Abstract

Mass spectrometry imaging (MSI) is a versatile tool for visualizing molecular distributions in complex biological specimens, but locating microscopic chemical features of interest can be challenging in samples that lack a well-defined anatomy. To address this issue, we developed a correlated imaging approach that begins with performing matrix-assisted laser desorption/ionization (MALDI) MSI to obtain low-resolution molecular maps of a sample. The resulting maps are then used to direct subsequent microscopic secondary ion mass spectrometry (SIMS) imaging and tandem mass spectrometry (MS/MS) experiments to examine selected chemical regions of interest. By employing MALDI undersampling, the sample surface is left mostly unperturbed and available for the SIMS analysis, while also generating an ablation array that can be used for navigation in SIMS. We validated this MALDI-guided SIMS approach using cultured biofilms of the opportunistic pathogen Pseudomonas aeruginosa; bioactive secondary metabolites, including rhamnolipids and quinolones, were detected and visualized on both macro- and microscopic size scales. MSI mass assignments were confirmed with in situ MALDI MS/MS and capillary electrophoresis–electrospray ionization MS/MS analysis of biofilm extracts. Two strains of P. aeruginosa were compared, wild type and a quorum sensing mutant, and differences in metabolite abundance and distribution were observed.
Mass spectrometry imaging (MSI) is an analytical approach that enables multiplexed, label-free, and nontargeted chemical imaging of sample surfaces.1−3 These features make it a useful tool for visualizing spatially heterogeneous chemical environments, especially for small molecules such as drugs and metabolites, which can otherwise be difficult to reliably label and distinguish from structurally similar compounds. MSI is commonly performed by scanning a microprobe over the sample surface and acquiring mass spectra at thousands of discrete positions in a rectangular array. Hundreds of unique ions are routinely detected per spectrum, and any of these signals may be used to generate an ion image representing the relative abundance of the ionized compound across the analyzed region.
Many different microprobes are available for MSI, including focused lasers for matrix-assisted laser desorption/ionization (MALDI)4,5 and focused ion beams for secondary ion mass spectrometry (SIMS).6−8 MALDI and SIMS are complementary techniques; MALDI affords excellent detection limits and a broad mass range extending to tens of kDa (in practice for MSI), and SIMS provides the highest lateral spatial resolution and nanometer scale depth resolution,9 but with a more limited (typically sub-kDa) mass range. MALDI requires chemical modification of the sample surface to enhance the yields of nonvolatile high molecular weight species to useful levels; this is often accomplished by applying a dissolved organic compound that subsequently dries and crystallizes on the surface, but alternative matrixes such as sputtered metal coatings10,11 are also used. SIMS does not require a matrix but can nevertheless benefit from similar treatments, including metal coatings.10−12 Methods that combine MALDI and SIMS imaging have been utilized recently for multiscale chemical mapping of nervous tissue,13,14 skin and kidney,15 single cultured neurons,13 bacterial biofilms,16 and a biofuel feedstock grass.12
Microscopic MSI allows multiplexed visualization of unlabeled analytes at cellular and subcellular sizes,17 but it also involves trade-offs: sampled surface area (per pixel) and detected ions decrease quadratically with probe diameter, while pixel count, and therefore data file size and acquisition time, increase quadratically with spatial resolution. In practice, the increase in experimental time may even be larger than quadratic because one often needs to acquire a greater number of laser/ion shots to make up for the smaller amount of material present in the smaller pixel size. These issues equate to practical limitations; large samples are not normally imaged entirely at high resolution given the time that this takes, and so a microscopic region of interest (ROI) must first be specified by some other means. Optical image correlation is often used for MSI of tissue sections, but this approach depends on having a sample with a well-understood anatomy and obvious morphological features, e.g., white and gray matter of brain tissue. Immunohistological staining has been applied prior to MSI in order to enhance the information content of the optical image,18 but this requires having a specific marker of interest, and also involves immersion of the sample in liquid washes and stain(s), which results in chemical modification and analyte delocalization on the microscopic scale. Thus, for samples that lack a visible and well-defined anatomy, new imaging techniques are needed.
To address the concerns outlined above, we developed a MALDI-guided SIMS approach wherein a full-sample chemical map is first acquired by MALDI MS. This map is then used as a guide in performing high resolution SIMS imaging of microscopic ROIs. Metal-assisted sample preparation is utilized to ensure compatibility between both ion imaging methods, and MALDI undersampling is used in order to leave an unperturbed sample area for subsequent SIMS imaging. Laser ablation spots are visualized in the SIMS images and used to precisely specify microscopic ROIs. The larger laser ablation volume of MALDI yields substantially higher ion counts, enabling in situ ion identification with tandem mass spectrometry (MS/MS) on the same sample subsequent to imaging. Additionally, mutual detection of a given molecular compound by both ionization techniques serves as cross-validation of the mass assignments.
