Abstract
To better understand the relationship between primate adeno-associated viruses (AAVs) and those of other mammals, we have cloned and sequenced the genome of an AAV found as a contaminant in two isolates of bovine adenovirus that was reported to be serologically distinct from primate AAVs. The bovine AAV (BAAV) genome has 4,693 bp, and its organization is similar to that of other AAV isolates. The left-hand open reading frame (ORF) and both inverted terminal repeats (ITRs) have the highest homology with the rep ORF and ITRs of AAV serotype 5 (AAV-5) (89 and 96%, respectively). However, the right-hand ORF was only 55% identical to the AAV-5 capsid ORF; it had the highest homology with the capsid ORF of AAV-4 (76%). By comparing the BAAV cap sequence with a model of an AAV-4 capsid, we mapped the regions of BAAV VP1 that are divergent from AAV-4. These regions are located on the outside of the capsid and are partially located in exposed loops. BAAV was not neutralized by antisera raised against recombinant AAV-2, AAV-4, or AAV-5, and it demonstrated a unique cell tropism profile in four human cancer cell lines, suggesting that BAAV might have transduction activity distinct from that of other isolates. A murine model of salivary gland gene transfer was used to evaluate the in vivo performance of recombinant BAAV. Recombinant BAAV-mediated gene transfer was 11 times more efficient than that with AAV-2. Overall, these data suggest that vectors based on BAAV could be useful for gene transfer applications.
Adeno-associated virus (AAV) was first described as a contaminant of tissue culture-grown simian virus 15 (a simian adenovirus) that is dependent on adenovirus for efficient replication (reviewed in reference 17). This description resulted in its name as well as its classification in the genus Dependovirus. AAV is a member of the Parvoviridae, a virus family characterized by a single-stranded linear DNA genome and a capsid with icosahedral symmetry measuring about 20 nm in diameter. AAV serotype 2 (AAV-2) is the best-characterized isolate and is discussed here as an AAV prototype.
The AAV-2 genome consists of a linear single strand of DNA that is 4,780 nucleotides long. Both polarities of DNA are encapsulated by AAV-2 with equal efficiency. The AAV-2 genome contains two open reading frames (ORFs). The rep ORF encodes the nonstructural proteins essential for viral DNA replication, packaging, and AAV targeted integration. The cap ORF encodes three related proteins that assemble to form the virus capsid, while the rep ORF is transcribed from promoters at map units P5 and P19. The rep transcripts contain an intron close to the 3′ end of the rep ORF that can be alternatively spliced. Thus, the rep ORF is expressed as four partially overlapping proteins, which are referred to according to their molecular weights: Rep78, Rep68, Rep52, and Rep40 (18, 20, 26). The cap ORF is expressed from a single promoter at P40. By alternative splicing and utilization of an alternative ACG start codon, the cap ORF produces three proteins (VP1 to VP3) ranging from 65 to 86 kDa. VP3 is the most abundant capsid protein and constitutes 90% of the capsid (4, 21, 27). All viral transcripts terminate at a common poly(A) signal at map unit 96 (32).
AAV infection appears to be common in humans, causing a steep rise in titers of antibodies against AAV-1 to AAV-3 early in life (6). AAV is readily isolated from anal and throat specimens from children (5), whereas isolation from adults is less common. Thus, it appears that AAV spreads primarily among the young (17). In Europe, Brazil, and Japan, the prevalences of antibodies against AAV are similar, suggesting a global spread of AAV (12). Infection with AAV in tissue culture does not cause permanent cytotoxic effects and appears to be benign in humans and laboratory animals. To date, no disease has been associated with AAV infections.
AAV, a common contaminant of adenovirus preparations, has been isolated from human virus samples (AAV-2, AAV-3, AAV-5, and AAV-6), simian virus 15 (AAV-1 and AAV-4), and stocks of canine, ovine, and avian adenoviruses (AAAV) (7-9, 22, 28, 29, 33). DNAs spanning the entire rep-cap ORFs of AAV-7 and AAV-8 have been amplified by PCR from cardiac tissue of rhesus monkeys (15). AAV also is detected in strains of bovine adenovirus (BAV) types 1, 2, and 3. Additionally, it can be purified from lysates of BAV-infected cells by CsCl gradient centrifugation and bands at a density of 1.37 to 1.40 g/cm3 (10, 19, 23). The bovine AAV (BAAV) isolated from BAV type 1 was found to be serologically distinct from AAV-1 to -4 (19) but serologically related to AAV contaminations in BAV types 2 and 3. Neutralizing antibodies have not been reported for humans (9). The diameter of the BAAV capsid, as estimated by electron microscopy, is 22 nm. Vectors based on new AAV serotypes might have different cell tropisms and immunological properties, thus allowing more-efficient transduction in certain cell types in the presence of neutralizing antibodies to other common AAV serotypes. In addition, characterization of new serotypes may aid in the identification of viral elements required for altered tissue tropism. Since BAAV is serologically distinct from other primate AAVs, in a gene therapy application BAAV would allow for transduction of a patient who already possesses neutralizing antibodies to primate isolates through either natural immunological defense or prior exposure to other AAV vectors.
