Abstract
Replication of the influenza A virus virion RNA (vRNA) requires the synthesis of full-length cRNA, which in turn is used as a template for the synthesis of more vRNA. A “corkscrew” secondary-structure model of the cRNA promoter has been proposed recently. However the data in support of that model were indirect, since they were derived from measurement, by use of a chloramphenicol acetyltransferase (CAT) reporter in 293T cells, of mRNA levels from a modified cRNA promoter rather than the authentic cRNA promoter found in influenza A viruses. Here we measured steady-state cRNA and vRNA levels from a CAT reporter in 293T cells, directly measuring the replication of the authentic influenza A virus wild-type cRNA promoter. We found that (i) base pairing between the 5′ and 3′ ends and (ii) base pairing in the stems of both the 5′ and 3′ hairpin loops of the cRNA promoter were required for in vivo replication. Moreover, nucleotides in the tetraloop at positions 4, 5, and 7 and nucleotides forming the 2-9 base pair of the 3′ hairpin loop were crucial for promoter activity in vivo. However, the 3′ hairpin loop was not required for polymerase binding in vitro. Overall, our results suggest that the corkscrew secondary-structure model is required for authentic cRNA promoter activity in vivo, although the precise role of the 3′ hairpin loop remains unknown.
Influenza A virus has a segmented, single-stranded RNA genome of negative polarity. The eight segments of influenza virus viral RNA (vRNA) are transcribed and replicated by the viral RNA-dependent RNA polymerase in the host cell nucleus. vRNAs are transcribed into mRNA and are also copied into cRNA molecules, which in turn are used as templates to generate more vRNA. The influenza virus RNA polymerase is a heterotrimeric complex of three subunits: PB1, PB2, and PA. All three subunits, in association with nucleoprotein (NP), are required for transcription and replication of the viral RNA genome (reviewed in references 8 and 18).
All eight genomic RNA segments of influenza A virus have 12 and 13 conserved nucleotides at their 3′ and 5′ termini, respectively, that constitute the vRNA promoter. Significant progress has been made in defining the sequence and secondary structural requirements of the vRNA promoter (reviewed in reference 8). These conserved 3′- and 5′-terminal nucleotides, together with two or three segment-specific nucleotides, show inverted partial complementarity. Interaction between the vRNA 5′ and 3′ ends, through base pairing, is required for promoter activity (11, 12, 24). A “corkscrew” secondary structural model for the vRNA promoter (7) is supported by several more recent studies both in vitro and in vivo (2, 6, 7, 19, 20). The main features of the corkscrew model are two short hairpin loop structures, each with a stem of 2 bp and a tetraloop, formed by residues near the 5′ and 3′ termini. In vitro experiments suggest that a hairpin loop at the 5′ end is required for ApG-primed transcription (11) and for polyadenylation of mRNA (29), whereas hairpin loops at both the 5′ and 3′ ends of the vRNA are required for endonuclease activity (19, 20) and for stability (2). However, Rao et al. (30) have recently reported that when the capped RNA substrate contains a CA cleavage site, the 3′ end of vRNA functions only as a template for mRNA synthesis and does not require a hairpin loop structure.
The mechanism by which transcription is initiated by cap primers and terminated to form a polyadenylated mRNA is reasonably well defined. Initiation of viral mRNA transcription from a vRNA template involves a “cap-snatching” mechanism. Caps are cleaved from host pre-mRNAs by the endonuclease activity of the RNA polymerase to generate short 9- to 17-nucleotide capped RNAs that are then used as primers by the influenza virus RNA polymerase complex. Viral mRNA synthesis is dependent on binding of the influenza virus RNA polymerase to both the 5′ and 3′ ends of the vRNA template, indicating that the vRNA promoter acts as an essential cofactor in the production of capped primers (16, 22). Earlier models (4, 22) suggested that the 5′ end of the vRNA promoter was sufficient to stimulate cap-binding activity by the polymerase complex. However, Lee et al. (21) have recently proposed that both the 5′ and 3′ ends of the vRNA promoter are needed for efficient cap binding. Subsequently, polymerase bound to the 5′ and 3′ ends of the vRNA activates an endonuclease activity that results in the cleavage of host pre-mRNAs (14, 19, 20). However, another recent report suggests that when the capped RNA substrate contains a CA cleavage site, the 5′ end of vRNA alone is apparently sufficient for endonuclease activation (30). Termination of transcription and polyadenylation of mRNA occur at a poly(A) signal sequence of 5 to 7 U residues located about 16 nucleotides from the 5′ end of the vRNA template (26).
Replication, unlike transcription, is primer independent (15, 31), but the mechanisms controlling transcription and replication have remained elusive (reviewed in reference 27). Recently, however, our laboratory has proposed a “cRNA stabilization model” to explain the switch from primary transcription to replication (F. Vreede and T. Jung, personal communication). This model suggests that cRNA accumulates only when sufficient newly synthesized polymerase subunits and NP are available to protect cRNA from degradation by host nucleases. It follows from this model that cRNA promoter activity becomes evident only if a stable cRNA-ribonucleoprotein complex forms intracellularly. The cRNA promoter is complementary to the vRNA promoter, and recent data suggest that it, like the vRNA promoter, adopts a corkscrew secondary structure (Fig. 1) (1). However, Azzeh et al. (1) analyzed a modified cRNA promoter containing 3′C→U, 8′G→A, and 5′A→G mutations in the 5′ end of the cRNA promoter, rather than the wild-type promoter. This modified cRNA promoter might not adopt the same RNA secondary structure as the authentic wild-type promoter. Moreover, these authors used a conventional chloramphenicol acetyltransferase (CAT) reporter assay to study replication. This assay measures transcription, which is only an indirect measure of the synthesis of rRNA from cRNA during replication. Thus, further investigation of the validity of the corkscrew model of the authentic cRNA promoter is needed.
