Abstract
Complement plays a pivotal role in the regulation of innate and adaptive immunity. It has been shown that the binding of C1q, a natural ligand of gC1qR, on T cells inhibits their proliferation. Here, we demonstrate that direct binding of the hepatitis C virus (HCV) core to gC1qR on T cells leads to impaired Lck/Akt activation and T-cell function. The HCV core associates with the surface of T cells specifically via gC1qR, as this binding is inhibited by the addition of either anti-gC1qR antibody or soluble gC1qR. The binding affinity constant of core protein for gC1qR, as determined by BIAcore analysis, is 3.8 × 10−7 M. The specificity of the HCV core-gC1qR interaction is confirmed by reduced core binding on Molt-4 T cells treated with gC1qR-silencing small interfering RNA and enhanced core binding on GPC-16 guinea pig cells transfected with human gC1qR. Interestingly, gC1qR is expressed at higher levels on CD8+ than on CD4+ T cells, resulting in more severe core-induced suppression of the CD8+-T-cell population. Importantly, T-cell receptor-mediated activation of the Src kinases Lck and ZAP-70 but not Fyn and the phosphorylation of Akt are impaired by the HCV core, suggesting that it inhibits the very early events of T-cell activation.
Hepatitis C virus (HCV) is a serious and growing threat to human health, having infected more than 170 million people worldwide. A remarkable feature of HCV is its ability to establish chronic infection. Indeed, the virus persists in >85% of patients following acute infection. These individuals carry an increased risk of developing various liver diseases, including cirrhosis and hepatocellular carcinoma (9). Unfortunately, no vaccine or effective treatment for HCV is currently available, and the mechanism(s) for the establishment of persistent HCV infection remains elusive.
CD4+- and CD8+-T-cell dysfunction may be a mechanism by which persistent HCV infection is established, because early and sustained CD4+- and CD8+-T-cell responses appear to be crucial for controlling HCV infection (7, 11, 21). The different clinical outcomes of HCV infection are associated with the relative strengths of antiviral cytotoxic T-lymphocyte responses (14, 17, 29, 39, 40). For patients with chronic hepatitis C, however, the frequency and magnitude of T-cell responses are dramatically lower than those for patients with self-limited infection (5, 34, 42). Correspondingly, the Th1-type cytokines are severely diminished in the periphery of patients with chronic HCV infection (20). This finding suggests that insufficient T-cell responses may be responsible for the establishment of persistent HCV infection. However, the mechanism of impaired T-cell function observed in patients with chronic HCV infection has yet to be defined. It is likely that a gene product(s) encoded by HCV directly affects T-cell functions crucial for limiting virus replication.
Various investigators previously demonstrated the immunomodulatory role of the HCV core in the inhibition of T-lymphocyte responsiveness (18, 19, 44, 45, 46). Furthermore, dendritic cell maturation and, correspondingly, their CD4+-T-cell-priming ability are impaired by the HCV core, leading to a defect in the induction of anti-HCV T cells (36, 37). Importantly, free core protein (non-virion associated) is secreted from infected cells and is detectable in the bloodstream of HCV-infected patients, possibly providing the virus with an indirect means of affecting host immunity (2, 25, 26, 43). This free core protein has been shown to interfere with both proliferation and effector activities of human T cells through its interaction with a complement receptor, gC1qR, in a mixed-lymphocyte reaction (18, 44, 45, 46). However, it is not clear whether the inhibition of T-cell function by the HCV core results directly from core interactions with T cells and/or indirectly from core-induced effects on antigen-presenting cells. Notably, treatment of T cells with C1q, a natural ligand for gC1qR, can inhibit their proliferative responses to mitogenic stimulation, suggesting that gC1qR may play a role in fine-tuning cellular immune responses by bridging innate and adaptive immunity (12).
C1q is part of the C1 complex, which is the first component in the classical pathway of complement activation, and thus plays a crucial role in the early defense against pathogens, including viruses and bacteria. The C1q receptor is a heterodimer consisting of a 33-kDa glycoprotein, gC1qR, and a 60-kDa calreticulin homologue, cC1qR. Although gC1qR lacks a transmembrane domain and is expressed mainly inside cells, it is also found on the surface of immune cells, such as macrophages and T cells, where it may be anchored through its association with β1-integrin (10, 12). In addition to the HCV core, gC1qR has been shown to bind a number of pathogen-derived proteins, including human immunodeficiency virus type 1 Rev (23), adenovirus core protein V (27), Epstein-Barr virus EBNA-1 (41), herpes simplex virus open reading frame protein P (4), Listeria monocytogenes internalin B (3), and Staphylococcus aureus protein A (31); these data suggest that many different pathogens exploit a similar strategy to subvert the host immune response. The underlying mechanism for the role of gC1qR in the inhibition of T-cell function and its possible role in HCV persistence have yet to be elucidated.
To address this issue, we examined the ability of the HCV core to bind gC1qR displayed on the T-cell surface as well as the functional impact of this core-gC1qR interaction on CD4+- and CD8+-T-cell subsets. Here, we show that the HCV core directly binds gC1qR on T lymphocytes, resulting in the inhibition of both their activation and their proliferation. gC1qR is expressed at higher levels on CD8+ T cells and, correspondingly, the core elicits more severe suppression on CD8+ T cells than on CD4+ T cells. In addition, the T-cell receptor (TCR)-mediated activation of Lck and ZAP-70 and the subsequent phosphorylation of Akt in both CD4+ and CD8+ subsets are impaired by the HCV core-gC1qR interaction on the surface of T cells. This study indicates that core-induced inhibition of T cells may accommodate the establishment of persistent infection, a finding which provides a target for potential HCV therapies.
MATERIALS AND METHODS
PBMC isolation and culture.
Human peripheral blood mononuclear cells (PBMC) were isolated from the peripheral blood of donors at Virginia Blood Services by Ficoll density centrifugation with Lympholyte-H (Cedarlane Labs, Hornby, Ontario, Canada). Purified cells were washed two times and cultured with RPMI 1640 (Life Technologies, Gaithersburg, Md.) containing 10% (vol/vol) fetal bovine serum (FBS) (Life Technologies), penicillin-streptomycin (100 μg/ml for each drug; Life Technologies), l-glutamine (2 mM), and 2-mercaptoethanol (5.5 × 10−5 M; Life Technologies) at 37°C with 5% CO2 in a humidified atmosphere.
T-cell proliferation analysis.
Subsets of CD4+ or CD8+ T cells were purified directly from whole blood by adding RosetteSep antibody cocktail before density gradient centrifugation in accordance with the manufacturer's instructions (StemCell Technologies Inc., Vancouver, British Columbia, Canada). To activate naive human T cells, anti-CD3/CD28 antibodies (1 μg/ml; BD Pharmingen, San Diego, Calif.) or concanavalin A (ConA) (2 μg/ml; Sigma, St. Louis, Mo.) was used. Recombinant core protein with six histidine residues fused at the N terminus (His6-core; amino acids 2 to 191) was expressed and purified under native conditions in our laboratory. An irrelevant control protein, His6-dihydrofolate reductase (DHFR), was prepared in the same manner.
For T-cell proliferation analysis, various concentrations of His6-core, His6-DHFR, or C1q (Advanced Research Technologies, San Diego, Calif.) were added to purified CD4+ or CD8+ T cells (2 × 105 cells/200 μl/well) activated as described above. The cultures were incubated in a U-bottom 96-well tissue culture plate (Corning Glass, Corning, N.Y.) for 5 days and pulsed with [3H]thymidine (specific activity, 6.7 Ci/mmol; Amersham Pharmacia Biotech Inc., Piscataway, N.J.) at 1 μCi/well for 18 h. The cells were harvested by using semiautomated cell harvester 96 (TOMTEC, Hamden, Conn.), and the amount of [3H]thymidine incorporated was measured by using a Wallac MicroBeta liquid scintillation counter (Trilux, Turku, Finland). To examine whether gC1qR mediates core-induced T-cell suppression, various concentrations of anti-gC1qR polyclonal antibody (PAb), rabbit control serum (1:10 and 1:100 [vol/vol] dilutions; QED Bioscience Inc., San Diego, Calif.), or soluble His6-gC1qR or His6-DHFR (2, 4, and 8 μg/ml; prepared in our laboratory) were coincubated with 1 μg of His6-core/ml, and T-cell proliferation was assessed as described above.
Flow cytometry.