We used MALDI-guided SIMS to interrogate cultured biofilms of the opportunistic pathogen Pseudomonas aeruginosa in order to visualize secondary metabolites that are involved in biofilm growth and cellular signaling. These compounds were detected as intact pseudomolecular ions for the first time using SIMS, and confirmed with in situ MS/MS as well as capillary electrophoresis–electrospray ionization (CE–ESI) MS/MS of bulk biofilm extracts. Although MALDI MSI has yielded informative “colony scale” images19,20 and SIMS imaging has been applied to obtain exquisite cell scale chemical information21−24 in previous microbiological studies, the unique sequential combination of MALDI and SIMS utilized here guides high-resolution imaging systematically and also enables this information to be placed within the macroscopic spatial context of the biofilm.
Experimental Section
Materials and Chemicals
Mixed rhamnolipid “R-95 Rhamnolipid” (95%, AldrichCPR), 2-heptyl-3-hydroxy-4(1H)-quinolone (Pseudomonas quinolone signal, PQS), and 2-heptyl-4-quinolone (HHQ) standards were purchased from Sigma-Aldrich (St Louis, MO) and dissolved in methanol for MS experiments. The substrates for biofilm growth were created by scoring silicon (100) wafers (4 in. diameter, Silicon, Inc., Boise, ID) with a diamond scribe, and breaking them to yield 2 × 2 cm tiles before use. Solvents and buffer ingredients for CE–MS were purchased from Sigma-Aldrich and the mixtures prepared in lab.
Biofilm Growth
Two P. aeruginosa strains were used in this work: wild type (WT) (ATCC 15692, ATCC, Manassas, VA) and the quorum sensing (QS) ΔlasIΔrhlI mutant,25 which is deficient in homoserine lactone production, i.e., has disabled QS. Cell suspensions were grown at 30 °C overnight in fastidious anaerobe broth culture medium26 with a filter-sterilized glucose carbon source (150 μL of 1.2 M glucose solution per 6 mL of culture) and then transferred to Petri dishes. Cell suspension in the Petri dishes was diluted 50× with culture medium, allowed 1 min for initial cell attachment, and then the silicon tiles were fully submerged in the mixture. Static biofilms were permitted to grow for 72 h at 30 °C. Growth medium was then removed from the Petri dishes by pipette and the biofilms were allowed to air-dry completely prior to preparation for MSI analysis.
Mass Spectrometry Imaging
Sputter coating was performed using a Desk II TSC sputter coater equipped with a gold target (Denton Vacuum, Moorestown, NJ) operated at 64 mTorr Ar pressure and 40% power with 6 s of application time. MALDI MS was performed on an UltrafleXtreme MALDI-time-of-flight (TOF)/TOF mass spectrometer (Bruker Daltonics, Billerca, MA) in positive ion mode, reflectron enabled, acquiring m/z 20–1000, and equipped with a frequency-tripled Nd:YAG laser focused to an ∼100 μm probe diameter and triggered at 1000 Hz. Quadratic mass calibration was performed with K+ and Aun+ cluster ions detected from the prepared sample. MS/MS (TOF/TOF) was performed by postsource decay without CID (collision-induced dissociation), adjusting the laser power to optimize fragmentation for individual ions of interest. Data were processed using FlexAnalysis v3.4 and FlexImaging v3.0 (Bruker Daltonics); ion filters were set to m/z ± 0.25 and images normalized to total ion count. SIMS imaging was performed on a TRIFT III (Physical Electronics, Chanhassen, MN) TOF mass spectrometer using a 22 keV Au+ primary ion source yielding an ∼ 2-nA sample current and a 500 nm diameter probe spot. Images were acquired in the static SIMS regime, i.e., with a primary beam flux of <1× 1012 primary ions/cm2. Control and data processing were performed using WinCadence v4.4 software (Physical Electronics, Eden Prairie, MN). Calibration was performed with H+, Na+, K+, and Aun+ cluster ions. Optical images of samples were acquired with a Perfection V300 Photo flatbed scanner (Epson, Suwa, Nagano, Japan).
For the MALDI-guided SIMS experiments, bacterial biofilms were first cultured and dried on silicon as described above. A thin layer of gold (∼ 2 nm) was applied to the sample and then an optical reference image was acquired. We affixed the silicon pieces onto steel plates with double-sided conductive copper-backed tape; the steel plates match the size required by the Bruker MTP ABI plate adapter that holds older ABI MALDI TOF targets. The steel plates were milled to produce a recessed area matching the height of the mounted sample (silicon substrate plus tape) in order to level the sample surface with the MALDI target surface; this was critical for obtaining high quality MS spectra. The entire sample surface was then imaged by MALDI MS at a 500 or 1000 μm pitch, 500 shots per position. The MALDI ion images were inspected and used to select ROIs for microscopic SIMS analysis. The sample was transferred to the SIMS instrument, with care taken to maintain orientation. A SIMS “survey” was then acquired in mosaic mapping mode in which a relatively large (typically 4 × 4 mm) area was imaged quickly at low resolution (16 × 16 μm pixel size) by stitching multiple small and fast (1 s accumulation time) ion images together. This yielded images with low signal and contrast, but sufficient to visualize the MALDI ablation marks that were used for sample navigation and correlation with MALDI images. High-resolution SIMS images (typically a 150 × 150 μm field of view and 0.6 × 0.6 μm pixel size) were then acquired at ROIs specified from the MALDI map and subsequent SIMS survey. At least three each of the WT and QS mutant biofilms were prepared and imaged in this manner; physical and chemical features (and therefore imaging results) varied slightly between batches, so the results presented here are representative of consistent observations.