We therefore subcloned and sequenced BAAV from BAV type 1 (ATCC VR-313) and BAV type 2 (ATCC VR-314) samples obtained from the American Type Culture Collection (ATCC). Afterwards, we generated recombinant BAAV particles in order to analyze the tropism of BAAV and study its utility as a vector for in vivo gene transfer.
MATERIALS AND METHODS
Cell culture and virus propagation.
293T (human kidney) and HeLa (human cervix) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. SkOV3 (human ovary) and SNB75 (human glioblastoma) cells were cultured in RPMI 1640 medium supplemented with 5% fetal bovine serum. MDBK (bovine kidney) cells were propagated in Dulbecco's modified Eagle's medium supplemented with 5% horse serum. All media contained 2 mM l-glutamine, 100 U of penicillin/ml, and 0.1 mg of streptomycin/ml. Cells were maintained at 37°C under a 5% CO2 humidified atmosphere.
BAV type 1 (ATCC VR-313) and BAV type 2 (ATCC VR-313) were obtained from the ATCC. For virus propagation, MDBK cells were infected with ATCC VR-313 or ATCC VR-314 and cultured for 5 days, after which cytopathic effects were evident. Cells were washed with phosphate-buffered saline and lysed by three freeze-thaw cycles.
Isolation, cloning, sequencing, and sequence analysis of BAAV DNA from BAV samples.
Viral DNA was isolated from BAV type 1 (ATCC VR-313)- and BAV type 2 (ATCC VR-314)-infected MDBK cells by using the High Pure viral nucleic acid kit (Roche). These DNA samples were subjected to PCR amplification using degenerate PCR primers, which amplified a fragment containing sequences of the rep and cap ORFs of all known AAV serotypes (17a) and generated a 1.4-kb amplification product. The amplicon was then cloned by using the TOPO TA cloning kit (Invitrogen) and sequenced with an ABI Prism 3100 genetic analyzer and FS dye-terminator chemistry (both from Applied Biosystems Inc.). The sequences were compared to sequences in GenBank by using BLAST (the basic local alignment search tool, available at http://www.ncbi.nlm.nih.gov/BLAST/). Sequence information from the amplicon was used to design a second set of PCR primers that bind the minus strand of the BAAV rep ORF (5′-TGTTCAAAGGTCGTGGAGTTCCCATC-3′) and the plus strand of the BAAV cap ORF (5′-GGTGATTGGCATTGCGATTCC-3′). PCR using these primers and DNAs isolated from ATCC VR-313- and ATCC VR-314-infected MDBK cells by use of the Plasmid minikit (Qiagen) as a template resulted in amplification of a fragment spanning the area from the cap ORF through the inverted terminal repeat (ITR) to the rep ORF. The PCR amplification products were subsequently cloned by using the TA cloning kit (Invitrogen), and 10 clones were sequenced. ITRs of two clones were sequenced by isothermal noncycling sequencing chemistry using radiolabeled dCTP (Epicentre). Protein sequence and DNA alignments were performed by using the ClustalW multiple sequence alignment tool of the Biology Workbench web-based software (SDSC) and MacVector (version 7; Oxford Molecular).
Cloning of a BAAV packaging plasmid and generation of recombinant BAAV particles.
For generation of recombinant particles, a BAAV packaging plasmid was constructed by PCR amplification of a BAAV fragment containing the complete rep and cap ORFs that used DNA isolated from ATCC VR-314 samples and primers 5′-CTGGACTGCTAGAGGACCCTC-3′ and 5′-CGGTTTATTGAGGGTATGCAAC-3′. Insertion of this fragment into an expression plasmid under the control of the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) promoter resulted in plasmid pMMTV-BAAV. Rep expression in this construct is controlled by the MMTV promoter. Sequencing of pMMTV-BAAV revealed that the plasmid differed from the BAAV consensus sequence by one nucleotide change. The sequence of pMMTV-BAAV was changed to the consensus sequence by using the QuikChange kit (Clontech). The AAV-5 ITR-containing vector pAAV5-LacZ (8), expressing the nucleus-localized β-galactosidase gene under the control of a Rous sarcoma virus (RSV) promoter, was used to generate recombinant BAAV-lacZ (rBAAV-lacZ) by triple transfection of 293T cells with pAAV5-lacZ, pMMTV-BAAV, and the adenovirus helper plasmid pRS449B (30). rAAV2-lacZ, rAAV4-lacZ, and rAAV5-lacZ were generated as described previously (8, 9). Recombinant vectors were purified by fractionation with CsCl gradient centrifugation. DNase-resistant genome copy titers of the vector preparations were determined by quantitative real-time PCR using the TaqMan system (Applied Biosystems) with probes specific to the RSV promoter. Titration of recombinant AAVs was performed in 293T, HeLa, SkOV3, and SNB75 cells. In a 96-well plate, 7 × 103 cells were plated per well. Half of the wells were infected with adenovirus type 5 (at a multiplicity of infection [MOI] of 10) followed by recombinant AAV in 10-fold serial dilutions (ranging from 1:103 to 1:109) of stock virus 2 h later. Forty-eight hours after infection, cells were fixed and stained for β-galactosidase activity with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Gold BioTechnology, Inc., St. Louis, Mo.). Transduced cells were visually counted by using a light microscope. The results were used to calculate the number of transduced cells per 108 DNase-resistant AAV particles.
