Abstract
The saimiri transforming protein (STP) oncogene of Herpesvirus saimiri subgroup A strain 11 (STP-A11) is not required for viral replication but is required for lymphoid cell immortalization in culture and lymphoma induction in primates. We previously showed that STP-A11 interacts with cellular Src kinase through its SH2 binding motif and that this interaction elicits Src signal transduction. Here we demonstrate that STP-A11 interacts with signal transducer and activator of transcription 3 (Stat3) independently of Src association and that the amino-terminal short proline-rich motif of STP-A11 and the central linker region of Stat3 are necessary for their interaction. STP-A11 formed a triple complex with Src kinase and Stat3 where Src kinase phosphorylated Stat3, resulting in the nuclear localization and transcriptional activation of Stat3. Consequently, the constitutively active Stat3 induced by STP-A11 elicited cellular signal transduction, which ultimately induced cell survival and proliferation upon serum deprivation. Furthermore, this activity was strongly correlated with the induction of Fos, cyclin D1, and Bcl-XL expression. These results demonstrate that STP-A11 independently targets two important cellular signaling molecules, Src and Stat3, and that these proteins cooperate efficiently to induce STP-A11-mediated transformation.
Signal transducer and activator of transcription 3 (Stat3) protein belongs to a seven-member family of latent cytoplasmic transcription factors that contribute to signal transduction initiated by cytokines, hormones, and growth factors (13). Stat3 proteins control fundamental cellular processes, including survival, proliferation, and differentiation. Upon stimulation with interferons, interleukin-6, granulocyte-macrophage colony-stimulating factor, epidermal growth factor, or platelet-derived growth factor, Stat3 protein becomes activated by phosphorylation on a single tyrosine (Y705) and dimerizes through a reciprocal interaction between the SH2 domain of one monomer and the phosphorylated tyrosine of the other (29). The dimers accumulate in the nucleus, recognize specific DNA elements, and activate transcription. Given the critical roles of Stat3 protein, it is hypothesized that inappropriate or aberrant activation of Stat3 contributes to cellular transformation and, in particular, leukemogenesis (6, 19, 20). In fact, the constitutive activation of Stat3 has been detected in many cancer cells and tissues, including those of multiple myeloma, leukemia, lymphoma, breast, lung, and head and neck cancer (6, 19, 20). Furthermore, it has been shown that Stat3 exerts its growth-deregulating activity by activating the expression of cellular genes that are involved in cell cycle progression such as fos, cyclin D1, myc, and pim-1 and by activating antiapoptotic processes such as Bcl-2 and Bcl-XL (3, 5, 7, 17, 35). Thus, activation of the Stat3 signal transduction pathway is likely a common strategy used by various viruses to transform a normal cell to a cancerous cell (8, 38). In fact, viral Src has been shown to interact with Stat3 and to activate Stat3 to induce cell growth transformation (4, 34, 37).
Herpesvirus saimiri (HVS) belongs to the gamma subfamily of herpesviruses (Gammaherpesvirinae). HVS naturally infects the squirrel monkey (Saimiri sciureus), a common South American primate, but with no apparent disease association. However, HVS infection of marmosets, owl monkeys, and other species of New World primates results in rapidly progressing fulminant lymphomas, lymphosarcomas, and leukemias of T-cell origin (18, 26). HVS can be further subclassified into three subgroups (subgroups A, B, and C) on the basis of the extent of DNA sequence divergence at the left end of coding DNA (32). Subgroups A and C are highly oncogenic and are able to immortalize common marmoset T lymphocytes to interleukin 2-independent growth in vitro (16, 36). Subgroup-C strains are further capable of immortalizing human, rabbit, and rhesus monkey lymphocytes into continuously proliferating T-cell lines (1, 2).
Mutational analyses have demonstrated that the leftmost open reading frame in the coding sequence of subgroup A strain 11 is not required for viral replication but is required for immortalization of common marmoset T lymphocytes in vitro and for lymphoma induction in vivo (14, 15). This open reading frame is termed STP-A11, for saimiri transformation-associated protein of subgroup A strain 11 (33). At a position and an orientation equivalent to those of the STP-A11 reading frame, the highly oncogenic HVS subgroup C strain 488 contains a distantly related reading frame termed STP-C488 (2, 28). Despite limited sequence similarity, STP-A11 and STP-C488 seem to be organized similarly in terms of the presence and localization of basic structural elements (2, 28). Both proteins are predicted to have a highly acidic amino terminus and collagen-like repeats in the central region. The primary amino acid sequence of STP-A11 has 9 repeats of a collagen-like motif (Gly-X-Y, where X and/or Y is proline), and in STP-C488 this motif is directly repeated 18 times (2, 28). The STP-A11 and STP-C488 proteins also contain a hydrophobic stretch at their carboxyl termini sufficient for a membrane-spanning domain (27). Both STPs have transforming and tumor-inducing activities independent of the rest of the herpesvirus genome (28). Specifically, both can transform rodent fibroblast cells, resulting in apparent loss of contact inhibition, formation of foci, growth at reduced serum concentrations, and formation of invasive tumors in nude mice (28).
To understand the structural and functional properties of STP-A, we analyzed the primary amino acid sequences of six different subgroup-A isolates (31). This analysis revealed that STP-A contains interesting structural and functional elements, including the 60PVQES64 binding motif for tumor necrosis factor (TNF) receptor-associated factors (TRAFs) and the 115YAEV118 SH2 binding motif for Src family kinases. Indeed, biochemical analysis has demonstrated that STP-A is capable of interacting with TRAF and Src kinase through the 60PVQES64 and 115YAEV118 motifs, respectively. While the role of TRAF interaction has not been well characterized, Src interaction has been shown to induce the tyrosine phosphorylation of STP-A11 as well as of other cellular proteins (30, 31).
In this report, we further demonstrate that STP-A11 interacts with Stat3 independently of TRAF and Src, and that Src kinase associated with STP-A11 phosphorylates Stat3, resulting in its nuclear localization and transcriptional activation. Consequently, the constitutive activation of Stat3 induced by STP-A11 leads to cell survival and proliferation upon serum deprivation. Thus, the STP-A11 oncoprotein targets multiple cellular signaling molecules to elicit cell growth transformation, which ultimately contributes to T-cell transformation induced by HVS subgroup-A strains.