To determine core binding, various concentrations (0.5, 1, 2, 4, and 8 μg/ml) of His6-core protein were incubated with 106 human PBMC at 37°C for 2 h. The cells were washed three times and resuspended with 1 μg of anti-HCV core monoclonal antibody (MAb) (ABR Inc., Golden, Colo.)/100 μl of fluorescence-activated cell sorting (FACS) medium (RPMI 1640 supplemented with 10% FBS and 0.1% NaN3) on ice for 1 h. The cells were washed three times and resuspended in 100 μl of FACS medium containing 1 μg of fluorescein isothiocyanate (FITC)-polyclonal anti-mouse immunoglobulin (BD Pharmingen) at 4°C for 1 h in the dark. The cells were washed three times, fixed in 0.1% paraformaldehyde in phosphate-buffered saline, and analyzed by flow cytometry (Becton Dickinson, San Jose, Calif.).
To determine gC1qR expression on lymphocytes, 106 PBMC were incubated with 1 μg of anti-gC1qR MAb (University of Virginia Hybridoma Center)/100 μl of FACS medium at 4°C for 1 h. The cells were washed three times and resuspended in 100 μl of FACS medium containing 1 μg of phycoerythrin (PE)-polyclonal anti-mouse immunoglobulin at 4°C for 1 h in the dark. The cells were analyzed by flow cytometry gated on the lymphocyte population. To determine gC1qR expression on CD4+- and CD8+-T-cell subsets, the cells were stained first with FITC-anti-CD4 or FITC-anti-CD8 conjugate (BD Pharmingen) and then with PE-gC1qR as described above.
To determine CD69 expression on activated T cells, 106 PBMC were stimulated with ConA and simultaneously treated with either His6-core or His6-DHFR at 1 μg/ml for various times. The cells were washed three times in FACS medium, spun at 200 × g for 5 min at 4°C, resuspended in 100 μl of FACS medium containing 1 μg of PE-anti-CD69 conjugate (BD Pharmingen), incubated at 4°C for 1 h in the dark, and subjected to FACS analysis. To determine CD69 expression on CD4+- and CD8+-T-cell subsets, the cells were stained first with FITC-anti-CD4 or FITC-anti-CD8 conjugate and then with PE-anti-CD69 conjugate as described above.
siRNA silencing of gC1qR expression.
To generate a double-stranded RNA template for gC1qR, a primer pair containing the T7 RNA polymerase promoter (underlined) was synthesized: sense, 5′-GCGTAATACGACTCACTATAGGGAGACGGCACCCCGGCTGTGCACC-3′; and antisense, 5′-GCGTAATACGACTCACTATAGGGAGACTACTGGCTCTTGACAAAAC-3′. Total RNA was isolated from 5 × 106 human PBMC with TRIzol reagent (Life Technologies) by following the manufacturer's protocol. A total of 1 μg of RNA was treated with DNase, and cDNA was synthesized by using a cDNA synthesis kit (PE Applied Biosystems, Foster City, Calif.). PCR was performed with a total reaction volume of 50 μl containing 10 μM deoxynucleoside triphosphates, 40 pM primers, and 2.5 U of AmpliTaq gold polymerase (PE Applied Biosystems). After an initial step at 95°C for 5 min, 45 cycles of amplification were conducted at 95°C for 40 s, 55°C for 30 s, and 72°C for 40 s, followed by a final step at 72°C for 10 min. After Geneclean (Qbiogene Inc., Carlsbad, Calif.) treatment of the gC1qR PCR product, double-stranded RNA was transcribed from the gC1qR DNA template, and small interfering RNA (siRNA) was generated by using recombinant DICER enzyme (Gene Therapy Systems Inc., San Diego, Calif.).
For T-cell transfection, 2 μg of siRNA in 50 μl of diluent was incubated with 10 μl of GeneSilencer reagent in 40 μl of diluent at room temperature for 30 min. The siRNA-GeneSilencer complex was gently added to 5 × 105 Molt-4 T cells in 500 μl of serum-free lymphocyte medium (Life Technologies). At 4 h after transfection, 500 μl of RPMI 1640 containing 20% FBS was added, and the cells were harvested at various times after transfection. Total RNA was extracted from transfected and untransfected cells, and reverse transcription (RT)-PCR was performed as described above but with a different gC1qR primer pair: sense, 5′-GGCTGCGGCTCGCTGCACACCGACGG-3′; and antisense, 5′-CTACTGGCTCTTGACAAAACTCTTGAG-3′. The specificity of the gC1qR-siRNA effect was verified by measuring the amount of β-actin cDNA in each sample. The levels of gC1qR expression and HCV core binding on the surface of Molt-4 T cells were detected by FACS analysis as described above.
Stable transfection of GPC-16 cells with Hu-gC1qR.
GPC-16 cells, a guinea pig cell line of epithelial origin (American Type Culture Collection, Manassas, Va.), were transfected with plasmid pCI:neo encoding human gC1qR (Hu-gC1qR) by using DMRIE-C (Life Technologies). Transfected cells were cultured with selection medium containing 400 μg of G418 (Life Technologies)/ml. The expression of gC1qR was verified by Western blot or FACS analysis with gC1qR PAbs (QED Bioscience) or MAbs (University of Virginia Hybridoma Center). Transfected and untransfected cells were also assessed for their ability to bind the HCV core as described above.
For sequence comparisons of gC1qR from guinea pigs and gC1qR from humans, RNA isolated from GPC-16 cells was treated with DNase, and RT-PCR was performed as described above. The PCR products so obtained were cloned into a TA cloning vector (Invitrogen, Carlsbad, Calif.). The DNA purified from positive clones was sequenced and analyzed by using DNA Strider 1.2.
GST pull-down analysis.
Glutathione S-transferase (GST) and GST-core (amino acids 1 to 124) fusion protein were expressed in Escherichia coli DH5α after transformation of E. coli with plasmid pGEX4T or pGEX:C1-124. GST and GST-core fusion protein were purified by using glutathione-agarose beads (Sigma) in accordance with the supplier's recommendations. To perform in vitro transcription-translation, Hu-gC1qR and guinea pig gC1qR were subcloned from the TA cloning vector into the pCI:neo vector by using restriction enzymes NheI and NotI. gC1qR labeled with [35S]methionine was generated by using a coupled transcription-translation system (TNT) from Promega Corp. with pCI:gC1qR as a template.
To perform GST pull-down analysis, GST (200 ng) or GST-core fusion protein coupled to 30 μl of glutathione-agarose beads (50% slurry) was incubated for 1 h at 4°C with [35S]methionine-labeled gC1qR in 500 μl of a buffer solution containing 40 mM HEPES-KOH (pH 7.5), 150 mM KCl, 0.5 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 0.1% Nonidet P-40. To minimize potential bead loss during subsequent washes, the buffer was mixed with glutathione-agarose beads adjusted to a total bead volume of 30 μl per reaction. After the incubation, the beads were washed five times with the same buffer. The bound proteins were eluted with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer and analyzed by SDS-PAGE and autoradiography.
BIAcore analysis.
gC1qR was covalently coupled to a BIAcore CM5 carboxymethylated dextran chip. Briefly, the chip surface was first activated with 10 mM sodium acetate (pH 4.0) at a flow rate of 10 μl/min for 10 min. gC1qR (50 μg/ml) was injected into the system at 10 μl/min for 10 min. The chip surface was deactivated for 7 min with ethanolamine-HCl (pH 8.5). To test the relative binding affinities of C1q, His6-core, and His6-DHFR, various concentrations of these proteins were injected at a flow rate of 50 μl/min for 1 min, after which they were dissociated for 2 min in a buffer containing 0.01 M HEPES (pH 7.4), 0.15 M NaCl, 3 mM EDTA, and 0.005% (vol/vol) Surfactant P20 (Biacore Inc., Piscataway, N.J.). The chip was regenerated in 10 mM glycine (pH 3.0) for 2.5 min. Kinetic parameters were defined by using the BIA analysis computer program.
Western blot analysis.
CD4+ and CD8+ T cells were purified from human PBMC by incubation with FITC-anti-CD4 and FITC-anti-CD8 conjugate, followed by positive isolation with anti-FITC magnetic beads (Miltenyi Biotec, Auburn, Calif.) in accordance with the manufacturer's instructions. A total of 2 × 106 purified CD4+ and CD8+ T cells were activated with anti-CD3/CD28 antibodies and treated with His6-core or His6-DHFR for 16 h. Cell lysates were prepared for 30 min at 4°C with a lysis buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium vanadate, 1 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride, and 5 μM okadaic acid. The cell lysates were sonicated three times for 1 min each time. Cellular debris was pelleted by centrifugation at 16,000 × g; supernatants were collected and frozen at −80°C.
