Abstract
The use of the PER.C6 adenovirus packaging cell line in combination with a designated vector plasmid system, whereby the cell line and vector with E1 deleted have no sequence overlap, eliminates the generation of replication-competent adenovirus during vector production. However, we have found cytopathic effect (CPE)-inducing particles in 2 out of more than 40 large-scale manufacturing lots produced in PER.C6 cells. The CPE inducer was detected at a frequency of 1 event in 7.5 × 1012 vector particles. Despite amplification, it was not readily purified, indicating that the agent itself is replication deficient and requires the parental recombinant adenovirus serotype 5 (rAd5) vector for replication and packaging. Therefore, we designated the agent as a helper-dependent E1-positive region containing viral particle (HDEP). Here, we report the molecular structure of the HDEP genome, revealing an Ad comprised of E1 sequences derived from PER.C6 cells flanked by inverted terminal repeat, packaging signal, and transgene sequences. These sequences form a palindromic structure devoid of E2, E3, E4, and late genes. Since only 5 bp were shared between E1 sequences in the PER.C6 genome and viral vector sequences, the data strongly suggested that insertion of genomic DNA into an adenoviral genome had occurred essentially via nonhomologous recombination. HDEPs have been found in unrelated virus batches and appear to share a common structure that may explain their mechanism of generation. This finding allowed development of an HDEP assay to screen batches of rAd5 produced on the PER.C6 cell line and resulted in detection of seven HDEP agents from four different transgene-virus vector constructs in separate batches of Ad.
Replication-deficient gene therapy vectors based on adenovirus serotype 2 (Ad2) or Ad5 and with E1 deleted have been routinely propagated on a permissive human cell line, HEK-293 (4). HEK-293 cells were established via stable insertion into primary embryonic kidney cells of a 4.3-kb Ad5 genome fragment containing the E1 region flanked on both sides by additional viral sequences (13). This cell line thus complements for E1 deficiencies and allows propagation of replication-defective adenoviral vectors. Due to homology between vector sequences and E1 flanking sequences in the HEK-293 cell line, homologous recombination via double crossover events can occur and often results in the formation of replication-competent Ad (RCA) (12, 21). The occurrence of RCA is undesired; and therefore, current U.S. Food and Drug Administration guidelines stipulate that a batch should contain less than 1 RCA per 3 × 1010 virus particles (Biological Response Modifiers Advisory Committee meeting, 5 April 2001). To eliminate RCA formation, a new cell line named PER.C6 was created by immortalization of human embryonic retina cells with the Ad5 E1 gene sequence. This cell line is used in combination with a vector plasmid system (pClip and pAdapt) that ensures absence of overlapping sequences and, thus, no sequences are available for homologous recombination between the cell line and the vector (3). Many recombinant Ad batches have been produced using the PER.C6 cell line, and RCA have not been detected. We have previously demonstrated the importance of using the appropriate vector-plasmid system in combination with PER.C6 cells, showing that a 177-bp sequence homology at one end of the E1 region is sufficient for recombination between cells and vector (14). In that study we showed that the single crossover event observed gave rise to a helper-dependent, replication-incompetent virus with a low frequency (less than 1 in 1011 particles). To accommodate the extra E1 sequences, the recombined genomes had deleted a significant portion of the viral backbone. This resulted in a cytopathic effect (CPE)-inducing agent that was completely dependent on the presence of the parental recombinant virus for replication. Since the CPE-inducing agent could not be propagated in the absence of recombinant vector, the term helper-dependent-E1-containing particle, or HDEP, was coined (14). We reported that the emergence of both RCA and these HDEPs could be circumvented, using the tailored vector that was specifically adapted for use with PER.C6 cells, after producing 20 production lots in which no CPE-inducing events could be detected. However, after having produced and tested more than 40 production lots of recombinant Ad5 vector using the tailored vector-plasmid system, we identified two virus lots in which CPE-like events were detected using a sensitive RCA assay. The CPE-inducing agent proved to be dependent for its replication on the presence of the Ad5 recombinant vector, thus qualifying as an HDEP. In this report, we describe the in vitro growth properties of these HDEPs together with the genomic characterization studies to identify their structure. Subsequently, more HDEPs were recovered in independent studies using other recombinant vectors containing different transgenes. We describe common structural features for these HDEP entities, which leads to a proposal of a mechanism of HDEP formation and replication and strongly suggests that nonhomologous recombination between vector sequences and genomic DNA has occurred.
MATERIALS AND METHODS
Vector construction and production.
An Ad5 vector carrying the cDNA encoding fibroblast growth factor 4 (Ad5.1FGF-4) was constructed by inserting a 1.2-kb EcoRI-digested FGF-4 cDNA (17) into plasmid pClip (6). The resulting pClip.1FGF-4 plasmid was digested with SalI and PacI, thereby liberating the Ad sequence. This DNA was cotransfected with PacI-digested cosmid pWE/AdAfIII-rITR (6) into PER.C6 cells (3) using Lipofectamine and the protocol provided by the manufacturer (Life Technologies). Virus propagation and purification have been described previously (14). For each production lot, a separate aliquot from a master virus bank was used for the initial infection. Particle counts were determined by an analytical ion-exchange high-performance liquid chromatography method similar to that described by Huyghe et al. (9).
RCA bioassay.
A summary of this bioassay has been presented previously (14). In brief, Ad5.1FGF-4 virus samples were used to infect adherent cultures of HeLa cells at 2,000 virus particles/cell. Typically, 109 particles were tested on 500,000 cells plated in a T25 flask with 5 ml of medium, whereas larger flasks and higher cell densities (5 × 106 to 5 × 107 cells) allowed for testing of particle inputs on the order of 1010 to 1011 virus particles per assay. In some instances, more than 30 flasks were tested to achieve testing of 1012 particles. Freeze-thawed lysates of the HeLa cells were used to infect adherent cultures of A549 cells. Flasks were monitored up to 15 days postinfection for visual signs of CPE. Lysates from flasks showing signs of CPE were analyzed by a PCR amplification procedure for the presence of E1A sequences.
HDEP enrichment protocol.
Material recovered from the RCA assay was used to infect 3 × 108 A549 cells at a multiplicity of infection (MOI) of 5 infectious units as determined by an end point infectivity assay on HEK-293 cells. Adsorption and internalization of the virus was allowed to proceed at 37°C for 2 h in a total of 4.5 ml per 15-cm culture dish. Fresh infection medium made of RPMI 1640 (Gibco-Life Technologies, Grand Island, N.Y.) with 1% fetal bovine serum (HyClone, Logan, Utah) was then added to the cells. The infection was continued for 72 h, at which point the first signs of CPE were observed. The medium was discarded, and the cells were recovered in infection medium and assayed for infectious particle count in a cell-based enzyme-linked immunosorbent assay on HEK-293 and A549 cells. Five additional amplification rounds were carried out similarly except that more cells were infected, and these infection cycles only required a period of 48 h. Depending on the round of amplification, the target MOI varied between 0.1 and 10 based on the titer obtained on A549 cells.