Serological analysis.
Total-serum samples from AAV2-lacZ-, AAV4-lacZ-, AAV5-lacZ-, or BAAV-lacZ-transduced BALB/c mice, which were isolated 4 weeks after direct injection of rAAVs into the salivary duct (H. Katano, M. R. Kok, S. Yamano, A. Cotrim, and J. A. Chiorini, unpublished data), were assayed for neutralizing antibodies against rBAAV and rAAV-4. Exponentially growing COS cells were plated at a density of 7 × 103/well in a flat-bottom 96-well plate. Twenty-four hours after seeding, cells were infected with adenovirus type 5 at an MOI of 10. Heat-inactivated serum samples from AAV2-lacZ-, AAV4-lacZ-, AAV5-lacZ-, or BAAV-lacZ-infected mice were serially diluted from 1:800 to 1:12,800 in RPMI medium containing 1% fetal calf serum (FCS). Forty transducing units of BAAV-lacZ or AAV4-lacZ was added to the diluted serum samples or 1:32-diluted FCS and incubated for 1 h at 37°C. Subsequently, the virus-serum mixture was added to COS cells. The cells were assayed for β-galactosidase expression by X-Gal staining (Gold BioTechnology, Inc.) 24 h after rAAV infection. Transduced cells were counted under a light microscope. The percentage of rAAV-4 or rBAAV neutralization was calculated by the following formula: 100 × [1 − (transducing titers of serum-incubated rAAV/transducing titers of untreated rAAV)].
Evaluation of BAAV vectors for gene transfer into submandibular glands.
We injected 1010 particles of AAV2-lacZ or BAAV-lacZ into submandibular glands of BALB/c mice by retrograde ductal instillation as previously described (34). Four weeks after transduction, blood was collected from these mice via a retro-orbital plexus bleed. Submandibular glands were excised, homogenized, and lysed in 500 μl of Galact-light lysis solution (100 mM potassium phosphate [pH 7.8], 0.2% Triton X-100) (Applied Biosystems). Phenylmethylsulfonyl fluoride and leupeptin were added to final concentrations of 0.2 mM and 5 μg/ml, respectively. The lysate was cleared by centrifugation at 10,000 × g for 5 min. Genomic DNA was extracted from a 100-μl aliquot by using the Wizard DNA extraction kit (Promega) according to the manufacturer's instructions. DNA concentrations were determined by spectrophotometry. Detection and quantification of genome copies of the AAV vectors were performed by quantitative real-time PCR on a TaqMan system (Applied Biosystems) with probes specific to the RSV promoter. The protein concentrations of the lysates were determined by using the BCA protein assay kit (Pierce), and the β-galactosidase expression of 3 mg of lysate was quantified with the β-Gal ELISA kit (Roche Molecular Biochemicals).
Nucleotide sequence accession number.
The assembled DNA sequence of the complete BAAV genome has been submitted to GenBank (accession no. AY388617).
RESULTS
BAV type 1 (ATCC VR-313) and BAV type 2 (ATCC VR-314) samples distributed through ATCC were reported by ATCC to be contaminated with an AAV. This uncharacterized virus genome was partially amplified by PCR from BAV type 2-infected MDBK cells by using a universal AAV PCR primer set which was shown to amplify a 1.4-kb fragment containing sequences of the rep and cap ORFs of all known AAV serotypes (17a). Sequence analysis of the PCR amplicon from BAV type 2 did not match that of previous isolates but was homologous with AAV-4 (68%) and AAV-5 (80%). The complete sequence of BAAV was obtained by PCR amplification, subcloning, and sequencing (GenBank accession no. AY388617). We propose the name bovine adeno-associated virus (BAAV). PCR amplification and sequencing of the BAAV cap ORFs from ATCC VR-313 and ATCC VR-314 revealed no differences between the two BAAVs (data not shown).