MATERIALS AND METHODS
Cell culture and transfection.
Human embryonic kidney (HEK) 293T cells, Cos-1 cells, and NIH 3T3 cells were cultured in Dulbecco's modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. Cells were plated at 106 per 100-mm-diameter plate or 2 × 105 per 64-mm-diameter plate 24 h before transfection. Cells were transfected with 8 to 15 μg of DNA by using the calcium phosphate precipitation method (Clontech, Palo Alto, Calif.) or Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. NIH 3T3 cells stably expressing STP-A11 or its mutants were maintained in DMEM containing 10% FBS and 5.0 μg of puromycin/ml (Sigma, St. Louis, Mo.).
Plasmid construction.
The STP-A11 gene (31) was amplified by PCR using a 5′ primer containing the hemagglutinin (HA) tag or the AU1 tag and a 3′ gene-specific primer. The gene was subcloned into the pEF6 vector (Invitrogen). To generate STP-A11 deletion mutants, two-step PCR mutagenesis was performed (21). Wild-type (wt) STP-A11 and its mutants were completely sequenced to verify 100% agreement with the authentic or expected mutant sequence by the CEQ 8000 genetic analysis system (Beckman Coulter, Fullerton, Calif.). To construct glutathione S-transferase (GST) fusion constructs, STP-A11 DNA fragments and Stat3 DNA fragments were amplified by PCR and inserted in frame into the pGEX4T-1 vector (Amersham, Piscataway, N.J.). The pBUD vector (Invitrogen) carrying two separate promoters was used to express both Src and STP-A11.
Immunoprecipitation and immunoblotting.
Cells were harvested and lysed with lysis buffer (0.15 M NaCl, 1% Nonidet P-40, 50 mM Tris [pH 7.5]) containing 0.1 mM Na2VO3, 1 mM NaF, and protease inhibitors (leupeptin, aprotinin, phenylmethylsulfonyl fluoride, and bestatin). For immunoblotting, polypeptides from whole-cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Immunoblot detection was performed with a 1:2,000 dilution of primary antibodies. For immunoprecipitation, cells were harvested after 36 to 48 h of transfection, and cell debris was removed by centrifugation at 10,000 × g for 10 min at 4°C. Cell lysates were precleared with 25 μl of protein A/G agarose and incubated with appropriate primary antibodies and protein A/G agarose for 2 h at 4°C. After three washes with the lysis buffer, the precipitates were loaded on SDS-PAGE gels and analyzed by immunoblotting with the appropriate antibodies. The AU1 antibody was purchased from Covance (Richmond, Calif.), and Fos, cyclin D1, Bcl-XL, Src, Stat3, and phosphospecific Stat3 antibodies were purchased from Santa Cruz Biotech (Santa Cruz, Calif.). The protein was visualized with a chemical luminescent detection reagent (Pierce, Rockford, Ill.) and detected by a Fuji Phosphor Imager.
In vitro GST pull-down assay.
The GST-STPA11 and GST-Stat3 fusion proteins were purified from Escherichia coli BL21 (Invitrogen) with glutathione Sepharose 4B beads as recommended by the manufacturer (Amersham). At 48 h after transfection with STP-A11 or Stat3 expression plasmids, 293T cells were lysed with binding buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1% protease inhibitor cocktail solution [Sigma]) and mixed with 10 μg of the GST fusion protein for 2 h at 4°C. Then glutathione Sepharose 4B beads were extensively washed and subjected to SDS-10% PAGE, followed by an immunoblot assay.
Stat3 luciferase reporter assay.
HEK 293T cells were transfected with wild-type STP-A11 or its mutant expression vectors together with the Stat3-dependent reporter vector pLucTKS3 (39) by the calcium phosphate transfection procedure. To normalize transfection efficiency, the pGK-βgal vector, which expresses β-galactosidase from a phosphoglucokinase promoter, was included in the transfection mixture. At 48 h posttransfection, cells were washed with cold phosphate-buffered saline (PBS) and lysed in lysis solution (25 mM Tris [pH 7.8], 2 mM EDTA, 2 mM dithiothreitol, 10% glycerol, and 1% Triton X-100). Luciferase activity was measured with a luminometer by using a luciferase assay kit (Promega, Madison, Wis.) and was normalized to β-galactosidase activity.
Immunofluorescence.
Cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with cold acetone for 15 min, blocked with 10% goat serum in PBS for 30 min, and reacted with 1:100 to 1:2,000 dilutions of primary antibody in PBS for 30 min at room temperature. After incubation, cells were washed extensively with PBS, incubated with a 1:100-diluted Alexa 488- or Alexa 568-conjugated anti-rabbit or anti-mouse antibody (Molecular Probes, Eugene, Oreg.) in PBS for 30 min at room temperature, and washed three times with PBS. Confocal microscopy was performed using a TCS SP laser-scanning microscope (Leica Microsystems, Exton, Pa.) fitted with a 40× Leica objective (PL APO, 1.4 NA), and using Leica imaging software. Images were collected at a resolution of 512 by 512 pixels. The stained cells were optically sectioned in the z axis, and the images in the different channels (photomultiplier tubes) were collected simultaneously. The step size in the z axis was varied from 0.2 to 0.5 μm to obtain 30 to 50 slices/imaged file. The images were transferred to a Macintosh G4 computer (Apple Computers, Cupertino, Calif.), and NIH Image software (version 1.61) was used to render the images.
Cell proliferation assay.
NIH 3T3 cells (5 × 103) stably expressing wt STP-A11 or its mutant were plated into a 96-well plate and incubated with different serum concentrations (0.1, 0.2, or 1%) for 0, 24, 48, or 72 h. Cell proliferation was measured by using a modified MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrasodium bromide tetrazolium] assay (Promega).
RESULTS
Expression of both STP-A11 and Src induces Stat3 transcription activation.