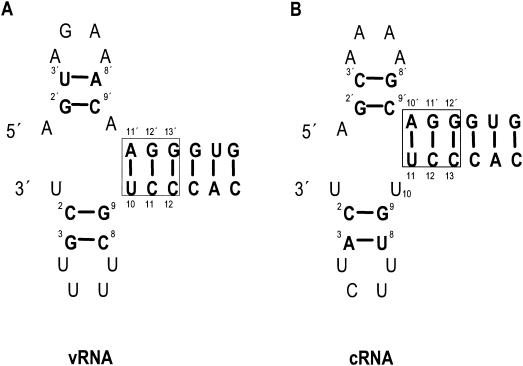
FIG. 1.
Influenza virus vRNA promoter (A) and cRNA promoter (B), shown in the corkscrew conformation. Base pairs are boldfaced and joined by lines. Conserved base-pairs in the double-stranded region, involving both the 5′ and 3′ ends of the promoter, are boxed. The ′ notation is used to identify nucleotides of the 5′ end of the promoter.
In this study, we conducted a systematic mutagenic analysis of both the 5′ and 3′ strands of the authentic wild-type cRNA promoter and examined the effects in vivo by a primer extension assay to directly measure the steady-state levels of vRNA, mRNA, and cRNA. To further understand the effects of mutations caused in vivo, we also performed a UV cross-linking assay to detect polymerase binding activity in vitro. Our in vivo results extended the previous data and validated the corkscrew model for the authentic cRNA promoter. Moreover, we show that the specific identities of the nucleotides in the tetraloop (residues 4, 5, and 7) and the base pair at positions 2 and 9 of the 3′ hairpin loop secondary structure are important for cRNA promoter activity. The in vitro polymerase binding assay further confirms that a hairpin loop structure is required at the 5′ end of the cRNA promoter. However, no direct evidence was obtained in vitro for the presence of a hairpin loop at the 3′ end of the cRNA promoter. Possible reasons for this discrepancy between the in vivo and in vitro results for the 3′ end of the promoter are discussed.
MATERIALS AND METHODS
Plasmids.
The protein expression plasmids pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-PA-His6, and pcDNA-NP and the pUC18-based plasmid pPOLI-cCAT-RT have been described previously (9). Plasmid pPOLI-cCAT-RT contains the 5′ and 3′ noncoding regions of the NS cRNA segment of influenza A/WSN/33 virus and a CAT open reading frame in positive sense. Modifications of the pPOLI-cCAT-RT plasmid were prepared by site-directed mutagenesis; sequences of the mutagenic primers are available upon request. All modified plasmid constructs were verified by sequencing.
293T cell transfection and RNA isolation.
Approximately 106 293T cells were transfected in suspension in 35-mm-diameter dishes by using 5 μl of LipofectAMINE 2000 (Gibco BRL) with 1 μg of plasmids pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and pPOLI-cCAT-RT or pPOLI-cCAT-RT mutant plasmids. Cells were harvested at about 48 h posttranfection, and total RNA was isolated by using TRIzol reagent (Gibco BRL).
Primer extension assay.
Primer extension reactions were performed essentially as published previously (9). Briefly, approximately 5 μg of total RNA was mixed with an excess of two CAT-specific 32P-labeled primers, 5′-CGCAAGGCGACAAGGTGCTGA-3′ (to detect vRNA) and 5′-ATGTTCTTTACGATGCGATTGGG-3′ (to detect mRNA and cRNA), in 6 μl of water. The mixture was denatured by heating at 95°C for 5 min, followed by cooling on ice, and was then incubated at 45°C. Primer extensions were performed at 45°C for 90 min after addition of 100 U of SuperScript reverse transcriptase (Gibco BRL) in the enzyme reaction buffer provided. Transcription products were denatured at 95°C for 5 min, separated on 6% polyacrylamide gels containing 7 M urea in Tris-borate-EDTA buffer, and detected by autoradiography. Primer extension reactions for each mutation were performed at least twice with independently transfected cells.
Preparation of partially purified recombinant His-tagged influenza A virus polymerase.
Recombinant His-tagged influenza A virus polymerase was prepared essentially as published previously (2, 9). Briefly, approximately 3 × 106 293T cells were transfected in 15-cm-diameter dishes by using 90 μl of LipofectAMINE 2000 (Gibco BRL) with 20 μg of pcDNA-PB1, pcDNA-PB2, and pcDNA-PA-His6 or with 20 μg of pcDNA-PB1 and pcDNA-PB2 as a negative extract control. Cells were harvested at about 48 h posttranfection, washed twice with 10 ml of ice-cold phosphate-buffered saline, and resuspended in 0.9 ml of lysis buffer (50 mM sodium phosphate [pH 7.5], 200 mM NaCl, 25% glycerol, 0.5% Nonidet P-40, 1 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride, and 1 Complete Mini EDTA-free protease inhibitor cocktail tablet [Roche] per 10 ml), as described previously (9). Preparations were incubated on ice for 15 min, and all subsequent steps were performed at 4°C. The cell lysate was centrifuged at 16,000 × g for 15 min, and the supernatant was transferred to a fresh 1.5-ml tube. A 100-μl volume of nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) in lysis buffer was added to the cell lysate. Imidazole was added to a final concentration of 5 mM, and the preparation was mixed gently for 2 h. The Ni-NTA agarose was collected by centrifugation at 10,000 × g for 2 min and then washed twice with lysis buffer containing 10 mM imidazole. Proteins were eluted from the Ni-NTA agarose in lysis buffer containing 100 mM imidazole and were stored at −20°C.
UV cross-linking assay.