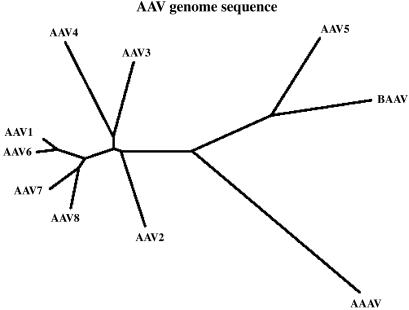
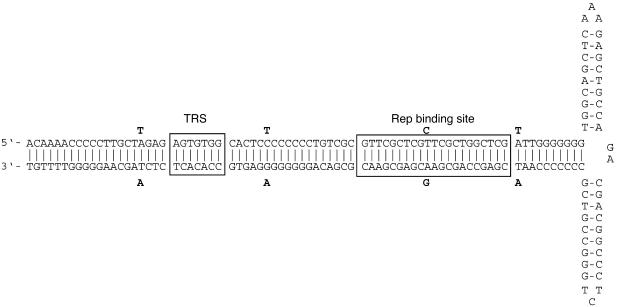
The genome of BAAV is composed of 4,693 nucleotides, and its organization is similar to that of other AAVs. BAAV encodes two ORFs flanked by ITRs. ClustalW alignments of the entire BAAV genome displayed 54 to 79% nucleotide homology with other known AAVs, and from them we generated a phylogenetic tree (Fig. 1). The highest homology was observed with AAV-5 (79%), and the lowest was observed with AAAV (54%). The ITRs of BAAV are 172 nucleotides long and form a characteristic T-shaped palindromic structure (Fig. 2). The BAAV ITR is 96% identical to that of AAV-5 and contains a terminal resolution site (TRS) identical to that of AAV-5 as well as a putative Rep-binding element consisting of a (GAGY)4 repeat.
FIG. 1.
Evolutionary relationship between BAAV and primate AAVs or AAAV. The nucleotide sequence of the entire BAAV genome was aligned to the corresponding sequences of primate AAVs and AAAV with ClustalW. A phylogenetic tree graph representing the sequence relationships was generated based on the homology between the sequences.
FIG. 2.
BAAV ITR. The sequence of the ITR is shown in a hairpin conformation. The putative Rep-binding site and TRS element are boxed. Sequence differences from the AAV-5 ITR are annotated in boldface either above or below the BAAV sequence.
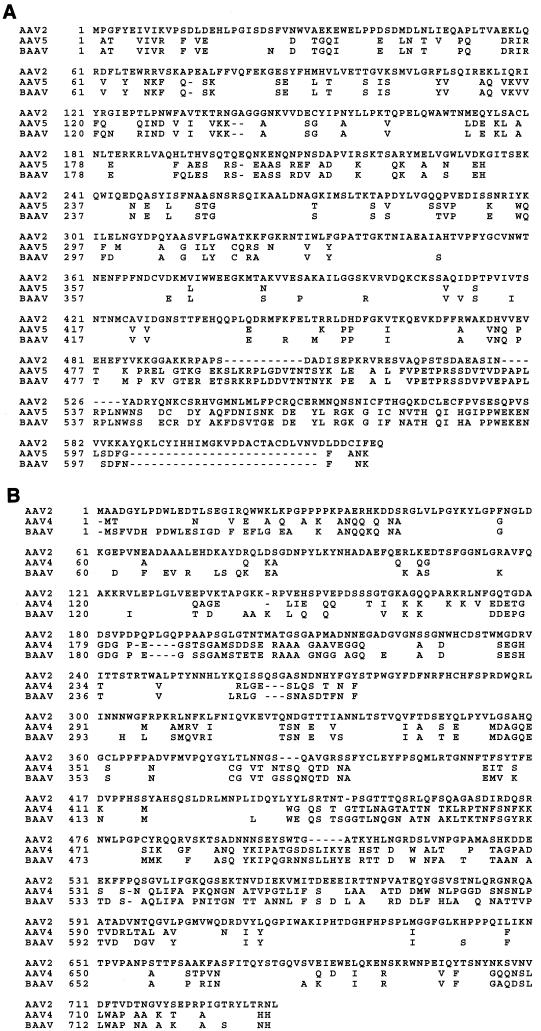
The rep ORF of BAAV displays 48 to 89% amino acid homology with the other AAVs, with most of the diversity clustered at the amino termini (Fig. 3A). High homology (89%) was found with AAV-5 Rep. In contrast, the Rep proteins of AAV-2 and AAAV were only 60 and 52% homologous, respectively. The amino-terminal region of Rep78 (amino acids 1 to 251), which in the case of AAV-2 is sufficient for binding to the ITR and cleavage of a single-stranded TRS substrate (11, 24), was 96% identical to that of AAV-5. The BAAV rep ORF contains a number of sequence motifs associated with DNA replication that also are found in the Rep proteins of other AAV isolates. These include sequence motifs 2 and 3 of the rolling-circle replication superfamily (16), the consensus helicase- and ATPase-associated motif I [GXXXXGK(T/S), where X represents any amino acid] and motif II [UUUU(D/E)(D/E), where U represents any hydrophobic amino acid] (31), and a proposed zinc finger motif in the carboxy-terminal region, all of which are conserved in AAV-2, AAV-5, and BAAV.
FIG. 3.
Comparison of Rep78 and VP1. Amino acid sequences of Rep78 from AAV-2, AAV-5, and BAAV (A) and of the capsid protein VP1 from AAV-2, AAV-4, and BAAV (B) were compared by multiple alignments using the ClustalW algorithm in MacVector.