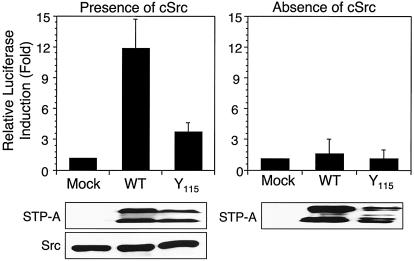
The Stat3 signal transduction pathway has been shown to be a common target for virus-induced cell transformation (8, 38). To investigate the potential effect of STP-A11 on Stat3 signal transduction, we examined Stat3-dependent transcriptional activity upon STP-A11 expression. 293T cells were transfected with the STP-A11 expression vector with or without the Src expression vector. The STP-A11 Y115A mutant, which was incapable of binding to Src kinase (31), was included as a control. At 48 h posttransfection, cells were harvested in order to measure Stat3 transcription factor activity by using the Stat3-dependent reporter vector pLucTKS3 (39). STP-A11 induced Stat3 transcriptional activity strongly (approximately 15-fold) in the presence of Src kinase but at only a minimal level in the absence of Src kinase (Fig. 1). Furthermore, STP-A11 Y115A mutant expression induced Stat3 transcription activity weakly in the presence of Src kinase but not at all in its absence (Fig. 1). This result indicates that STP-A11 expression strongly induces Stat3 transcription activity only in the presence of Src expression and that the interaction of STP-A11 with Src kinase is likely necessary for efficient activation of Stat3 transcription activity.
FIG. 1.
Activation of Stat transcription activity by STP-A11 and Src expression. 293T cells were transfected with a Stat3 luciferase reporter vector (pLuc-TKS3) and an expression vector containing either wt STP-A11 or the Y115A mutant either alone or together with a Src expression vector. At 48 h posttransfection, cells were harvested, and luciferase activity was measured. Transfection efficiency was normalized with the β-galactosidase reporter vector pGK-βgal. Results are averages from three independent experiments. Error bars, standard deviations. Gels at the bottom show the expression of STP-A11, its mutant, and Src in whole-cell lysates.
STP-A11 specifically interacts with Stat3.
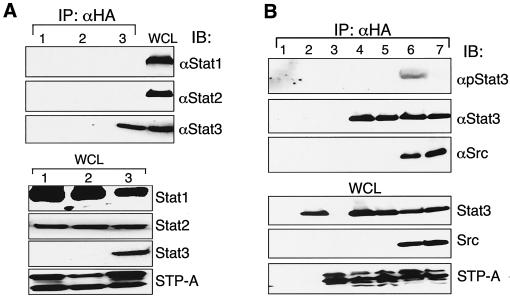
To further investigate the effect of STP-A11 on the Stat signal pathway, we examined the potential interaction between STP-A11 and Stat transcription factors. Stat1 and Stat2, but not Stat3, were found to be expressed at high levels in 293T cells (data not shown). Therefore, 293T cells were transfected with the amino-terminal HA-tagged STP-A11 expression vector with or without with the Stat3 expression vector. At 48 h posttransfection, cell lysates were immunoprecipitated with an anti-HA antibody, followed by immunoblotting with an anti-Stat1, anti-Stat2, or anti-Stat3 antibody. The results showed that STP-A11 interacted specifically with Stat3 but not with Stat1 and Stat2 (Fig. 2A). Stat1, Stat2, and Stat3 were expressed at equivalent levels (Fig. 2A).
FIG. 2.
Specific interaction of STP-A11 with Stat3. (A) STP-A11 interacts with Stat3 but not with Stat1 or Stat2. At 48 h after transfection with the carboxy-terminal HA-tagged STP-A11 expression vector alone (lanes 1 and 2) or together with the Stat3 expression vector (lane 3), 293T cells were lysed and used for immunoprecipitation (IP) with an anti-HA (αHA)antibody, followed by immunoblotting with an anti-Stat1, anti-Stat2, or anti-Stat3 antibody. To demonstrate the expression of endogenous Stat1 and Stat2 and transfected Stat3, whole-cell lysates (WCL) were used for immunoblotting with an anti-Stat1, anti-Stat2, or anti-Stat3 antibody and an anti-HA antibody. (B) Stat3 interacts with STP-A11 and is phosphorylated at Y705 by Src kinase associated with STP-A11. At 48 h after transfection with various expression vectors, 293T cells were lysed and used for immunoprecipitation with an anti-HA antibody, followed by immunoblotting with an anti-Stat3 antibody, an anti-Stat3 Y705 phosphospecific antibody, or an anti-Src antibody. Lane 1, vector only; lane 2, Stat3; lane 3, STP-A11; lane 4, Stat3 plus STP-A11; lane 5, Stat3 plus STP-A11 Y115; lane 6, Stat3 plus STP-A11 plus Src; lane 7, Stat3Y705A plus STP-A11 plus Src.
Since Src interaction was necessary for STP-A11-induced activation of Stat3 transcription activity, we next tested whether it was also necessary for Stat3 interaction. 293T cells were transfected with HA-STP-A11, Stat3 expression vectors, and Src expression vectors in various combinations. The HA-tagged STP-A11 Y115A mutant was also included as a control. At 48 h posttransfection, cell lysates were used for immunoprecipitation with an anti-HA antibody, followed by immunoblotting with an anti-Src antibody, an anti-Stat3 antibody, or the anti-phospho-Stat3 antibody, which reacts only with Y705-phsophorylated Stat3. Stat3 protein was readily detected in wt STP-A11 and mutant STP-A11 Y115A immune complexes in the presence or absence of Src expression, indicating that STP-A11 binds to Stat3 independently of Src interaction (Fig. 2B, lanes 4, 5, and 6). However, the Y705 phosphorylation of Stat3 was very strong in the wt STP-A11 complex (Fig. 2B, lane 6) but was only very weakly detected in the STP-A11 Y115A immune complex under the same conditions (data not shown). Furthermore, when the Stat3 Y705A mutant was expressed, its phosphorylation was not detected in the complex of wt STP-A11 (Fig. 2B, lane 7), indicating that Src kinase associated with STP-A11 phosphorylates the Y705 residue of Stat3. These results indicate that while Src interaction of STP-A11 is not necessary for its binding to Stat3, it is required for the efficient phosphorylation of the Stat3 Y705 residue in the STP-A11 complex.