The UV cross-linking assay was performed essentially as described previously (9) except that reaction mixtures included 5 ng of tRNA (Sigma)/μl. For cross-linking of the polymerase to the 5′ end of the wild-type influenza A virus cRNA promoter (5′-AGCAAAAGCAGGC-3′) (wild-type cRNA promoter sequences are underlined) or mutant 5′-end cRNA, we used an existing library of chemically synthesized RNA molecules (28). For cross-linking of the polymerase to the 3′ end of the cRNA promoter, short synthetic oligonucleotides corresponding to the 5′ end (5′-AGCAAAAGCAGGCC-3′) or the 3′ end (5′-GGCCUUGUUUCUACU-3′) of the wild-type influenza A virus cRNA promoter or to the mutant 3′ end of the cRNA promoter were purchased from Dharmacon Inc. RNAs were 5′ end labeled by using T4 polynucleotide kinase and [γ-32P]ATP. A 2-μl volume of partially purified His-tagged RNA polymerase (see above) was mixed with approximately 1 pmol (100,000 cpm) of 32P-labeled RNA and incubated at 30°C for 15 min. The reaction mixture was transferred to a U-bottom 96-well plate and irradiated on ice for 10 min in a UV Stratalinker (Stratagene) equipped with a 254-nm (G8T5) bulb. Cross-linked products were denatured at 95°C for 5 min, separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (7% acrylamide), and analyzed by autoradiography.
Immunoprecipitation of polymerase proteins.
Rabbit polyclonal antibodies specific for influenza virus polymerase proteins PB1 and PB2 were kindly provided by S. C. Inglis (5). An anti-His monoclonal antibody (Qiagen) was used to immunoprecipitate His-tagged PA. Immunoprecipitation of the polymerase complex after cross-linking was performed essentially as published previously (13). Briefly, the polymerase complex (after cross-linking to the 32P-labeled 3′ end of the cRNA promoter in the presence of the unlabeled 5′ cRNA promoter) was disrupted with 1% SDS at 95°C for 2 min, followed by dilution in immunoprecipitation buffer A (10 mM Tris-HCl [pH 7.5], 100 mM NaC1, 2 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 1 μM leupeptin, 1 μM pepstatin, 1 μM Pefabloc [Roche]) to 0.1% SDS, and 5 μl of a specific antiserum was added. After incubation on ice for 2 h, 100 μl of 10% protein A-Sepharose (Pharmacia) in buffer A was added to the antibody-cross-linked product mixture, and the mixture was rotated at 4°C for 1 h. Then buffer B (13) was used to wash Sepharose-bound material twice. Finally, the immunoprecipitates were boiled in 15 μl of SDS-PAGE sample buffer for 5 min. Proteins were separated by SDS-7% PAGE, and after drying they were detected by autoradiography.
RESULTS
Base pairing between the 5′ and 3′ ends of the cRNA promoter is required for replication in vivo.
The present corkscrew model of the cRNA promoter (1) proposes base-pairing between the 5′ residues 10′ to 12′ and the 3′ residues 11 to 13 of the cRNA promoter (Fig. 1B) (nucleotide positions at the 5′ end of the promoter are designated with a prime as well as a number to distinguish them from positions at the 3′ end). To test whether this base pairing was required for replication, a series of pPOLI-cCAT-RT mutants with substitutions at positions 10′, 11′, and 12′ of the 5′ strand were constructed to disrupt one or two potential base pairs. To re-form alternative base pairs at these positions, additional double mutations at positions 11, 12, and 13 of the 3′ strand were made. Primer extension assays were performed (see Materials and Methods) to specifically measure the levels of steady-state vRNA, mRNA, and cRNA in transfected 293T cells expressing the influenza virus polymerase and NP and either a wild-type or a mutant cRNA-like CAT reporter RNA. Two primers, one specific for the negative-sense CAT RNA (detecting vRNA) and the other specific for the positive-sense CAT RNAs (detecting mRNA and cRNA), were used in the same primer extension reaction (9). Due to the possible difference in priming efficiency by the two primers, vRNA levels might be underestimated in this assay, since it is expected that vRNA levels would vastly exceed cRNA levels in transfected cells.
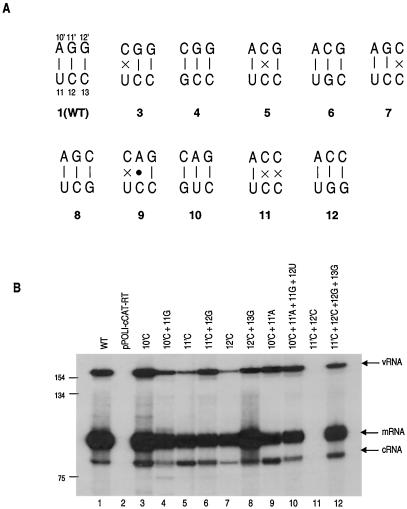
It can be seen (Fig. 2) that disruption of any one of the base pairs at positions 10′, 11′, and 12′ (Fig. 2, lanes 3, 5, and 7) in the 5′ strand was still compatible with the synthesis of vRNA, cRNA, and mRNA (compared with that for the wild type [lane 1]). The mutant with the change at position 10′ showed activity similar to that of the wild type, indicating that disruption of the 10′-11 base pair is not crucial for polymerase activity. A similar result was also reported for the vRNA promoter (17). With a mutation at position 11′ or 12′, the vRNA level was significantly lower than that for the wild type (Fig. 2; compare lanes 5 and 7 with lane 1). Disruption of the two adjacent base pairs at positions 11′ and 12′, however, resulted in no detectable RNA (Fig. 2, lane 11), whereas disrupting two other adjacent base pairs at positions 10′ and 11′ did not reduce vRNA and cRNA levels significantly, although mRNA levels decreased (Fig. 2, lane 9). This result may be explained by an A · C base pair formation (25) at positions 11′ and 12 and the observation that base pairing at positions 10′ and 11 is not essential for polymerase activity. Thus, this double mutant is behaving as a point mutant, like the mutant of Fig. 2, lane 3, rather than the double mutant of lane 11. Overall, therefore, these results suggest that disruption of 1 base pair is still compatible with replication, usually at a reduced level. However, disruption of 2 base pairs at positions 11′ and 12′ blocks replication completely.
FIG. 2.
