Abstract
The structure and genetic diversity of a California Cucumber mosaic virus (CMV) population was assessed by single-strand conformation polymorphism and nucleotide sequence analyses of genomic regions 2b, CP, MP, and the 3′ nontranslated region of RNA3. The California CMV population exhibited low genetic diversity and was composed of one to three predominant haplotypes and a large number of minor haplotypes for specific genomic regions. Extremely low diversity and close evolutionary relationships among isolates in a subpopulation suggested that founder effects might play a role in shaping the genetic structure. Phylogenetic analysis indicated a naturally occurring reassortant between subgroup IA and IB isolates and potential reassortants between subgroup IA isolates, suggesting that genetic exchange by reassortment contributed to the evolution of the California CMV population. Analysis of various population genetics parameters and distribution of synonymous and nonsynonymous mutations revealed that different coding regions and even different parts of coding regions were under different evolutionary constraints, including a short region of the 2b gene for which evidence suggests possible positive selection.
RNA viruses are intrinsically heterogeneous, partly because of the error-prone nature of RNA replication (14, 20). In addition, genetic exchange (either by recombination or by reassortment) can be another major evolutionary force which is able to rapidly increase variation and is proposed to have evolved to offset fitness losses due to the accumulation of deleterious mutations through the effect known as Muller's ratchet (6). Other evolutionary factors such as selection, genetic drift, and bottleneck effects can serve to decrease diversity. However, in spite of the high potential for variability of RNA viruses, most RNA virus populations analyzed so far are genetically stable with relatively low diversity (17).
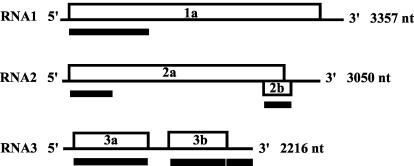
Cucumber mosaic virus (CMV) is a tripartite, positive-sense plant RNA virus (Fig. 1). CMV occurs naturally worldwide and has perhaps the widest host range among all plant viruses, including some monocotyledonous and a great number of dicotyledonous plant hosts (26). RNA1 encodes the 1a protein, which together with the RNA2-encoded 2a protein forms the viral component of the replicase complex (19). RNA2 also encodes a second protein, 2b. The 2b coding region overlaps the coding region for the C-terminal portion of the 2a protein but is in a different reading frame register. The CMV 2b protein functions in host-specific long-distance movement (12, 13) and as a virulence determinant by suppressing posttranscriptional gene silencing (3). RNA3 encodes two proteins. The 3a protein is a cell-to-cell movement protein (MP), and the 3b protein is the capsid protein (CP), which is also involved in cell-to-cell movement and aphid-mediated CMV transmission from plant to plant (5, 27, 28).
FIG. 1.
Genome organization of CMV. Numbers of nucleotides (nt) are given for the Fny strain. Open boxes, open reading frames. Solid boxes correspond to the genomic regions analyzed here.
Various methods have been used to analyze the variability among different CMV isolates. These include the host range assay (26), PCR-based methods, restriction mapping (31), coat protein peptide mapping (15), dot blot hybridization (32), serological analyses (21), RNase protection assays (16), and nucleotide sequence analysis (8, 9, 33, 34, 39). Most of these studies were phylogenetically oriented and allowed the subdivision of CMV isolates from throughout the world into three subgroups: IA, IB, and II (26, 33, 34). The genetic structure of natural populations of CMV from Spain was analyzed (16), but genetic diversity based on genetic distances between haplotypes and polymorphism patterns of the CMV population was not. Studies of genetic structure and diversity would be important in helping to understand the evolutionary mechanisms that generate and/or maintain variation in viral populations and their evolution; thus, such studies may facilitate the development of strategies for the control of viral diseases.
In a previous study, we analyzed the biological and molecular variation of California CMV isolates by single-strand conformation polymorphism (SSCP) and by sequence and phylogenetic analyses of the CP gene (22). Here we extend the analyses to other genomic regions (1a, 2a, 2b, MP, and the 3′ nontranslated region [3′ NTR]) and analyze the genetic structure, diversity, and various population genetics parameters for a California CMV population.
MATERIALS AND METHODS
Virus isolates.
The biological characteristics, geographic origins, and collection years of the 81 California CMV isolates used here have been reported previously (22). These isolates represent two groups; group α contains 63 isolates collected from cucurbit plants in two growing seasons at the Kearney Agricultural Center, Parlier, Calif., in 1999, and group β contains 18 isolates collected from various hosts and different California locations during 1985 to 1994.
RT-PCR amplification.
Total RNA extraction, reverse transcriptase PCR (RT-PCR), and PCR were carried out as described previously (22). Primers used for amplifying different genomic regions of CMV, designed according to CMV strain Fny (GenBank accession numbers for RNA1, RAN2, and RNA3 are D00356, D00355, and D10538, respectively), are listed in Table 1.
TABLE 1.
Primers designed for RT-PCR amplification of different genomic regions of CMV
| Primera | Genomic region | Positionsb | Sizec (nt) | Nucleotide sequence |
|---|---|---|---|---|
| 1a-forward | 1a | 1-22 | 1,089 | 5′-GTTTATTTACAAGAGCGTACGG-3′ |
| 1a-reverse | 1070-1089 | 5′-TGTCGAATGAGTTCGGGTGG-3′ | ||
| 2a-forward | 2a | 1-24 | 653 | 5′-GTTTATTTACAAGAGCGTACGCTT-3′ |
| 2a-reverse | 633-653 | 5′-AGACGTTCGGGACCACGGTTC-3′ | ||
| 2b-forward | 2b | 2418-2438 | 370 | 5′-TTATGGAATTGAACGTAGGTG-3′ |
| 2b-reverse | 2769-2787 | 5′-ACAGAAAACCGGAGGGAGA-3′ | ||
| M1-forward | MP | 118-138 | 842 | 5′-CATGGCTTTCCAAGGTACCAG-3′ |
| M2-reverse | 938-959 | 5′-CTAAAGACCGTTAACCACCTGC-3′ | ||
| F4-forward | CP | 1245-1266 | 678 | 5′-TTGAGTCGAGTCATGGACAAATC-3′ |
| F3-reverse | 1902-1922 | 5′-AACACGGAATCAGACTGGGAG-3′ | ||
| F2-forward | 3′ NTR (RNA3) | 1902-1922 | 315 | 5′-CTCCCAGTCTGATTCCGTGTT-3′ |
| F1-reverse | 2197-2216 | 5′-TGGTCTCCTTTTGGAGGCCC-3′ |
“Forward” indicates positive polarity; “reverse” indicates negative polarity.