Viral titer determinations.
HEK-293 or A549 cells, seeded in 96-well plates (5 × 104 cells/well), were infected with 100 μl of a wild-type Ad5 (wtAd5) standard starting at an MOI of 1 that was subsequently reduced by twofold serial dilutions. Since a priori the concentration of the experimental samples was not known, multiple initial dilutions to target an MOI of 1 were prepared, followed by twofold step dilutions. Infection was allowed to proceed at 37°C for 44 h, after which the medium was discarded and cells were fixed with 95% ethanol-5% acetic acid (200 μl/well) for 15 min at −20°C. Cells were washed with phosphate-buffered saline and then blocked for 1 h using 200 μl of Superblock (Pierce Chemical, St. Louis, Mo.)/well while shaking at room temperature. The primary antibody (rabbit polyclonal anti-Ad5; Access Biomed, San Diego, Calif.) was added (diluted 1:1,000 in Tris-buffered saline-1.35% normal goat serum; Pierce Chemical) in a total volume of 100 μl/well, and binding was allowed to proceed for 1 h at room temperature on a shaker. The primary antibody was removed by three wash steps using Tris-buffered saline-0.1% Tween 20 and secondary antibody (goat anti-rabbit alkaline phosphatase; Pierce Chemical) diluted 1:1,000 was added followed by an incubation period of 1 h. The secondary antibody was removed by washing before the paranitrophenylphosphate substrate was added, according to the protocol supplied by the manufacturer (Pierce Chemical). The reaction was stopped after 5 min by the addition of 100 μl of 2 N NaOH. The plates were analyzed at 405 nm on a Vmax plate reader (Molecular Devices, Sunnyvale, Calif.) using the Softmax endpoint dilution protocol. Titers for the unknown samples were extrapolated from the standard curve obtained with the wtAd5 standard.
CsCl equilibrium density centrifugation.
CsCl was added to a final concentration of 0.51 g/ml to samples that were enriched for HDEPs. Subsequently, the CsCl mixture was centrifuged at 25,000 rpm at 15°C for 25 h in a Sorvall RC80 ultracentrifuge using a Sorvall AH-629 swinging bucket rotor. After centrifugation, a broad, fuzzy band was visible. A hole was made in the centrifuge tube below the level of the visible band, 35 fractions spanning the region of the virus band were collected and dialyzed twice using 10 mM Tris-HCl (pH 8)-10 mM MgCl2-5% sucrose, and materials were stored at −20°C until analysis by PCR.
Molecular biology analyses.
For TaqMan PCR analyses, E1A-specific primers and probe were designed (upstream primer, 5′-AGAGAGCCTTGGGTCCGGT-3′; downstream primer, 5′-TTCATCCTCGTCGTCACTGG-3′; probe, 5′-car-boxyfluorescein-ATCGATCTTACCTGCCACGAGGCTGG-carboxytetrameth-ylrhodamine-3′), as well as Ad5.1FGF-4 primers recognizing the Ad5 hexon gene sequence (upstream primer, 5′-CTTCTCTACGCCAACTCCGC-3′; downstream primer, 5′-GCAGGTACACGGTTTCGATGA-3′; probe, 5′-carboxy-fluorescein-TTGAGGTGGATCCCATGGACGAGC-carboxytetramethylrhod-amine-3′). The assay was run using universal PCR Master Mix reagents from Applied Biosystems (Foster City, Calif.). Final concentrations of primers and probe in the reaction mixtures were 400 and 200 nM, respectively. As template, 5 μl of each fraction was used, in a total reaction volume of 50 μl. To standardize the TaqMan assay, wtAd5 was used for both PCR assays. The TaqMan protocol consisted of 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Adenoviral DNA preparation, PCR amplification, field inversion gel electrophoresis (FIGE), biotinylated probe preparation, and Southern blotting were performed as described previously (14).
RESULTS
Detection of HDEPs.
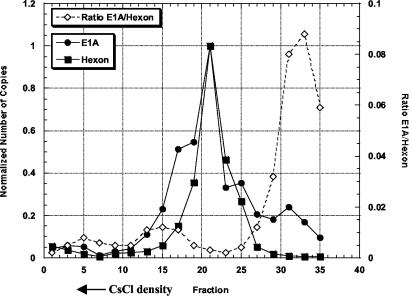
Unexpectedly, we detected slow-developing CPE events in the 25th and 33rd manufacturing lots of recombinant Ad5.1FGF-4 vector produced in PER.C6 cells, using a sensitive cell-based RCA assay. CPE was just visible at day 13, whereas the typical RCA (from HEK-293 packaging cells) is normally evident by day 5. These observations provided a first hint that the CPE-causing agent was not the typical RCA. Cumulative RCA testing (more than 40 production batches) of 1.5 × 1013 virus particles detected only two separate CPE events and suggested a frequency of 1 CPE event per 7.5 × 1012 virus particles. In an effort to enrich and possibly purify the CPE-causing entity, five consecutive culture rounds on A549 or HEK-293 cells were performed. From titration data (Table 1) it could be concluded that, although the titer of the CPE-causing agent could be amplified by approximately a factor of 100 during the cell culture rounds on A549 cells, the ratio of HDEP versus Ad5.1FGF-4 vector did not change significantly. Thus, we concluded that replication of the CPE-causing agent is dependent upon the presence of the Ad5.1FGF-4 vector. Based on these data, the CPE-causing agent appeared to be helper dependent and was thus categorized as HDEP, analogous to our previous report (14). Our inability to purify the HDEPs via propagation on selective cell lines suggested that we turn to physical separation methods in seeking to understand whether the genome size of the HDEP is sufficiently different to aid in its fractionation. CsCl fractions were collected and analyzed for the presence of Ad5-E1A DNA (HDEP) and the Ad5 hexon DNA (parental Ad5.1FGF-4) by quantitative PCR. The PCR on Ad5 hexon DNA showed one clear peak with a maximum in fraction 21, indicative of a single Ad5.1FGF-4 genome size (Fig. 1). The PCR on E1A showed two distinct peaks (fractions 21 and 31) and two partially resolved shoulders off the main peak (fractions 15 to 17 and 25). Thus, at least two, but possibly four, different HDEP species are present in the analyzed HDEP sample together with a single Ad5.1FGF-4 species. These data also suggest that the HDEP entities present in fractions 17 and 31 (and perhaps 25) contain little or no Ad5 hexon sequence. For example, by plotting the ratio of E1A signal to hexon signal, fraction 31 could be identified as significantly enriched (more than 17-fold) for HDEP (Fig. 1). Since the predominant HDEP species (fraction 21) coeluted with the Ad5.1FGF-4 vector, indicating a genome size similar to that of Ad5.1FGF-4 (35.6 kb), it appears that density gradient separation is not an effective method to purify the predominant HDEP species.