CE–ESI MS/MS Biofilm Extract Analysis
CE–ESI MS/MS was performed as reported previously27 using both an Impact HD or a maXis 4G Qq-ToF mass spectrometer (Bruker Daltonics) operated in positive and negative ion modes. Biofilm extracts were prepared by collecting ∼1 mg of dried biofilm from the silicon substrate with a clean razor blade, depositing the film into a microcentrifuge vial, adding 20 μL of extraction solution (50/50 (v/v) MeOH:H2O + 0.5% AcOH), shaking vigorously for 2 min, then centrifuging for 5 min at 2000g. For each run, 6 nL of supernatant was loaded into a capillary (65–70 cm long) and a separation potential of 15 kV applied. For negative ion mode analysis, CE was performed using a background electrolyte composed of 20 mM ammonium bicarbonate, and a sheath liquid of 60% (v/v) isopropyl alcohol and 200 μM ammonium bicarbonate, delivered at 600 nL per minute. Instrument calibration was performed using sodium acetate clusters in negative ion mode. Molecular features were assigned with high confidence through matching of the tandem mass spectral data from the endogenous substances with those found at publicly available mass spectral databases (METLIN28).
Results
MALDI-directed SIMS imaging enables the specification of ROIs for high-resolution MSI of specimens that lack a characteristic anatomy, as in the bacterial biofilms studied here. A schematic of the overall workflow is shown in Figure 1 and described below.
Figure 1.

Workflow schematic of a MALDI-guided SIMS experiment, illustrating: (A) sample preparation to meet vacuum MS requirements (must be flat, dry, and mounted to a conductive or semiconductive substrate), (B) surface chemical treatment for ion signal enhancement, (C) undersampling MALDI MS image acquisition across the specimen surface, (D) specification of ROIs for SIMS analysis based on the MALDI chemical map, and (E) SIMS imaging at microscopic ROIs.
Dried biofilm samples were first sputter-coated with a 1–2 nm thick Au film for metal-assisted MS, providing signal enhancement for small molecules (effective to <1000 Da) with multiple additional advantages: mutual compatibility with both laser desorption/ionization and SIMS, no reduction in lateral resolution due to diffusion of analytes during solvent application or due to matrix particle size, minimal low-mass chemical background, monoisotopic Aun+ peaks for internal calibration, improved sample conductance to mitigate sample charging effects in SIMS, and optical transparency, facilitating sample navigation and optical imaging after application. The thickness of the Au coating, optimized using mixed rhamnolipid standard spotted on a silicon wafer, was found to provide an over 10-fold signal enhancement for [M + Na]+ ions (shown in Figure S1, Supporting Information). Typical SIMS and MALDI MS profiles are shown in Figure S2 (Supporting Information).
MS microprobes are inherently destructive, so a challenge in MALDI-guided SIMS is to acquire the MALDI image while leaving most of the sample surface unperturbed for subsequent SIMS imaging. This was accomplished by undersampling, i.e., setting pixel size and array pitch to a length much larger than the microprobe diameter. The effect of undersampling is apparent in Figure 2, showing selected results from the analysis of a WT P. aeruginosa biofilm.
Figure 2.
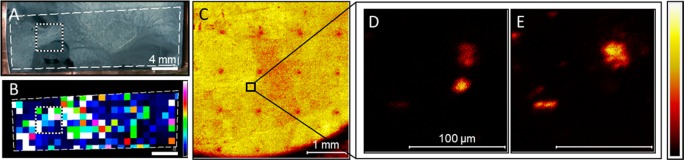
Selected results from MALDI-guided SIMS of a WT P. aeruginosa biofilm. (A) Optical and (B) MALDI ion images of quinolone PQS (m/z 260.17) are acquired, and an ROI (dotted white square) is selected for further analysis (scale bars = 4 mm). (C) Mosaic-mode SIMS total ion image reveals laser ablation marks in the sample, and these positions are correlated with the MALDI ion image to select a microscopic ROI (black square, scale bar = 1 mm). High-resolution SIMS imaging at the ROI reveals micrometer scale localization of (D) PQS (m/z 260.17) and (E) HHQ (m/z 244.17) on the biofilm (scale bars = 100 μm).