Nucleotide positions of the primers in the genomic RNAs of CMV Fny. The 1a protein is encoded by RNA1; 2a and 2b are encoded by RNA2; MP, CP, and the 3′ NTR are encoded by RNA3.
Expected size of the resulting RT-PCR product.
Cloning and sequencing.
RT-PCR products were purified by using the QIAquick PCR extraction kit (QIAGEN, Valencia, Calif.). The purified PCR products of the 2b, MP, and CP genes and of the RNA3 3′ NTR were directly sequenced, and those of the 1a and 2a genes were ligated into pGEM-T (Promega, Madison, Wis.) according to the manufacturer's instructions, followed by transformation into Escherichia coli DH5α. Recombinant colonies were screened by PCR using the same conditions described above. Nucleotide sequences were determined in both directions by means of a model 377 ABI PRISM DNA sequencer (Perkin-Elmer, Fremont, Calif.) in the Automated DNA Sequencing Facility of the University of California—Davis. Three colonies for each isolate were used for sequence analysis, and the consensus sequences were used for phylogenetic analysis. Sequences of oligonucleotide primers were excluded for nucleotide sequence comparisons.
SSCP analysis.
RT-PCR products or bacterial-colony PCR products were used for SSCP analysis. For the MP gene, PCR products were first digested by KpnI in order to obtain smaller fragments and greater accuracy in SSCP. The DNAs were denatured and subjected to electrophoresis by the methods described previously (22). All SSCP analyses were repeated at least twice, and only samples within the same gel were compared.
Sequence analysis.
Multiple nucleotide sequence alignments were performed by using CLUSTAL W (40). Alignments were manually adjusted by using MacClade, version 4.0 (24). All the sequence alignments used in this paper are available upon request. Genetic diversity (the average number of nucleotide substitutions per site between two sequences) within and between populations and the degree of population subdivision were calculated by following the method of Lynch and Crease (23). Population genetics parameters with respect to the total number of mutations, the statistic θ○ from the number of segregating sites (S), the number of nonsynonymous mutations, the average number of nucleotide differences between two random sequences in a population (π), the average number of synonymous and nonsynonymous nucleotide substitutions, and synonymous codon usage bias were calculated by the DnaSP program, version 3.99 (35). The distribution of synonymous and nonsynonymous substitutions along the coding regions was analyzed by using the SNAP program (available at http://hiv-web.lanl.gov). Synonymous codon usage bias was measured by quantifying the “effective” number of codons (ENC) (41) that are used in a gene. For the nuclear universal genetic code, the value of ENC ranges from 20 (if only one codon is used for each amino acid, i.e., the codon bias is maximum) to 61 (if all synonymous codons for each amino acid are equally used, i.e., no codon bias).
Phylogenetic relationships were inferred by using the PAUP* 4.0b10.0 program with the maximum parsimony optimality criterion (38). For all analyses, a limit of 100,000 trees was imposed. To avoid reaching the tree limit on the first replicate, a maximum of 100 trees were saved per replicate (“nchuck” option in effect). Gaps were treated as a fifth character state. Heuristic searches were performed by using 1,000 replicates of random taxon addition and tree bisection-reconnection branch swapping. Bootstrap analyses were performed by using 1,000 replicates, each with 10 replicates of random taxon addition heuristic search. All branches with bootstrap values of <70% were collapsed. Two other members of the genus Cucumovirus, Peanut stunt virus (PSV) and Tomato aspermy virus (TAV), were included as outgroups. The GenBank accession numbers of 13 CMV isolates with full-length nucleotide sequences and of PSV (strain ER), used as reference isolates, have been given by Roossinck (33). The GenBank accession numbers of TAV (strain V) are D10044 (RNA1), L79972 (RNA2), and AJ277268 (RNA3).
RESULTS
Genetic structure of a California CMV population.
RT-PCR products corresponding to genomic regions 2b and MP and the RNA3 3′ NTR of 81 CMV isolates were analyzed by SSCP. Each SSCP pattern was considered to be a distinct haplotype. Sixteen haplotypes were observed for the 2b gene. One was predominant (haplotype C) and corresponded to 52 isolates, all belonging to group α (Fig. 2A). For the MP gene, we first used KpnI to cleave the PCR product into three smaller DNA fragments of 321, 502, and 18 bp (only the SSCP bands of the 321- and 502-bp fragments are shown on the gels). KpnI failed to digest PCR products for 23 isolates; therefore, AccI was used for these 23 isolates to yield two fragments of 331 and 510 bp. However, PCR products for three isolates were not digested by AccI, and therefore these were directly used for SSCP analysis. Considering the SSCP analysis and restriction endonuclease digestion results, 21 haplotypes were observed for the MP gene. Haplotype C was predominant, corresponding to 35 isolates, of which 34 were from group α and 1 was from group β. Haplotype P, corresponding to 13 group α isolates, was the second most frequent, and haplotype E, found for 9 isolates (5 from group α and 4 from group β), was the third most frequent (Fig. 2B). For the RNA3 3′ NTR, 16 haplotypes were found. Haplotypes A and B were the two predominant haplotypes, corresponding to 35 (33 from group α and 2 from group β) and 26 (20 from group α and 6 from group β) isolates, respectively (Fig. 2C). Collectively, these results showed that the California CMV population comprised one to three predominant haplotypes, depending on the genomic region analyzed, and a number of minor haplotypes with low frequency.
FIG. 2.
Genetic structure of the California CMV population based on SSCP analyses of different genomic regions. (A) The CMV 2b gene; (B) the CMV MP gene; (C) the RNA3 3′ NTR. Each lane contains a distinct SSCP pattern, which is considered a distinct haplotype, designated by the letter above the lane. Below each lane, the number of samples with that SSCP pattern is given (upper numbers), as well as the haplotype frequencies for group α and β isolates, respectively (lower numbers) (e.g., 0 + 1 means that no isolates in group α and 1 isolate in group β have pattern A). For the MP gene, those isolates without a KpnI site were digested by AccI; however, three isolates were digested neither by KpnI nor by AccI (lanes T and U).