A total of 80 μg of protein, as determined by bicinchoninic acid analysis (Pierce, Rockford, Ill.), was denatured with sample loading buffer at 100°C for 5 min and resolved by SDS-PAGE; these steps were followed by semidry transfer (Amersham Pharmacia Biotech) to a Hybond-P membrane (Amersham Biosciences, Arlington Heights, Ill.). After blocking in Blotto-Tween 20 (10 mM Tris, 0.9% NaCl, 0.1% Tween 20, 5% nonfat dry milk) at room temperature for 1 h, the membrane was probed with MAbs (Santa Cruz Biotechnology, Santa Cruz, Calif.) to p56 Lck (1:500), p59 Fyn (1:500), and ZAP-70 (1:1,000) or rabbit PAbs (Cell Signaling Technology, Beverly, Mass.) to phospho-Akt (1:1,000), Akt (1:1,000), and β-actin (1:500). After several 5-min washes with Tris-buffered saline-Tween and Tris-buffered saline, the membrane was incubated with horseradish peroxidase-conjugated donkey anti-mouse or anti-rabbit immunoglobulin G (IgG) secondary antibodies (1:5,000) and subsequently developed by enhanced chemiluminescence (ECL-plus; Amersham Biosciences) on X-OMAT-LS X-ray film (Kodak, Rochester, N.Y.).
Kinase assay.
A total of 3 × 106 purified CD4+ and CD8+ T cells were activated with anti-CD3/CD28 antibodies and treated with His6-core or His6-DHFR for 18 h. The cells were lysed in 500 μl of Brij 98 lysis buffer (1% Brij 98, 25 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1 mM Pefabloc, 5 mM iodoacetamide, 1 mM Na3VO4, 1 mM NaF). A total of 100 μg of total cell lysate was immunoprecipitated with 10 μl (2 μg) of anti-Lck antibody (Santa Cruz Biotechnology) and 50 μl of protein G/A-plus agarose (Santa Cruz Biotechnology) in 500 μl of phosphate-buffered saline at 4°C for 2 h with rotation. Lck precipitates were washed two times with Brij 98 lysis buffer and kinase assay buffer (0.1% Brij 98, 25 mM HEPES [pH 7.4]), respectively, before incubation at 30°C for 20 min with 30 μl of assay buffer, containing 10 mM MnCl2, 5 μCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham Biosciences), and acidified enolase (5 μg; Sigma-Aldrich). After the addition of 30 μl of 2× sample buffer and boiling at 100°C for 5 min, Lck kinase activity was detected by SDS-15% PAGE, followed by phosphorimager analysis (Molecular Dynamics).
RESULTS
HCV core directly binds to gC1qR on the surface of human T cells.
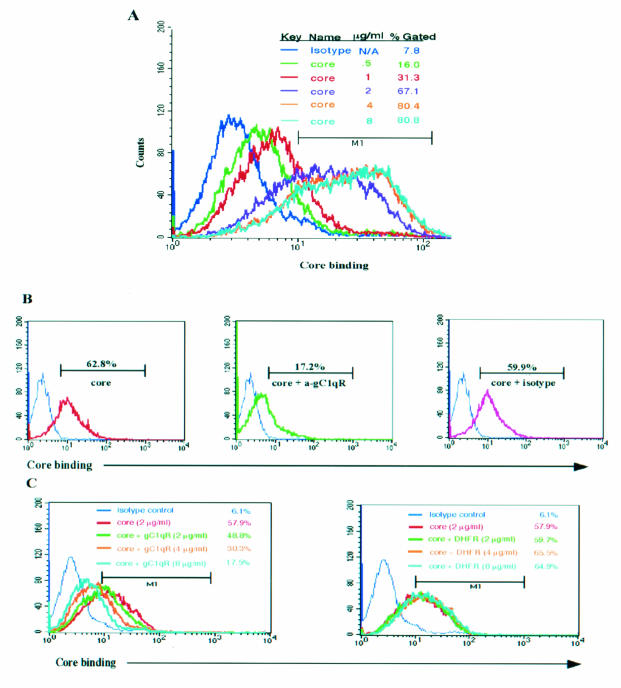
It was previously demonstrated that the interaction of the HCV core with gC1qR inhibits T-cell proliferation in a mixed-lymphocyte reaction (18, 44). To determine whether the HCV core-gC1qR interaction is responsible for the inhibition of T-cell function, we examined the requirement of gC1qR for core binding on the surface of T cells. We first analyzed core binding to lymphocytes incubated with recombinant His6-core or a control protein, His6-DHFR, by flow cytometry. As shown in Fig. 1A, the HCV core was found to bind to the surface of lymphocytes in a dose-dependent manner. Importantly, HCV core binding to the cell surface was saturated at core concentrations above 4 μg/ml. This result suggests that the binding sites for the core on these cells are fully occupied and that the binding of the core to the cell surface is mediated through a specific receptor. In the absence of the core protein or in the presence of His6-DHFR, the fluorescence profile was identical to that of the isotype control, suggesting that the anti-core MAb used in this assay is specific for the core (data not shown).
FIG. 1.
The HCV core specifically binds to gC1qR displayed on the cell surface of T lymphocytes. (A) Dose-dependent core binding on lymphocytes. A total of 106 PBMC were incubated with various concentrations (0.5, 1, 2, 4, and 8 μg/ml) of the HCV core at 37°C for 2 h. After washing of the residual core protein, core binding was examined by FACS analysis with an anti-core MAb followed by FITC-labeled goat anti-mouse IgG. N/A, not applicable. (B) Anti-gC1qR antibody (a-gC1qR) blocks core binding on lymphocytes. Cells were treated with 2 μg of HCV core/ml in the presence of either 1:10 (vol/vol) anti-gC1qR PAb or prebleed serum at 37°C for 2 h. Core binding was measured as described above. (C) Soluble gC1qR prevents core binding on lymphocytes. Cells were incubated at 37°C for 2 h with 2 μg of HCV core/ml in the presence of various concentrations (2, 4, and 8 μg/ml) of soluble gC1qR (or DHFR) equivalent to core-gC1qR molar ratios of 1:2, 1:4, and 1:8, respectively. Core binding was determined as described above. The results were reproducible in two independent experiments. M1, gated on lymphocyte populations based on isotype control.
To determine whether the binding of the HCV core to the surface of T cells is specifically mediated by its interaction with gC1qR, we first examined the ability of anti-gC1qR antibody to prevent the core from associating with these cells. To this end, T cells were incubated with the HCV core and either anti-gC1qR antibody or a control antibody, and core binding was assessed as described above. As shown in Fig. 1B, core binding to T lymphocytes was blocked by anti-gC1qR antibody, while binding was unaffected by control antibody. Additionally, increasing concentrations of soluble gC1qR, corresponding to core/gC1qR molar ratios of 1:2 to 1:8, were able to inhibit core binding, as measured by flow cytometry, suggesting that soluble gC1qR is able to compete for core binding with surface gC1qR (Fig. 1C). There were, however, no detectable changes in the ability of the core to bind to lymphocytes in the presence of DHFR. Correspondingly, coincubation of PBMC with the HCV core and various concentrations of either anti-gC1qR antibody or soluble gC1qR was able to dose dependently restore the core-induced suppression of T-cell proliferation (data not shown). Interestingly, a MAb which is directed against the N terminus of the HCV core and which recognizes an epitope distinct from that used for the above assays was able to block the binding of the core to lymphocytes (data not shown).
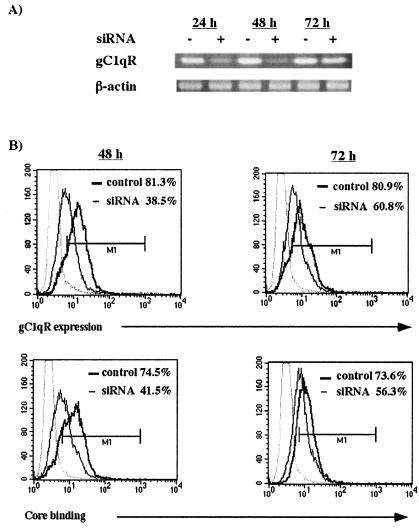
Silencing of gC1qR expression reduces HCV core binding on the surface of T cells.