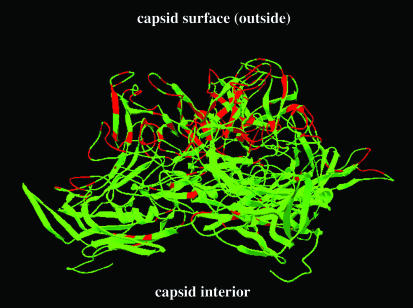
Comparison of the capsid proteins of BAAV along with the primate and avian dependoviruses revealed 55 to 76% homology with other known AAVs (Fig. 3B). AAV-4 showed the highest homology to BAAV (76%), while AAAV was the most divergent (55%). A pseudoatomic structure for AAV-4 has been proposed based on low-resolution cryoelectron microscopy experiments (E. Padron, V. Bowman, N. Kaludov, L. Govindasamy, R. McKenna, J. A. Chiorini, T. S. Baker, and M. Agbandje-McKenna, unpublished data). To identify the location of the 16% sequence divergence between BAAV and AAV-4, we superimposed the BAAV VP1 sequence onto a pseudoatomic structure for AAV-4 (Fig. 4). Divergent regions in the capsid ORF are clustered mainly on the exposed surface loops that comprise the threefold axis of symmetry, suggesting that despite the homology with AAV-4, the tropism of BAAV may differ from that of AAV-4 and other AAVs.
FIG. 4.
Structure of AAV-4 capsid subunits and comparison with BAAV. A ribbon model generated in SwissPDB Viewer of three VP molecules shows the capsid side view of the mounds at the threefold axis of symmetry. The AAV-4 VP sequence is shown in green. Red indicates amino acids where the BAAV sequence differs from that of AAV-4. The structure is based on a pseudoatomic model of the AAV-4 capsid at 22 Å.
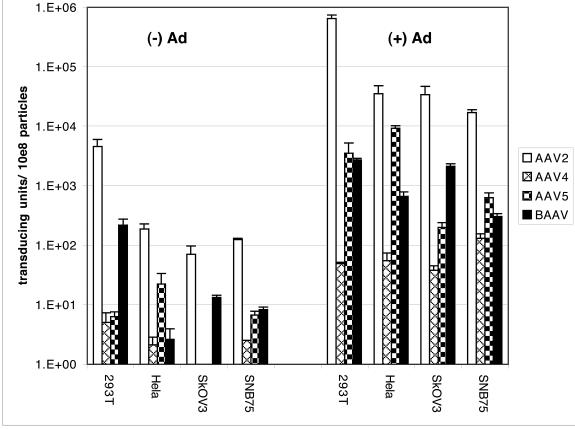
To study BAAV tropism, we generated rBAAV expressing β-galactosidase. We constructed a BAAV packaging plasmid expressing BAAV Rep under the control of an MMTV promoter and Cap from the native P40 promoter. Based on the high homology between the ITR sequences of BAAV and AAV-5, we hypothesized that BAAV Rep proteins could replicate and package an AAV-5 ITR-containing vector. This assumption was confirmed in initial experiments. pAAV5-lacZ, a vector plasmid containing a β-galactosidase expression cassette flanked by AAV-5 ITRs, was packaged with equal efficiency in an AAV-5 and a BAAV capsid (data not shown). Therefore, AAV-5 ITR-containing vector plasmids were used for all subsequent studies to produce rBAAV. rAAVs derived from BAAV, AAV-2, AAV-4, and AAV-5 expressing nucleus-localized β-galactosidase under the control of an RSV promoter were generated by triple transfection of 293T cells with an AAV vector, AAV packaging, and adenovirus helper plasmids. Recombinant vectors were purified by CsCl isopycnic density gradient centrifugation, and particle titers were measured by quantitative PCR. A panel of the following cells was transduced with serial dilutions of rAAVs: human kidney (293T), human cervix (HeLa), human ovary (SkOV3), and human glioblastoma (SNB75) cells. Cells were stained with X-Gal for detection of β-galactosidase activity 48 h after transduction. Each of the AAV serotypes analyzed demonstrated a unique transduction profile (Fig. 5). rAAV-2 had the highest transduction titers in every cell line analyzed. rBAAV transduced 293T and SkOV3 cells more efficiently than rAAV-4 and rAAV-5. However, rBAAV transduction titers were similar to those of AAV4 but lower than those of rAAV-5 in HeLa cells. Coinfection with adenovirus increased the transduction titers of rAAV-2, rAAV-4, and rAAV-5 (8, 9, 13, 14). Coinfection of rBAAV with wild-type adenovirus type 5 increased the rBAAV transduction efficiency 114-fold on average over that of rBAAV alone. The largest increase was observed in HeLa cells (250-fold), and the lowest was in 293T cells (12-fold). Adenovirus effects were dependent on both cell type and AAV serotype. Adenovirus coinfection increased rAAV-2 transduction titers 235-fold on average, ranging from a 476-fold increase in transduction titers in SkOV3 cells to a 135-fold increase in SNB75 cells. The unique transduction profile of BAAV suggests that BAAV uptake might be dependent on cellular factors that differ from factors required for AAV-2, AAV-4, and AAV-5.
FIG. 5.
Transduction titers of rAAV-2, rAAV-4, rAAV-5, and rBAAV-lacZ. Human cell lines were incubated with rAAV-2, rAAV-4, rAAV-5, or rBAAV expressing LacZ in serial dilutions and were either coinfected with advenovirus type 5 at an MOI of 10 [(+) Ad] or mock infected [(−) Ad]. Columns represent β-galactosidase-positive transducing units per 108 DNase-resistant rAAV particles. Values are means from three experiments; error bars, standard deviations.