Genetic diversity of the California CMV population.
The nucleotide sequences of the 2b and MP regions and the RNA 3 3′ NTR for one isolate of each haplotype were determined from RT-PCR products in order to estimate genetic diversity. Genetic diversity (average nucleotide substitutions per site for pairs of randomly selected haplotypes) was estimated based on the haplotype frequencies (obtained from SSCP analysis) and nucleotide distances of the 2b, MP, 3′ NTR, and CP regions. Data obtained from SSCP and sequence analyses of the CP gene described previously (22) were used in the estimation of genetic diversity here. Among the four genomic regions analyzed, genetic diversity was significantly lower for group α (collected from cucurbit plants, same location and year) than for group β (collected from various locations, host plants, and years) (P = 0.01) (Table 2). Curiously, the 2b gene had the highest genetic diversity for group β isolates but the lowest for group α isolates (Table 2). For the California CMV population, genetic diversity ranged from 0.01323 ± 0.00275 to 0.02186 ± 0.00607 according to the genomic region analyzed; this gave a mean genetic diversity of 0.01648 ± 0.00366 (Table 2).
TABLE 2.
Genetic diversity within the California CMV population
| Isolatesa | Genetic diversityb of genomic region:
|
Mean genetic diversity | |||
|---|---|---|---|---|---|
| 2b | MP | CP | 3′ NTR | ||
| All | 0.02186 (0.00607) | 0.01428 (0.00216) | 0.01323 (0.00275) | 0.01653 (0.00367) | 0.01648 (0.00366) |
| Group α | 0.00182 (0.00092) | 0.00956 (0.00144) | 0.00526 (0.00155) | 0.00759 (0.00245) | 0.00606 (0.00159) |
| Group β | 0.06770 (0.01472) | 0.02592 (0.00591) | 0.03178 (0.00498) | 0.03974 (0.00897) | 0.04129 (0.00865) |
All, the California population containing 81 isolates. Group α isolates were collected from cucurbit plants in 1999 at the Kearney Agricultural Center, and group β isolates were collected from various host species and different areas during 1985 to 1994 (see Lin et al. [22]).
Average nucleotide substitutions per site for pairs of random haplotypes within populations. Numbers in parentheses are the standard errors.
Phylogenetic relationship among CMV isolates.
Phylogenetic analyses of genomic regions 2b, MP, and 3′ NTR were performed by using nucleotide sequences of isolates corresponding to the different haplotypes (Fig. 3) in order to clarify the evolutionary relationships among these CMV isolates. For the 2b gene, various isolates of group α (CK31, CK35, CK13, CK5, and CK27) formed a clade supported by a bootstrap value of 87%. The group β isolates were much more dispersed and fell into distinct clades (e.g., 113B, 190A, MD284, and SJ91B) (Fig. 3A). For the MP gene, four group α isolates (CK7, CK5, CK58, and CK27) formed a small clade with a supporting value of 92%, which fell within a large clade composed of most isolates. The remaining group α isolates were unresolved, forming polytomy clades (Fig. 3B). However, as for the 2b phylogeny, group β isolates appeared to be more diverse than group α isolates. The same trend was also observed in the phylogenetic tree based on the 3′ NTR (data not shown). These results showed that group α isolates were closely related.
FIG. 3.
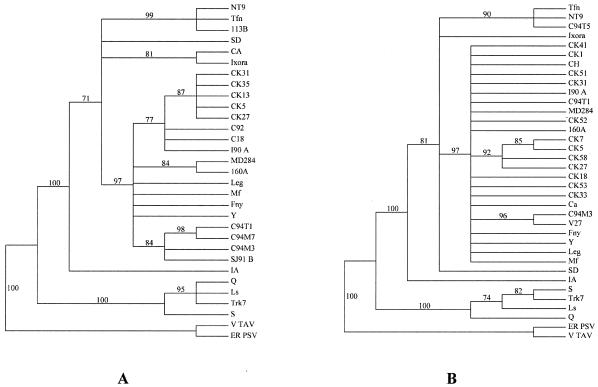
Bootstrap majority rule (70%) consensus trees of the CMV 2b (A) and MP (B) genes obtained by using California CMV isolates corresponding to the respective haplotypes of the regions analyzed (see Fig. 1) and reconstructed by maximum parsimony heuristic searches. Bootstrap values are given above the branches. The GenBank accession numbers of the reference CMV isolates used here are as follows: Fny, D10538; IA, AB042294; Ixora, U20219; Leg, D16405; Ls, AF127976; Mf, AJ276481; NT9, D28780; Q, M21464; S, AF063610; SD, AB008777; Tfn, Y16926; Trk 7, L15336; Y, D12499. Outgroups are ER-PSV (U15730) and V-TAV (L79972). Standard subgroup II CMV isolates are Q, Ls, S, and Trk7. Standard subgroup IB isolates are NT9, Tfn, IA, Ixora, and SD. Standard subgroup IA isolates are Fny, Mf, Y, and Leg.
Inter- and intrasubgroup reassortment revealed by phylogenetic analysis.