MALDI imaging was performed in this experiment with a 1 mm pitch and 100 μm diameter laser microprobe, obtaining a coarse chemical map for multiple biologically related ions, including PQS (MH+ at m/z 260.17), shown in Figure 2B. The laser ablation spots are clearly visible in the SIMS total ion image shown in Figure 2C as a decrease in overall signal, likely due to removal of the signal-enhancing metal coating, and these features form an array that can be used to navigate the sample and select ROIs for SIMS imaging, precisely positioned in relation to the MALDI data. In the ROI selected here, microscopic SIMS images show differential distribution of PQS (Figure 2D) and HHQ (MH+ at m/z 244.17) (Figure 2E). In addition to the MS/MS analyses discussed below, the assignment of these ions was confirmed by colocalization of ion fragments that are characteristic of their respective quinolone molecular classes (as shown in Figure S3, Supporting Information).29
Although mass profiles varied between and within individual biofilms, up to nine mono- and di-rhamnolipids, and seven quinolone compounds, were consistently detected in the WT P. aeruginosa biofilms. All nine putative rhamnolipids were confirmed by CE–ESI-MS/MS analysis of bulk WT biofilm extract (results shown in Figure S4, Supporting Information), and six of the nine could be confirmed directly from the biofilm using in situ MALDI MS/MS, as we reported recently,30 in comparison with standards and in agreement with previously published fragmentation analysis.31 Rhamnolipids were predominantly detected as sodium adducts ([M + Na]+ and sometimes [M – H + 2Na]+) by both MALDI and SIMS, and as deprotonated pseudomolecular ions by ESI, consistent with previous work.32 Similarly, all seven putative quinolones were confirmed by CE–ESI MS/MS (shown in Figure S4, Supporting Information); HHQ, PQS, and NHQ were also confirmed by in situ MALDI MS/MS (spectra shown in Figure S5, Supporting Information). Quinolones were detected as primarily MH+ ions (and [M – H]− ions with ESI), which is consistent with previous ESI MS work,29 and formation of these ions in SIMS was also confirmed with quinolone standards (PQS and HHQ; others were not available), both of which yielded primarily MH+ ions after metal enhancement (as shown in Figure S6, Supporting Information). Several pairs of known quinolones are isobaric, including PQS and 4-hydroxy-2-heptylquinoline-N-oxide (HQNO), but these were discerned by MS/MS fragmentation patterns.29 In the case of PQS and HQNO at m/z 260, in situ MALDI MS/MS yielded predominantly PQS-related fragments (m/z 175, 188) and little or no contribution from HQNO (m/z 159, 172). Using CE–MS/MS, detectable quantities of HQNO were observed from a biofilm extract; our lack of detection of this compound by MALDI and SIMS may indicate that HQNO is localized below the MALDI and SIMS sampling depth within the biofilm, or that low levels (below our MSI detection limits) are uniformly distributed on the surface. Table 1 compiles the MALDI MS, SIMS, and CE–ESI MS data acquired from the WT P. aeruginosa biofilms. The standard rhamnolipid notation used here is detailed elsewhere.33
Table 1. Combined MALDI MS, SIMS, and CE–ESI MS Results for Rhamnolipid and Quinolone Species Detected from the WT P. aeruginosa Biofilma.
| CE-ESI
MS |
MALDI
MS |
SIMS |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| compound ID | molecular formula | [M + H]+ observed | [M – H]− observed | ΔM (neg) | [M + H]+ observed | [M + Na]+ observed | [M – H + 2Na]+ observed | ΔM | [M + H]+ observed | [M + Na]+ observed | [M – H + 2Na]+ observed | ΔM |
| HHQ (2-heptyl-4-quinolone) | C16H22NO | 244.170 | 242.152 | –0.003 | 244.18 | 0.01 | 244.17 | 0.00 | ||||
| PQS (2-heptyl-3-hydroxyquinolone) | C16H21NO2 | 260.164 | 258.148 | –0.002 | 260.17 | 0.01 | 260.16 | –0.01 | ||||
| HQNO (4-hydroxy-2-heptylquinolone-N-oxide) | C16H21NO2 | 260.165 | 258.148 | –0.002 | 260.17 | 0.01 | 260.16 | –0.01 | ||||
| C9:1-NHQ (2-nonenyl-4-quinolone) | C18H23NO | 268.168 | –0.002 | 270.19 | 0.01 | |||||||
| NHQ (2-nonyl-4-quinolone) | C18H25NO | 272.202 | 270.184 | –0.002 | 272.21 | 0.01 | 272.20 | 0.00 | ||||