Effects of mutations in the double-stranded region of the cRNA promoter on CAT RNA levels. (A) Wild-type (WT) (pattern 1) and mutant (patterns 3 to 12) sets of base pairs tested. Solid dot indicates mismatched base pair. Pattern numbers correspond to lane numbers in panel B. (B) Primer extension analysis of the effects of mutations to disrupt and re-form predicted base pairs on CAT RNA levels. 293T cells were transfected with pPOLI-cCAT-RT only (pPOLI-cCAT-RT) or with pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and WT or mutant pPOLI-cCAT-RT, as indicated above lanes. Size standards of the 32P-labeled 1-kb DNA ladder (Gibco BRL) are shown on the left. Positions of vRNA, mRNA, and cRNA signals are indicated on the right.
Re-forming alternative base pairs at positions 11′ and 12 and positions 12′ and 13 “rescued” wild-type vRNA levels (Fig. 2; compare lane 5 with lane 6 and lane 7 with lane 8). The double mutant that re-formed the two base pairs at positions 11′ and 12′ also rescued RNA to wild-type levels (Fig. 2; compare lanes 11 and 12). On the other hand, re-forming base pairs at positions 10′ and 11′ (Fig. 2, lane 10) did not fully restore the wild-type pattern (compare lanes 1 and 10); mRNA levels were still reduced. Overall, these results confirmed that base pairing between the 5′ and 3′ ends of the cRNA promoter is required for replication, although not all base pairs are equally critical.
Mutagenic analysis of nucleotides in the proposed 5′ and 3′ hairpin loop of the cRNA promoter in vivo.
We next extended the mutagenic analysis of pPOLI-cCAT-RT to the proposed short 5′ hairpin loop (residues 1′ to 9′) and the proposed 3′ hairpin loop (residues 1 to 10) of the corkscrew model of the cRNA promoter (Fig. 1B). Our aim was to establish if these secondary-structure motifs were present in the authentic wild-type cRNA promoter. A set of single-substitution mutations at each nucleotide from position 1′ to 9′ in the 5′ end and position 1 to 10 in the 3′ end of the authentic cRNA promoter, flanking the CAT reporter gene, was therefore constructed for analysis by the in vivo primer extension assay.
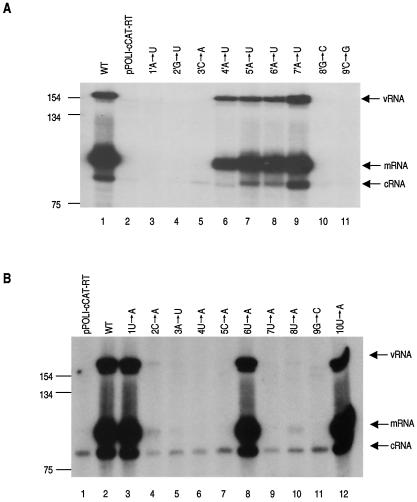
Significant levels of vRNA, cRNA, and mRNA, compared to that of the wild type, were detected with a mutation at nucleotide position 4′, 5′, 6′, or 7′ in the 5′ end of the cRNA promoter (Fig. 3A, lanes 6 to 9), indicating that these positions are not critical for replication. However, RNA levels were essentially at background (Fig. 3A, lanes 3 to 5, 10, and 11) when the 5′ end of the cRNA promoter contained a mutation at position 1′, 2′, 3′, 8′, or 9′, suggesting that either the nucleotides themselves are essential for cRNA to vRNA synthesis or they are involved in base pairs with other residues, forming essential secondary structures. However, the results for the nucleotide at position 1′ must be treated with caution, because we noticed that the input cRNA signal was not detectable. Possibly the mutation decreased POLI promoter efficiency.
FIG. 3.
Effects of point mutations in the 5′ and 3′ termini of the influenza virus cRNA promoter on CAT RNA synthesis. (A) Point mutations from positions 1′ to 9′ of the 5′ strand of the cRNA promoter; (B) point mutations at positions 1 to 10 of the 3′ strand of the cRNA promoter. RNA was analyzed by a primer extension assay (see Materials and Methods). 293T cells were transfected either with pPOLI-cCAT-RT only (pPOLI-cCAT-RT) or with pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and wild-type (WT) or mutant pPOLI-cCAT-RT as indicated above the lanes. Size standards of the 32P-labeled 1-kb DNA ladder (Gibco BRL) are shown on the left. Positions of the vRNA, mRNA, and cRNA signals are indicated on the right. To show the input cRNA signals, the mRNA and vRNA signals had to be somewhat overexposed in panel B. The input cRNA signals are also visible in panel A in all lanes, with the exception of lane 2, after overexposure (data not shown). The signal in lane 2 was not detected, possibly because the mutation at position 1′ affected POLI promoter efficiency.
In comparison with the 5′ end of the cRNA promoter, only a mutation at nucleotide position 1, 6, or 10 in the 3′ end of the cRNA promoter was compatible with significant levels of the different RNA species (Fig. 3B, lanes 3, 8, and 12). A mutation at position 2, 3, 4, 5, 7, 8, or 9 (Fig. 3B, lanes 4 to 7 and 9 to 11) resulted in no or minimal RNA. Therefore, the 3′ end of the cRNA promoter contained fewer positions at which point mutations were tolerated than the 5′ end. This finding suggests that either the 3′ end of the cRNA promoter has a different arrangement of base pairs and secondary structure than the 5′ end or the specific identities of most of the nucleotides in the 3′ end of the promoter are critical for function.