RNA interference (RNAi) is a useful tool for the study of gene function because it allows for sequence-specific gene suppression in a variety of mammalian cell types. This mechanism of gene silencing involves targeted mRNA degradation, which results from the introduction of 21- to 25-nucleotide siRNAs. In order to confirm whether gC1qR is required for the core to bind to the surface of T cells, gC1qR-specific RNAi was used. Briefly, following transient transfection of Molt-4 T cells with gC1qR-specific interfering RNA, gC1qR expression and core binding ability were determined by RT-PCR and FACS analysis, respectively. As shown in Fig. 2A, gC1qR mRNA expression was markedly diminished as early as 24 h, with the strongest suppression being observed 48 h after transfection. In contrast, no effect was observed in untransfected cells or β-actin expression. Correspondingly, gC1qR expression on the surface of transfected T cells was also inhibited, consistent with the observed mRNA levels (Fig. 2B). In addition, the ability of the HCV core to associate with these cells was also decreased. Importantly, both gC1qR expression and core binding to the surface of these cells were most inhibited at 48 h and recovered by 72 h posttransfection, suggesting that the ability of the HCV core to bind to the cell surface strongly depends on gC1qR expression.
FIG. 2.
RNAi-mediated gC1qR silencing inhibits core binding on the T-cell surface. (A) Inhibition of gC1qR expression following siRNA transfection of T cells. Molt-4 T cells were transiently transfected with gC1qR-specific siRNAs. At 24, 48, and 72 h after transfection, gC1qR-specific mRNAs from siRNA-transfected (+) and untransfected (−) cells were analyzed by RT-PCR. β-Actin was used as a control. (B) Inhibition of gC1qR cell surface expression as well as core binding on siRNA-transfected T cells. (Upper panels) gC1qR cell surface expression in siRNA-transfected and untransfected Molt-4 T cells was determined at 48 and 72 h after transfection by FACS analysis. The percentages of gC1qR-positive cells in siRNA-transfected cells (thin line) and untransfected cells (thick line) are shown. (Lower panels) HCV core binding was determined as described above. The percentages of transfected cells (thin line) and untransfected cells (thick line) that were positive for HCV core binding at 48 and 72 h after transfection are shown. Gating of cells was based on an anti-mouse IgG isotype control (light gray line). The results were reproducible in two independent experiments. M1, gated on Molt-4 T cells based on isotype control.
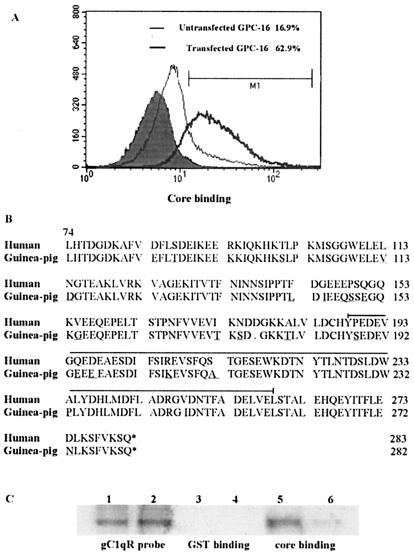
Transfection of GPC-16 cells with Hu-gC1qR enhances core binding.
To further characterize the specificity of the interaction between the HCV core and gC1qR, we compared the relative abilities of the HCV core to bind to Hu-gC1qR-transfected GPC-16 cells and untransfected cells. GPC-16 cells are a guinea pig cell line to which the HCV core binds less efficiently than to human cells, such as Molt-4 cells and PBMC (data not shown). To this end, GPC-16 cells were stably transfected with a plasmid encoding Hu-gC1qR, and the increased expression of gC1qR in this transfected cell line was confirmed by FACS and Western blot analyses (data not shown). The cells then were incubated with the HCV core and assessed for core binding as described above. As shown in Fig. 3A, core binding was significantly higher on Hu-gC1qR-transfected GPC-16 cells than on untransfected cells.
FIG. 3.
HCV core binding on GPC-16 cells is increased upon transfection with Hu-gC1qR. (A) HCV core binding on GPC-16 cells. GPC-16 cells, both untransfected (thin line) and stably transfected with Hu-gC1qR (thick line), were incubated with 2 μg of HCV core/ml at 37°C for 2 h. Core binding was determined by FACS analysis as described above. The percentages of cells that were positive for HCV core binding, relative to the isotype control (filled area), are shown. (B) Alignment of the cDNA-derived gC1qR sequences from humans and guinea pigs. Amino acid residues that diverge in humans and guinea pigs are underlined. A deleted amino acid at position 177 (D) is notated with a dot. The HCV core binding region of gC1qR is bracketed. This alignment was constructed by using DNA Strider. (C) Comparison of the relative HCV core binding abilities of Hu-gC1qR and guinea pig gC1qR by GST pull-down analysis. GST alone (lanes 3 and 4) or GST-core (lanes 5 and 6) was incubated with in vitro-translated Hu-gC1qR (lanes 1, 3, and 5) or guinea pig gC1qR(lanes 2, 4, and 6). After a pull-down reaction with glutathione-agarose beads, samples were separated by SDS-PAGE, and [35S]Met-labeled gC1qR was detected by autoradiography.
We next sought to explore whether the differences in core binding observed above might result from sequence variations between Hu-gC1qR and guinea pig gC1qR. Because the mature form of Hu-gC1qR is comprised of residues 74 to 283, DNA sequence comparisons of gC1qR from transfected and untransfected cells were performed within this region. As shown in Fig. 3B, the cDNA-derived amino acid sequences demonstrated 90% similarity between Hu-gC1qR and guinea pig gC1qR. This finding supports a previously reported significant degree of cross-species conservation for this molecule (24). While differences between the two sequences are observed throughout, most occur in the N-terminal region. In addition to 23 amino acid substitutions, the guinea pig sequence contains a deletion at position 177. Interestingly, 12 of these substitutions are conserved in both the mouse and the rat, and 10 of these 12 are identical to those observed in the guinea pig. Importantly, within the core binding region of gC1qR (amino acids 188 to 259, as denoted in Fig. 3B), only 7 amino acids diverge in guinea pigs and humans.
To determine whether these divergent amino acids are critical for the differences in the core binding abilities of Hu-gC1qR and guinea pig gC1qR, we used [35S]methonine-labeled gC1qR probes in a GST pull-down analysis. Guinea pig gC1qR bound GST-core much less efficiently (Fig. 3C, lane 6) than did Hu-gC1qR (lane 5). Both Hu-gC1qR and guinea pig gC1qR failed to bind GST alone (Fig. 3C, lanes 3 and 4, respectively). Based on these data, it is likely that the 7 residues which are found to diverge in guinea pigs and humans and which fall within the core binding domain of gC1qR play a critical role in the HCV core-gC1qR interaction.
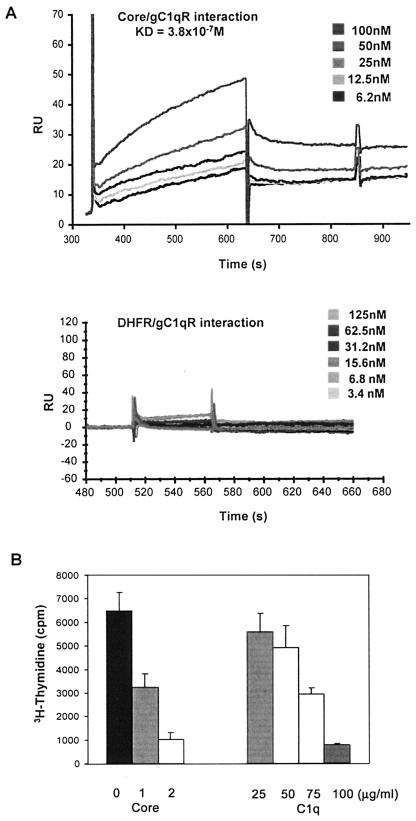
HCV core binds gC1qR with an affinity similar to that of C1q but delivers a stronger inhibitory signal to T cells.
To examine the affinity of binding of the HCV core to gC1qR, we used the BIAcore system, which allows real-time measurement of protein-protein interactions under physiological salt and pH conditions. We covalently coupled gC1qR (50 μg/ml) to a BIAcore dextran chip. Buffer containing various concentrations of His6-core or His6-DHFR was passed over the immobilized gC1qR at a flow rate of 50 μl/min. His6-core (Fig. 4A, upper panel) bound gC1qR in a dose-dependent manner, with an apparent Kd of 3.8 × 10−7 M, a value which falls within the normal range for protein-protein interactions. Importantly, the Kd for the C1q-gC1qR interaction has been reported to be 2.4 × 10−7 M (15), suggesting that the HCV core and C1q bind gC1qR with similar affinities. In contrast, the control protein, His6-DHFR (Fig. 4A, lower panel), failed to demonstrate gC1qR binding.