| C9:1-PQS (2-nonenyl-3-hydroxyquinolone) C9:1-NQNO (4-hydroxy-2-nonenylquinolone-N-oxide) | C18H23NO2 | 286.180 | 284.161 | –0.005 | 286.19 | 0.01 | 286.18 | 0.00 | ||||
| C9-PQS (2-nonyl-3-hydroxyquinolone) NQNO (4-hydroxy-2-nonylquinolone-N-oxide) | C18H25NO2 | 286.178 | –0.003 | 288.20 | 0.01 | 288.19 | 0.00 | |||||
| C11:1-UHQ (2-undecenyl-4-quinolone) | C20H27NO | 298.21 | –0.01 | |||||||||
| Rha-C10 | C16H30O7 | 335.208 | 333.187 | –0.005 | 357.21 | 379.19 | 0.02 | 379.16 | –0.01 | |||
| Rha-C8-C10 | C24H44O9 | 475.289 | –0.002 | 499.31 | 0.02 | 499.28 | –0.01 | |||||
| Rha-Rha-C10 | C22H40O11 | 481.256 | 479.247 | –0.002 | 503.27 | 525.25 | 0.02 | |||||
| Rha-C10-C10 | C26H48O9 | 503.321 | –0.001 | 527.34 | 549.33 | 0.02 | 527.30 | –0.02 | ||||
| Rha-C10-C12:1 | C28H50O9 | 529.337 | –0.001 | 553.38 | 0.04 | 553.33 | –0.01 | |||||
| Rha-C10-C12 | C28H52O9 | 531.355 | 0.002 | 555.38 | 0.03 | 555.33 | –0.02 | |||||
| Rha-Rha-C8-C10 | C30H54O13 | 621.351 | 0.002 | 645.38 | 0.03 | 645.34 | –0.01 | |||||
| Rha-Rha-C10-C10 | C32H58O13 | 649.379 | –0.001 | 673.42 | 695.41 | 0.04 | 673.40 | 0.02 | ||||
| Rha-Rha-C10-C12 | C34H62O13 | 677.411 | –0.001 | 701.46 | 723.44 | 0.05 | 701.40 | –0.01 | ||||
Comparison of WT and QS mutant P. aeruginosa biofilms
WT and QS mutant P. aeruginosa biofilms were compared using MALDI-guided SIMS to characterize spatiochemical differences arising from a disabled QS system, with the results shown in Figure 3. These biofilms were gold-coated and imaged by MALDI in a single experiment to minimize preparation- and instrument-related variations.
Figure 3.
MALDI-guided SIMS images reveal multiscale compositional differences between the biofilms of WT and QS mutant P. aeruginosa. (Top row) Arrows in the optical (top image) and MALDI ion (bottom image) images indicate ROIs where the (middle and bottom rows) SIMS images were acquired. MALDI and SIMS ion image false color scales are shown on the right; for each specific ion, the SIMS color scale range was set identically in QS Mutant and WT ion images: Rha-Rha-C10-C10 ([M + Na]+m/z 673.4), 0–5 counts (cts); Rha-C10-C10 ([M + Na]+m/z 527.3), 0–8 cts; HHQ (MH+m/z 244.2), 0–40 cts; PQS (MH+m/z 260.2), 0–10 cts; NHQ (MH+m/z 272.2), 0–100 cts. WT and QS mutant biofilms were imaged in a single experiment by MALDI and the color scales are the same for each ion in MALDI images as well. Optical/MALDI scale bars =2 mm, SIMS scale bars =100 μm. Regions of WT biofilm where tape was adhered to the surface were excluded.
The MALDI images in Figure 3 show a high abundance of the major rhamnolipids Rha-Rha-C10-C10 and Rha-C10-C10 (both detected as [M + Na]+ at m/z 673.4 and 527.3, respectively), distributed uniformly across the WT biofilm but completely absent in the QS mutant biofilm. The SIMS images reflect these observations on the microscopic scale; rhamnolipids are uniformly distributed in the WT biofilm, but they are not seen in the QS mutant biofilm. In contrast, quinolones were highly heterogeneous within and between both samples. HHQ shows comparable overall abundance between biofilms in the MALDI image, though a macroscopic region of relatively high abundance can be seen to follow the growth contour of the QS mutant. The HHQ SIMS image at this feature likewise shows higher overall signal as well as “hot spots” of intense localization in aggregated clusters of <10 μm features, whereas the WT biofilm SIMS image shows lower overall abundance and smaller, less intense localizations without the aggregate features. Another prominent quinolone, 2-nonyl-4-quinolone (NHQ, MH+ at m/z 272.2), exhibits sparse localizations in the MALDI images of both biofilms, and appears to colocalize with HHQ in the microscopic aggregates. Lower resolution mosaic SIMS images acquired around the specified ROIs on these biofilms (shown in Figure S7, Supporting Information) indicate that these aggregates are abundant across the surface of the QS mutant biofilm in the selected area, while not observed at all with the WT. In contrast, PQS shows higher abundance on the WT biofilm in the MALDI image, and this is also reflected in the selected SIMS ROIs, which show less PQS on the QS mutant relative to the other quinolones.