Identification of the binding regions of STP-A11 and Stat3.
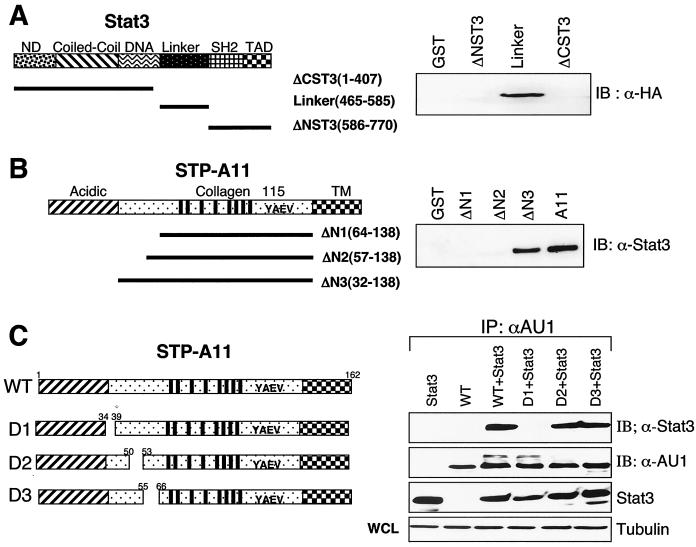
To further define the regions of STP-A11 required for Stat3 interaction, we generated various deletion mutants of both Stat3 and STP-A11. The Stat3 protein consists of an amino-terminal domain, a coil-coiled domain, a DNA binding domain, a linker, an SH2 binding domain, and a carboxyl-terminal activation domain (13, 24, 25). We generated three GST-Stat3 fusion constructs: GST-ΔCST3, in which the carboxy-terminal domain was deleted; GST-Link, containing the central linker region; and GST-ΔNST3, in which the amino-terminal domain was deleted. Lysates of 293T cells containing HA-tagged STP-A11 were used for a pull-down assay with bacterially purified GST-Stat3 fusion proteins, followed by immunoblotting with an anti-HA antibody. This experiment showed that the central linker region of Stat3 was capable of interacting with STP-A11, whereas the amino-terminal and carboxy-terminal regions of Stat3 were not capable of doing so under the same conditions (Fig. 3A). Thus, the central linker region of Stat3 is sufficient for interacting with STP-A11.
FIG. 3.
Identification of the regions of Stat3 and STP-A11 necessary for their interaction. (A) The central linker region of Stat3 is sufficient for interacting with STP-A11. Bacterially purified GST or GST fusion proteins containing the amino-terminal region (ΔCST3), the central linker region, or the carboxyl-terminal region (ΔNST3) of Stat3 were mixed with lysates of 293T cells transfected with the HA-tagged STP-A11 expression vector. Polypeptides present in GST beads were used for immunoblotting (IB) with an anti-HA (α-HA) antibody. Similar amounts of each GST fusion protein were used in this assay (data not shown). ND, N-terminal domain; TAD, transcriptional activation domain. (B) The amino-terminal region of STP-A11 is necessary for interacting with Stat3. Bacterially purified GST or GST fusion proteins containing the amino-terminal regions of STP-A11 were mixed with lysates of 293T cells transfected with the Stat3 expression vector. Polypeptides present in GST beads were used for immunoblotting with an anti-Stat3 antibody. Similar amounts of each GST fusion protein were used in this assay (data not shown). TM, transmembrane domain. (C) The amino-terminal proline-rich motif of STP-A11 is necessary for interacting with Stat3. At 48 h after transfection with a Stat3 expression vector together with an expression vector containing the HA-tagged wt STP-A11 or its mutant, 293T cells were lysed and used for immunoprecipitation with an anti-HA antibody, followed by immunoblotting with an anti-Stat3 antibody.
The STP-A11 protein contains three distinct regions: an amino-terminal acidic region, a central collagen-like region, and a carboxyl transmembrane region (Fig. 3B). To identify the region of STP-A11 required for Stat3 interaction, we generated GST-STP-A11 fusion constructs containing serial deletions of STP-A11 from the amino terminus. These deletion constructs were GST-ΔN1 (amino acids 64 to 138 deleted), GST-ΔN2 (amino acids 57 to 138 deleted), and GST-ΔN3 (amino acids 32 to 138 deleted). Lysates of 293T cells transfected with a Stat3 expression vector were used for a pull-down assay with bacterially purified GST-STP-A11 fusion proteins, followed by immunoblotting with an anti-Stat3 antibody. The experiment showed that GST-ΔN3 was capable of interacting with Stat3, while GST-ΔN1 and GST-ΔN2 were not (Fig. 3B). Thus, the amino acid sequences between residues 32 and 57 of STP-A11 are necessary for Stat3 binding.
Precise inspection reveals that this region of STP-A11 contains three recognizable motifs: the proline-rich motifs 34PTPYLP38 and 50PYNP53 and a cysteine bridge between C56 and C65. To further investigate whether these motifs play a role in Stat3 interaction, we introduced mutations into full-length STP-A11. The D1 mutation contains a deletion of the 34PTPYLP38 proline-rich motif, the D2 mutation contains point mutations of two prolines to glycine in the 50PYNP53 proline-rich motif, and the D3 mutation contains a deletion from C56 to C65. 293T cells were cotransfected with the amino-terminally AU1-tagged STP-A11 wt and mutant forms together with a Stat3 expression vector. At 48 h posttransfection, cell lysates were used for immunoprecipitation with an anti-AU1 antibody, followed by immunoblotting with an anti-Stat3 antibody. The D1 mutation completely abolished Stat3 binding ability, whereas the D2 and D3 mutations did not affect Stat3 binding ability (Fig. 3C). wt STP-A11 and its mutants were expressed at equivalent levels (Fig. 3C).