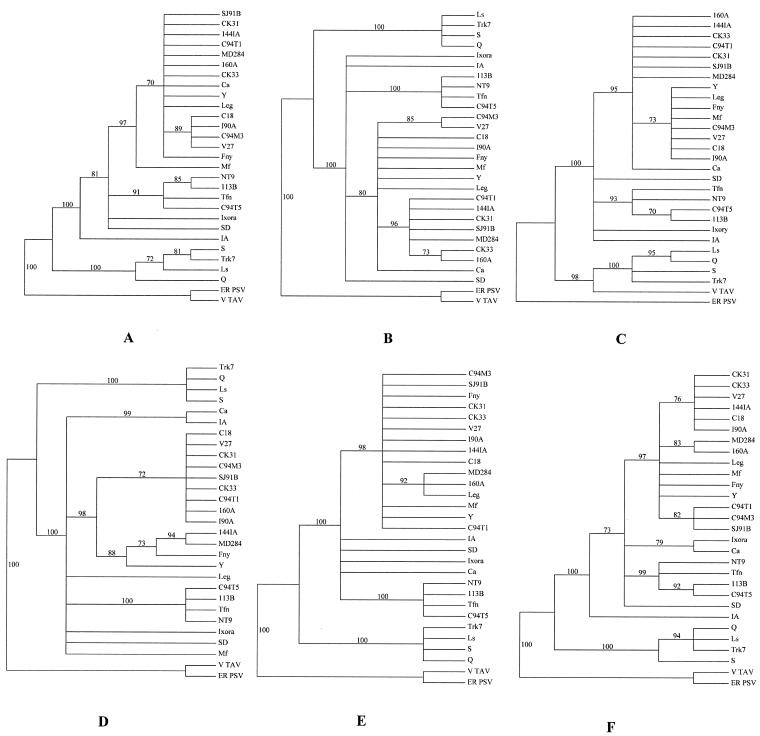
Genetic exchange and recombination also can be important factors affecting virus evolution and the resulting populations. To assess whether evidence suggested that genetic exchange events had occurred between California isolates, we performed phylogenetic analyses of sequence data sets for 14 California isolates corresponding to 2b (RNA2), MP and CP (RNA3), and the 3′ NTR of RNA3, as well as parts of the CMV coding regions for the 1a (RNA1) and 2a (RNA2) proteins. These 14 isolates (2 from group α and 12 from group β) were chosen as representative of the California population based on the nucleotide distances of isolates in the population. In addition, 13 CMV isolates whose complete genome sequences were in GenBank were used for reference. The bootstrap maximum-parsimony trees for these genomic regions are shown (Fig. 4). Strikingly, isolate Ca was assigned to subgroup IA based on phylogenetic analysis of the MP and CP regions and the 3′ NTR of RNA3 (Fig. 4A, B, and C) but to subgroup IB based on the 1a, 2a, and 2b genes (Fig. 4D, E, and F). In addition, trees for all three regions of RNA3 showed similar topologies, including the placement of isolate Ca, and trees for the two RNA2 genomic regions (2a and 2b) also showed similar topologies. Based on these congruent topologies, it seems unlikely that recombination events led to the origin of isolate Ca; rather, these topologies suggest that genome segment reassortment could have played a role. However, because only partial sequences of the 1a and 2a genes were used, phylogenetic analyses were performed in parallel by using complete 1a and 2a sequences for 13 CMV isolates (from GenBank) and also using partial sequences (corresponding to those obtained for our CMV isolates) for the same 13 isolates. These analyses were performed to assess the validity of our data; the results showed that, for the 2a gene, the two data sets gave essentially identical tree topologies, suggesting that the partial 2a sequence can be used for reconstruction of the evolutionary history of the 2a gene. In contrast, the topologies for the 1a gene in the two data sets were slightly different. Based on the complete sequence of 1a, isolate Leg was placed into a clade together with isolates Fny and Y, but Leg was unresolved with respect to isolates Fny and Y when the partial sequences were used (data not shown and Fig. 4D). In spite of this, our phylogenetic analyses still suggested that isolate Ca was a natural reassortant resulting from genetic exchange between subgroup IA and IB isolates. However, no potential ancestors of this isolate were identified among the 27 isolates analyzed here.
FIG. 4.
Bootstrap majority rule (70%) consensus trees of six genomic regions of CMV reconstructed by maximum-parsimony heuristic searches. Bootstrap values are given above branches. (A) MP; (B) CP; (C) 3′ NTR of RNA3; (D) 1a; (E) 2a; (F) 2b. See the legend to Fig. 3 for isolate designations.
Genetic exchange can result from mixed infections. Therefore, it is possible that the parental sequences could still be present in isolate Ca, but at a low proportion within the resulting hybrid isolate. It is possible that the minor sequence variants (or parental sequences) could outcompete the hybrid isolate in a different host species, thereby allowing them to be easily detected (16). To test this possibility, 30 clones each from RT-PCR products of the 1a, 2a, and MP genes amplified from small, CMV Ca-infected sugar pumpkin plants, and 30 clones each of the 2b and CP genes amplified from CMV Ca-infected Nicotiana benthamiana plants were analyzed first by SSCP and then by nucleotide sequence analysis of clones representing the different haplotypes as described above. However, no sequences corresponding to RNA3 subgroup IB or to RNA1 or RNA2 subgroup IA were found (data not shown).
Close examination of the topologies for subgroup IA isolates also suggested evidence for reassortment between some subgroup IA isolates. Examination of the trees for the RNA2 coding regions showed that isolates V27, CK31, and CK33 appeared to be closely related (Fig. 4E and F). However for RNA3, V27 was placed separate from CK31 and CK33 but closer to C94M3 (Fig. 4A, B, and C). CK31 and CK33 were placed with several isolates including C94T1. To help clarify their relationships, pairwise nucleotide distances among these isolates and a subgroup IB isolate (NT9) were calculated by using sequence data for the 2a, 2b, MP, and CP coding regions (Table 3). Assuming that C94T1-like and V27-like isolates were the parental strains, these data suggested that RNA3 of isolates CK31 and CK33 came from a C94T1-like isolate but RNA2 (the 2a and 2b coding regions) came from a V27-like isolate. For isolate SJ91B, these analyses suggested that RNA3 (based on the CP and MP regions) also originated from a C94T1-like isolate, but the origin of RNA2 (based on the 2a and 2b sequences) was not clear. RNA3 of isolate C94M3 could have originated from a V27-like isolate, but the parental isolate for RNA2 (based on the 2a and 2b sequences) was also unknown.
TABLE 3.