Discussion
MALDI-guided SIMS imaging enables visualization of molecular distributions on both the microscopic (cell) and macroscopic (population) size scales. The approach was validated with P. aeruginosa in order to visualize the chemical features associated with growth and cellular signaling. P. aeruginosa is an opportunistic pathogen known for organizing into biofilms, thin layers of bacteria adhered to a surface and embedded in a self-produced extracellular polymeric matrix, which imparts numerous advantages to ensure survival.34−36 Biofilm development is regulated via cell-to-cell chemical signaling, which includes quorum sensing (QS),37 utilizing several classes of secondary metabolites such as homoserine lactones,38 quinolones,39 rhamnolipids40 and phenazines.41 This chemical language is expansive, P. aeruginosa produces over 50 distinct quinolone compounds alone,29 and is also highly complex, as a single metabolite may play several distinct roles in biofilm development.39 Functional characterization is incomplete for most of these compounds, so there is a great deal yet to be elucidated about their roles in biofilm growth and homeostasis.
Several interesting observations were made about P. aeruginosa biofilm composition based on the optical, low-resolution MALDI, and high-resolution SIMS images obtained in this work. The MALDI MS images show heterogeneous macroscopic distributions of many biomolecules, including PQS (Figure 2B), which appear to follow biofilm growth contours, at least partially. PQS production varies with cell density and population maturity,42 thus the observed distribution may reflect temporal changes in production during biofilm growth and expansion. SIMS analysis directed at a region of high PQS abundance revealed a punctate microscopic distribution for PQS (Figure 2D), concentrated in micrometer scale features. These features could be clusters of cells exposed at the surface of the biofilm, or aggregations of membrane vesicles, which have been shown to contain ∼90% of the PQS produced by P. aeruginosa.43 Several other quinolones were typically also detected along with PQS, and interestingly, these quinolones were not always colocalized; Figure 2E shows a region where HHQ was observed to be similarly punctate but distributed differently. HHQ is the direct precursor to PQS,44 released by cells and taken back up by others for PQS production;29 therefore, the features observed in the HHQ SIMS image may represent pockets of secreted HHQ. HHQ is also an autoinducer,45 so local concentration around producing cells in the biofilm may increase production further via positive feedback. The differential distributions of HHQ and PQS observed here also raise the possibility of cell specialization within a biofilm population, a phenomenon known to occur with other biofilm-producing microbes such as Bacillus subtilis, but not yet reported with P. aeruginosa.46
Several of the differences observed between the WT and QS mutant P. aeruginosa strains studied here fit well with what is known about their genomes. The QS mutant is a ΔlasIΔrhlI strain, which is incapable of synthesizing N-3-oxo-dodecanoyl-l-homoserine lactone (HSL) (3-oxo-C12-HSL) and N-butyryl-l-HSL (C4-HSL), two “master regulator” signals controlling the P. aeruginosa QS systems, las and rhl, respectively.38 One expected effect of these mutations is a complete lack of rhamnolipid production,25 a change that is clearly visualized in both the MALDI and SIMS ion images of the rhamnolipids Rha-Rha-C10-C10 and Rha-C10-C10. Quinolone biosynthesis is also regulated by these signals indirectly and in a competitive manner; 3-oxo-C12-HSL promotes PQS production whereas C4-HSL suppresses it.38 Thus, the reduced PQS levels detected in the QS mutant biofilm may arise from loss of las promotion, with residual PQS produced by promotion from other signaling pathways, as the las system is not the dominant driver of PQS biosynthesis under certain growth conditions.47
The changes observed in the HHQ and NHQ distributions are not as easily explained, partly because they are more subtle, and partly because these quinolones are not as well-characterized as PQS, so the effect of the ΔlasIΔrhlI mutations on their abundance and distribution is less easily interpreted. The microscopic quinolone aggregations observed in the QS mutant biofilm could be due to a lack of rhamnolipid production; rhamnolipids act as surfactants, which can mobilize cells40 and solubilize quinolones in the biofilm,48 so secreted quinolones or cells may aggregate more extensively without them. Another possible explanation is that PQS is required for production of the membrane vesicles that ordinarily transmit quinolones and other metabolites between cells,43 so decreased PQS levels may result in reduced vesicular production and consequently, a local buildup of other quinolones around the cells of origin. Ongoing work is focused on elucidating the role of these quinolones and others in P. aeruginosa biofilms using MALDI-guided SIMS and Raman imaging.
Conclusions
We have developed a sequentially combined chemical imaging approach wherein an undersampled MALDI MS image is used to guide microscopic SIMS imaging experiments to target chemically interesting ROIs on the same sample. This combination of complementary microprobes conveys numerous advantages in an MSI experiment, including: (1) specification of microscopic ROIs from a chemical map rather than an optical image, (2) spatially registered macro- and microscopic chemical images of a single sample, (3) generation of a fiducial grid for sample navigation in SIMS, and (4) MALDI MS/MS capability for in situ ion characterization. Applied to bacterial biofilms of P. aeruginosa, MALDI-guided SIMS enabled the detection and visualization of multiple secondary metabolites, including rhamnolipid surfactants and quinolone signaling molecules that heretofore have not been reported using SIMS alone. The combination of SIMS and MALDI revealed both macroscopic and cell scale chemical heterogeneity across the biofilms for the analytes studied, and can be applied to other samples where similar multiscale complexity exists, such as tissue sections. Additionally, although a single metal-enhanced preparation was utilized in this work for compatibility with both MALDI and SIMS, future work will include applying traditional MALDI matrixes in a microdroplet array in order to extend the mass range to include detection of proteins and other macromolecules of interest.
