- and Gammaherpesvirinae subfamilies. MHV-68 ORF31 has identities of 31.3% to its KSHV homologue, 33.6% to herpesvirus saimiri, 24.4% to EBV, 21.3% to human cytomegalovirus, and 19.6% to HHV-6 (3, 8, 16, 24, 37). The product of ORF31 and its viral homologues has not yet been characterized. We showed here that the native MHV-68 ORF31 is a 27-kDa protein expressed during lytic replication. RNAi results suggested that ORF31 is critical for MHV-68 replication after transfection of virion DNA into 293T cells. Through functional analysis of an ORF31-null MHV-68(BAC) mutant (31STOP), we demonstrated that the ORF31 protein is essential for viral replication and that its deletion causes a depletion of late viral protein (ORF26, ORF45, and ORF65) expression. Finally, we found that the lesion in ORF31 in 31STOP mutant virus replication can be complemented in trans by MHV-68 ORF31 protein and, importantly, by the KSHV ORF31 homologue.
The cellular localization of ORF31 was examined by both indirect immunofluorescence and GFP fluorescence on cells transfected with FLAG-tagged ORF31 or GFP-ORF31 fusion protein expression plasmid. Both assays indicated that the MHV-68 ORF31 is distributed evenly in both the cytoplasm and nuclei of the transfected cells (Fig. 1C). Interestingly, the KSHV ORF31 had a similar cellular localization to MHV-68 ORF31 (data not shown). The relationship between the subcellular localization of ORF31 and its functional role is under further investigation.
RNAi is triggered by the presence of a double-stranded RNA in the cell, and results in the silencing of homologous gene expression. We showed that siRNA targeted against MHV-68 ORF31 inhibits ORF31 protein expression from plasmid transfection. Cotransfection of viral DNA with siRNA-ORF31 suppresses the production of MHV-68 viral progeny, indicating a critical role of ORF31 in viral replication. Since the RNAi experiment did not completely abolish the ORF31 protein expression, we generated an ORF31-null MHV-68(BAC) mutant by insertion of translation termination codons in ORF31 to confirm the functional importance of ORF31 in viral replication. The stop codon insertion in ORF31 ensured no functional protein would be expressed, which led to virus production below the detection level (a drop of over 6 log10 in virus titer). Providing the FLAG-tagged ORF31 or GFP-m31 fusion protein in trans can restore the defect in the 31STOP mutant, confirming the essential role of ORF31 in viral replication. Importantly, the lack of ORF31 in ORF31-null MHV-68(BAC) can also be rescued by its KSHV homologue (GFP-k31). Successful complementation of the MHV-68 ORF31 function by GFP-k31 suggests that ORF31 is not only genetically but also functionally conserved among gammaherpesviruses, which provides a strong argument that MHV-68 serves an appropriate model for investigating gene functions of KSHV.
Acknowledgments
We thank members of Ren Sun's laboratory for helpful discussions. We are grateful to Helen Brown and Eric Bortz for critical reading of the manuscript.
This study was supported by NIH grants CA91791, DE14153, and DE15752 and the Stop Cancer Foundation (R.S.).
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, et al. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 4.Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for murine herpesvirus 4. J. Gen. Virol. 84:111-113. [DOI] [PubMed] [Google Scholar]
- 5.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bortz, E., J. P. Whitelegge, Q. Jia, Z. H. Zhou, J. P. Stewart, T. T. Wu, and R. Sun. 2003. Identification of proteins associated with murine gammaherpesvirus 68 virions. J. Virol. 77:13425-13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty, P. C., R. A. Tripp, A. M. Hamilton-Easton, R. D. Cardin, D. L. Woodland, and M. A. Blackman. 1997. Tuning into immunological dissonance: an experimental model for infectious mononucleosis. Curr. Opin. Immunol. 9:477-483. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 12.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 13.Horsburgh, B. C., M. M. Hubinette, D. Qiang, M. L. MacDonald, and F. Tufaro. 1999. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 6:922-930. [DOI] [PubMed] [Google Scholar]
- 14.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia, Q., and R. Sun. 2003. Inhibition of gammaherpesvirus replication by RNA interference. J. Virol. 77:3301-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence, G. L., M. Chee, M. A. Craxton, U. A. Gompels, R. W. Honess, and B. G. Barrell. 1990. Human herpesvirus 6 is closely related to human cytomegalovirus. J. Virol. 64:287-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 18.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackett, M., J. P. Stewart, V. P. S. de, M. Chee, S. Efstathiou, A. A. Nash, and J. R. Arrand. 1997. Genetic content and preliminary transcriptional analysis of a representative region of murine gammaherpesvirus 68. J. Gen. Virol. 78:1425-1433. [DOI] [PubMed] [Google Scholar]
- 20.McGregor, A., and M. R. Schleiss. 2001. Recent advances in herpesvirus genetics using bacterial artificial chromosomes. Mol. Genet. Metab. 72:8-14. [DOI] [PubMed] [Google Scholar]
- 21.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mistrikova, J., and D. Blaskovic. 1985. Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol. 29:312-317. [PubMed] [Google Scholar]
- 23.Moore, P. S., and Y. Chang. 1995. Kaposi's sarcoma findings. Science 270:15. [DOI] [PubMed] [Google Scholar]
- 24.Moore, P. S., S. J. Gao, G. Dominguez, E. Cesarman, O. Lungu, D. M. Knowles, R. Garber, P. E. Pellett, D. J. McGeoch, and Y. Chang. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J. Virol. 70:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moorman, N. J., D. O. Willer, and S. H. Speck. 2003. The gammaherpesvirus 68 latency-associated nuclear antigen homolog is critical for the establishment of splenic latency. J. Virol. 77:10295-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 27.Pavlova, I. V., H. W. t. Virgin, and S. H. Speck. 2003. Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication. J. Virol. 77:5731-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajcani, J., D. Blaskovic, J. Svobodova, F. Ciampor, D. Huckova, and D. Stanekova. 1985. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 29:51-60. [PubMed] [Google Scholar]
- 29.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr Virus, p. 2575-2628. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins Co., Philadelphia, Pa.
- 30.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 34.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Virgin, H. W. t., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, T. T., L. Tong, T. Rickabaugh, S. Speck, and R. Sun. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 75:9262-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]