Abstract
During infection of both vertebrate and invertebrate cell lines, the alphanodavirus Nodamura virus (NoV) expresses two nonstructural proteins of different lengths from the B2 open reading frame. The functions of these proteins have yet to be determined, but B2 of the related Flock House virus suppresses RNA interference both in Drosophila cells and in transgenic plants. To examine whether the NoV B2 proteins had similar functions, we compared the replication of wild-type NoV RNA with that of mutants unable to make the B2 proteins. We observed a defect in the accumulation of mutant viral RNA that varied in extent from negligible in some cell lines (e.g., baby hamster kidney cells) to severe in others (e.g., human HeLa and Drosophila DL-1 cells). These results are consistent with the notion that the NoV B2 proteins act to circumvent an innate antiviral response such as RNA interference that differs in efficacy among different host cells.
Nodamura virus (NoV) is the type species of the genus Alphanodavirus of the Nodaviridae, a family of small riboviruses with bipartite, positive-strand RNA genomes that also includes Flock House virus (FHV). NoV is unique among alphanodaviruses in its ability to lethally infect both insects and mammals, including the mosquitoes Aedes aegypti, Aedes albopictus, and Toxorhynchites amboinensis (4, 36, 41), suckling mice, and suckling hamsters (17, 35, 36). NoV infects cultured mosquito cells from Aedes pseudoscutellaris, A. aegypti, and A. albopictus (1, 3, 41) and cultured baby hamster kidney BHK21 cells (3, 23, 30). When NoV genomic RNAs are introduced by transfection, they can replicate in a wide range of cultured cells (6).
The divided nodavirus genome naturally separates the replicative and packaging functions onto two different positive-sense RNA molecules, RNA1 and RNA2, respectively. These two genomic RNAs are copackaged into the same virion, and both are required for infectivity (24, 30, 38). RNA1 encodes protein A, the RNA-dependent RNA polymerase (RdRp) that catalyzes the replication of both genome segments. RNA2 encodes the viral capsid precursor protein, α. RNA2 and protein α are dispensable for RNA1 replication. Protein A also catalyzes the synthesis of a single subgenomic RNA3 from an RNA1 template. RNA3 is not packaged into virus particles. For FHV, RNA3 encodes two small proteins, B1 and B2, in overlapping reading frames. Protein B1 is in the same reading frame as protein A and thus represents its C-terminal fragment, whereas protein B2 is in the +1 reading frame relative to protein A (11). For NoV, the first and second AUG codons of RNA3 initiate the translation of two forms of B2 (B2-137 and B2-134) that differ only at the N terminus, whereas B1 initiates at the third AUG codon. As for FHV, the B2 proteins are in the +1 reading frame relative to the A/B1 open reading frame (ORF) (23). All three proteins are detected in cells transfected with NoV RNA1 (NoV1), which replicates autonomously and leads to the synthesis of RNA3 (23).
The functions of the nodavirus B1 and B2 proteins remain unclear. Both B1 and B2 are dispensable for FHV RNA replication in some mammalian cells, and B2 is dispensable for FHV RNA replication in yeast cells (5, 13, 33). However, a mutant FHV RNA1 unable to synthesize B2 exhibits phenotypic defects in RNA replication in cultured Drosophila cells (21, 27). Li et al. (27) showed that FHV B2 inhibits RNA interference (RNAi) in Drosophila cells and in transgenic plants, although its mechanism of action is as yet unknown.
The construction and characterization of full-length cDNA clones for NoV1 and NoV2 was recently described (23). When these clones were transcribed from T7 promoters in BSR T7/5 cells, which are BHK21 derivatives that constitutively express T7 RNA polymerase (7), the entire viral infectious cycle was reconstituted, including the replication of the genomic RNAs, the synthesis of subgenomic RNA3 and viral proteins, and the assembly of infectious NoV particles (23). In the present study, we used this reverse genetic system to investigate the function of the NoV B2 proteins in cultured mammalian and insect cells. Since the phenotypic consequences of mutating B2 were observed at the level of RNA replication, the RNA replication cycle was initiated by RNA transfection rather than viral infection except where otherwise noted. All of the RNA transfection experiments described here were performed in the absence of RNA2, which is not required for RNA1 replication.