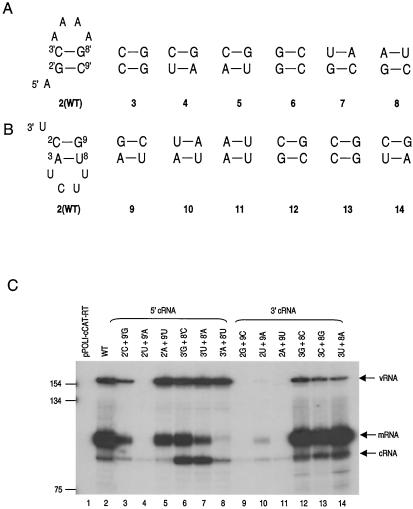
The main features of the corkscrew model, other than base pairing between the 5′ and 3′ strands of the cRNA promoter, are two short hairpin-loop structures, each with a stem of 2 bp and a tetraloop formed by residues close to the 3′ and 5′ termini (Fig. 1). We next tested whether base pairs 2-9 and 3-8 could be replaced in both the 5′ and 3′ strands of the cRNA promoter by alternative base pairs (Fig. 4). In the 5′ end of the cRNA promoter, replacing the wild-type putative GC base pair at position 2′-9′ with CG resulted in low levels of vRNA, replacing with UA resulted in undetectable cRNA, and replacing with AU produced wild-type levels of vRNA (Fig. 4, lanes 3 to 5). Thus, the nature of the alternative 2′-9′ base pair had a dramatic effect on the promoter activity observed. However, when alternative base pairs were introduced at positions 2 and 9 in the 3′ end of the cRNA promoter, essentially no activity was apparent (Fig. 4, lanes 9 to 11). Therefore, it appears that replication requires a C at position 2 and a G at position 9 in the 3′ end of the cRNA promoter.
FIG. 4.
Investigation of “rescue” mutations to re-form proposed base pairing in the cRNA 5′- and 3′-terminal stem-loop structures. (A) Proposed wild-type (WT) stem in the 5′ end of the cRNA promoter (pattern 2) and base pair mutants (patterns 3 to 8) tested. (B) Proposed wild-type stem in the 3′ end of the cRNA promoter (pattern 2) and base pair mutants (patterns 9 to 14) tested. (C) 293T cells were transfected either with pPOLI-cCAT-RT only (pPOLI-cCAT-RT) or with pcDNA-PB1, pcDNA-PB2, pcDNA-PA, pcDNA-NP, and wild-type or mutant pPOLI-cCAT-RT as indicated. Size standards of the 32P-labeled 1-kb DNA ladder (Gibco BRL) are shown on the left. Positions of the vRNA, mRNA, and cRNA signals are indicated on the right.
In contrast, significant RNA synthesis activity was detected when 3 alternative base pairs were introduced at positions 3′ and 8′ in the 5′ end, or at positions 3 and 8 in the 3′ end, of the cRNA promoter. In the 5′ end of the cRNA promoter, replacing the putative CG base pair at 3′ and 8′ with either GC, UA, or AU (Fig. 4, lanes 6, 7, and 8) resulted in wild-type levels of vRNA, suggesting that cRNA to vRNA synthesis was not significantly affected. Interestingly, these 3′-8′ mutations produced different effects on the levels of cRNA and mRNA. In vivo, mutation of the cRNA promoter will affect the vRNA promoter, because these are complementary in sequence. Introduction of a mutation in the 5′ end of the cRNA promoter will cause a corresponding mutation in the 3′ end of the vRNA promoter and potentially affect both vRNA to mRNA and vRNA to cRNA activity. Indeed, these alternative base pairs resulted in reduced (<50% [GC and UA]) or negligible (<5% [AU]) levels of mRNA compared to that for the wild type (Fig. 4; compare lane 2 with lanes 6 to 8). Furthermore, both the GC and UA base pairs increased the level of cRNA compared to that for the wild type (Fig. 4, lanes 6 and 7), although this was not seen for the AU mutation (lane 8). Thus, the 3′-8′ base pair mutations may be affecting cRNA and mRNA levels by affecting the 3′ end of the vRNA promoter. The introduction of all three possible alternative base pairs at positions 3 and 8 in the 3′ end of the cRNA promoter, on the other hand, resulted in slightly reduced levels of vRNA compared to that for the wild type (Fig. 4; compare lane 2 with lanes 12 to 14). All three base pair mutations resulted in CAT mRNA levels similar to that of the wild type but increased cRNA levels (Fig. 4, lanes 12 to 14). This indicates that these base pair substitutions are likely to have had an effect on cRNA to vRNA synthesis but are unlikely to have affected vRNA to mRNA synthesis. Overall, the data (Fig. 4) for the 3′-8′ and 3-8 base pair mutants contrast with those for the 2′-9′ and 2-9 mutants. With the 3′-8′ and 3-8 mutants, cRNA to vRNA replication occurs, whereas with some of the 2′-9′ and all the 2-9 mutants, replication is blocked.
The in vivo data described above—derived from point mutants (Fig. 3) and from “rescue” of base pair mutants at positions 3′ and 8′ in the 5′ end and positions 3 and 8 in the 3′ end (Fig. 4)-suggest that hairpin loop structures are required at both the 5′ and the 3′ end of the cRNA promoter. However, the data (Fig. 3 and 4) for the 2′-9′ and 2-9 mutations are less convincing. cRNA to vRNA synthesis was detected for two of the three base pair mutations made at positions 2′ and 9′ in the 5′ end but not for the third base pair mutant. Moreover, essentially no activity was detected for all three base pair mutations made at positions 2 and 9 in the 3′ end. This finding for 2-9 base pair mutations suggests that if base pairing occurs, as is likely from the 3′-8′ and 3-8 results, then sequence-specific effects must limit the mutations possible at positions 2 and 9 in the 3′ end of the cRNA promoter.
UV cross-linking of the polymerase to the 5′ and 3′ ends of the cRNA promoter.