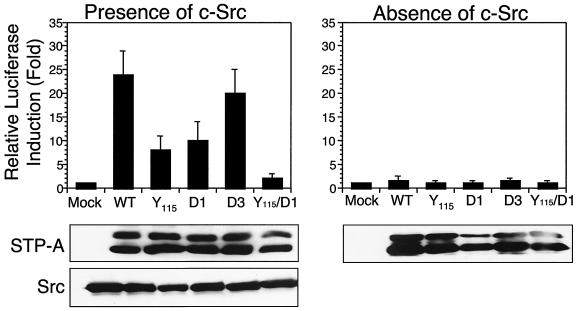
Finally, we tested whether Stat3 interaction was necessary for efficient STP-A11-induced activation of Stat3 transcriptional activity. 293T cells were transfected with a wt STP-A11 or STP-A11 D1, D2, or D3 mutant expression vector in the presence or absence of the Src expression vector. The STP-A11 Y115A mutant, which was incapable of binding to Src, and the STP-A11 Y115A/D1 mutant, which was incapable of binding to either Src or Stat3, were included in this assay. At 48 h posttransfection, cells were harvested to measure Stat3 transcription factor activity by using the Stat3-dependent reporter vector pLucTKS3 (39). wt STP-A11 and the STP-A11 D3 mutant strongly induced Stat3 transcriptional activity, whereas STP-A11 D1 and STP-A11 Y115A mutants exhibited significantly reduced levels of Stat3 transcriptional activation (Fig. 4). In contrast, the STP-A11 Y115A/D1 mutant induced little or no activation of Stat3 transcriptional activity (Fig. 4). These results indicate that the central linker region of Stat3 and the amino-terminal proline-rich motif of STP-A11 are responsible for their interaction and that both STP-A11 interaction with Stat3 and STP-A11 interaction with Src are necessary for the efficient activation of Stat3 transcriptional activity.
FIG. 4.
Effect of the interaction of STP-A11 with Stat3 and Src on the efficient activation of Stat3 transcriptional activity. 293T cells were transfected with a Stat3 luciferase reporter vector (pLuc-TKS3) and an expression vector containing wt STP-A11 or the Y115A, D1, D2, D3, or Y115A/D1 mutant in the presence or absence of the Src expression vector. At 48 h posttransfection, cells were harvested for measurement of luciferase activity. Transfection efficiency was normalized with the β-galactosidase reporter vector pGK-βgal. Results are averages from three independent experiments. Error bars, standard deviations. Gels at the bottom show expression of STP-A11, its mutants, and Src in whole-cell lysates.
STP-A11 and Src expression induces Stat3 nuclear localization.
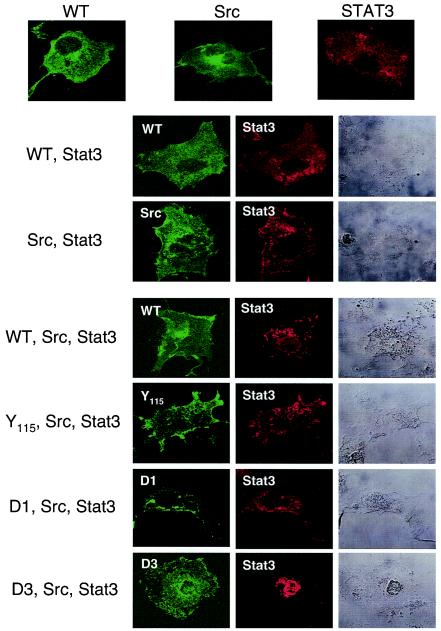
Upon stimulation with a cytokine, activated Stat3 forms a homodimer through the reciprocal interaction between its own SH2 domains. The dimer translocates into the nucleus, where it recognizes specific DNA elements and activates transcription. We used confocal immunofluorescence microscopy to examine the effect of STP-A11 on Stat3 localization. Cos-1 cells were transfected with an STP-A11, Src, or Stat3 expression vector individually or in combination as depicted in Fig. 5. As previously described (10, 29), Src and Stat3 proteins were primarily present in the cytoplasm (Fig. 5). Interestingly, unlike the STP-C488 oncoprotein of HVS subgroup C strain 488, which was primarily localized in the endoplasmic reticulum (27), STP-A11 was present in both the cytoplasm and the plasma membrane (Fig. 5). Interestingly, Stat3 was primarily located in the cytoplasmic region in the presence of either Src or STP-A11 expression, whereas it efficiently translocated into the nucleus upon expression of wt STP-A11 or the STP-A11 D3 mutant together with Src (Fig. 5). In contrast, expression of the Y115A or D1 mutant did not induce nuclear localization of Stat3 under the same conditions (Fig. 5). These results indicate that STP-A11 efficiently induces the nuclear localization of Stat3 and that both Src and Stat3 interactions with STP-A11 are necessary for this activity.
FIG. 5.
Nuclear localization of Stat3 upon STP-A11 and Src expression. Cos-1 cells were transfected with a Stat3 expression vector alone or together with pBud expression vectors carrying c-Src and/or wt or mutant STP-A11. At 48 h posttransfection, cells were fixed and reacted with anti-Stat3 (red), anti-STP-A11 (green), and/or anti-Src (green) antibodies, followed by incubation with an Alexa-488-conjugated anti-mouse immunoglobulin G antibody and/or an Alexa-568-conjugated anti-rabbit immunoglobulin G antibody. Immunofluorescence was examined with a Leica confocal microscope. Cells were visualized with Nomarski optics.
The activation of Stat3 activity induced by STP-A11 contributes to the alteration of cell growth.