Mutation of NoV B2 results in a wild-type (WT)-like phenotype in BSR T7/5 cells.
We examined the ability of four NoV B2 mutants to direct RNA1 replication and subgenomic RNA3 synthesis in plasmid-transfected BSR T7/5 cells. The NoV1 mutants used for this analysis are detailed in Table 1. Mutant 1 (B2−−) eliminated both B2 ORFs without affecting the overlapping A/B1 ORF (23). Mutant 2 (B2+−) synthesized only B2-137 (23), while mutant 6 (B2−+) synthesized only B2-134 (data not shown). Mutant 7 (RNA3−) eliminated the synthesis of RNA3, which encodes the B1 and B2 proteins. Similar mutations have previously been demonstrated to reduce or eliminate FHV RNA3 synthesis (13). The indicated changes in mutant 7 introduce two conservative coding changes in the overlapping protein A ORF (Table 1) without impairing its RdRp activity (Fig. 1A, lane 6). The NoV RNA1 mutations were generated by PCR-based circular mutagenesis and DpnI selection, as described previously (34). The regions of interest were confirmed by sequencing, and small DNA fragments carrying the desired mutations were ligated into the parental plasmid background by standard techniques (34).
TABLE 1.
NoV RNA1 mutants
| NoV1 | Phenotype | Genotypeb | Protein sequence corresponding toc:
|
|
|---|---|---|---|---|
| B2 ORFd | A/B1 ORFe | |||
| WTa | B2++ | WT | MTNMS | MRLR |
| Mutant 1a | B2−− | U2745C, U2754C, C2757G | T..T* | .... |
| Mutant 2a | B2+− | U2754C | M..T. | .... |
| Mutant 6 | B2−+ | U2745C | T..M. | .... |
| Mutant 7 | RNA3− | C2731A, G2732A, U2733A, G2734A | No RNA3 | R904K, V905I |
Construction of the WT NoV1 cDNA and mutants 1 and 2 has been previously described (23).
Nucleotide numbers refer to positions in NoV RNA1.
A period indicates identity to WT.
Protein B2 is in the +1 reading frame with respect to the overlapping protein A ORF. The first five positions are indicated. The asterisk indicates a stop codon.
Residue 1 of protein B1 corresponds to residue 908 of protein A; protein B1 is identical to the C terminus of protein A. The first four positions are indicated.
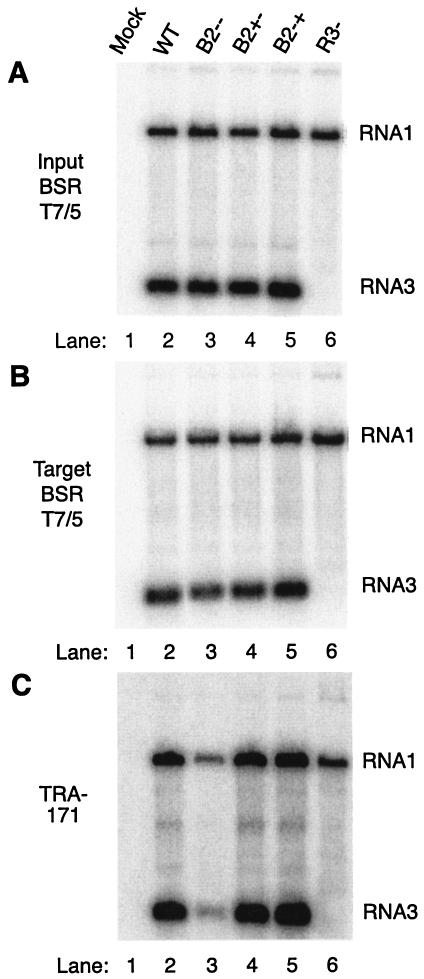
FIG. 1.