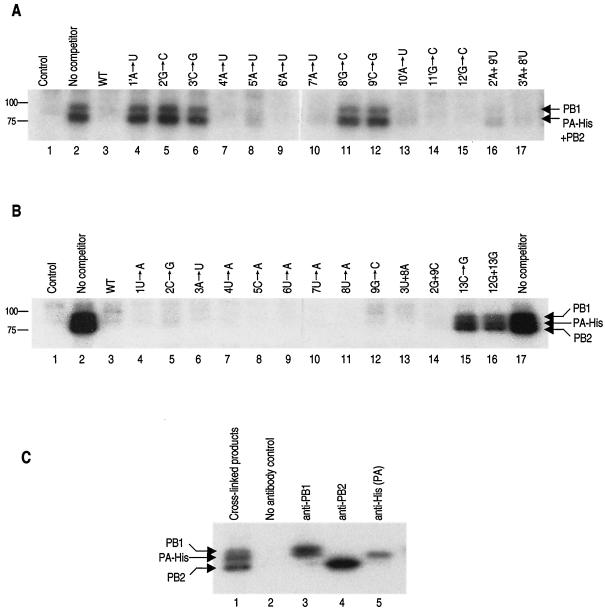
To determine whether a hairpin loop structure at the 5′ and 3′ ends of the cRNA promoter was required for RNA polymerase binding, we used a UV cross-linking protocol with recombinant influenza virus RNA polymerase (9). A competition assay was used to examine the specificity of cross-linking of 32P-labeled wild-type RNA (see Materials and Methods) in the presence of a >100-fold excess of unlabeled mutant competitor RNA. Mutant RNAs that could not bind the polymerase would not be expected to compete with the labeled wild-type probe for cross-linking. On the other hand, mutants that retained binding ability would compete and therefore would prevent the wild-type labeled probe from cross-linking. The results (Fig. 5A, lanes 4, 5, 6, 11, and 12) show that nucleotide substitutions at positions 1′, 2′, 3′, 8′, or 9′ in the 5′ end of the cRNA promoter failed to compete and must, therefore, have prevented polymerase binding. Moreover, double mutants in which the wild-type base pair at either positions 2′ and 9′ or 3′ and 8′ in the 5′ end of the cRNA promoter was replaced with an alternative base-pair (Fig. 5A, lanes 16 and 17) could effectively compete with the 32P-labeled wild-type probe. These results suggest that a hairpin loop structure is needed at the 5′ end of the cRNA promoter for polymerase binding.
FIG. 5.
Identification of nucleotide residues involved in RNA polymerase binding by competitive UV cross-linking. (A) Cross-linking of the 32P-labeled 5′ end of wild-type cRNA (see Materials and Methods). Lane 1, wild-type cRNA 5′ end with polymerase-negative extract (see Materials and Methods); lanes 2 to 17, cross-linking to RNA polymerase with no added competitor or a 100-fold excess of unlabeled competitor, as indicated above the lanes. (B) Cross-linking of the 32P-labeled 3′ end of cRNA in the presence of excess unlabeled 5′ strand (see Materials and Methods). Lane 1, wild-type cRNA 3′ end with polymerase-negative extract; lanes 2 to 14, cross-linking to RNA polymerase with no added competitor or a 100-fold excess of unlabeled competitor, as indicated above the lanes. Sizes of the protein standards (Bio-Rad) are given on the left (in kilodaltons). (C) Identification by immunoprecipitation of the products that were UV cross-linked to the 32P-labeled 3′ end of cRNA. Cross-linking was performed as for panel B, lane 2, followed by immunoprecipitation with an anti-PB1, anti-PB2, or anti-His antibody as indicated (see Materials and Methods). The pattern of the “triplet” of PB1, PA-His, and PB2 varies slightly in panels B and C, possibly because of slight variation in the conditions of electrophoresis.
In contrast, all mutants with point mutations in the 3′ hairpin loop (residues 1 to 9) of cRNA and all double base pair mutants in the stem of the 3′ end of the cRNA promoter could compete with the 32P-labeled 3′ strand of the wild-type probe (Fig. 5B, lanes 4 to 14). However, a point mutant at position 13 (Fig. 5B, lane 15) and a double mutant at positions 12 and 13 (lane 16) in the base-paired region of the 3′ end of the cRNA promoter were unable to compete. Thus, the cross-linking data provide no evidence that a hairpin loop is needed for polymerase binding to the 3′ strand of the cRNA promoter. However, they supported the evidence for base pairing between the 5′ and 3′ ends of the cRNA promoter.
DISCUSSION
There has been significant progress in understanding the sequence and secondary-structure requirements of the influenza virus vRNA promoter (reviewed in reference 8). In contrast, relatively little is known about the requirements of the cRNA promoter. In vivo and in vitro analysis of the vRNA promoter has led to a proposed corkscrew model for the vRNA promoter (Fig. 1A). In this model, two short hairpin loop structures are formed by residues in both the 5′ and 3′ termini of the vRNA (2, 7, 19, 20). The cRNA promoter is complementary to the vRNA promoter, and recent data have suggested that it may also adopt a corkscrew configuration (1). However, those authors (1) used a conventional CAT reporter assay system and performed only limited mutational analysis of a modified cRNA promoter containing 3′C→U, 8′G→A, and 5′A→G mutations in the 5′ strand. The aim of our study was to initially investigate the cRNA promoter requirements for replication by directly measuring the levels of cRNA, vRNA, and mRNA by a primer extension assay (9). The primer extension assay directly measures RNA levels, unlike the conventional CAT activity reporter assay, which only indirectly measures mRNA levels and gives no information on the steady-state levels of vRNA and cRNA.