TABLE 1.
Enrichment of RCA and HDEP by successive rounds of propagation on A549 cellsa
| Round of amplifi- cation | Titer on A549 cells (HDEP infectious units/ml) | Titer on HEK-293 cells (total virus infectious units/ml) | HDEP/ Ad5.1FGF-4 ratio |
|---|---|---|---|
| 1 | 4.8 × 107 | 3.6 × 109 | 1:75 |
| 2 | 1.7 × 108 | 2.5 × 1010 | 1:147 |
| 3 | 1.6 × 106 | 4.4 × 108 | 1:275 |
| 4 | 3.3 × 109 | 4.0 × 1011 | 1:121 |
| 5 | 7.4 × 109 | 2.56 × 1012 | 1:346 |
The CPE-positive, secondary culture supernatant from the RCA assay was subjected to five rounds of amplification on the nonpermissive cell line A549. Infectivity was measured using a cell-based enzyme-linked immunosorbent assay on both HEK-293 and A549 cells for the five cultures, as described in Materials and Methods.
FIG. 1.
Analysis of CsCl centrifugation fractions by TaqMan PCR. A sample containing both HDEP and Ad5.1FGF-4 was fractionated using CsCl gradient density centrifugation. Thirty-five fractions were collected (proceeding from greatest to lowest CsCl density, from fraction 1 to fraction 35), and alternative fractions starting with the first one were analyzed by TaqMan PCR either specific to the E1A sequence (closed circles) or to the Ad5 hexon gene sequence (closed squares). Quantitation was performed by assigning a value of 1 to the fraction with the highest copy number. The ratio of E1A to hexon signal is represented by open diamonds.
Determination of the origin of the E1 sequence present in the HDEP genome.
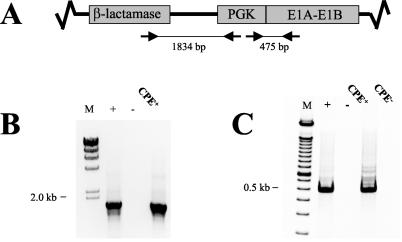
To identify whether the HDEP originates from a recombination event between Ad5.1FGF-4 and E1 sequences present in PER.C6cells, PCR analyses were performed amplifying (i) a 475-bp fragment encompassing the PGK promoter-E1A gene junction and (ii) an 1,834-bp fragment spanning the β-lactamase gene-PGK promoter sequence. Both junctions were present only on plasmid pIG.E1A.E1B (Fig. 2A), which was used for immortalization of primary human embryonic retinal cells, ultimately resulting in a cell line which is now marketed under the trademark PER.C6. Amplicons for both reactions were clearly detected (Fig. 2B and C, respectively), and together with additional PCR runs (data not shown) they indicated that the genome of the HDEP likely contains essentially the entire 7.3-kb pIG.E1A.E1B plasmid. As a consequence, if the HDEP genome originated by an insertion of 7.3 kb of plasmid DNA from the PER.C6 cell line into the Ad5.1FGF-4 vector, it must have then undergone deletions in the Ad5.1FGF-4 backbone, or else the genome would be too large to be packaged efficiently (1).
FIG. 2.
Confirmation of PER.C6-derived gene sequence by PCR. (A) Schematic drawing of part of plasmid pIG.E1A.E1B showing the locations of selected plasmid elements and oligonucleotide primers. (B) Oligonucleotides specific for the β-lactamase gene (5′-TTTATCCGCCTCCATCCAGTC-3′) and the PGK promoter (5′-GTGAAGAATGTGCGAGACCC-3′) were used to amplify an 1,834-bp PCR product. As a positive control (+), DNA extracted from PER.C6 cells (5-ng input) was used as template, whereas Tris-EDTA (TE) buffer served as the negative control (−). A CPE-positive culture from the RCA assay was diluted 10-fold before use as template. (C) Oligonucleotides specific for the PGK promoter (5′-GGCTCCCTCGTTCCGAAT-3′) and the E1A gene (5′-CGGTACAAGGTTTGGCATAGA-3′) were used to amplify a 475-bp fragment. As a positive control (+), DNA isolated from PER.C6 cells (5-ng input) was used as template, whereas TE buffer served as a negative control (−). A CPE-positive culture (RCA+) or CPE-negative culture (CPE−) from the RCA assay was diluted 10-fold before use as template.
Genomic organization of HDEP.
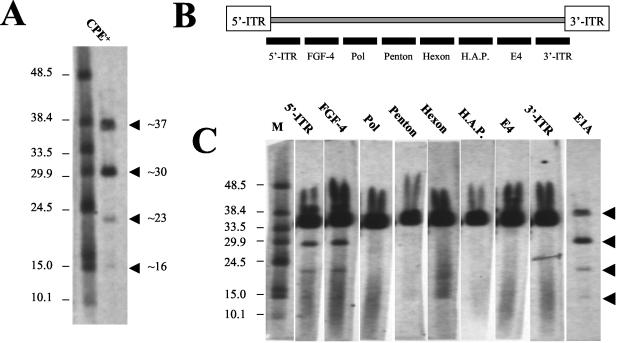
Using FIGE and subsequent Southern blotting analyses with an E1A probe, four distinct HDEP entities were detected (Fig. 3A) with genome sizes of approximately 37, 30, 23, and 16 kb. These results are consistent with multiple virus peaks seen after CsCl gradient separation. Next, a set of probes was designed recognizing distinct regions of the Ad5.1FGF-4 genome to determine which regions are retained in HDEP (Fig. 3B). Only those HDEPs smaller than 37 kb could be investigated in this experiment, since the presence of the Ad5.1FGF-4 genome obscures analysis of an HDEP of similar size. From the results, it could be concluded that the HDEP entities of 30 and 23 kb both retained the 5′ inverted terminal repeat (ITR) and the FGF-4 transgene region but had lost all sequence 3′ of the FGF-4 transgene, since no signal could be detected for any of the probes tested (Fig. 3C). The positive signal around 35.6 kb in all blots indicated the presence of the parental Ad5.1FGF-4.