Pairwise nucleotide distances among six California CMV isolates and a reference subgroup IB isolate for the 2a, 2b, MP, and CP genes
| Gene and isolate | Pairwise nucleotide distance between the indicated isolatesa | |||||
|---|---|---|---|---|---|---|
| 2a gene | C94M3 | C94T1 | SJ91B | CK33 | V27 | CK31 |
| C94M3 | ||||||
| C94T1 | 0.0243 | |||||
| SJ91B | 0.0167 | 0.0299 | ||||
| CK33 | 0.0242 | 0.0337 | 0.0222 | |||
| V27 | 0.0318 | 0.0414 | 0.0298 | 0.0111 | ||
| CK31 | 0.0242 | 0.0337 | 0.0222 | 0.0037 | 0.0111 | |
| NT9 | 0.0653 | 0.0754 | 0.0711 | 0.0671 | 0.0752 | 0.0632 |
| 2b gene | C94M3 | C94T1 | SJ91B | CK31 | CK33 | V27 |
| C94M3 | ||||||
| C94T1 | 0.0306 | |||||
| SJ91B | 0.0401 | 0.0338 | ||||
| CK31 | 0.0527 | 0.0526 | 0.0494 | |||
| CK33 | 0.0527 | 0.0526 | 0.0494 | 0.0000 | ||
| V27 | 0.0591 | 0.0526 | 0.0526 | 0.0060 | 0.0060 | |
| NT9 | 0.1505 | 0.1647 | 0.1502 | 0.1498 | 0.1498 | 0.1462 |
| CP gene | SJ91B | C94T1 | CK31 | CK33 | C94M3 | V27 |
| SJ91B | ||||||
| C94T1 | 0.0048 | |||||
| CK31 | 0.0048 | 0.0063 | ||||
| CK33 | 0.0079 | 0.0095 | 0.0095 | |||
| C94M3 | 0.0404 | 0.0421 | 0.0420 | 0.0387 | ||
| V27 | 0.0371 | 0.0388 | 0.0387 | 0.0354 | 0.0032 | |
| NT9 | 0.0640 | 0.0640 | 0.0623 | 0.0623 | 0.0589 | 0.0555 |
| MP gene | SJ91B | CK33 | CK31 | C94T1 | C94M3 | V27 |
| SJ91B | ||||||
| CK33 | 0.0092 | |||||
| CK31 | 0.0092 | 0.0131 | ||||
| C94T1 | 0.0105 | 0.0144 | 0.0118 | |||
| C94M3 | 0.0305 | 0.0264 | 0.0291 | 0.0251 | ||
| V27 | 0.0238 | 0.0198 | 0.0251 | 0.0238 | 0.0065 | |
| NT9 | 0.0682 | 0.0667 | 0.0653 | 0.0597 | 0.0611 | 0.0626 |
For specific isolate information, see Lin et al. (22).
Different evolutionary processes may have affected different genomic regions.
The rate of molecular evolution differs from one gene to another and even from one part of a gene to another part of that same gene. Due to the small size and high efficiency of RNA virus genomes, even synonymous mutations might be subjected to selective constraints due to codon usage bias or RNA structure. To provide insight into the evolutionary processes affecting different genomic regions of CMV, we examined different population genetics parameters for the California CMV population based on the 1a, 2a, 2b, MP, and CP coding regions of 20 randomly selected isolates.
π, the average number of nucleotide differences between two random sequences in a population (also called genetic diversity), and θ (S), the statistic of the number of segregating sites, were used here as two indicators to estimate genetic variation. For both estimations, the order of genetic variation, from greatest to least, was as follows: 2b, 2a, 1a, MP, and CP (Table 4). Thus, the 2b gene showed the most segregating sites and the highest frequency of mutations among the different genomic regions analyzed here. In terms of the number of nonsynonymous mutations, less than one-third of the mutations for 1a, MP, and CP were nonsynonymous, but the percentage was much higher (50% or higher) for the 2a and 2b regions, suggesting that the 2a and 2b genes were more flexible with regard to amino acid changes. To further understand the evolutionary constraints imposed on different coding regions, the ratio of the average number of nonsynonymous substitutions per nonsynonymous site (Ka) to the average number of synonymous substitutions per synonymous site (Ks) was estimated. The Ka/Ks ratios for all coding regions analyzed here were less than 1.0, indicating that they were all subjected to negative selection (Table 4). The 1a, MP, and CP genes showed low Ka/Ks ratios, suggesting high selective pressure, whereas the Ka/Ks ratios for the 2a and 2b regions were 5 to 36 times higher (Table 4), suggesting that the proteins encoded by these two genes were considerably more tolerant of amino acid changes.
TABLE 4.
Population genetics parameters of different coding regions of CMV
| Region | Positiona | Sites (total)b | πc | θ (S)d | % Nonsynonymous mutationse | Ka/Ks ratiof | ENCg |
|---|---|---|---|---|---|---|---|
| 1a | 95-1069 | 975 | 0.01945 | 0.03068 | 21.7 (23/106) | 0.06972 (0.00482/0.06913) | 58.185 |
| 2a | 87-632 | 546 | 0.02153 | 0.03820 | 52.7 (39/74) | 0.25909 (0.01275/0.04921) | 55.684 |
| 2b | 2419-2751 | 333 | 0.03248 | 0.04741 | 50.0 (29/58) | 0.31170 (0.02161/0.06933) | 59.916 |
| MP | 159-926 | 768 | 0.01841 | 0.02760 | 17.3 (13/75) | 0.04564 (0.00303/0.06639) | 55.405 |
| CP | 1269-1901 | 633 | 0.01489 | 0.02494 | 12.5 (7/56) | 0.05206 (0.00258/0.04956) | 59.696 |
Nucleotide positions of the regions in the genomic RNAs of CMV Fny.
Total nucleotides of the given genomic region used for analysis.
Average number of nucleotide difference in terms of total sites analyzed.
Statistic θ from segregating sites.
Percentage of total mutations that are nonsynonymous. Values in parentheses are the number of nonsynonymous mutations/total number of mutations.
Values in parentheses are Ka (average number of nonsynonymous substitutions per nonsynonymous site)/Ks (average number of synonymous substitutions per synonymous site). Ka and Ks are estimated by the method described by Nei and Gojobori (25).
ENC, effective number of codons.