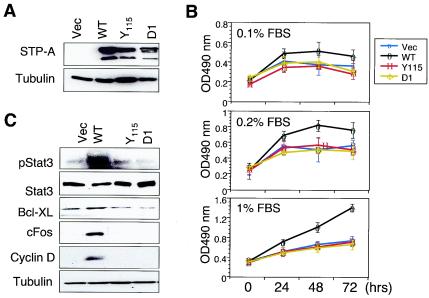
Inappropriate or aberrant activation of Stats has been shown to contribute to cell growth transformation through induction of cell cycle progression and antiapoptotic pathways (3, 5, 7, 17, 35). We hypothesized that Src kinase recruited by STP-A11 activated the Stat 3 transcription factor, which led to alteration of cell growth. To test this hypothesis, we examined cell growth upon serum deprivation. NIH 3T3 cells were transfected with the pEF-IRES-puro vector (23) containing wt or Y115A or D1 mutant STP-A11 and were selected for puromycin resistance. STP-A11 and its mutants were expressed at equivalent levels in puromycin-resistant NIH 3T3 cells (Fig. 6A). These cells were incubated in the presence of 0.1, 0.2, or 1% serum, and their growth rates were then measured by an MTT assay. Control NIH 3T3 cells carrying vector alone and NIH 3T3 cells expressing the STP-A11 Y115A or D1 mutant exhibited almost no growth or drastically reduced growth under all serum conditions (Fig. 6B). By striking contrast, NIH 3T3 cells expressing wt STP-A11 showed detectable levels of growth in the presence of 0.1 or 0.2% serum and almost a normal growth rate in the presence of 1% serum (Fig. 6B). These results indicate that STP-A11 expression significantly alters cell growth, which allows cells to survive and/or grow upon serum deprivation.
FIG. 6.
Contribution of STP-A11-induced Stat3 activation to enhanced cell growth. (A) Expression of wt STP-A and its mutants in stable NIH 3T3 cells. Lysates from NIH 3T3-puro (Vec), NIH 3T3-wt STP-A11 (WT), NIH 3T3-STP-A11 Y115A (Y115), and NIH 3T3-STP-A11 D1 (D1) cells were prepared and blotted with an anti-AU1 antibody for detection of STP-A. Tubulin was used for loading equivalent amount of proteins from each sample. (B) Cell growth rate upon serum deprivation. NIH 3T3-puro, NIH 3T3-wt STP-A11, NIH 3T3-STP-A11 Y115A, and NIH 3T3-STP-A11 D1 cells were incubated with 0.1, 0.2, or 1% serum. Cell proliferation rates of these NIH 3T3 cells were measured by a modified MTT assay. Results are averages from three independent experiments. Error bars, standard deviations. (C) Y705 phosphorylation of Stat3 and enhanced expression of Fos, cyclin D1, and Bcl-XL upon STP-A11 expression. Equivalent amounts of polypeptides from NIH 3T3-puro, NIH 3T3-wt STP-A11, NIH 3T3-STP-A11 Y115A, and NIH 3T3-STP-A11 D1 cells were used for immunoblotting with an anti-Stat3 antibody, an anti-Stat3 Y705 phosphospecific antibody, an anti-Fos antibody, an anti-cyclin D1 antibody, or an anti-Bcl-XL antibody.
To further delineate the effect of STP-A11 on cell growth control, we tested the level of Stat3 Y705 phosphorylation by immunoblotting. Results showed that the Stat3 Y705 residue was highly phosphorylated in NIH 3T3 cells expressing wt STP-A11 relative to phosphorylation in NIH 3T3 cells expressing vector alone or STP-A11 mutants, indicating that STP-A11 expression induces an endogenous Stat3 activation (Fig. 6C). Stat3 activation has been shown to provide a survival signal by activating the expression of cellular genes that are involved in cell cycle progression, such as Fos and cyclin D1, and antiapoptotic genes, such as Bcl-XL (3, 5, 7, 17, 35). We examined whether the STP-A11-induced activation of Stat3 transcription affected the expression of cellular Fos, cyclin D1, and Bcl-XL. Equivalent amounts of polypeptides from NIH 3T3 cells containing a vector, wt STP-A11, or mutant STP-A11 were used for immunoblotting with anti-Fos, anti-cyclin D1, and anti-Bcl-XL antibodies. An anti-tubulin antibody was included as a control. This experiment showed that wt STP-A11 considerably enhanced the levels of Fos, cyclin D1, and Bcl-XL protein expression (Fig. 6C). These results suggest that the constitutive activation of Stat3 induced by STP-A11 contributes to the alteration of cell growth through the induction of cell cycle and antiapoptotic signaling pathways.
DISCUSSION
A growing body of evidence shows that inappropriate or aberrant activation of Stat3 contributes to cellular transformation and, in particular, leukemogenesis (6, 19, 20). Here we report that STP-A11 expression induces the activation of Stat3 transcription activity, which upregulates cellular Fos, cyclin D1, and Bcl-XL expression and thereby cell growth transformation. Furthermore, this activation requires the specific interactions of STP-A11 with the Src kinase and the Stat3 transcription factor. These results indicate that STP-A11 targets two important signaling molecules, Src and Stat3, to deregulate cell growth control and facilitate virus-induced oncogenesis.
The Stat3 protein consists of an amino-terminal domain, a coil-coiled domain, a DNA binding domain, a linker, an SH2 binding domain, and a carboxy-terminal activation domain (13, 24, 25). We found that STP-A11 efficiently interacted with Stat3 but not with Stat1 or Stat2. Furthermore, our mapping results showed that the central linker region of Stat3 was sufficient for binding to STP-A11 and that the amino-terminal 34PTPYLP38 proline-rich motif of STP-A11 was necessary for binding to Stat3. In addition, despite extensive sequence variation of the central region of STP-A, this proline-rich motif appears to be well conserved among six different isolates (31). Furthermore, several STP-B isolates that are distant relatives of STP-A also show considerable conservation of this proline-rich motif in their amino-terminal domains, reflecting the important role of this motif in Stat 3 interaction (9). Recently, hepatitis C virus core protein (38) has been shown to interact with the central linker region of Stat3. Interaction and activation of Stat3 by the HCV core also resulted in rapid cell growth and upregulation of Bcl-XL and cyclin D1. Furthermore, exogenous expression of Stat3 in HCV core-expressing cells led to anchorage-independent growth and tumorigenesis (38). Thus, because of its important role in cell growth control, Stat3 is likely a common target for viral oncoproteins. It should be noted that the amino-terminal proline-rich motif of STP-A11 also shows a great resemblance to the binding motifs to SH3 and WW domains. In fact, we have found that STP-A11 is able to interact with the WW domain of NEDD4 ubiquitin ligase (unpublished data). This suggests that the central linker region of Stat3 may potentially mimic the SH3 and/or WW domains of cellular signaling molecules for ligand interaction.