FIG. 3.
Determination of HDEP genome size and overall HDEP structure. (A) A sample containing both HDEP and Ad5.1FGF-4 (RCA+) was loaded on a FIGE gel and probed with an E1A probe. The E1A probe was generated with PCR using the oligonucleotides 5′-TCCTAGCCATTTTGAACCAC-3′ and 5′-CGGTACAAGGTTTGGCATAG-3′, which amplified an E1A fragment corresponding to nt 661 to 910 (wtAd5 genome). (B) Schematic drawing of Ad5.1FGF-4 vector genome and approximate location of PCR-generated probes used to determine HDEP structure. HAP, hexon-associated protein. The 5′-ITR probe and 3′-ITR probe are specific, since they hybridize just downstream of the 5′ ITR or just upstream of the 3′ ITR, respectively. (C) A sample containing both HDEP and Ad5.1FGF-4 was loaded on a FIGE gel and probed with diverse probes. All probes were generated by PCR using biotinylated oligonucleotides. Probe sequences corresponded to wtAd5 sequence nt 91 to 389 (5′ ITR), nt 7522 to 7874 (DNA polymerase gene), nt 14957 to 15281 (penton gene), nt 20653 to 20983 (hexon gene), nt 256546 to 25977 (hexon-associated protein), nt 32219 to 32584 (E4 region), and nt 35188 to 35548 (3′ ITR).
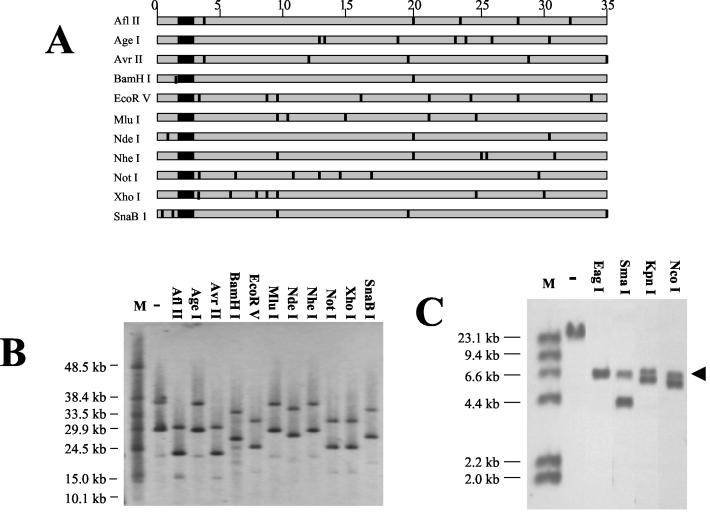
Since an adenoviral genome requires an ITR at both ends for replication (20), we surmised that the 3′ ITR of the HDEP might have been replaced by duplication of the 5′ ITR (and packaging sequence). Such an entity has previously been shown to have normal replicative ability (8). To confirm a duplication of the 5′ ITR and to investigate in more detail what sequence of the Ad5.1FGF-4 vector remained in the HDEP entities, viral DNA was digested with a panel of restriction enzymes known to cut at least once in the Ad5.1FGF-4 vector DNA but not in the pIG.E1A.E1B plasmid (Fig. 4A). Upon FIGE and Southern analyses using the E1A probe, a characteristic three-band pattern seen in the uncut sample was unchanged, with each fragment reduced in size by a constant amount after each digestion (Fig. 4B). Restriction sites such as MluI and NheI had apparently been lost in HDEPs, consistent with the preceding hybridization results showing loss of several viral genes. These results suggest that all viral sequences 3′ to the 10-kb reference point are absent in HDEPs. As a result, the HDEP genomes should all share common 5′ sequence derived from Ad5.1FGF-4. We believe this indicates that the differences in genome lengths of the different HDEP entities are caused by differences in the lengths of PER.C6-derived inserted sequences. By example, the predominant HDEP entity (30 kb) seen in the uncut sample showed a maximum difference in size of approximately 7 kb following digestion with enzymes cutting within viral sequences but not within plasmid pIG.E1A.E1B, which suggested that the uncut 23 kb of the HDEP genome is derived from PER.C6 cells. This finding, together with previous PCR analyses suggesting that most if not all of the pIG.E1A.E1B sequence is present within HDEP, was taken to imply the existence of multiple copies of the 7.3-kb pIG.E1A.E1B in the HDEP genome. Such an insertion is consistent with a single recombination event, as PER.C6 cells have been previously shown to contain approximately 15 copies of pIG.E1A.E1B as tandem repeats (the orientation of individual repeats are unknown) at a single site on chromosome 14 (15). Additional evidence for the presence of multiple copies of pIG.E1A.E1B was provided by the Southern blot assay shown in Fig. 4C, in which viral DNA was digested with a panel of restriction enzymes which each had one site in the pIG.E1A.E1B sequence. Most digests showed two hybridizing fragments, indicative of at least two copies of the plasmid in the HDEP genome. Significantly, all digestions yielded a hybridizing fragment of approximately 7 kb, roughly the size of pIG.E1A.E1B, suggesting that the plasmid copies were in a tandem, head-to-tail orientation.
FIG. 4.
Restriction enzyme digestion of HDEP genome. (A) Schematic representation of Ad5.1FGF-4 genome with approximate positions of restriction enzyme sites in the genome. The large black square represents the FGF-4 cDNA region. DNA size (in kilobases) is listed above in steps of 5 kb. (B) A sample containing both HDEP and Ad5.1FGF-4 was loaded on a FIGE gel either uncut (−) or cut with a panel of restriction enzymes as listed. All chosen enzymes had no recognition sites in pIG.E1A.E1B. Lane M, molecular weight markers. The blot was probed with the same E1A probe that was used for Fig. 3A. (C) A sample containing both HDEP and Ad5.1FGF-4 was loaded on a 1% agarose gel either uncut (−) or cut with restriction enzymes as listed. All chosen enzymes had only one recognition site in pIG.E1A.E1B. The blot was probed with the same E1A probe as that used for Fig. 3A. The black arrow denotes a common 7-kb hybridizing fragment.
The data from Fig. 4B further support the hypothesis that a duplication of the 5′ ITR has occurred. Digestion with either AflII or AvrII resulted in a total HDEP genome reduction of approximately 7 kb. This was unexpected, because the enzymes did not have similarly situated 3′ sites (nucleotides [nt] 32807 and 35166, respectively), although they had closely situated sites in the 5′-Ad5.1FGF-4 sequence (nt 3236 and 3204, respectively). Furthermore, all enzymes that yielded similar reductions in size of the main band had closely situated 5′-Ad5.1FGF-4 sites, and the reduction in DNA fragment size expected from cleavage at the 5′ sites was approximately half of the observed reduction (for AflII, cleavage at nt 3236 should result in a decrease of 3.2 kb, instead of the observed 7 kb).