Acknowledgments
This work was funded by the Department of Energy Office of Biological and Environmental Research through grant DE SC0006642 and NIH through grant U54GM093342. SIMS imaging was performed in the Center for Microanalysis of Materials, University of Illinois at Urbana–Champaign, partially supported by the U.S. Department of Energy through grant DEFG02-91-ER45439. The authors are grateful to Dr. Vanessa Phelan and Prof. Pieter Dorrestein for stimulating scientific discussions.
Supporting Information Available
Supplemental figures described in the text, including MS/MS spectra and analysis of standards. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Rubakhin S.; Sweedler J. In Mass Spectrometry Imaging; Rubakhin S. S.; Sweedler J. V., Eds.; Humana Press: New York, 2010; pp 21–49. [Google Scholar]
- Chughtai K.; Heeren R. M. A. Chem. Rev. 2010, 110, 3237–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setou M.Imaging Mass Spectrometry: Protocols for Mass Microscopy; Springer: Tokyo, 2010. [Google Scholar]
- Gusev A. I.; Vasseur O. J.; Proctor A.; Sharkey A. G.; Hercules D. M. Anal. Chem. 1995, 67, 4565–4570. [Google Scholar]
- Caprioli R. M.; Farmer T. B.; Gile J. Anal. Chem. 1997, 69, 4751–4760. [DOI] [PubMed] [Google Scholar]
- Castaing R.; Slodzian G. J. Microsc. (Paris) 1962, 1, 395–410. [Google Scholar]
- Galle P. Ann. Phys. Biol. Med. 1970, 42, 83–94. [Google Scholar]
- Boxer S. G.; Kraft M. L.; Weber P. K. Annu. Rev. Biophys. 2009, 38, 53–74. [DOI] [PubMed] [Google Scholar]
- Mao D.; Brenes D. A.; Lu C.; Wucher A.; Winograd N. Surf. Interface Anal. 2013, 45, 65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altelaar A. F. M.; Klinkert I.; Jalink K.; de Lange R. P. J.; Adan R. A. H.; Heeren R. M. A.; Piersma S. R. Anal. Chem. 2005, 78, 734–742. [DOI] [PubMed] [Google Scholar]
- Delcorte A.; Bour J.; Aubriet F.; Muller J. F.; Bertrand P. Anal. Chem. 2003, 75, 6875–6885. [DOI] [PubMed] [Google Scholar]
- Li Z.; Bohn P. W.; Sweedler J. V. Bioresour. Technol. 2010, 101, 5578–5585. [DOI] [PubMed] [Google Scholar]
- Yang H.-J.; Sugiura Y.; Ishizaki I.; Sanada N.; Ikegami K.; Zaima N.; Shrivas K.; Setou M. Surf. Interface Anal. 2010, 42, 1606–1611. [Google Scholar]
- Monroe E. B.; Annangudi S. P.; Hatcher N. G.; Gutstein H. B.; Rubakhin S. S.; Sweedler J. V. Proteomics 2008, 8, 3746–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touboul D.; Roy S.; Germain D. P.; Chaminade P.; Brunelle A.; Laprévote O. Int. J. Mass spectrom. 2007, 260, 158–165. [Google Scholar]
- Debois D.; Hamze K.; Guerineau V.; Le Caer J. P.; Holland I. B.; Lopes P.; Ouazzani J.; Seror S. J.; Brunelle A.; Laprevote O. Proteomics 2008, 8, 3682–3691. [DOI] [PubMed] [Google Scholar]
- Lanni E. J.; Rubakhin S. S.; Sweedler J. V. J. Proteomics 2012, 75, 5036–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurand P.; Schwartz S. A.; Billheimer D.; Xu B. J.; Crecelius A.; Caprioli R. M. Anal. Chem. 2004, 76, 1145–1155. [DOI] [PubMed] [Google Scholar]
- Blaze M. T. M.; Aydin B.; Carlson R. P.; Hanley L. Analyst 2012, 137, 5018–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J.; Roach P.; Alexandrov T.; Heath B. S.; Yang J. Y.; Kersten R. D.; van der Voort M.; Pogliano K.; Gross H.; Raaijmakers J. M.; Moore B. S.; Laskin J.; Bandeira N.; Dorrestein P. C. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, E1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekas A. E.; Poretsky R. S.; Orphan V. J. Science 2009, 326, 422–426. [DOI] [PubMed] [Google Scholar]