It has previously been shown that STP-A11 interacts with and activates Src kinase through its SH2 binding motif (31). Here we have also demonstrated that STP-A11 interacts with and activates the Stat3 transcription factor through its proline-rich motif. Furthermore, we have shown that Src kinase in association with STP-A11 phosphorylates the Y705 residue of Stat3, which induces its nuclear localization and transcriptional activation. The STP-A Y115A mutant, defective in Src interaction, and the D1 mutant, defective in Stat3 interaction, both showed diminished levels of Stat3 transcriptional activation, whereas the STP-A11 Y115A/D1 mutant, defective in both Stat3 and Src interaction, showed little or no activation of Stat3 transcriptional activity. Furthermore, neither the Y115A nor the D1 mutant of STP-A11 displayed any effect on cell growth control. This suggests that while STP-A11 interaction either with Src or with Stat3 weakly induces Stat3 transcriptional activation, it is not sufficient to allow cells to proliferate upon serum deprivation, indicating that both Src interaction and Stat3 interaction are necessary for inducing the full strength of STP-A11-mediated cell growth transformation. This also suggests that STP-A11 functions as an adaptor to link Src kinase and the Stat3 transcription factor: STP-A11 recruits Stat3 in the vicinity of Src kinase to allow Stat3 tyrosine phosphorylation and thereby Stat3 transcriptional activation.
A striking feature of gammaherpesviruses is that they contain a distinct open reading frame at the end of their genome; each of these open reading frames has characteristic transforming ability (11, 12, 26). These include the Epstein-Barr virus (EBV) latent membrane protein 1 (LMP-1) and HVS STP. LMP-1 acts like a permanently activated receptor of the TNF receptor superfamily and is absolutely required for B-cell immortalization by EBV. Gires et al. (22) have shown that the LMP-1 oncoprotein in B cells interacts with Jak3 kinase through its carboxyl-terminal proline-rich sequence, leading to the enhanced tyrosine auto/transphosphorylation of Jak3 kinase and thereby the activation of Stat transcription factor activity. Furthermore, Chen et al. (8) have also shown that LMP-1 expression in nasopharyngeal carcinoma cells induces a significant increase in the tyrosine-phosphorylated forms of Stat3 and Stat5 and allows the normally cytoplasmic Stat proteins to enter the nucleus and bind to their recognition sequences in responsive promoters. However, it should be noted that despite its important role in cell growth control, the specific role of Stat activation induced by EBV LMP-1 and HVS STP-A11 has not been elucidated. Nevertheless, although there is no discernible homology between STP-A11 and LMP-1, these two oncoproteins utilize similar but distinct motifs to target Stat transcription factors and activate their transcriptional activity, which ultimately elicits cellular activation and cell proliferation. Future experiments will seek to understand the detailed molecular nature of STP-A11/Stat3/Src-mediated activation of downstream signal transduction pathways.
Acknowledgments
We especially thank Ganes Sen for providing the Stat expression vector and J. Macke for manuscript editing.
This work was partly supported by U.S. Public Health Service grants CA31363, AI38131, and RR00168. P. Feng and J. Jung are Leukemia & Lymphoma Society Fellow and Scholar, respectively.
REFERENCES
- 1.Biesinger, B., I. Muller-Fleckenstein, B. Simmer, G. Lang, S. Wittmann, E. Platzer, R. C. Desrosiers, and B. Fleckenstein. 1992. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc. Natl. Acad. Sci. USA 89:3116-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesinger, B., J. J. Trimble, R. C. Desrosiers, and B. Fleckenstein. 1990. The divergence between two oncogenic Herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology 176:505-514. [DOI] [PubMed] [Google Scholar]
- 3.Bowman, T., M. A. Broome, D. Sinibaldi, W. Wharton, W. J. Pledger, J. M. Sedivy, R. Irby, T. Yeatman, S. A. Courtneidge, and R. Jove. 2001. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc. Natl. Acad. Sci. USA 98:7319-7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bromberg, J. F., C. M. Horvath, D. Besser, W. W. Lathem, and J. E. Darnell, Jr. 1998. Stat3 activation is required for cellular transformation by v-src. Mol. Cell. Biol 18:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bromberg, J. F., M. H. Wrzeszczynska, G. Devgan, Y. Zhao, R. G. Pestell, C. Albanese, and J. E. Darnell, Jr. 1999. Stat3 as an oncogene. Cell 98:295-303. [DOI] [PubMed] [Google Scholar]
- 6.Catlett-Falcone, R., W. S. Dalton, and R. Jove. 1999. STAT proteins as novel targets for cancer therapy. Signal transducer and activator of transcription. Curr. Opin. Oncol. 11:490-496. [DOI] [PubMed] [Google Scholar]
- 7.Catlett-Falcone, R., T. H. Landowski, M. M. Oshiro, J. Turkson, A. Levitzki, R. Savino, G. Ciliberto, L. Moscinski, J. L. Fernandez-Luna, G. Nunez, W. S. Dalton, and R. Jove. 1999. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 10:105-115. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., L. Hutt-Fletcher, L. Cao, and S. D. Hayward. 2003. A positive autoregulatory loop of LMP1 expression and STAT activation in epithelial cells latently infected with Epstein-Barr virus. J. Virol. 77:4139-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, J. K., S. Ishido, and J. U. Jung. 2000. The collagen repeat sequence is a determinant of the degree of herpesvirus saimiri STP transforming activity. J. Virol. 74:8102-8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courtneidge, S. A., and J. M. Bishop. 1982. Transit of pp60v-src to the plasma membrane. Proc. Natl. Acad. Sci. USA 79:7117-7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damania, B., J. K. Choi, and J. U. Jung. 2000. Signaling activities of gammaherpesvirus membrane proteins. J. Virol. 74:1593-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damania, B., and J. U. Jung. 2001. Comparative analysis of the transforming mechanisms of Epstein-Barr virus, Kaposi's sarcoma-associated herpesvirus, and herpesvirus saimiri. Adv. Cancer Res. 80:51-82. [DOI] [PubMed] [Google Scholar]