These results suggested that the AflII site (nt 3236) was the most 3′ site that was clearly present in the HDEP genome, while the XhoI site (nt 5491) was the most 5′ site in the adenoviral backbone (excluding the polylinker) that was clearly absent.
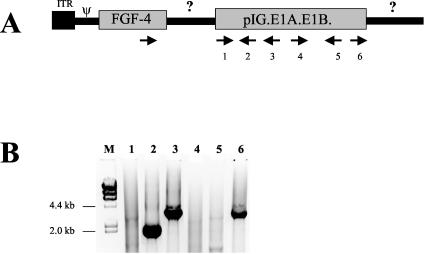
Using a preliminary HDEP genome schematic (Fig. 5A), we sought to identify the precise recombination junction between the Ad5.1FGF-4 vector genome and plasmid pIG.E1A.E1B sequence within PER.C6 cells. Primers were designed to amplify the HDEP region downstream of the FGF-4 transgene region by PCR using a FGF-4 forward primer, in combination with a panel of reverse primers specific for pIG.E1A.E1B in either orientation (Fig. 5A). As seen in Fig. 5B, several reactions gave clear, single amplification products, which were subsequently isolated and subjected to DNA sequencing. Nucleotide sequence analyses on amplicons 2 and 3 demonstrated that a recombination had occurred, resulting in the junction sequence (5′…ATGTAGCTTAGAAGGCAGTGGAATT…3′) whereby 10 bases of Ad5.1FGF-4-specific sequence located in the pIX region of the Ad5 genome (italic font) were followed by 10 bases of pIG.E1A.E1B-specific sequence. Both sequences have 5 nt in common (bold font). The PCR product obtained using reverse primer 6 in combination with the FGF-4 transgene forward primer was intriguing, as this primer was opposite in orientation to primers 2 and 3 (both of which generated amplicons which contained the same Ad5.1FGF-4-pIG.E1A.E1B junction described above). Priming from a second copy of pIG.E1A.E1B (oriented antiparallel to the first copy) would support polymerization, but the amplicon predicted would have been over 8 kb as it traversed an entire copy of pIG.E1A.E1B. This was not consistent with the 4.1-kb amplicon detected. An alternative explanation is that a second copy of FGF-4 exists (antiparallel to the first one) 3′ to the pIG.E1A.E1B depicted in Fig. 5A. Nucleotide sequencing of the 4.1-kb amplicon revealed that this PCR product contained an unusual pIG.E1A.E1B-inverted pIG.E1A.E1B junction (5′-GCTTCTAAGGCCGTATCGTA-3′ [inverted sequence is shown in bold type]) representing a fusion of E1B and β-lactamase sequences.
FIG. 5.
Mapping of the recombination site between Ad5.1FGF-4 and pIG.E1A.E1B. (A) Schematic drawing of the 5′ end of the Ad5.1FGF-4 genome, with location and orientation of oligonucleotides used in a PCR to amplify the genome region between the FGF-4 cDNA and pIG.E1A.E1B. (B) Ethidium bromide-stained gel of PCR products. Numbers 1 to 6 above the lanes correspond with oligonucleotides used in combination with the forward FGF-4 primer. M, molecular weight markers, from bacteriophage λ DNA digested with HindIII. Primers used were AGCAAGGGCAAGCTCTATG (FGF-4), TCTTGGACTCCCAGCAATG (oligonucleotide 1), ATGATTACGCCAAGCTAATTC (2), CTCACCAGTCACAGAAAAGCA (3),TTTATCCGCCTCCATCCAGTC (4), CTGCCCATCCTCTGTAATTG (5), and CGCTAATGAGCTTGATCTGC (6).
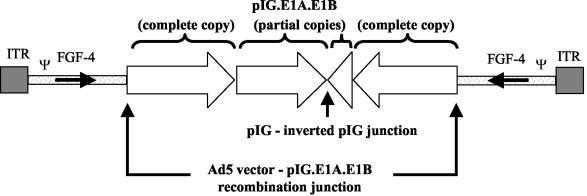
From these molecular analyses, a theoretical schematic HDEP genome was assembled (Fig. 6). This HDEP model contains sequence derived from the 5′ end of Ad5.1FGF-4 on both ends, joined to repeated pIG.E1A.E1B sequences derived from the PER.C6 genome. The Ad5.1FGF-4-derived sequence includes the entire FGF-4 transgene but no additional viral genes except pIX (nt 3312 to 3731 in Ad5.1FGF-4). We interpret this to mean that the plasmid pIG.E1A.E1B-specific sequence in HDEP includes one complete and one partial copy of pIG.E1A.E1B on each side of a specific pIG.E1A.E1B-inverted pIG.E1A.E1B junction. The predicted size of this HDEP genome would be 30.4 kb.
FIG. 6.
Schematic drawing of HDEP genome structure. In the drawing, the Ad5.1FGF-4-pIG.E1A.E1B and pIG.E1A.E1B-inverted pIG.E1A.E1B junction are denoted, as well as the orientation of the E1 coding sequence. See text for further details.
Common HDEP structure and model for replication.