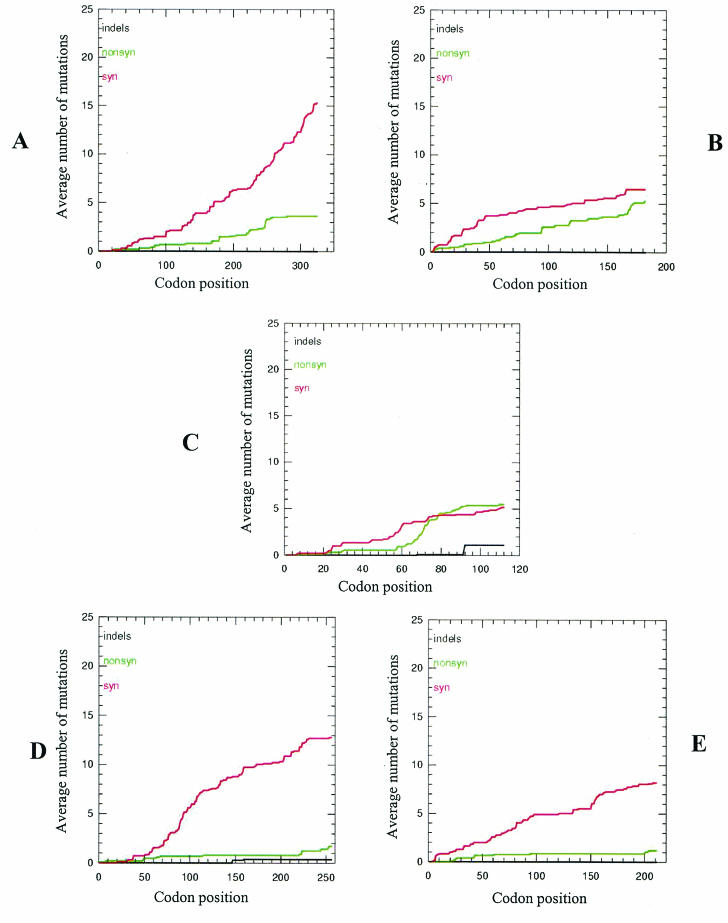
To compare in detail the selective constraints across coding regions 1a, 2a, 2b, MP, and CP, we also analyzed the distributions of both synonymous and nonsynonymous mutations. For 1a, the curve of synonymous mutations showed a steady increase with a generally consistent slope, suggesting that the synonymous mutations were relatively evenly distributed across the region and thus were selectively neutral. However, most of the nonsynonymous mutations in the 1a coding region were clustered around codon positions 180 to 260 (Fig. 5A). The cumulative incidences of the synonymous mutations for the MP and CP coding regions were similar to each other. Accumulation of only a few synonymous mutations was observed, but some areas showed very few mutations. For example, mutations occurred at only four and three positions within positions 159 to 198 of the MP region and positions 98 to 151 of the CP coding region, respectively (Fig. 5D and E). The nearly flat lines showing the distribution of nonsynonymous mutations indicated very strong negative selection against amino acid changes across the entire MP and CP regions (Fig. 5D and E). For 2a, the cumulative incidences of both synonymous and nonsynonymous mutations increased steadily across the sequence, showing consistent slopes, suggesting that there was little bias in the distribution of these mutations (Fig. 5B). The cumulative behavior of 2b was distinct from that of other regions (Fig. 5C). For the region overlapping 2a (positions 1 to 80), the distributions of both synonymous and nonsynonymous mutations were uneven. Mutations were few within positions 1 to 20 and 30 to 53 but frequent within positions 55 to 80. Interestingly, within a short section (codons 81 to 93) of the nonoverlapping region of 2b (codons 81 to 111), the average number of nonsynonymous mutations was greater than the number of synonymous mutations (Fig. 5C). This region corresponds to nucleotide positions 2659 to 2697 of Fny RNA2. Further analysis showed that 87.5% of the mutations in this area were nonsynonymous, and the Ka/Ks ratio was estimated as 5.21709 (0.03052/0.00585), suggesting that this area might be under positive selection.
FIG. 5.
Cumulative incidences of synonymous and nonsynonymous substitutions in coding regions 1a (A), 2a (B), 2b (C), MP (D), and CP (E). The x axis represents the position of the codon, and the y axis represents the average cumulative number of synonymous or nonsynonymous mutations estimated at a specific codon position. Red, green, and black curves, synonymous, nonsynonymous, and indel mutations, respectively. See Fig. 1 for diagrams of the genomic regions analyzed.
Synonymous codon usage bias was calculated by using the ENC. The value of ENC for these coding regions ranged from 55.405 to 59.696, suggesting that these coding regions had only a slight bias in codon choice (Table 4).
DISCUSSION
Based on the SSCP profiles of the 2b and MP regions and the 3′ NTR of RNA3 in this study (Fig. 1), and on that of the CP gene in a previous study (22), we found that these genomic regions for the isolates comprising the California CMV population were composed of one to three predominant haplotypes and a large number of minor haplotypes for the specific genomic regions analyzed. The population also exhibited overall low genetic diversity, averaging 0.01648 ± 0.00366 (Table 2). Similar genetic structure and diversity have been found in populations of most plant viruses studied so far (19). Among those California CMV isolates analyzed here, group β isolates showed significantly higher genetic diversity (mean, 0.04129 ± 0.00865) whereas group α isolates showed very low diversity (mean, 0.00606 ± 0.00159), irrespective of the genomic region used for analysis (Table 2). This result is not very surprising for the group β isolates, because they were collected from different host plants and locations and in different years. Phylogenetic analyses using nucleotide sequences of CMV isolates of the different haplotypes revealed that group α isolates were closely related (Fig. 3). Together with the low diversity among group α isolates, these data suggest that group α isolates were most likely derived via a founder event from a common ancestor and that they only recently colonized and spread within the area. However, we cannot rule out the possibility that the low diversity and close evolutionary relationship among isolates in group α could also be due, at least in part, to selection by the host plant, since all the group α isolates were collected from cucurbit plants (although of different cultivars [22]).
Phylogenetic analyses of different CMV genomic regions revealed natural reassortment between subgroup IA and IB isolates and potential reassortment between subgroup IA isolates but yielded no evidence for recombination (Fig. 4). Our results are seemingly in agreement with the hypothesis that the purpose of a multipartite viral genome is to favor genetic exchange through reassortment (7, 30). However, conflicting data arguing against this hypothesis have been provided in a report for a Spanish CMV population in which both recombination and reassortment were infrequent, and reassortment was not more frequent than recombination (16). Thus, the absence of detectable recombinants in our study may also be explained by two alternative scenarios: either (i) recombination events occurred between two closely related isolates and are difficult to detect by the methods we used here or (ii) recombination events did occur, but the resulting recombinants were not favored and subsequently were selected out. Indeed, recombination events in the CMV genome and CMV satellite RNA have been described recently in both experimental systems and natural populations (1, 2, 4, 10, 16, 18, 37).