NoV1 B2 mutants exhibit defects in RNA replication in TRA-171 but not in BSR T7/5 cells. (A) BSR T7/5 cells, which constitutively express T7 RNA polymerase (7), were mock transfected (lane 1) or transfected with WT (lane 2) or mutant (lanes 3 through 6) versions of NoV RNA1 cDNA clones. After 48 h, total cellular RNAs were isolated, and 500 ng of each RNA was analyzed by Northern blot hybridization using 32P-labeled probes specific for the positive strands of RNA1 and RNA3, as described in the text. (B and C) Each RNA preparation shown in panel A (500 ng) was used to transfect fresh monolayers of BSR T7/5 cells (B) or TRA-171 cells (C). After 24 h at 28°C, total RNAs were isolated, quantitated, and subjected to Northern blot hybridization as described above. Panel C was exposed 16 times longer than panels A and B. Lanes: 1, mock transfection; 2, WT NoV1; 3, mutant 1 (B2−−); 4, mutant 2 (B2+−); 5, mutant 6 (B2−+); 6, mutant 7 (RNA3−).
We transfected confluent monolayers of hamster BSR T7/5 cells with WT or mutant versions of the pNoV1 cDNA plasmid by using Lipofectamine 2000 (2, 23) and incubated the cells for 48 h at 28°C. Plasmid DNA (2.5 μg) was used to transfect 2 × 106 cells. Total cellular RNAs were isolated with guanidinium isothiocyanate (10), quantitated by spectrophotometry, and analyzed by Northern blot hybridization using 32P-labeled probes specific for the positive strands of RNA1 and RNA3, as described previously (23). Five hundred nanograms of each RNA sample was analyzed, and results were visualized with a Molecular Dynamics PhosphorImager digital radioactivity imaging system.
We observed the accumulation of WT RNA1 and subgenomic RNA3 (Fig. 1A, lane 2). No decrease in RNA1 accumulation relative to WT levels was observed for the four mutants (Fig. 1A, compare lanes 3 through 6 and lane 2). Mutation of the RNA1 region corresponding to the 5′ end of RNA3 (mutant 7; Table 1) reduced the synthesis of RNA3, which serves as a message for the translation of B2, to below the level of detection by this method (Fig. 1A, lane 6). The two mutations in the overlapping RdRp ORF (Table 1) did not impair the ability of RNA1 to replicate in these cells (Fig. 1A, compare lanes 6 and 2). In fact, we observed a slight increase in RNA1 synthesis for mutant 7 (Fig. 1A and B, lanes 6, and Fig. 2B), possibly because RNA3 was no longer present to compete with RNA1 for the RdRp.
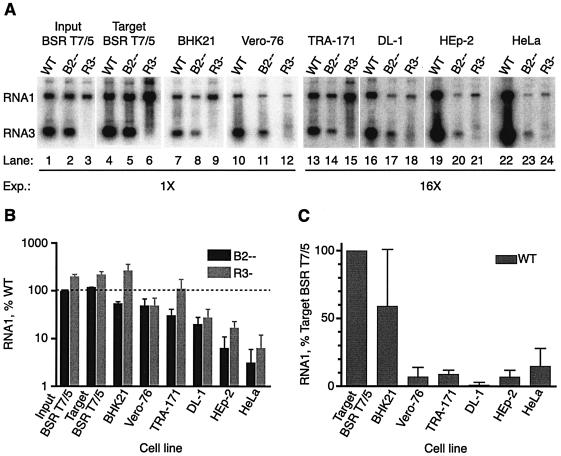
FIG. 2.