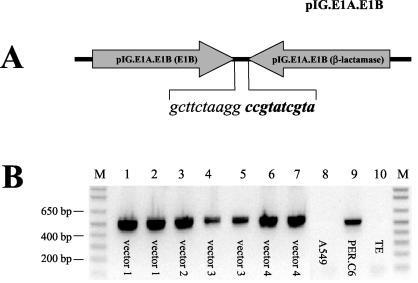
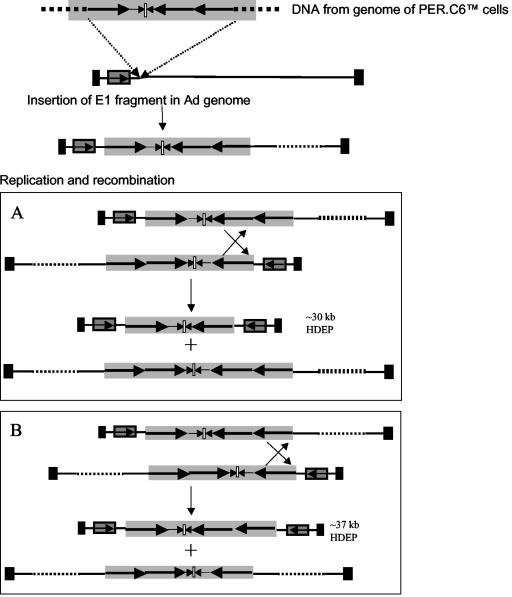
The identification of a unique pIG.E1A.E1B-inverted pIG.E1A.E1B junction in the HDEP analyzed suggested a possible means to specifically detect HDEP. Thus, primers were designed such that the unique inverted junction could be specifically amplified (Fig. 7A). These primers were used to amplify DNA isolated from a total of seven independent HDEP isolates (involving four unrelated vectors with different transgenes) as well as chromosomal DNA isolated from PER.C6 cells. The results, shown in Fig. 7B, demonstrated that the same specific inversion of pIG.E1A.E1B present in PER.C6 cells was also seen in all HDEP isolates studied thus far. Thus, these results suggest that HDEP formation is strongly linked to the presence of a specific head-to-head pIG.E1A.E1B arrangement found in PER.C6 chromosomal DNA and is independent of vector transgene. This arrangement may also be key to explaining the mechanism for HDEP replication. The largest HDEP molecule observed is ∼37 kb and likely contains a third copy of pIG.E1A.E1B. This suggests a model starting with an insertion of a genomic fragment from PER.C6 cells containing three copies of the entire E1 plasmid (see Fig. 8, below). Consequently, a double-stranded Ad is created that can replicate by virtue of the two ITRs but that is too large to be packaged into a virion. However, if multiple double-stranded copies are present, intermolecular recombination mediated by the inverted repeat sequences in the E1 fragment could give rise to the HDEP structures that were identified in this paper. Depending on where the E1 copies recombine, a 30- or 37-kb HDEP could arise. Secondary recombination events between copies of the 30- or 37-kb HDEP genomes may explain the 23- and 16-kb HDEP genomes detected by Southern blotting. These smaller HDEP species would likely contain, stepwise, fewer copies of pIG.E1A.E1B plasmid.
FIG. 7.
Determination of common PER.C6-derived structure elements of HDEP entities. (A) Schematic representation of the head-to-head orientation of pIG.E1A.E1B, with a unique junction sequence indicated. Sequence shown in italics is located in E1B, and sequence shown in bold italic type is present in the β-lactamase gene, thus offering a unique sequence for primer design. (B) Ethidium bromide gel with PCR results using oligonucleotide primers (E1B, 5′-TTAGCTTAATGACCAGACACC-3′; β-lactamase, 5′-GCACCTATCTCAGCGATCTG-3′) for detection of the pIGE1A.E1B-inverted pIG.E1A.E1B vector junction present in HDEP and PER.C6 cells. Both primers have the same orientation in pIG.E1A.E1B and thus amplify only material derived from PER.C6 cells, where two copies of the vector are next to each other in a head-to-head orientation. PCRs were run according to standard procedures, in the presence of 3% dimethyl sulfoxide and at an annealing temperature of 59°C. Lanes 1 to 7 show the PCR results with seven different HDEP-containing samples derived from four independent studies using four different recombinant Ad5 vectors (different transgenes), labeled in the figure. Lane 8 represents the PCR result with DNA isolated from human A549 cells. Lane 9 represents the PCR result with DNA isolated from PER.C6 cells. Lane 10 represents the PCR result using TE buffer as template. Lane M, marker.
FIG. 8.
Theoretical model for HDEP formation. A specific head-to-head E1 coding sequence present in PER.C6 cells is incorporated into the Ad5 recombinant vector with E1 deleted via a nonhomologous recombination mechanism (top). The light gray arrow represents the FGF-4 coding sequence, and solid black boxes represent the ITRs. The genomic fragment from PER.C6 cells (shaded box) contains three full copies of the E1 plasmid (long black arrows in shaded box) and two partial E1 plasmid sequences (small black arrows in shaded box). The location of the sequence inversion point is represented by a small white box. Recombination events, illustrated by the crosses, can result in HDEP genomes of 37 or 30 kb, depending on where recombination takes place. The reciprocal molecules are too large to be packaged.
DISCUSSION
Due to the complete absence of any sequence homology between the tailored vector-plasmid system and the PER.C6 cell line, RCA-like events were not expected during production of Ad5.1FGF-4 virus, even when testing very high concentrations of vector. Despite initial success in producing virus batches apparently free of RCA particles (more than 3.2 × 1012 particles per tested dose), we subsequently detected unexpected CPE-inducing events in later-production samples, albeit at very low frequency. Because these CPE-inducing particles could not result from homologous recombination, we set out to analyze these events to better understand the underlying mechanism and possible consequences. Growth analyses, starting from a sample enriched via propagation on human A549 cells, demonstrated that the CPE-causing agent could not be selectively amplified or isolated by limiting dilution. We have described this feature previously (14) and, hence, the CPE-causing agent was termed HDEP. Subsequent attempts to physically separate the recombinant vector from HDEP (by CsCl gradient centrifugation or by serial passage) have proven unsuccessful. While isolation of a pure, well-characterized helper entity would confirm our explanation of this unusual recombination event, the molecular analyses of HDEP demonstrated its complete lack of any late genes and, therefore, it is totally dependent for packaging on either the packaging cell or the recombinant Ad5 vector. Since the cell line only contains early genes, the source of the late genes can only be derived from the parental Ad5 recombinant vector. Considering our explanation of partial functionality of the HDEP genome, it is then apparent that, unlike conventional RCA, the HDEP would never be able to overgrow the parental virus since it lacks the viral structural genes needed for packaging and amplification.
There appear to be limited options to achieve production of pure HDEP by creating the mutant genome in vitro (itself not an easy task), where such an effort could require (i) construction of a new cell line able to provide all viral structural genes that would support the independent amplification of HDEPs without need of other virus help or (ii) coinfection of HDEP DNA plus a virus with E1 deleted into an E1-containing cell line, where the outcome would be a mixed population of virus such as we described in this report. Future work with HDEPs would benefit from efforts such as these.