FIG. 4.
The HCV core, which binds gC1qR with an affinity similar to that of C1q, elicits a stronger inhibitory signal on T cells. (A) Kinetics of the HCV core-gC1qR interaction and its dissociation constant (Kd) determined by BIAcore analysis. Various concentrations of the HCV core or DHFR were passed over gC1qR (50 μg/ml) covalently coupled to a dextran chip at a flow rate of 50 μl/min. The Kd for the HCV core-gC1qR interaction, as determined by using the BIA analysis computer program, is shown. Results are presented as RU/s, where 1 RU = 1 pg/mm2. (B) Dose-dependent inhibition of T-cell proliferation by the HCV core or C1q. A total of 2 × 105 ConA-stimulated PBMC were incubated with various concentrations of either the HCV core or C1q for 5 days at 37°C. [3H]thymidine was added to the cultures 18 h prior to harvesting, and incorporation was determined by using a MicroBeta liquid scintillation counter. Error bars indicate standard deviations. The results were reproducible in three independent experiments.
Since both C1q (6, 13) and the HCV core (18, 44) have been shown to inhibit T-cell proliferation, we next sought to compare their relative antiproliferative abilities. To this end, we treated ConA-stimulated PBMC with either the HCV core or C1q at various concentrations for 5 days and examined T-cell proliferation by measuring [3H]thymidine incorporation. As shown in Fig. 4B, both the HCV core and C1q induced a dose-dependent inhibition of ConA-stimulated T-cell proliferation. The HCV core, however, had a stronger inhibitory effect on T-cell proliferation than did C1q. Specifically, 1 μg of core protein/ml inhibited ∼50% of T-cell proliferation, while the same molar concentration of C1q (25 μg/ml) failed to induce an inhibitory effect. In fact, subphysiological concentrations (i.e., below 50 μg/ml) of C1q also failed to elicit significant inhibition. T-cell proliferation was inhibited by 50% in the presence of C1q at a concentration of 75 μg/ml, which is almost three times the molar concentration of the core protein required to elicit the same inhibitory effect.
CD8+ T cells exhibit higher levels of gC1qR and are more susceptible to HCV core-mediated inhibition than are CD4+ T cells.
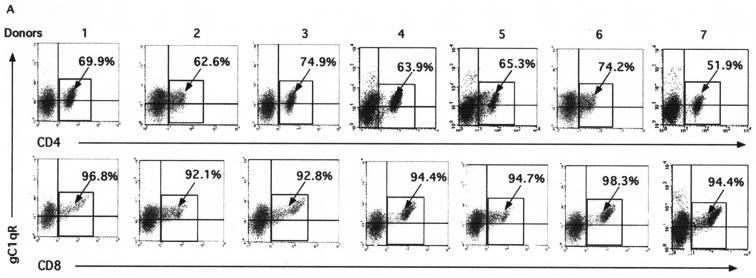
gC1qR is expressed on most cell types, with the exception of erythrocytes (12), and it is well established that PBMC from various donors may display different receptor expression patterns. Based on this information, we examined PBMC from seven blood donors to determine whether the gC1qR expression patterns on human lymphocytes are consistent from donor to donor. The cell surface expression of gC1qR on these lymphocytes was comparable in the seven individuals examined, ranging from 59 to 68% (data not shown). We next determined whether gC1qR is expressed at similar levels on specific CD4+- and CD8+-T-cell subsets by flow cytometry. As shown in Fig. 5A, in all seven donors examined, the level of expression of gC1qR was higher on the CD8+-T-cell population (i.e., 94.8% ± 2.4% [mean and standard deviation] of CD8+ cells expressed gC1qR) than on the CD4+-T-cell population (i.e., 66.1% ± 4.6% of CD4+ cells expressed gC1qR).
FIG. 5.
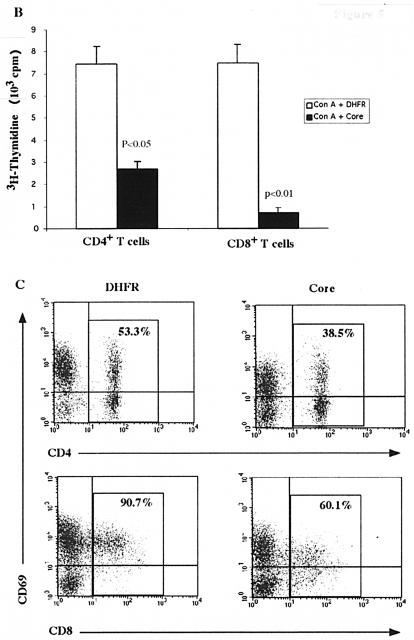
gC1qR expression and core-induced inhibitory effects are more profound on CD8+ than on CD4+ T cells. (A) gC1qR expression on CD4+ and CD8+ T cells. A total of 106 human PBMC were doubly stained with either FITC-conjugated anti-CD8 or anti-CD4 and anti-gC1qR antibody, followed by PE-conjugated anti-mouse IgG antibody. Cells subjected to FACS analysis were gated on the lymphocyte population. The percentages of CD4+ cells (upper panel) and CD8+ cells (lower panel) that were positive for gC1qR are shown. (B) Proliferation of CD4+ and CD8+ T cells in the presence of the core. Purified CD4+ and CD8+ T cells were activated by ConA in the presence of the HCV core (2 μg/ml) or DHFR (2 μg/ml) for 5 days at 37°C in a 5% CO2 atmosphere. [3H]thymidine was added to the cultures 18 h prior to harvesting. The uptake of [3H]thymidine was measured by using a liquid scintillation counter. P values determined by the Student t test are shown. Error bars indicate standard deviations. The results were reproducible in three independent experiments. (C) CD69 expression on activated CD4+ and CD8+ T cells in the presence of the core. PBMC were activated with ConA in the presence of either the HCV core (right panels) or DHFR (left panels) at 37°C for 24 h in a 5% CO2 atmosphere. CD69 expression on CD4+ T cells (upper panels) and CD8+ T cells (lower panels) was determined by FACS analysis with PE-conjugated anti-CD69 antibody and either FITC-conjugated anti-CD4 or FITC-conjugated anti-CD8 MAb. The percentages of CD4+ and CD8+ T lymphocytes that were positive for CD69 are shown. The results were reproducible in three independent experiments.
Because of the higher level of gC1qR expression on CD8+ T cells than on CD4+ T cells, we next sought to determine whether this disparity influences the HCV core-induced antiproliferative effect. To this end, CD4+ and CD8+ T cells were purified from PBMC by negative selection and subjected to proliferation analysis as described above. Figure 5B shows that the HCV core inhibited ConA-stimulated CD4+- and CD8+-T-cell proliferation, while DHFR failed to elicit such an effect. Interestingly, core-treated CD8+ T cells exhibited a more significant reduction in [3H]thymidine uptake than did the CD4+ subset, a result which correlates with the higher levels of gC1qR detected on CD8+ T cells. These results suggest a dependence of HCV core-induced inhibition of T-cell proliferation on gC1qR expression.
To further examine the role of the HCV core-gC1qR interaction in the inhibition of T-cell function, we next analyzed the surface expression of CD69, an early T-cell activation marker. We found diminished CD69 expression in core-treated cells as early as 6 h and sustained for 5 days after treatment (data not shown). To determine the levels of CD69 expression on CD4+ and CD8+ subpopulations, ConA-stimulated PBMC were treated with the core or DHFR for 24 h, stained for both CD69 and either CD4 or CD8, and analyzed by flow cytometry. As shown in Fig. 5C, CD69 down-regulation was observed for both CD4+ and CD8+ T cells in the presence of the HCV core, with more significant inhibition in the CD8+ subpopulation. CD69 expression on CD4+ and CD8+ T cells changed from 53.3 and 90.7% in the absence of the core to 38.5 and 60.1% in the presence of the core (i.e., mean decreases of 14.8 and 30.6%), respectively (Fig. 5C). These results are in concordance with the above data indicating that the HCV core inhibits the proliferation of the CD8+ subset more significantly than it does that of the CD4+ subset.
The HCV core-gC1qR interaction impairs the activation of Lck, ZAP-70, and Akt.