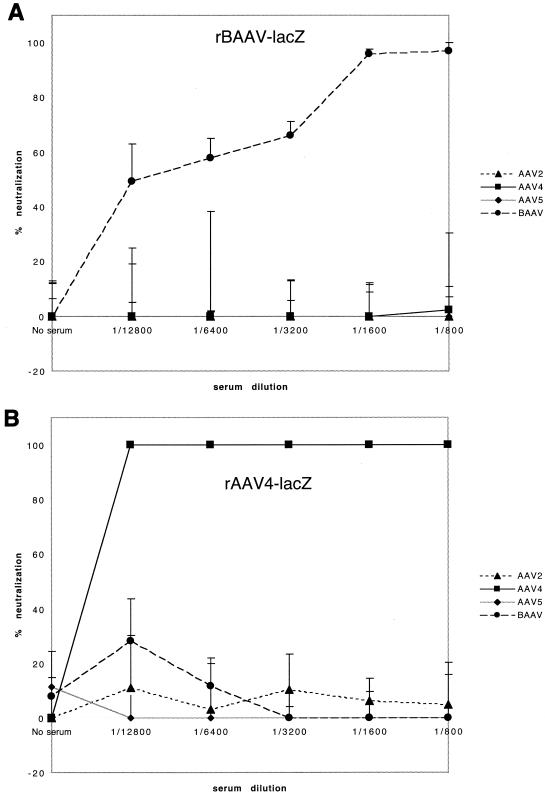
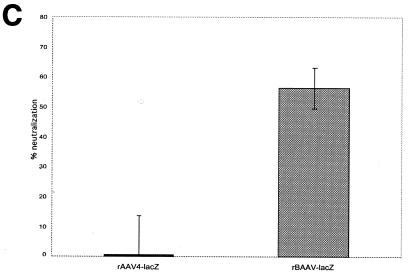
Classical differentiation of viruses relies on a lack of cross-reacting antibodies in serological studies. Therefore, to test for serological identity, rAAV-4 and rBAAV were titered on COS cells in the presence of sera raised against rAAV-2, rAAV-4, rAAV-5, and rBAAV. rBAAV was efficiently inhibited by sera from rBAAV-lacZ-transduced BALB/c mice. In contrast, sera from rAAV-2-, rAAV-4-, or rAAV-5-transduced animals did not inhibit rBAAV-mediated gene transfer (Fig. 6A). Despite the high homology between BAAV and AAV-4, sera from rBAAV-transduced animals did not affect AAV-4 transduction (Fig. 6B). Thus, regardless of the sequence homology between BAAV and AAV-4, BAAV is serologically distinct from other characterized AAV serotypes. Therefore, it should be considered a new AAV serotype. Because most cells used in the experiments were grown in FCS, we also tested whether FCS could inhibit BAAV transduction. Three lots of FCS were tested for neutralization activity against rBAAV, and each demonstrated some inhibitory activity. In contrast, rAAV-4 transduction was unaffected, a result that may suggest existing immunity against BAAV in bovines (Fig. 6C).
FIG. 6.
Neutralization assay. rBAAV-lacZ (A) or rAAV4-lacZ (B) was incubated with serial dilutions of a polyclonal mouse serum against rAAV-2, rAAV-4, rAAV-5, or rBAAV or with a 1:32 dilution of three randomly selected lots of FCS (C). COS cells coinfected with adenovirus type5 (at an MOI of 10) were incubated with the virus-serum mixture. The percentage of neutralization was calculated by the formula 100 × [1 − (transducing titers of serum-incubated rAAV/transducing titers of untreated rAAV)]. Neutralization values that were calculated to be less than zero were adjusted to zero. Values shown are means from three experiments; error bars, standard deviations.
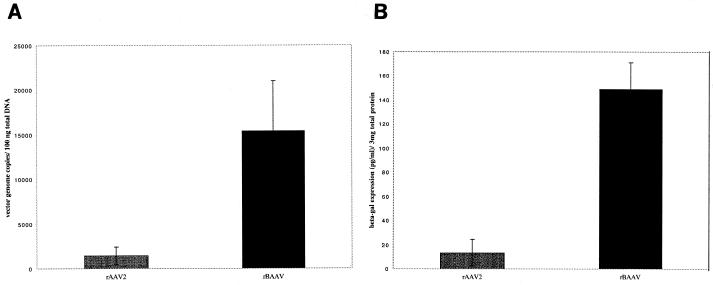
The utility of rBAAV as a vector for in vivo gene transfer was evaluated in a murine model of salivary gland gene transfer (Fig. 7). Salivary glands, a natural secretory organ, are a potent depot organ for the production of proteins either in the blood or in saliva (reviewed in references 2 and 3). Submandibular glands of BALB/c mice were cannulated and injected with 1010 particles of rAAV2-lacZ or rBAAV-lacZ. Four weeks posttransduction, blood was collected and submandibular glands were excised, homogenized, and assayed for vector genome transfer by quantitative PCR and for β-galactosidase activity by an enzyme-linked immunosorbent assay (Fig. 7A and B, respectively). Quantitative PCR analysis indicated that gene transfer was 11-fold higher with rBAAV vectors than with rAAV-2 vectors (Fig. 7A). A similar increase in β-galactosidase activity also was detected in glands from rBAAV-transduced animals relative to that with AAV-2 (Fig. 7B). The results suggest that rBAAV is an efficient gene transfer system for submandibular glands.