Subsequent molecular analyses showed that the E1 sequence in HDEP clearly originates from the PER.C6 packaging cell line, since it contains the engineered PGK promoter present in plasmid pIG.E1A.E1B that was used to make the PER.C6 cell (3). The molecular evidence also demonstrates that distinct HDEP agents can coexist in one sample. Although the number of HDEPs available for analysis is low due to the low frequency of generation, it appears that HDEP formation is favored by the presence of a unique head-to-head orientation of pIG.E1A.E1B within the PER.C6 cells. Based on this finding, a model is proposed for HDEP formation and replication. Formation of packaged HDEP apparently occurs within PER.C6 cells via nonhomologous recombination between the adenoviral genome and pIG.E1A.E1B sequence. At present it seems most likely that such nonhomologous recombination might initiate via intracellular interaction of viral DNA with intact chromosomes of PER.C6 cells; however, one might consider that there may be interactions with exogenous PER.C6 genomic fragments present in the vector crude lysate and taken up by the cells. Indications that host cell DNA can be incorporated, and even covalently linked with (incomplete) Ad genomes, in virions were obtained early in Ad research (5, 18). Host cell contamination was typically found in defective virions of lower densities in CsCl gradients than in the complete virions. Similar findings were reported for other DNA viruses, like simian virus 40 and polyomavirus (10, 11). In one case (2), a recombinant between Ad12 and host cell DNA was described that appeared to have the same mirror-image structure as has been described here. In most of these studies, the presence of host cell DNA in viral genomes was found when crude virus lysates were serially passaged at high MOIs. Experimental evidence for one or the other mechanism described above is important, because if recombination were to occur preferentially in combination with exogenous DNA, then DNase treatment or altered propagation conditions could perhaps prevent formation of HDEP. While we cannot exclude alternate theories of HDEP formation, it is apparent that the key step in HDEP assembly occurs within PER.C6 cells.
The scientific rationale to propose that the sole recombination event between the Ad vector and the genome of PER.C6 cells is not site specific is based on (i) the lack of substantial DNA homology at the site of recombination and (ii) the finding that the E1 genome fragment derived from the genome of PER.C6 cells can be incorporated into the Ad5 genome at different sites (based on data from other transgene constructs [results not shown]). The possible sites of insertion, however, are likely to be limited by the total length of the resulting HDEP, in that the duplicated left end and E1-containing insert together must not exceed the maximum length for virus formation, i.e., approximately 105% of the wtAd5 genome (1). The next step in the proposed HDEP replication model encompasses the creation of a complete HDEP genome mediated by the presence of the inverted repeat sequences within the incorporated E1 region. The characteristic duplication of the left-end sequences to the right end, as seen in the HDEP genomes, can be mediated by intermolecular recombination. Evidence that such events take place during Ad replication of genomes with inverted repeats was reported earlier (16). In this study, repeat sequences, in the former E1 region, were placed on different viruses. When both viruses were mixed in complementing cells, deleted viruses were formed that contained the inverted repeat on one (helper-dependent) genome, the structure of which resembles the HDEP structure identified here.
While these experiments provide proof for the occurrence of intermolecular recombination, they do not exclude replication of the genomes with inverted repeat sequences via a process of intramolecular hybridization. In this case, the displaced single-stranded molecule resulting from replication gives rise to a stem-loop structure with a double-stranded ITR via intramolecular recombination and elongation on the displaced strand. Replication of this molecule then results in a double-stranded genome with the characteristic left-end duplication. Such intramolecular interactions are thought to occur during Ad replication when the ITRs in single-stranded intermediates form a so-called pan-handle structure (7, 19). However, in vitro studies have shown that the Ad DNA-binding protein, when present in sufficient amounts, inhibits intramolecular renaturation and enhances intermolecular renaturation (22). The inhibitory effect was attributed to the rigid structure of single-stranded DNA bound to DNA-binding protein and which might, therefore, be weaker on longer templates. In theory, upon intramolecular renaturation, a molecule resulting from end-joining of a left-end fragment derived from a recombinant Ad and a genomic fragment containing inverted repeat sequences could result in the observed structure without the presence of two ITRs in the initial molecule. The model presented in Fig. 8 shows the intermolecular recombinations that form HDEP genomes of approximately 37 and 30 kb in length. Similarly, the observed smaller genomes of approximately 23 and 16 kb could be formed that still contain at least one full copy of the E1 plasmid. However, whether these constitute the smaller genomes observed in Fig. 3 was not further investigated, since these smaller HDEP agents may be formed in secondary culture rounds in the RCA assay (on the human HeLa or A549 cells).
Besides suggesting a hypothetical replication model, the finding of the common inversion sequence present in all HDEP agents can be exploited to develop a detection assay for HDEP. With such an assay the frequency, both in terms of number of batches contaminated as well as number of HDEP entities per number of viral particles, could be established more easily. Moreover, such a detection assay is a prerequisite when designing and testing strategies to prevent HDEP formation. This PCR assay was designed to detect HDEP containing the common inversion sequence and will not identify any HDEPs which have picked up E1 sequence not containing the inversion. However, as this sequence has been found in all HDEP isolates detected to date, historical evidence suggests that this assay should be able to detect most E1 recombinants produced in PER.C6 cells. Finally, with regard to this assay, as the amplicon is common to both HDEP and PER.C6 genomic DNA, residual host cell DNA in a vector preparation could generate a false-positive result. In our experience, this has not been a problem, as only highly purified Ad preparations containing little host cell DNA are tested. Further, the selective amplification of HDEP through multiple passages on nonpermissive cells in the RCA bioassay, prior to PCR analysis, also serves to dilute any residual PER.C6 DNA.
It should be noted that if the formation of HDEPs also occurs in other Ad packaging cell lines, they may go undetected due to the appearance of classical RCA (an independently lytic entity present at much higher frequency). HEK-293 cells have been screened for the number, orientation, and location of E1 copies, demonstrating that the Ad5 region was located in the pregnancy-specific beta-glycoprotein 4 present at chromosome 19 (19q13.2) as a single colinear insertion of viral DNA with no rearrangements (13). Thus, if indeed a head-to-head E1 inversion present in a packaging cell line is pivotal for HDEP formation, the phenomenon is not likely to occur in HEK-293 cells.
When considering safety aspects of HDEP, it should be noted that HDEP is replication deficient, since it lacks the necessary viral genes for autonomous replication and, thus, HDEPs will not independently disseminate in a host. Replication and spread may occur only in the presence of a recombinant vector within the same cell. We expect the clinical influence of HDEPs to be effectively limited for the following reasons: (i) the low frequency of HDEP formation, (ii) the low ratio of HDEPs versus recombinant vector particles (<0.1 HDEP per dose), (iii) the typical effective human immune response to circulating virus, (iv) the lack of evidence regarding in vivo rescue, and (v) simple dilution over time. However, the presence of a mirror image of an expression cassette carrying DNA of human origin (FGF-4 transgene or PGK promoter) within some HDEP agents could potentially result in a more efficient interaction with human genomic DNA, thereby inserting the E1 sequences, located between the expression cassettes of the cellular genome through homologous recombination. In this case, host cell immortalization could theoretically occur in the absence of recombinant Ad5 vector and concomitant cell lysis. Thus, the sensitive RCA detection assay that has detected all HDEPs to date, in conjunction with the HDEP-specific PCR test, will be most helpful in screening Ad vector batches to exclude HDEP-positive vector batches from ongoing clinical studies.