The observed defect in RNA replication is not specific for insect cells. (A) Phosphorimage of representative Northern blot. Five hundred nanograms of each of the RNA preparations (WT, B2−−, or RNA3−) shown in Fig. 1A (Fig. 1A lanes 2, 3, and 6, respectively) was transfected into fresh monolayers of the following cells: hamster BSR T7/5 (lanes 4 through 6), hamster BHK21 (lanes 7 through 9), monkey Vero-76 (lanes 10 through 12), mosquito TRA-171 (lanes 13 through 15), Drosophila DL-1 (lanes 16 through 18), human HEp-2 (lanes 19 through 21), and human HeLa cells (lanes 22 through 24). Lanes 1 through 3 contain the indicated input BSR T7 RNAs shown in Fig. 1A. Total cellular RNAs were isolated and analyzed by Northern blot hybridization as described in the legend to Fig. 1. Lanes 13 to 24 were exposed 16 times longer than lanes 1 to 12, as indicated. Detection of RNA1 and RNA3 from DL-1 cells (lanes 16 through 18) also required the loading of 5 times more RNA per lane (2.5 μg). WT, WT NoV1; B2−−, mutant 1; R3−, mutant 7. (B) Histogram of mutant RNA1 (B2−− or R3−) accumulation in each cell line, expressed as a percentage of WT RNA1 accumulation, which was set at 100% (indicated by a dotted line). (C) Histogram of WT RNA1 accumulation in the various cell lines. For each cell type, individual WT values were expressed as a percentage of the mean WT RNA1 accumulation detected in BSR T7/5 target cells, and the mean percentage was determined. In panels B and C, the relative RNA levels are shown as mean values ± standard deviations.
Transfection of BSR T7/5 and TRA-171 cells with WT and mutant NoV1 RNAs.
Previous reports have shown that FHV mutants that are unable to synthesize B2 exhibit WT-like phenotypes in mammalian and yeast cells (5, 13, 33). However, FHV B2− mutants showed defects in RNA replication in cultured Drosophila cells (21, 27). Therefore, we examined the phenotypes of the NoV mutants in insect as well as mammalian cells. For these studies, we used a line of cultured cells from T. amboinensis mosquitoes, TRA-171 (25), which can be productively infected with WT NoV (data not shown).
We isolated total cellular RNAs from WT or mutant NoV1 plasmid-transfected BSR T7/5 cells and quantitated them as described above. This pool of RNA contained cellular RNAs and viral RNA species, including RNA1 and RNA3 of both polarities. These RNAs (roughly 0.4% of the harvest) were transfected into fresh monolayers containing 5 × 105 BSR T7/5 or TRA-171 cells, where they initiated new replication cycles. Following 24 h of incubation at 28°C, total cellular RNA was isolated and analyzed by Northern blot hybridization with RNA1 and RNA3 probes as described above (Fig. 1B and C). The overall pattern of RNA accumulation in RNA-transfected BSR T7/5 cells (Fig. 1B, lanes 1 through 6) was identical to that seen in plasmid-transfected BSR T7/5 cells (Fig. 1A, lanes 1 through 6) and confirmed that the mutants exhibited WT-like phenotypes upon passage in BSR T7/5 cells. However, in TRA-171 cells, mutants 1 and 7 (B2−− and RNA3−, respectively) exhibited defects in the accumulation of viral RNAs (Fig. 1C, lanes 3 and 6). A similar phenotype was observed in TRA-171 cells when the replicative cycle was initiated by infection with B2−− mutant viruses (data not shown), suggesting that the phenotype was independent of the RNA passage protocol. The B2+− and B2−+ mutants (mutants 2 and 6, respectively; Table 1) synthesized near-WT levels of RNA1 and RNA3 (Fig. 1C, lanes 4 and 5, respectively), suggesting that the two forms of B2 were functionally interchangeable in TRA-171 cells.
To determine the extent to which the input RNA replicated in the target cells, we measured the input RNA signal by Northern blot hybridization of total HeLa cell RNA isolated 3 h after transfection with the WT, B2−−, or RNA3− RNA (see below) and extrapolated the results to the other cell types. We had observed that the kinetics of RNA replication in all of the cell types tested were similar (data not shown) and were consistent with previous direct measurements of the kinetics of NoV RNA replication in transfected BHK21 cells (6). Based on these observations, we expected to find no detectable RNA replication products 3 h posttransfection and therefore assumed that all of the RNA measured at 3 h was input RNA. These results showed that the input RNA could have contributed no more than 20% of the TRA-171 cell signal and therefore does not influence our interpretation. In target BSR T7/5 cells, the input RNA contributed 0.1 to 0.3% of the signal for WT and mutant RNAs (data not shown).
NoV B2−− phenotype in other mammalian and insect cells.
Previous reports of FHV B2− mutants (21, 27) suggested to us that the observed B2− phenotype, observed in Drosophila cells but not in mammalian or yeast cells (5, 13, 33), might be due to a difference between cells from insects and those from other sources. We examined this possibility by comparing the replication abilities of NoV B2−− and RNA3− mutant RNAs in a variety of other mammalian and insect cell types.