FIG. 7.
Comparison of rAAV-2 and rBAAV transduction of salivary glands. By retrograde ductal instillation, 1010 particles of AAV2-RnlacZ or BAAV-RnlacZ were injected into the submandibular glands of BALB/c mice. Four weeks after infection, glands were removed and analyzed for the presence of vector genome DNA by real-time PCR (A) and for expression of the bacterial β-galactosidase reporter by an enzyme-linked immunosorbent assay (B). Values are means of data from seven animals; error bars, standard deviations.
DISCUSSION
As early as 1970, AAV was detected in BAV strains (19). This virus has been studied by serological methods and found to be distinct from AAV-1 to -4. Until now, it had never been cloned or sequenced; thus, its true identity and relationship with other AAVs were unknown (10, 19, 23). Our goals were to subclone, sequence, and initially characterize AAV contaminants detected in BAV types 1 and 2, which we have termed BAAV. To characterize the tropism of this virus, we generated recombinant particles and demonstrated both in vitro and in vivo the distinct transduction activity of BAAV.
To aid in our cloning of this new serotype, we developed a novel strategy to subclone a complete AAV genome in two steps by PCR amplification. Initially, we amplified a 1.4-kb fragment from the core region of AAV by using degenerative PCR primers which bind conserved regions of the AAV genome. The sequence obtained from this region was used to design a second set of PCR primers, which amplified a BAAV fragment spanning the region from the cap through the ITR to the rep ORF. We based the design of the second PCR on the assumption that BAAV forms circular monomers or concatemers in a head-to-tail orientation during either integration or replication. Thus, the whole genome of an unknown AAV isolate could be cloned in two steps. The BAAV genome had the highest homology with AAV-5, and we have demonstrated that the Rep proteins of BAAV can be used to package an AAV-5 genome. Another unique feature of AAV-5, a dimer of polyadenylation sites at the 3′ end of the rep78 ORF, is also conserved in BAAV (data not shown) (25). At the protein level, BAAV Rep78 and VP1 have the highest homology with AAV-5 Rep78 and AAV-4 VP1, respectively. While this could be the result of a recombination event between AAV-5 and AAV-4, alternatively BAAV, AAV-4, and AAV-5 might have a common ancestor and the differences observed today might have arisen due to diverse evolution in different hosts. A structural map of the capsids of BAAV and AAV-4 indicates that the sequence differences are localized to exposed loops on the surfaces of the particles and suggests that BAAV might be serologically distinct from AAV-4. This hypothesis also was confirmed in neutralizing antibody assays.
rBAAV appears to have several attributes that make it an attractive vector for gene transfer: unique serological identity and cell tropism, and efficient gene transfer in vivo. However, additional testing and characterization of its tropism in vivo will be necessary to better define its optimal utility.
BAAV is the second dependovirus of nonprimate origin to be cloned and sequenced. The high homology between the BAAV and AAV-5 Rep proteins, along with the biochemically distinct mechanisms of replication for these two viruses vis-à-vis other mammalian AAVs, suggest that BAAV and AAV-5 might form a distinct group within the Dependovirus genus. Thus, BAAV and human AAV-5 might be the result of a cross-species transmission such as has been proposed for the evolutionary history of parvoviruses (1).
Acknowledgments
We thank Beverly Handelman for excellent technical assistance, Karen Knight and Roberta Knox for administrative assistance, and Robert Kotin for critical discussion of the manuscript. I.B. identified the AAV-contaminated adenovirus isolates used in the study.