It was previously demonstrated that the TCR-dependent activation of ERK/MEK mitogen-activated protein kinase and the subsequent induction of interleukin 2 and interleukin 2 receptor α-chain gene expression were impaired in core-treated T cells (44). These findings suggested that the HCV core may inhibit an early event(s) in T-cell signaling. The earliest signaling events coupled to TCR ligation are the expression and activation of several protein tyrosine kinases (PTK), including Lck and Fyn, two members of the Src family expressed in T cells. Either one or both of these enzymes phosphorylate multiple immunoreceptor tyrosine-based activation motifs located in cytosolic domains of TCR ζ chains. Each immunoreceptor tyrosine-based activation motif contains two tyrosine residues which, upon phosphorylation, serve as binding sites for the tandem SH2 domains of ZAP-70, a Syk family PTK. Lck or Fyn then phosphorylates bound ZAP-70, which in turn activates downstream events in T-cell signaling cascades.
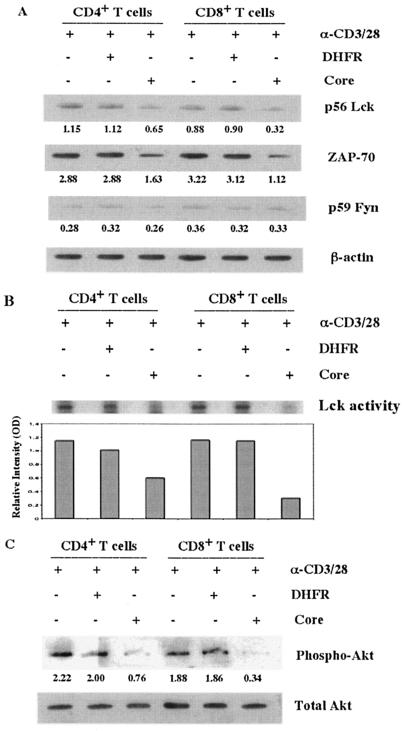
To test the possibility that the HCV core inhibits T-cell signaling at this early stage, we examined its effect on the expression of Lck, Fyn, and ZAP-70 after TCR stimulation. To this end, purified CD4+ and CD8+ T cells were activated by anti-CD3/CD28 antibodies in the presence of the HCV core or DHFR, and cell lysates were subjected to Western blot analysis. As shown in Fig. 6A, the expression of Lck (CD4+, 56%, and CD8+, 36%, relative to anti-CD3/CD28) and ZAP-70 (CD4+, 56%; CD8+, 35%) but not of Fyn (CD4+, 93%; CD8+, 92%) was inhibited in HCV core-treated T cells compared to those in the presence of DHFR (CD4+, 97%; CD8+, 100%) or anti-CD3/CD28 antibodies alone (CD4+ and CD8+, 100%). Interestingly, the HCV core suppressed the expression of Lck and ZAP-70 more profoundly in CD8+ than in CD4+ T cells, consistent with our earlier results showing enhanced gC1qR expression and core-induced suppressive effects in this population. For analysis of Lck activation, we immunoprecipitated Lck from TCR-activated CD4+ and CD8+ T cells in the presence of the HCV core or DHFR, and its PTK activity was detected by a kinase assay with enolase as a substrate. As shown in Fig. 6B, the relative intensity of the phosphorylated enolase-specific band, examined by [γ-32P]ATP labeling and phosphorimager quantification, was decreased by treatment with the HCV core (CD4+, 51%, and CD8+, 21%, relative to anti-CD3/CD28), most significantly in CD8+ T cells compared to those treated with DHFR or anti-CD3/CD28 antibodies alone.
FIG. 6.
The HCV core inhibits the activation of Src family protein kinases and Akt in T cells. (A) Src family kinase expression in CD4+ and CD8+ T cells in the presence of the HCV core. Totals of 2 × 106 purified CD4+ and CD8+ T cells were activated with anti-CD3/CD28 antibodies (α-CD3/28) (1 μg/ml) in the presence of the HCV core (2 μg/ml) or DHFR (2 μg/ml) for 16 h. Cell lysates were subjected to Western blotting for the detection of p56 Lck, p59 Fyn, and ZAP-70. β-Actin was used as a loading control. The optical density, relative to β-actin, is shown below each band. The data were reproducible in four independent experiments. (B) Lck kinase activity in CD4+ and CD8+ T cells in the presence of the HCV core. Totals of 3 × 106 purified CD4+ and CD8+ T cells were activated with anti-CD3/CD28 antibodies (1 μg/ml) in the presence of the HCV core (2 μg/ml) or DHFR (2 μg/ml) for 18 h. Lck tyrosine kinase activity was detected by a kinase immunocomplex assay. The relative intensities of phosphorylated enolase bands determined by phosphorimager analysis are shown. OD, optical density. (C) Phosphorylation of Akt in CD4+ and CD8+ T cells in the presence of the HCV core. Purified CD4+ and CD8+ T cells were activated and treated as described above. Phosphorylated Akt was determined by Western blot analysis; the total Akt protein level was used as a loading control. These results were reproducible in three independent experiments.
TCR-mediated activation of Lck has been linked to the induction of phosphatidylinositol 3′-kinase and Akt activities, which are associated with cell cycle progression and the propagation of an antiapoptotic signal (8). In addition, it was previously shown that the HCV core suppresses cell cycle progression from G1 to S phase through the stabilization of p27kip1 (45). It is possible that the HCV core inhibits Lck-mediated Akt phosphorylation, resulting in cell cycle arrest. To test this possibility, we treated anti-CD3/CD28 antibody-activated CD4+ and CD8+ T cells as described above and detected the phosphorylation of Akt by Western blot analysis. As shown in Fig. 6C, the phosphorylation of Akt was down-regulated in T cells treated with the HCV core (CD4+, 34%, and CD8+, 18%, relative to anti-CD3/CD28), most significantly in the CD8+ population compared to that treated with DHFR or anti-CD3/CD28 antibodies alone.
DISCUSSION
In this study, we demonstrated the specific binding of the HCV core to gC1qR on T cells and explored the pivotal role of this interaction in the core-induced inhibition of CD4+- and CD8+-T-cell functions, respectively. We found that HCV core binding to and inhibition of T cells depended solely on gC1qR expression, in that increasing concentrations of soluble gC1qR or anti-gC1qR antibody significantly diminished both. Importantly, silencing of gC1qR expression by RNAi decreased core binding on the surface of T cells. In addition, the inefficient binding of the core to guinea pig GPC-16 cells was greatly enhanced upon stable transfection of these cells with Hu-gC1qR. gC1qR was expressed at higher levels on CD8+ T cells and, correspondingly, the core elicited a more profound inhibitory effect on these cells than on CD4+ T cells. The HCV core was also found to suppress the activation of the Src family kinases Lck and ZAP-70 and of Akt, all of which are important during the early events of T-cell activation. These results suggest that the core-gC1qR interaction may play a crucial role in the impaired T-cell responses observed during HCV infection.
Despite low viral titers throughout the course of HCV infection, chronic HCV is associated with impaired T-cell function, as manifested by increased susceptibility to secondary microbial infections (1, 16, 22, 30, 32, 38). These data indicate that the virus may produce one or more factors that counteract or modulate host immune response. Studies on T-cell activity during the acute phase of HCV infection have suggested that the resolution of viral infection is associated with strong and multispecific T-cell responses (7, 11, 14, 17, 21, 29, 39, 40). However, the magnitude of T-lymphocyte responses is dramatically lower in patients with chronic HCV infection than in patients with acute HCV infection (5, 20, 34, 42). The immunomodulatory function of the HCV core protein, as described in this report, may contribute to the inactivation of HCV-specific T cells by interfering with proliferative and cytolytic activities through its interaction with gC1qR. Thus, the HCV core-induced inhibition of T-cell function may be responsible for dampening adaptive immunity during the early phase of viral infection, thus facilitating the establishment of persistent infection.