- 13.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 14.Desrosiers, R. C., A. Bakker, J. Kamine, L. A. Falk, R. D. Hunt, and N. W. King. 1985. A region of the Herpesvirus saimiri genome required for oncogenicity. Science 228:184-187. [DOI] [PubMed] [Google Scholar]
- 15.Desrosiers, R. C., R. L. Burghoff, A. Bakker, and J. Kamine. 1984. Construction of replication-competent herpesvirus saimiri deletion mutants. J. Virol. 49:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desrosiers, R. C., D. P. Silva, L. M. Waldron, and N. L. Letvin. 1986. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J. Virol. 57:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epling-Burnette, P. K., B. Zhong, F. Bai, K. Jiang, R. D. Bailey, R. Garcia, R. Jove, J. Y. Djeu, T. P. Loughran, Jr., and S. Wei. 2001. Cooperative regulation of Mcl-1 by Janus kinase/Stat and phosphatidylinositol 3-kinase contribute to granulocyte-macrophage colony-stimulating factor-delayed apoptosis in human neutrophils. J. Immunol. 166:7486-7495. [DOI] [PubMed] [Google Scholar]
- 18.Fickenscher, H., and B. Fleckenstein. 2001. Herpesvirus saimiri. Philos. Trans. R. Soc. Lond. B 356:545-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia, R., T. L. Bowman, G. Niu, H. Yu, S. Minton, C. A. Muro-Cacho, C. E. Cox, R. Falcone, R. Fairclough, S. Parsons, A. Laudano, A. Gazit, A. Levitzki, A. Kraker, and R. Jove. 2001. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene 20:2499-2513. [DOI] [PubMed] [Google Scholar]
- 20.Garcia, R., C. L. Yu, A. Hudnall, R. Catlett, K. L. Nelson, T. Smithgall, D. J. Fujita, S. P. Ethier, and R. Jove. 1997. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ. 8:1267-1276. [PubMed] [Google Scholar]
- 21.Gibbons, I. R., D. J. Asai, N. S. Ching, G. J. Dolecki, G. Mocz, C. A. Phillipson, H. Ren, W. J. Tang, and B. H. Gibbons. 1991. A PCR procedure to determine the sequence of large polypeptides by rapid walking through a cDNA library. Proc. Natl. Acad. Sci. USA 88:8563-8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobbs, S., S. Jitrapakdee, and J. C. Wallace. 1998. Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1α promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem. Biophys. Res. Commun. 252:368-372. [DOI] [PubMed] [Google Scholar]
- 24.Horvath, C. M., Z. Wen, and J. E. Darnell, Jr. 1995. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 9:984-994. [DOI] [PubMed] [Google Scholar]
- 25.Jove, R. 2000. Preface: STAT signaling. Oncogene 19:2466-2467. [DOI] [PubMed] [Google Scholar]
- 26.Jung, J. U., J. K. Choi, A. Ensser, and B. Biesinger. 1999. Herpesvirus saimiri as a model for gammaherpesvirus oncogenesis. Semin. Cancer Biol. 9:231-239. [DOI] [PubMed] [Google Scholar]
- 27.Jung, J. U., and R. C. Desrosiers. 1994. Distinct functional domains of STP-C488 of herpesvirus saimiri. Virology 204:751-758. [DOI] [PubMed] [Google Scholar]
- 28.Jung, J. U., and R. C. Desrosiers. 1991. Identification and characterization of the herpesvirus saimiri oncoprotein STP-C488. J. Virol. 65:6953-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kisseleva, T., S. Bhattacharya, J. Braunstein, and C. W. Schindler. 2002. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285:1-24. [DOI] [PubMed] [Google Scholar]
- 30.Lee, H., J. K. Choi, M. Li, K. Kaye, E. Kieff, and J. U. Jung. 1999. Role of cellular tumor necrosis factor receptor-associated factors in NF-κB activation and lymphocyte transformation by herpesvirus saimiri STP. J. Virol. 73:3913-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, H., J. J. Trimble, D. W. Yoon, D. Regier, R. C. Desrosiers, and J. U. Jung. 1997. Genetic variation of herpesvirus saimiri subgroup A transforming protein and its association with cellular src. J. Virol. 71:3817-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medveczky, P., E. Szomolanyi, R. C. Desrosiers, and C. Mulder. 1984. Classification of herpesvirus saimiri into three groups based on extreme variation in a DNA region required for oncogenicity. J. Virol. 52:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murthy, S. C., J. J. Trimble, and R. C. Desrosiers. 1989. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J. Virol. 63:3307-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiner, S. J., A. P. Schiavone, and T. E. Smithgall. 2002. Activation of STAT3 by the Src family kinase Hck requires a functional SH3 domain. J. Biol. Chem. 277:45680-45687. [DOI] [PubMed] [Google Scholar]
- 35.Sinibaldi, D., W. Wharton, J. Turkson, T. Bowman, W. J. Pledger, and R. Jove. 2000. Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: role of activated STAT3 signaling. Oncogene 19:5419-5427. [DOI] [PubMed] [Google Scholar]
- 36.Szomolanyi, E., P. Medveczky, and C. Mulder. 1987. In vitro immortalization of marmoset cells with three subgroups of herpesvirus saimiri. J. Virol. 61:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turkson, J., T. Bowman, R. Garcia, E. Caldenhoven, R. P. De Groot, and R. Jove. 1998. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol. Cell. Biol. 18:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida, T., T. Hanada, T. Tokuhisa, K. Kosai, M. Sata, M. Kohara, and A. Yoshimura. 2002. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J. Exp. Med. 196:641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, Y., J. Turkson, C. Carter-Su, T. Smithgall, A. Levitzki, A. Kraker, J. J. Krolewski, P. Medveczky, and R. Jove. 2000. Activation of Stat3 in v-Src-transformed fibroblasts requires cooperation of Jak1 kinase activity. J. Biol. Chem. 275:24935-24944. [DOI] [PubMed] [Google Scholar]