Estimation of various population genetics parameters showed that coding regions on RNA2 (2a and 2b) were more variable, suggesting that the resultant proteins were more tolerant to amino acid changes than were regions on RNA1 (1a) and RNA3 (MP and CP) (Table 4). Among all the coding regions analyzed here, the 2b gene appeared to be the most flexible. Furthermore, a short region in the 2b gene, corresponding to nucleotide positions 2659 to 2697 of RNA2, might be subjected to positive selection. It is noteworthy that the extent of selection pressure imposed on different coding regions seems to be correlated with the functions of the proteins they encode and/or their interactions with the host. This correlation, inferred from phylogenetic analyses of 15 CMV isolates with full-length sequences in GenBank, has been proposed by Roossinck (33). It is of interest to consider the 2b protein and its role(s) in host interactions. The 2b protein is related to long-distance movement and virulence and is a suppressor of RNA silencing (3, 13). Unlike the conserved 1a-membrane, MP-plasmodesma, and CP-RNA and -aphid vector interactions (11, 29), the 2b-host interaction has been suggested to be host specific. The fact that the 2b protein is essential for long-distance virus movement within cucumber plants but not for systemic spread in Nicotiana spp. is evidence in support of this hypothesis (13). This host-specific function of the 2b protein was believed to provide a genetic basis for the extremely wide host range of CMV (13), thereby theoretically allowing 2b to be considerably more tolerant to nucleotide and amino acid changes. Additionally, the 2b gene is present in all members of the genus Cucumovirus but in only one other genus of the family Bromoviridae, the genus Ilarvirus, and is proposed to be a novel, naturally occurring hybrid gene (13, 42). One might argue that since approximately 72% of the 2b gene is embedded within the 2a gene, its high variability might be affected by its overlapping nature. However, high variability was also observed in a short, nonoverlapping region of 2b (codon positions 81 to 93 [Fig. 5C]). Considering these facts together, it is reasonable to postulate that positive selection might still be apparent in the 2b gene and that this gene is still evolving. Further analysis for positive selection in the 2b gene will require the use of more-sophisticated tools and additional data sets.
Analysis of the distribution of synonymous and nonsynonymous mutations revealed that different coding regions exhibit different cumulative behaviors of mutations (Fig. 5), indicating that different parts of these coding regions are under different evolutionary constraints. This notion is consistent with the idea that different coding regions are under different selection pressures, as revealed by the estimation of Ka/Ks ratios (Table 4). The lack of synonymous mutations in some coding regions, i.e., codon positions 1 to 20 and 30 to 50 in the 2b coding region and positions 98 to 151 in the CP coding region (Fig. 5C and E) also suggests that these regions might be subjected to negative selection, possibly due to codon usage bias and/or maintenance of RNA structures (primary, secondary, and tertiary) important for RNA-RNA or RNA-protein interactions. It is also possible that the sample size in our study was insufficient for accurate analysis of these regions. The first scenario seems less likely, since an estimation of codon usage bias based on the ENC suggested that there was only slight bias in codon usage in these coding regions (Table 4). Interestingly, CP codon positions 98 to 151, corresponding to nucleotide positions 1564 to 1721 of Fny RNA3, were also found to have only a few mutations in quasispecies populations recovered from various host plants infected by a genetically identical CMV population (36).
Acknowledgments
We are grateful to Fernando Garcia-Arenal for critical reading of the manuscript and helpful suggestions and to Brian Foley and Bette T. Korber for valuable discussions.
This work was supported in part by the Biotechnology Risk Assessment Research Grants Program (award 99-33120-8293 to B.W.F.) of the U.S. Department of Agriculture and by the University of California. H.-X.L. was partly supported by the China Scholarship Council of the Ministry of Education, People's Republic of China.
REFERENCES
- 1.Aaziz, R., and M. Tepfer. 1999. Recombination between genomic RNAs of two cucumoviruses under conditions of minimal selection pressure. Virology 263:282-289. [DOI] [PubMed] [Google Scholar]
- 2.Aranda, M. A., A. Fraile, J. Dopazo, J. M. Malpica, and F. Garcia-Arenal. 1997. Contribution of mutation and RNA recombination to the evolution of a plant pathogenic RNA. J. Mol. Evol. 44:81-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brigneti, G., O. Voinnet, W.-X. Li, L.-H. Ji, S.-W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Canto, T., S. K. Choi, and P. Palukaitis. 2001. A subpopulation of RNA 1 of Cucumber mosaic virus contains 3′ termini originating from RNAs 2 or 3. J. Gen. Virol. 82:941-945. [DOI] [PubMed] [Google Scholar]
- 5.Canto, T., D. A. M. Prior, K.-H. Hellwald, K. J. Oparka, and P. Palukaitis. 1997. Characterization of cucumber mosaic virus. IV. Movement protein and coat protein are both essential for cell-to-cell movement of cucumber mosaic virus. Virology 237:237-248. [DOI] [PubMed] [Google Scholar]
- 6.Chao, L. 1997. Evolution of sex and the molecular clock in RNA viruses. Gene 205:301-308. [DOI] [PubMed] [Google Scholar]
- 7.Chao, L. 1988. Evolution of sex in RNA viruses. J. Theor. Biol. 133:99-112. [DOI] [PubMed] [Google Scholar]
- 8.Chaumpluk, P., Y. Sasaki, N. Nakajima, H. Nagano, I. Nakamura, K. Suzuki, K. Mise, N. Inouye, T. Okundo, and I. Furusawa. 1996. Six new subgroup I members of Japanese cucumber mosaic virus as determined by nucleotide sequence analysis of RNA3’s cDNAs. Ann. Phytopathol. Soc. Jpn. 62:40-44. (In Japanese.) [Google Scholar]
- 9.Chen, Y. K., A. F. L. M. Derks, S. Langeveld, R. Goldbach, and M. Prins. 2001. High sequence conservation among cucumber mosaic virus isolates from lily. Arch. Virol. 146:1631-1636. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y.-K., R. Goldbach, and M. Prins. 2002. Inter- and intramolecular recombinations in the cucumber mosaic virus genome related to adaptation to alstroemeria. J. Virol. 76:4119-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cillo, F., I. M. Roberts, and P. Palukaitis. 2002. In situ localization and tissue distribution of the replication-associated proteins of Cucumber mosaic virus in tobacco and cucumber. J. Virol. 76:10654-10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, S.-W., B. J. Anderson, H. R. Haase, and R. H. Symons. 1994. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198:593-601. [DOI] [PubMed] [Google Scholar]
- 13.Ding, S. W., W. X. Li, and R. H. Symons. 1995. A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 14:5762-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingo, E., and J. J. Holland. 1994. Mutation rates and rapid evolution of RNA viruses, p. 161-184. In S. S. Mores (ed.), The evolutionary biology of viruses. Raven Press, New York, N.Y.