Intriguingly, the HCV core protein is capable of being secreted from infected cells and is therefore detectable in the bloodstream of HCV-infected patients (25, 26). Free core protein, which is not associated with immune complexes or with virus particles, is especially detectable in the serologically negative window phase; the detection of total core protein (i.e., core-immune complex) in serum or plasma serves as an indirect marker for HCV replication and viremia (2, 43). The amount of free core protein does not seem to be large in chronically infected patients and may not be sufficient for eliciting the immunosuppressive effect. However, because of the limited availability of acute-phase patient samples, the actual amount of free core protein during the early phase of viral infection has not yet been determined. Given vigorous HCV replication in the early phase of infection, it is likely that a large quantity of core protein is released from infected cells and circulates in the bloodstream of infected patients during the acute phase. In addition, it is likely that the amount of free core protein is larger in the local liver environment than in the bloodstream, as HCV replication occurs in hepatocytes. We are currently in the process of quantifying free core protein present during the acute phase of infection. Nonetheless, the presence of the circulating core in patient serum during the early phase of infection may contribute to its multiple immunopathological effects and the anergic state of the host immune response (33). Interestingly, these effects are conserved among other hepatotropic viruses, such as hepatitis B virus (HBV). In HBV infection, soluble HBV core antigen is secreted into the blood and has been implicated in mediating neonatal T-cell tolerance (28) and altering the reactivity of HBV-specific CD8+ T cells (35).
gC1qR was initially identified by its ability to bind the globular “heads” of C1q. The binding of C1q to gC1qR is both specific and saturable, with an approximate Kd of 2.4 × 10−7 M (15). The interaction of C1q with gC1qR induces a wide range of biological responses, including the inhibition of T-cell proliferation (6, 13). Here, we found that the HCV core binds to gC1qR with an affinity similar to that of C1q, a finding which is fundamentally important. However, why the HCV core elicits a stronger inhibitory signal for T cells than does C1q remains unknown. While the BIAcore system allows for real-time measurements of protein-protein interactions in vitro, gC1qR covalently coupled to a chip likely is present in a conformation different from that associated with the surface of cells in vivo, a scenario which might artificially skew the binding ability of the HCV core. Additionally, it is possible that while both the HCV core and C1q cause diminished T-cell proliferation, this effect may occur through different signaling pathways, accounting for variances in potency.
gC1qR is ubiquitously expressed in most cell types, both intracellularly and on the plasma membrane, although how gC1qR is anchored to the surface of cells remains unclear. Indeed, gC1qR is devoid of a typical hydrophobic transmembrane-spanning region or a consensus site for glycosylphophatidylinositol anchoring. It has been reported that gC1qR is anchored to the cell surface through its association with β1-integrin (10). The investigation of another possible docking protein(s) complexed with gC1qR as well as the signaling pathway for the gC1qR-mediated inhibition of T-cell function is currently under way. Ligation of gC1qR may provide a signal to turn off or raise the threshold for T-cell activation. Thus, complement activation can regulate T-cell responses, providing a bridge between innate immunity and adaptive immunity. Importantly, the binding of the HCV core to gC1qR displayed on the surface of T cells leads to the impaired activation of Lck and ZAP-70, which are critical during the early stages of T-cell activation. However, the mechanism of this core-gC1qR-induced suppression of these Src kinases remains unknown. It is likely that negative regulatory receptors, such as PD-1, CTLA-4, and Fas, cytoplasmic inhibitory proteins, such as Csk, SHP-1, and SHIP-2, and suppressors of cytokine signaling (SOCS) play roles in inhibiting the early events of T-cell activation. Importantly, SOCS family members interact with proteins critical for T-cell activation (e.g., ZAP-70, Gfi-1, and calcineurin). We are currently investigating the possibility that these negative regulators are induced by core-gC1qR engagement.
It is important to point out that both CD4+ and CD8+ T-cell responses are impaired in patients with chronic HCV infection. Although we cannot rule out the possibility that impaired T-cell function also arises from core-induced effects on antigen-presenting cells, such as dendritic cells, we demonstrated here that the suppression of mitogen-stimulated T-cell activation and proliferation by the HCV core is directly dependent on the surface expression of gC1qR. Interestingly, CD8+ T cells were found to express gC1qR at higher levels and to exhibit a higher level of inhibition in the presence of HCV core than CD4+ T cells. While these findings may explain why we observed a stronger inhibitory effect of the core on the CD8+-T-cell population, it does not exclude other potential mechanisms; i.e., different cell subsets may possess distinct molecular characteristics that can alter a subsequent intracellular signaling pathway(s).
Here, we demonstrated that the core-gC1qR interaction is required for inhibiting T-cell function. The interaction between the HCV core and gC1qR may provide the virus with a means of host immune evasion, which would contribute to the development of persistent infection. Thus, an factor that can intervene in the HCV core-gC1qR interaction may offer a potential rationale for designing therapeutic agents to prevent persistent HCV infection.
Acknowledgments
We thank our colleagues for constructive criticism and comments. We also greatly appreciate the outstanding technical support of Travis Lillard and Susan Landes.
This work was supported by an American Association for the Study of Liver Diseases/Schering Advanced Hepatology fellowship (to Z.Q.Y.) and by Public Health Service grant DK066754 (to Y.S.H.).
REFERENCES
- 1.Blanton, R. E., E. A. Salam, H. C. Kariuki, P. Magak, L. K. Silva, E. M. Muchiri, F. Thiongo, I. E. Abdel-Meghid, A. E. Butterworth, M. G. Reis, and J. H. Ouma. 2002. Population-based differences in Schistosoma mansoni- and hepatitis C-induced disease. J. Infect. Dis. 185:1644-1649. [DOI] [PubMed] [Google Scholar]
- 2.Bouvier-Alias, M., K. Patel, H. Dahari, S. Beaucourt, P. Larderie, L. Blatt, C. Hezode, G. Picchio, D. Dhumeaux, A. U. Neumann, J. G. McHutchison, and J. M. Pawlotsky. 2002. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology 36:211-218. [DOI] [PubMed] [Google Scholar]
- 3.Braun, L., B. Ghebrehiwet, and P. Cossart. 2000. gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J. 19:1458-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruni, R., and B. Roizman. 1996. Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc. Natl. Acad. Sci. USA 93:10423-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, K. M., B. Rehermann, J. G. McHutchison, C. Pasquinelli, S. Southwood, A. Sette, and F. V. Chisari. 1997. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J. Clin. Investig. 100:2376-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, A., S. Gaddipati, Y. Hong, D. J. Volkman, E. I. B. Peerschke, and B. Ghebrehiwet. 1994. Human T cells express specific binding sites for C1q. Role in T cell activation and proliferation. J. Immunol. 153:1430-1440. [PubMed] [Google Scholar]
- 7.Cooper, S., A. L. Erickson, E. J. Adams, J. Kansopon, A. J. Weiner, D. Y. Chien, M. Houghton, P. Parham, and C. M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity 10:439-449. [DOI] [PubMed] [Google Scholar]
- 8.Cuevas, B., Y. Lu, S. Watt, R. Kumar, J. Zhang, K. A, Siminovitch, and G. B. Mills. 1999. SHP-1 regulates Lck-induced phosphatidylinositol 3-kinase phosphorylation and activity. J. Biol. Chem. 274:27583-27589. [DOI] [PubMed] [Google Scholar]
- 9.Di Bisceglie, A. M. 1998. Hepatitis C. Lancet 351:351-355. [DOI] [PubMed] [Google Scholar]
- 10.Feng, X., M. G. Tonnesen, E. I. Peerschke, and B. Ghebrehiwet. 2002. Cooperation of C1q receptors and integrins in C1q-mediated endothelial cell adhesion and spreading. J. Immunol. 168:2441-2448. [DOI] [PubMed] [Google Scholar]
- 11.Gerlach, J. T., H. M. Diepolder, M. C. Jung, N. H. Gruener, W. W. Schraut, R. Zachoval, R. Hoffmann, C. A. Schirren, T. Santantonio, and G. R. Pape. 1999. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology 117:933-941. [DOI] [PubMed] [Google Scholar]