- Lechene C. P.; Luyten Y.; McMahon G.; Distel D. L. Science 2007, 317, 1563–1566. [DOI] [PubMed] [Google Scholar]
- Thiel V.; Toporski J.; Schumann G.; Sjövall P.; Lausmaa J. Geobiology 2007, 5, 75–83. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan S.; Fletcher J. S.; Goodacre R.; Lockyer N. P.; Micklefield J.; Vickerman J. C. Anal. Chem. 2008, 80, 1942–1951. [DOI] [PubMed] [Google Scholar]
- Shrout J. D.; Chopp D. L.; Just C. L.; Hentzer M.; Givskov M.; Parsek M. R. Mol. Microbiol. 2006, 62, 1264–1277. [DOI] [PubMed] [Google Scholar]
- Heydorn A.; Nielsen A. T.; Hentzer M.; Sternberg C.; Givskov M.; Ersbøll B. K.; Molin S. Microbiology 2000, 146, 2395–2407. [DOI] [PubMed] [Google Scholar]
- Nemes P.; Rubakhin S. S.; Aerts J. T.; Sweedler J. V. Nat. Protoc. 2013, 8, 783–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A.; O’Maille G.; Want E. J.; Qin C.; Trauger S. A.; Brandon T. R.; Custodio D. E.; Abagyan R.; Siuzdak G. Ther. Drug Monit. 2005, 27, 747–751. [DOI] [PubMed] [Google Scholar]
- Lépine F.; Milot S.; Déziel E.; He J.; Rahme L. G. J. Am. Soc. Mass Spectrom. 2004, 15, 862–869. [DOI] [PubMed] [Google Scholar]
- Masyuko R. N.; Lanni E. J.; Driscoll C. M.; Shrout J. D.; Sweedler J. V.; Bohn P. W.. Analyst, in press, DOI: 10.1039/C4AN00435C. [DOI] [Google Scholar]
- de Koster C. G.; Vos B.; Versluis C.; Heerma W.; Haverkamp J. Biol. Mass. Spectrom. 1994, 23, 179–185. [DOI] [PubMed] [Google Scholar]
- Price N. P. J.; Ray K. J.; Vermillion K.; Kuo T.-M. Carbohydr. Res. 2009, 344, 204–209. [DOI] [PubMed] [Google Scholar]
- Abdel-Mawgoud A.; Lépine F.; Déziel E. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H.-C.; Wingender J. Nat. Rev. Microbiol. 2010, 8, 623–633. [DOI] [PubMed] [Google Scholar]
- Costerton J. W.; Stewart P. S.; Greenberg E. P. Science 1999, 284, 1318–1322. [DOI] [PubMed] [Google Scholar]
- Drenkard E. Microbes Infect. 2003, 5, 1213–1219. [DOI] [PubMed] [Google Scholar]
- Davies D. G.; Parsek M. R.; Pearson J. P.; Iglewski B. H.; Costerton J. W.; Greenberg E. P. Science 1998, 280, 295–298. [DOI] [PubMed] [Google Scholar]
- Juhas M.; Eberl L.; Tümmler B. Environ. Microbiol. 2005, 7, 459–471. [DOI] [PubMed] [Google Scholar]
- Huse H.; Whiteley M. Chem. Rev. 2010, 111, 152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J.; Richardson A.-P.; Lépine F.; Déziel E. Environ. Microbiol. 2007, 9, 2622–2630. [DOI] [PubMed] [Google Scholar]
- Dietrich L. E. P.; Price-Whelan A.; Petersen A.; Whiteley M.; Newman D. K. Mol. Microbiol. 2006, 61, 1308–1321. [DOI] [PubMed] [Google Scholar]
- Diggle S. P.; Winzer K.; Chhabra S. R.; Worrall K. E.; Cámara M.; Williams P. Mol. Microbiol. 2003, 50, 29–43. [DOI] [PubMed] [Google Scholar]
- Mashburn L. M.; Whiteley M. Nature 2005, 437, 422–425. [DOI] [PubMed] [Google Scholar]
- Diggle S. P.; Cornelis P.; Williams P.; Cámara M. Int. J. Med. Microbiol. 2006, 296, 83–91. [DOI] [PubMed] [Google Scholar]
- Diggle S. P.; Matthijs S.; Wright V. J.; Fletcher M. P.; Chhabra S. R.; Lamont I. L.; Kong X.; Hider R. C.; Cornelis P.; Cámara M.; Williams P. Chem. Biol. 2007, 14, 87–96. [DOI] [PubMed] [Google Scholar]
- López D.; Kolter R. FEMS Microbiol. Rev. 2010, 34, 134–149. [DOI] [PubMed] [Google Scholar]
- Schafhauser J.; Lépine F.; McKay G.; Ahlgren H. G.; Khakimova M.; Nguyen D. J. Bacteriol. 2014, 196, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfee M. W.; Shelton J. G.; McCubrey J. A.; Pesci E. C. Infect. Immun. 2005, 73, 878–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