Theoretically it has been proposed that, unlike transcription, replication could occur without the involvement of the 5′ end of the RNA template (reviewed in reference 8). Our results (Fig. 2) with an in vivo primer extension assay do not support this hypothesis; rather, they confirm the findings of Azzeh et al. (1) suggesting that replication from a cRNA template requires a double-stranded region, formed between the 5′ and 3′ ends of the cRNA termini. Interestingly, however, disruption of one of the base pairs at either position 10′, 11′, or 12′ in this double-stranded region was still compatible with replication. The pattern of bands observed for the 10′ mutants in primer extension did not differ markedly from the wild-type pattern, but with mutations at position 11′ or 12′, subtle differences were observed. Of particular interest was the point mutation at position 11′ in the 5′ arm of the cRNA promoter, which produced an increase in cRNA levels, a significant decrease in vRNA levels, and a slight decrease in mRNA levels. This indicates that cRNA-to-vRNA synthesis is affected, while vRNA-to-cRNA transcription is unaffected or even increased. When the base pair at this position was restored, all three species of RNA appeared to return to wild-type levels. Interestingly, the mutation of two adjacent nucleotides, to disrupt two potential base pairs, gave different results. The double mutation at positions 11′ and 12′ resulted in no detectable RNA, whereas the double mutation at positions 10′ and 11′ resulted in significant levels of cRNA and vRNA but low levels of mRNA. This is best explained by the formation of an A · C mismatch base pair (25) at positions 11′ and 12 and the observation that base pairing at positions 10′ and 11 is not essential for polymerase activity. We conclude that single-base-pair mutations are compatible with replication but mutation of two adjacent base pairs may not be. These results are consistent with those of Fodor et al. (10) and Catchpole et al. (3), who showed that base pair mutations of the duplex region of vRNA (which would have inevitably affected cRNA) could be rescued into viable virus. Our results (Fig. 2) also showed that the single rescue mutation at position 10′ (lane 4) and double rescue mutations at positions 10′ and 11′ (lane 10) resulted in reduced cRNA levels relative to those for their corresponding base pair disruption mutants (lanes 3 and 9). This suggested that the nature of the nucleotide at position 11 of the cRNA promoter affected vRNA-to-cRNA synthesis, in agreement with previous results (17).
We also conducted a systematic mutagenic analysis of the 5′ and 3′ ends of the authentic cRNA promoter by using the same primer extension assay to investigate whether hairpin loop structures were required. A single-nucleotide substitution at position 1′, 2′, 3′, 8′, or 9′ in the 5′ end, or at position 2, 3, 4, 5, 7, 8, or 9 in the 3′ end, of the cRNA promoter led to low or undetectable levels of CAT RNA species. Re-forming alternative base pairs at positions 3′ and 8′ in the 5′ strand of the cRNA promoter with either 3′G-8′C, 3′U-8′A, or 3′A-8′U rescued cRNA-to-vRNA replication (Fig. 4). However, at positions 2′ and 9′ in the 5′ strand of the cRNA promoter, cRNA-to-vRNA synthesis activity was detected for only two (2′C-9′G and 2′A-9′U) of the three base pair mutations (Fig. 4). This suggests that there may also be some sequence-specific effects at the 2′-9′ base pair. A purine residue at position 2′ and a pyrimidine residue at position 9′ might be required. In contrast, all three alternative base pair substitution at positions 3 and 8 in the 3′ end of the cRNA promoter rescued cRNA-to-vRNA synthesis, although vRNA levels were much lower than wild-type vRNA levels (Fig. 4). These results suggest that these mutations affected cRNA-to-vRNA synthesis, a conclusion in agreement with the findings of Azzeh et al. (1). Strikingly, all three alternative base pair mutants with mutations at positions 2 and 9 in the 3′ end of the cRNA promoter were still inactive in replication (Fig. 4). Taken together, our results suggest that a hairpin loop is required in both the 5′ end and the 3′ end of the cRNA promoter. However, the identities of the nucleotides at positions 2 and 9 are more critical than those at positions 3 and 8, particularly in the 3′ strand, as previously found (6). These 2-9 or 2′-9′ base pairs are presumably recognized structurally by the polymerase complex.
The predicted tetraloop of the hairpin loop structure is composed of nucleotides 4′ to 7′ of the 5′ end and nucleotides 4 to 7 of the 3′ end of the cRNA promoter. Transversions at positions 4′ to 7′ in the 5′ end of the cRNA promoter showed significant levels of vRNA, mRNA, and cRNA compared to those for the wild type. In contrast, transversions at positions 4, 5, and 7 in the 3′ end of the cRNA promoter showed essentially background levels of all three specific influenza virus transcripts. Because transversions (pyrimidine to purine) could have interfered with stacking interactions in the hairpin loop region, a set of transitions was also made at these positions. No RNA synthesis activity was detected when either a 5C→U or a 7U→C transition was introduced in the 3′ end of the cRNA promoter (data not shown). But the transitions 4U→C and 6U→C resulted in significant replication and transcription activity (data not shown). Thus, whether transitions or transversions were studied, no replication was observed at positions 5 and 7, in agreement with earlier results (23). In agreement with our results, mutation of the corresponding residues at positions 4′, 5′, and 7′ in the 5′ end of the vRNA promoter also severely inhibited transcription and replication in the primer extension assay (data not shown), findings consistent with those of Flick et al. (7). Therefore, the nature of the nucleotides at positions 4, 5, and 7, but not 6, in both the 5′ end of the vRNA promoter and the 3′ end of the cRNA promoter is critical for replication. Invariable unpaired nucleotide positions may represent positions of direct RNA-protein interaction.
An in vitro UV cross-linking assay was subsequently performed to test whether mutations of the 5′ and 3′ strands of the cRNA promoter affected polymerase binding activity. We found that point mutations at positions 1′, 2′, 3′, 8′, and 9′ in the 5′ end of the cRNA promoter resulted in decreased cross-linking activity, whereas mutants with point mutations at positions 4′ to 7′ or double-base-pair mutations at positions 2′ and 9′ and positions 3′ and 8′ retained significant polymerase binding activity. These in vitro results were consistent with the in vivo results obtained in the primer extension assay (Fig. 3A and 4). They strongly suggested that a hairpin loop structure is required for the 5′ end of the cRNA promoter. However, the UV cross-linking data provided no evidence to support a hairpin loop structure in the 3′ strand of the cRNA promoter (Fig. 5B). We found that NP did not influence the UV cross-linking results with the 3′ strand of the cRNA promoter (data not shown). Further confirmation that such a hairpin loop structure was not required for the initial RNA polymerase binding and initial events in replication was obtained from the results of a limited ApG-primed replication assay (2) in vitro, in which a 15-nucleotide transcript was synthesized (data not shown). However, these in vitro results may differ from the in vivo results because essential cofactors may be absent in the partially purified polymerase used in vitro.