- 15.Edwards, M. C., and D. Gonsalves. 1983. Grouping of seven biologically defined isolates of cucumber mosaic virus by peptide mapping. Phytopathology 73:1117-1120. [Google Scholar]
- 16.Fraile, A., J. L. Alonso-Prados, M. A. Aranda, J. J. Bernal, J. M. Malpica, and F. Garcia-Arenal. 1997. Genetic exchange by recombination or reassortment is infrequent in natural populations of a tripartite RNA plant virus. J. Virol. 71:934-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Arenal, F., A. Fraile, and J. M. Malpica. 2001. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 39:157-186. [DOI] [PubMed] [Google Scholar]
- 18.Graves, M. V., and M. J. Roossinck. 1995. Characterization of defective RNAs derived from RNA 3 of the Fny strain of cucumber mosaic cucumovirus. J. Virol. 69:4746-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes, R. J., and K. W. Buck. 1990. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63:363-368. [DOI] [PubMed] [Google Scholar]
- 20.Holland, J. J., K. Spindler, F. Horodyski, E. Grabau, S. Nichol, and S. VandePol. 1982. Rapid evolution of RNA genomes. Science 215:1577-1582. [DOI] [PubMed] [Google Scholar]
- 21.Hsu, H. T., L. Barzuna, Y. H. Hsu, W. Bliss, and K. L. Perry. 2000. Identification and subgrouping of Cucumber mosaic virus with mouse monoclonal antibodies. Phytopathology 90:615-620. [DOI] [PubMed] [Google Scholar]
- 22.Lin, H. X., L. Rubio, A. Smythe, M. Jiminez, and B. W. Falk. 2003. Genetic diversity and biological variation among California isolates of Cucumber mosaic virus. J. Gen. Virol. 84:249-258. [DOI] [PubMed] [Google Scholar]
- 23.Lynch, M., and T. J. Crease. 1990. The analysis of population survey data on DNA sequence variation. Mol. Biol. Evol. 7:377-394. [DOI] [PubMed] [Google Scholar]
- 24.Maddison, D. R., and W. P. Maddison. 1999. MacClade 4.0. Analysis of phylogeny and character evolution. Sinauer, Sunderland, Mass. [DOI] [PubMed]
- 25.Nei, M., and T. Gojobori. 1986. Simple methods of estimating the number of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 26.Palukaitis, P., M. J. Roossinck, R. G. Dietzgen, and F. I. B. Francki. 1992. Cucumber mosaic virus. Adv. Virus Res. 41:281-348. [DOI] [PubMed] [Google Scholar]
- 27.Perry, K. L., L. Zhang, and P. Palukaitis. 1998. Amino acid changes in the coat protein of cucumber mosac virus differentially affect transmission by the aphids Myzus persicae and Aphis gossypii. Virology 242:204-210. [DOI] [PubMed] [Google Scholar]
- 28.Perry, K. L., L. Zhang, M. H. Shintaku, and P. Palukaitis. 1994. Mapping determinants in cucumber mosaic virus for transmission by Aphis gossypii. Virology 205:591-595. [DOI] [PubMed] [Google Scholar]
- 29.Power, A. G. 2000. Insect transmission of plant viruses: a constraint on virus variability. Curr. Opin. Plant Biol. 3:336-340. [DOI] [PubMed] [Google Scholar]
- 30.Pressing, J., and D. C. Reaney. 1984. Divided genomes and intrinsic noise. Mol. Evol. 20:135-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizos, H., L. V. Gunn, R. D. Pares, and M. R. Gillings. 1992. Differentiation of cucumber mosaic virus isolates using the polymerase chain reaction. J. Gen. Virol. 73:2099-2103. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Alvarado, G., G. Kurath, and J. A. Dodds. 1995. Heterogeneity in pepper isolates of cucumber mosaic virus. Plant Dis. 79:450-455. [Google Scholar]
- 33.Roossinck, M. J. 2002. Evolutionary history of Cucumber mosaic virus deduced by phylogenetic analyses. J. Virol. 76:3382-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roossinck, M. J., L. Zhang, and K.-H. Hellwald. 1999. Rearrangements in the 5′ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J. Virol. 73:6752-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an intergrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 36.Schneider, W. L., and M. J. Roossinck. 2001. Genetic diversity in RNA virus quasispecies is controlled by host-virus interactions. J. Virol. 75:6566-6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, M., T. Hibi, and C. Masuta. 2003. RNA recombination between cucumoviruses: possible role of predicted stem-loop structures and an internal subgenomic promoter-like motif. Virology 306:77-86. [DOI] [PubMed] [Google Scholar]
- 38.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer, Sunderland, Mass.
- 39.Szilassy, D., K. Salanki, and E. Balazs. 1999. Molecular evidence for the existence of two distinct subgroups in cucumber mosaic cucumovirus. Virus Genes 18:221-227. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright, F. 1990. The “effective number of codons” used in a gene. Gene 87:23-29. [DOI] [PubMed] [Google Scholar]
- 42.Xin, H. W., L. H. Ji, S. W. Scott, R. H. Symons, and S. W. Ding. 1998. Ilarviruses encode a Cucumovirus-like 2b gene that is absent in other genera within the Bromoviridae. J. Virol. 72:6956-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]