REFERENCES
- 1.Badgett, M. R., A. Auer, L. E. Carmichael, C. R. Parrish, and J. J. Bull. 2002. Evolutionary dynamics of viral attenuation. J. Virol. 76:10524-10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum, B. J., and B. C. O'Connell. 1999. In vivo gene transfer to salivary glands. Crit. Rev. Oral Biol. Med. 10:276-283. [DOI] [PubMed] [Google Scholar]
- 3.Baum, B. J., R. B. Wellner, and C. Zheng. 2002. Gene transfer to salivary glands. Int. Rev. Cytol. 213:93-146. [DOI] [PubMed] [Google Scholar]
- 4.Berns, K. I. 1990. Parvovirus replication. Microbiol. Rev. 54:316-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blacklow, N. R., M. D. Hoggan, and W. P. Rowe. 1967. Isolation of adenovirus-associated viruses from man. Proc. Natl. Acad. Sci. USA 58:1410-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blacklow, N. R., M. D. Hoggan, and W. P. Rowe. 1968. Serologic evidence for human infection with adenovirus-associated viruses. J. Natl. Cancer Inst. 40:319-327. [PubMed] [Google Scholar]
- 7.Bossis, I., and J. A. Chiorini. 2003. Cloning of an avian adeno-associated virus (AAAV) and generation of recombinant AAAV particles. J. Virol. 77:6799-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiorini, J. A., F. Kim, L. Yang, and R. M. Kotin. 1999. Cloning and characterization of adeno-associated virus type 5. J. Virol. 73:1309-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiorini, J. A., L. Yang, Y. Liu, B. Safer, and R. M. Kotin. 1997. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J. Virol. 71:6823-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coria, M. F., and H. D. Lehmkuhl. 1978. Isolation and identification of a bovine adenovirus type 3 with an adenovirus-associated virus. Am. J. Vet. Res. 39:1904-1906. [PubMed] [Google Scholar]
- 11.Davis, M. D., J. Wu, and R. A. Owens. 2000. Mutational analysis of adeno-associated virus type 2 Rep68 protein endonuclease activity on partially single-stranded substrates. J. Virol. 74:2936-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erles, K., P. Sebokova, and J. R. Schlehofer. 1999. Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV). J. Med. Virol. 59:406-411. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher, K. J., G. P. Gao, M. D. Weitzman, R. DeMatteo, J. F. Burda, and J. M. Wilson. 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol. 70:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman, A. B., D. R. Ronning, R. M. Kotin, and F. Dyda. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus. Rep. Mol. Cell 10:327-337. [DOI] [PubMed] [Google Scholar]
- 17.Hoggan, M. D. 1970. Adenovirus associated viruses. Prog. Med. Virol. 12:211-239. [PubMed] [Google Scholar]
- 17a.Katano, H., S. Afione, M. Schmidt, and J. A. Chiorini. 2004. Identification of adeno-associated virus contamination in cell and virus stocks by PCR. Biotechniques 36:1-4. [DOI] [PubMed] [Google Scholar]
- 18.Laughlin, C. A., H. Westphal, and B. J. Carter. 1979. Spliced adenovirus-associated virus RNA. Proc. Natl. Acad. Sci. USA 76:5567-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luchsinger, E., R. Strobbe, G. Wellemans, D. Dekegel, and S. Sprecher-Goldberger. 1970. Haemagglutinating adeno-associated virus (AAV) in association with bovine adenovirus type 1. Brief report. Arch. Gesamte Virusforsch. 31:390-392. [DOI] [PubMed] [Google Scholar]
- 20.Lusby, E. W., and K. I. Berns. 1982. Mapping of the 5′ termini of two adeno-associated virus 2 RNAs in the left half of the genome. J. Virol. 41:518-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muralidhar, S., S. P. Becerra, and J. A. Rose. 1994. Site-directed mutagenesis of adeno-associated virus type 2 structural protein initiation codons: effects on regulation of synthesis and biological activity. J. Virol. 68:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muramatsu, S., H. Mizukami, N. S. Young, and K. E. Brown. 1996. Nucleotide sequencing and generation of an infectious clone of adeno-associated virus 3. Virology 221:208-217. [DOI] [PubMed] [Google Scholar]
- 23.Myrup, A. C., S. B. Mohanty, and F. M. Hetrick. 1976. Isolation and characterization of adeno-associated viruses from bovine adenovirus types 1 and 2. Am. J. Vet. Res. 37:907-910. [PubMed] [Google Scholar]
- 24.Owens, R. A., M. D. Weitzman, S. R. Kyostio, and B. J. Carter. 1993. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J. Virol. 67:997-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu, J., R. Nayak, G. E. Tullis, and D. J. Pintel. 2002. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J. Virol. 76:12435-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redemann, B. E., E. Mendelson, and B. J. Carter. 1989. Adeno-associated virus Rep protein synthesis during productive infection. J. Virol. 63:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, J. A., J. V. Maizel, Jr., J. K. Inman, and A. J. Shatkin. 1971. Structural proteins of adenovirus-associated viruses. J. Virol. 8:766-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samulski, R. J., K. I. Berns, M. Tan, and N. Muzyczka. 1982. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. USA 79:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, R. H., S. A. Afione, and R. M. Kotin. 2002. Transposase-mediated construction of an integrated adeno-associated virus type 5 helper plasmid. BioTechniques 33:204-206, 208, 210-211. [DOI] [PubMed] [Google Scholar]
- 31.Smith, R. H., and R. M. Kotin. 1998. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J. Virol. 72:4874-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao, W., N. Chirmule, S. C. Berta, B. McCullough, G. Gao, and J. M. Wilson. 1999. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 73:3994-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamano, S., L. Y. Huang, C. Ding, J. A. Chiorini, C. M. Goldsmith, R. B. Wellner, B. Golding, R. M. Kotin, D. E. Scott, and B. J. Baum. 2002. Recombinant adeno-associated virus serotype 2 vectors mediate stable interleukin 10 secretion from salivary glands into the bloodstream. Hum. Gene Ther. 13:287-298. [DOI] [PubMed] [Google Scholar]