In summary, we have detected and characterized CPE-inducing agents that form at a very low frequency and that are dependent for replication and packaging on the presence of the parental recombinant Ad5 vector. HDEP genomes are formed in the absence of significant sequence overlap, suggesting that E1 coding sequences can be picked up via nonhomologous recombination. All HDEP agents isolated and analyzed thus far share a common structural motif based upon which a replication model for HDEP has been proposed.
REFERENCES
- 1.Bett, A. J., L. Prevec, and F. L. Graham. 1993. Packaging capacity and stability of human adenovirus type 5 vectors. J. Virol. 67:5911-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deuring, R., G. Klotz, and W. Doerfler. 1981. An unusual symmetric recombinant between adenovirus type 12 DNA and human cell DNA. Proc. Natl. Acad. Sci. USA 78:3142-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fallaux, F. J., A. Bout, I. van der Velde, D. J. M. van den Wollenberg, K. M. Hehir, J. Keegan, C. Auger, S. J. Cramer, H. van Ormondt, A. van der Eb, D. Valerio, and R. C. Hoeben. 1998. New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 9:1909-1917. [DOI] [PubMed] [Google Scholar]
- 4.Graham, F. L., J. Smiley, W. C. Russel, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 5.Hammarskjold, M. L., G. Winberg, E. Norrby, and G. Wadell. 1977. Isolation of incomplete adenovirus 16 particles containing viral and host cell DNA. Virology 82:449-461. [DOI] [PubMed] [Google Scholar]
- 6.Havenga, M. J. E., A. A. C. Lemckert, J. M. Grimbergen, R. Vogels, L. G. M. Huisman, D. Valerio, A. Bout, and P. H. A. Quax. 2001. Improved adenovirus vectors for infection of cardiovascular tissues. J. Virol. 75:3335-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hay, R. T., N. D. Stow, and I. M. McDougall. 1984. Replication of adenovirus minichromosomes. J. Mol. Biol. 174:493-510. [DOI] [PubMed] [Google Scholar]
- 8.Hearing, P., R. J. Samulski, W. L. Wishart, and T. Shenk. 1987. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J. Virol. 61:2555-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huyghe, B. G., X. Liu, Sutjipto, S., B. J. Sugarman, M. T. Horn, H. M. Shepard, C. J. Scandella, and P. Shabram. 1995. Purification of a type 5 recombinant adenovirus encoding human p53 by column chromatography. Hum. Gene Ther. 6:1403-1416. [DOI] [PubMed] [Google Scholar]
- 10.Lavi, S., S. Rozenblatt, M. F. Singer, and E. Winocour. 1973. Acquisition of sequences homologous to host DNA by closed circular simian virus 40 DNA. II. Further studies on the serial passage of virus clones. J. Virol. 12:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavi, S., and E. Winocour. 1974. Accumulation of closed-circular polyoma DNA molecules containing host DNA during serial passage of the virus. Virology 57:296-299. [DOI] [PubMed] [Google Scholar]
- 12.Lochmuller, H., A. Jani, J. Huard, M. Prescott, M. Simoneau, B. Massie, G. Karpath, and G. Acsadi. 1994. Emergence of early region 1-containing replication-competent adenovirus in stocks of replication-defective adenovirus recombinants (dE1+dE3) during multiple passages in 293 cells. Hum. Gene Ther. 5:1485-1491. [DOI] [PubMed] [Google Scholar]
- 13.Louis, N., C. Evelegh, and F. L. Graham. 1997. Cloning and sequencing of the cellular-viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology 233:423-429. [DOI] [PubMed] [Google Scholar]
- 14.Murakami, P., E. Pungor, J. Files, L. Do, R. van Rijnsoever, R. Vogels, A. Bout, and M. McCaman. 2002. A single short stretch of homology between adenoviral vector and packaging cell line can give rise to cytopathic effect-inducing, helper-dependent E1-positive particles. Hum. Gene Ther. 13:909-920. [DOI] [PubMed] [Google Scholar]
- 15.Nichols, W. W., R. Lardenoije, B. J. Ledwith, K. Brouwer, S. Manam, R. Vogels, D. Kaslow, D. Zuijdgeest, A. J. Bett, L. Chen, M. v. d. Kaaden, S. M. Galloway, R. B. Hill, S. V. C. A. A. Machotka, J. Lewis, D. Martinez, J. Lebron, C. Russo, D. Valerio, and A. Bout. 2002. Propagation of adenoviral vectors: use of PER.C6 cells. Elsevier Science, New York, N.Y.
- 16.Steinwaerder, D. S., C. A. Carlson, and A. Lieber. 1999. Generation of adenovirus vectors devoid of all viral genes by recombination between inverted repeats. J. Virol. 73:9303-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taira, M., T. Yoshida, K. Miyagawa, H. Sakamoto, M. Terada, and S. Sugimura. 1987. cDNA sequence of human transforming gene hst and identification of the coding sequence required for transforming activity. Proc. Natl. Acad. Sci. USA 84:2980-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tjia, S., E. Fanning, J. Schick, and W. Doerfler. 1977. Incomplete particles of adenovirus type 2. III. Viral and cellular DNA sequences in incomplete particles. Virology 76:365-379. [DOI] [PubMed] [Google Scholar]
- 19.van der Vliet, P. C. 1995. Adenovirus DNA replication, p. 1-31. In P. B. W. Doerfler (ed.), The molecular repertoire of adenoviruses, vol. 2. Springer Verlag, Berlin, Germany.
- 20.Wolfson, J., and D. Dressler. 1972. Adenovirus DNA contains an inverted terminal repetition. Proc. Natl. Acad. Sci. USA 69:3054-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu, J., M. Grace, J. Casale, A. T. I. Chang, M. L. Musco, R. Bordens, R. Greenberg, E. Schaefer, and S. R. Indelicato. 1999. Characterization of replication-competent adenovirus isolates from large-scale production of a recombinant adenoviral vector. Hum. Gene Ther. 10:113-121. [DOI] [PubMed] [Google Scholar]
- 22.Zijderveld, D. C., M. H. Stuiver, and P. C. Vandervliet. 1993. The adenovirus DNA binding protein enhances intermolecular DNA renaturation but inhibits intramolecular DNA renaturation. Nucleic Acids Res. 21:2591-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]