The obvious difference in results between the in vivo primer extension assay and the in vitro polymerase binding studies for the 3′ strand of the cRNA promoter suggests the following scenario. RNA polymerase binding to the cRNA promoter requires both a 5′ hairpin loop structure and 5′-to-3′ base pairing but does not require a corkscrew structure. However, an unknown host protein, we speculate, is required in addition to the polymerase and binds the hairpin loop in the 3′ strand, acting as a chaperone for the RNA polymerase. This host protein, we suggest, is absent or inactivated in our in vitro studies. We conclude that a corkscrew structure probably exists for the authentic cRNA promoter at some stage in replication but that initial binding of the cRNA promoter to the RNA polymerase probably requires only the 5′ hairpin loop and base pairing between the two ends of the cRNA promoter.
Acknowledgments
We thank Ervin Fodor, Frank Vreede, Pierre Fechter, and Ruth Harvey for helpful discussions. We also thank D. Peiris and J. Robinson for DNA sequencing.
This work was supported by MRC program grant G9523972, awarded to G.G.B., and by an ORS (to T.D.).
REFERENCES
- 1.Azzeh, M., R. Flick, and G. Hobom. 2001. Functional analysis of the influenza A virus cRNA promoter and construction of an ambisense transcription system. Virology 289:400-410. [DOI] [PubMed] [Google Scholar]
- 2.Brownlee, G. G., and J. L. Sharps. 2002. The RNA polymerase of influenza A virus is stabilized by interaction with its viral RNA promoter. J. Virol. 76:7103-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catchpole, A., L. Mingay, E. Fodor, and G. Brownlee. 2003. Alternative base pairs attenuate influenza A virus when introduced into the duplex region of the conserved viral RNA promoter of either the NS or the PA gene. J. Gen. Virol. 84:507-515. [DOI] [PubMed] [Google Scholar]
- 4.Cianci, C., L. Tiley, and M. Krystal. 1995. Differential activation of the influenza-virus polymerase via template RNA-binding. J. Virol. 69:3995-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Digard, P., V. C. Blok, and S. C. Inglis. 1989. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology 171:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flick, R., and G. Hobom. 1999. Interaction of influenza virus polymerase with viral RNA in the ′corkscrew' conformation. J. Gen. Virol. 80:2565-2572. [DOI] [PubMed] [Google Scholar]
- 7.Flick, R., G. Neumann, E. Hoffmann, E. Neumeier, and G. Hobom. 1996. Promoter elements in the influenza vRNA terminal structure. RNA 2:1046-1057. [PMC free article] [PubMed] [Google Scholar]
- 8.Fodor, E., and G. G. Brownlee. 2002. Influenza virus replication, p. 1-29. In C. W. Potter (ed.), Influenza. Elsevier Science, New York, N.Y.
- 9.Fodor, E., and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor, E., P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1998. Attenuation of influenza A virus mRNA levels by promoter mutations. J. Virol. 72:6283-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1995. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol. 69:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor, E., B. L. Seong, and G. G. Brownlee. 1993. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding-site at residue-9 to residue-12 of the promoter. J. Gen. Virol. 74:1327-1333. [DOI] [PubMed] [Google Scholar]
- 14.Hagen, M., T. D. Chung, J. A. Butcher, and M. Krystal. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol. 68:1509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hay, A. J., J. J. Skehel, and J. McCauley. 1982. Characterization of influenza virus RNA complete transcripts. Virology 116:517-522. [DOI] [PubMed] [Google Scholar]
- 16.Honda, A., A. Endo, K. Mizumoto, and A. Ishihama. 2001. Differential roles of viral RNA and cRNA in functional modulation of the influenza virus RNA polymerase. J. Biol. Chem. 276:31179-31185. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H. J., E. Fodor, G. G. Brownlee, and B. L. Seong. 1997. Mutational analysis of the RNA-fork model of the influenza A virus vRNA promoter in vivo. J. Gen. Virol. 78:353-357. [DOI] [PubMed] [Google Scholar]
- 18.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1579. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 19.Leahy, M. B., D. C. Pritlove, L. L. Poon, and G. G. Brownlee. 2001. Mutagenic analysis of the 5′ arm of the influenza A virus virion RNA promoter defines the sequence requirements for endonuclease activity. J. Virol. 75:134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leahy, M. B., H. C. Dobbyn, and G. G. Brownlee. 2001. Hairpin loop structure in the 3′ arm of the influenza A virus virion RNA promoter is required for endonuclease activity. J. Virol. 75:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, M. T., K. Klumpp, P. Digard, and L. Tiley. 2003. Activation of influenza virus RNA polymerase by the 5′ and 3′ terminal duplex of genomic RNA. Nucleic Acids Res. 31:1624-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M. L., B. C. Ramirez, and R. M. Krug. 1998. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17:5844-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, X., and P. Palese. 1992. Mutational analysis of the promoter required for influenza virus virion RNA synthesis. J. Virol. 66:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann, G., and G. Hobom. 1995. Mutational analysis of influenza virus promoter elements in vivo. J. Gen. Virol. 76:1709-1717. [DOI] [PubMed] [Google Scholar]
- 25.Nowakowski, J., and I. Tinoco, Jr. 1997. RNA structure and stability. Semin. Virol. 8:153-165. [Google Scholar]
- 26.Poon, L. L., D. C. Pritlove, J. Sharps, and G. G. Brownlee. 1998. The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J. Virol. 72:8214-8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portela, A., and P. Digard. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 83:723-734. [DOI] [PubMed] [Google Scholar]
- 28.Pritlove, D. C., E. Fodor, B. L. Seong, and G. G. Brownlee. 1995. In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J. Gen. Virol. 76:2205-2213. [DOI] [PubMed] [Google Scholar]
- 29.Pritlove, D. C., L. L. Poon, L. J. Devenish, M. B. Leahy, and G. G. Brownlee. 1999. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J. Virol. 73:2109-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao, P., W. Yuan, and R. M. Krug. 2003. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 22:1188-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skehel, J. J., and A. J. Hay. 1978. Nucleotide sequences at the 5′ termini of influenza virus RNAs and their transcripts. Nucleic Acids Res. 5:1207-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]