- 12.Ghebrehiwet, B., B. L. Lim, R. Kumar, X. Feng, and E. I. Peerschke. 2001. gC1q-R/p33, a member of a new class of multifunctional and multicompartmental cellular proteins, is involved in inflammation and infection. Immunol. Rev. 180:65-77. [DOI] [PubMed] [Google Scholar]
- 13.Ghebrehiwet, B., G. S. Habicht, and G. Beck. 1990. Interaction of C1q with its receptor on cultured cell lines induces an anti-proliferative response. Clin. Immunol. Immunopathol. 54:148-160. [DOI] [PubMed] [Google Scholar]
- 14.Gruner, N. H., T. J. Gerlach, M. C. Jung, H. M. Diepolder, C. A. Schirren, W. W. Schraut, R. Hoffmann, R. Zachoval, T. Santantonio, M. Cucchiarini, A. Cerny, and G. R. Pape. 2000. Association of hepatitis C virus-specific CD8+ T cells with viral clearance in acute hepatitis C. J. Infect. Dis. 181:1528-1536. [DOI] [PubMed] [Google Scholar]
- 15.Herwald, H., J. Dedio, R. Kellner, M. Loos, and W. Muller-Esterl. 1996. Isolation and characterization of the kininogen-binding protein p33 from endothelial cells. Identity with the gC1q receptor. J. Biol. Chem. 271:13040-13047. [DOI] [PubMed] [Google Scholar]
- 16.Jardi, R., F. Rodriguez, M. Buti, X. Costa, M. Cotrina, R. Galimany, R. Esteban, and J. Guardia. 2001. Role of hepatitis B, C, and D viruses in dual and triple infection: influence of viral genotypes and hepatitis B precore and basal core promoter mutations on viral replicative interference. Hepatology 34:404-410. [DOI] [PubMed] [Google Scholar]
- 17.Kamal, S. M., J. W. Rasenack, L. Bianchi, A. Al Tawil, K. El Sayed Khalifa, T. Peter, H. Mansour, W. Ezzat, and M. Koziel. 2001. Acute hepatitis C without and with schistosomiasis: correlation with hepatitis C-specific CD4(+) T-cell and cytokine response. Gastroenterology 121:646-656. [DOI] [PubMed] [Google Scholar]
- 18.Kittlesen, D. J., K. A. Chianese-Bullick, Z. Q. Yao, T. J. Braciale, and Y. S. Hahn. 2000. Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J. Clin. Investig. 106:1239-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Large, M. K., D. J. Kittlesen, and Y. S. Hahn. 1999. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistence. J. Immunol. 162:931-938. [PubMed] [Google Scholar]
- 20.Lechmann, M., R. P. Woitas, B. Langhans, R. Kaiser, H. G. Ihlenfeldt, G. Jung, T. Sauerbruch, and U. Spengler. 1999. Decreased frequency of HCV core-specific peripheral blood mononuclear cells with type 1 cytokine secretion in chronic hepatitis C. J. Hepatol. 31:971-978. [DOI] [PubMed] [Google Scholar]
- 21.Lechner, F., D. K. H. Wong, P. R. Dunbar, R. Chapman, R. T. Chung, P. Dohenwend, G. Robbins, R. Phillips, P. Klenerman, and B. D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 191:1499-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liaw, Y. F. 1995. Role of hepatitis C virus in dual and triple hepatitis virus infection. Hepatology 22:1101-1108. [DOI] [PubMed] [Google Scholar]
- 23.Luo, Y., H. Yu, and B. M. Peterlin. 1994. Cellular protein modulates effects of human immunodeficiency virus type 1 Rev. J. Virol. 68:3850-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch, N. J., K. B. M. Reid, R. H. van den Berg, M. R. Daha, L. A. E. Leigh, B. Ghebrehiwet, W. B. L. Lim, and W. J. Schwaeble. 1997. Characterisation of the rat and mouse homologues of gC1qBP, a 33 kDa glycoprotein that binds to the globular ′heads' of C1q. FEBS Lett. 418:111-114. [DOI] [PubMed] [Google Scholar]
- 25.Maillard, P., J. Nitkiewicz, K. Krawczynski, C. Bronnert, M. Sidorkiewicz, P. Gounon, J. Dubuisson, G. Faure, R. Crainic, and A. Budkowska. 2001. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J. Virol. 75:8240-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masalova, O. V., S. N. Atanadze, E. I. Samokhvalov, N. V. Petrakova, T. I. Kalinia, V. D. Smirnov, Y. E. Khudyakov, H. A. Fields, and A. A. Kushch. 1998. Detection of hepatitis C virus core protein circulating within different virus particle populations. J. Med. Virol. 55:1-6. [DOI] [PubMed] [Google Scholar]
- 27.Matthews, D. A., and W. C. Russell. 1998. Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and the nucleus. J. Gen. Virol. 79:1677-1685. [DOI] [PubMed] [Google Scholar]
- 28.Milich, D. R., J. E. Jones, J. L. Hughes, J. Price, A. K. Raney, and A. McLachlan. 1990. Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc. Natl. Acad. Sci. USA 87:6599-6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Missale, G., R. Bertoni, V. Lamonaca, A. valli, M. Massari, C. Mori, M. G. Rumi, M. Houghton, F. Fiaccadori, and C. Ferrari. 1996. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J. Clin. Investig. 98:706-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohamed, A., A. Elsheikh, Z. Ghandour, and M. Al Karawi. 1998. Impact of hepatitis C virus infection on schistosomal liver disease. Hepatogastroenterology 45:1492-1496. [PubMed] [Google Scholar]
- 31.Nguyen, T., B. Ghebrehiwet, and E. I. Peerschke. 2000. Staphylococcus aureus protein A recognizes platelet gC1qR/p33: a novel mechanism for staphylococcal interactions with platelets. Infect. Immun. 68:2061-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellicano, R., N. Leone, M. Berrutti, M. A. Cutufia, M. Fiorentino, M. Rizzetto, and A. Ponzetto. 2000. Helicobacter pylori seroprevalence in hepatitis C virus positive patients with cirrhosis. J. Hepatol. 33:648-650. [DOI] [PubMed] [Google Scholar]
- 33.Ray, R. B., and R. Ray. 2001. Hepatitis C virus core protein: intriguing properties and functional relevance. FEMS Microbiol. Lett. 202:149-156. [DOI] [PubMed] [Google Scholar]
- 34.Rehermann, B., K. M. Chang, J. G. McHutchison, R. Kokka, M. Houghton, and F. V. Chisari. 1996. Quantitative analysis of the peripheral blood cytotoxic T lymphocyte response in patients with chronic hepatitis C virus infection. J. Clin. Investig. 98:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reignat, S., G. J. Webster, D. Brown, G. S. Ogg, A. King, S. L. Seneviratne, G. Dusheiko, R. Williams, M. K. Maini, and A. Bertoletti. 2002. Escaping high viral load exhaustion: CD8 cells with altered tetramer binding in chronic hepatitis B virus infection. J. Exp. Med. 195:1089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarobe, P., J. J. Lasarte, A. Zabaleta, L. Arribillaga, A. Arina, I. Melero, F. Borras-Cuesta, and J. Prieto. 2003. Hepatitis C virus structure proteins impair dendritic cell maturation and inhibit in vivo induction of cellular immune responses. J. Virol. 77:10862-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarobe, P., J. J. Larsarte, N. Casares, A. Lopez-Diaz de Cerio, E. Baixeras, P. Labarga, N. Garcia, F. Borras-Cuesta, and J. Prieto. 2002. Abnormal priming of CD4+ T cells by dendritic cells expressing hepatitis C virus core and E1 proteins. J. Virol. 76:5062-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spinzi, G., R. Pellicano, G. Minoli, N. Terreni, M. A. Cutufia, S. Fagoonee, M. Rizzetto, and A. Ponzetto. 2001. Helicobacter pylori seroprevalence in hepatitis C virus positive patients with cirrhosis. The Como cross-sectional study. Panminerva Med. 43:85-87. [PubMed] [Google Scholar]
- 39.Takaki, A., M. Wiese, G. Maertens, E. Depla, U. Seifert, A. Liebetrau, J. L. Miller, M. P. Manns, and B. Rehermann. 2000. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat. Med. 6:578-582. [DOI] [PubMed] [Google Scholar]
- 40.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinant of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 19:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, Y., J. E. Finan, J. M. Middeldrop, and S. D. Hayward. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236:18-29. [DOI] [PubMed] [Google Scholar]
- 42.Wedemeyer, H., X. S. He, M. Nascimbeni, A. R. Davis, H. B. Greenberg, J. H. Hoofnagle, T. J. Liang, H. Alter, and B. Rehermann. 2002. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J. Immunol. 169:3447-3458. [DOI] [PubMed] [Google Scholar]
- 43.Widell, A., V. Molnegren, F. Pieksma, M. Calmann, J. Peterson, and S. R. Lee. 2002. Detection of hepatitis C core antigen in serum or plasma as a marker of hepatitis C viraemia in the serological window-phase. Transfus. Med. 12:107-113. [DOI] [PubMed] [Google Scholar]
- 44.Yao, Z. Q., D. T. Nguyen, A. I. Hiotellis, and Y. S. Hahn. 2001. Hepatitis C virus core protein inhibits human T lymphocyte responses by a complement-dependent regulatory pathway. J. Immunol. 167:5264-5272. [DOI] [PubMed] [Google Scholar]
- 45.Yao, Z. Q., A. Eisen-Vandervelde, S. Ray, and Y. H. Hahn. 2003. HCV core/gC1qR interaction arrests T cell cycle progression through stabilization of the cell cycle inhibitor p27kip1. Virology 314:271-282. [DOI] [PubMed] [Google Scholar]
- 46.Yao, Z. Q., S. Ray, A. Eisen-Vandervelde, S. Waggoner, and Y. S. Hahn. 2001. Hepatitis C virus: immunosuppression by complement regulatory pathway. Viral Immunol. 14:277-295. [DOI] [PubMed] [Google Scholar]