We transfected an aliquot (500 ng) of total RNA from DNA-transfected BSR T7/5 cells (Fig. 2A, lanes 1 to 3) into several target cell types, including a fresh monolayer of BSR T7/5 cells, hamster BHK21 cells, African green monkey kidney Vero-76 cells (32), TRA-171 cells, Schneider's Drosophila melanogaster line 1 cells (DL-1; WR strain) (15, 37), human HEp-2 cells (8), and human HeLa cells (8). HeLa and HEp-2 cells were transfected with Lipofectin; all other cells were transfected with Lipofectamine 2000. Mammalian and TRA-171 cells were transfected at a density of 5 × 105 cells/2-cm2 well, and DL-1 cells were transfected at 1.5 × 106 cells/well. After 24 h of incubation at 28°C, total cellular RNAs were isolated, quantitated, and analyzed by Northern blot hybridization as described above. A representative Northern blot is shown in Fig. 2A. The detection of signals from TRA-171, DL-1, HEp-2, and HeLa cells required 16-times-longer exposures than the detection of signals from the rest; in DL-1 cells, the detection of mutant RNA1 and RNA3 also required analysis of 5 times more RNA.
The amount of RNA1 in each case was quantitated, using ImageQuant software (Amersham Biosciences Products), from phosphorimages of Northern blots in which samples from three independent RNA transfection experiments were analyzed, and the resulting three values were averaged. Only sample results falling within the linear range of the assays were included. The RNA samples were analyzed on a total of seven different Northern blots; as a standard, an identical sample was included on each gel, and the standard values were normalized to one another to allow direct comparison of RNA samples run on different gels. For each cell type, the level of WT RNA1 accumulation was set to 100% (Fig. 2B), and the relative amounts of RNA1 detected for the B2−− and RNA3− mutants were expressed as percentages of the WT levels on a logarithmic scale. RNA1 levels are shown in Fig. 2B as mean values ± standard deviations.
The ability of mutant 1 (B2−−) and 7 (RNA3−) RNAs to replicate did not correlate with the animal origin (mammal or insect) of the target cell (Fig. 2A and B). For mutant 1, RNA1 accumulation was greatly reduced in Drosophila DL-1 cells (Fig. 2A, lane 17). The reduction was even more dramatic for human HEp-2 cells (Fig. 2A, lane 20) and human HeLa cells (Fig. 2A, lane 23). The quantitative analysis confirmed that the defect in RNA1 accumulation was most severe in Drosophila DL-1 cells and in human HeLa and HEp-2 cells. In HEp-2 cells, mutant 1 accumulated only 10% of the WT level of RNA1, whereas it directed the accumulation of less than 5% of the WT level in HeLa cells (Fig. 2B). The B2−− mutant also exhibited a similar decrease in RNA3 synthesis in these cells (Fig. 2A and data not shown).
Mutant 7 exhibited a similar phenotype in DL-1, HEp-2, and HeLa cells (Fig. 2A, lanes 18, 21, and 24, respectively). In these cell types, the ability of this mutant to direct the accumulation of RNA1 was severely impaired relative to that of the WT, although it consistently accumulated more RNA1 than did mutant 1 (Fig. 2B). This apparent ability of mutant 7 to synthesize more RNA1 than mutant 1 may be a consequence of the lack of competition by RNA3 for RdRp. As demonstrated above for BSR T7/5 and TRA-171 cells (Fig. 1), mutant 7 failed to synthesize detectable levels of RNA3 in any of the cell types tested (Fig. 2A). However, the reduced severity of the defect in RNA1 accumulation of the RNA3− mutant raises the possibility that this mutant might synthesize tiny amounts of RNA3 that are sufficient to direct the synthesis of low levels of B2.
To determine the extent to which the input RNA replicated in these target cells, we analyzed the accumulation of BSR T7/5-derived RNA1 as a function of time in DL-1 and HeLa cells (data not shown). We found that WT RNA1 was amplified over a 24-h time course in both of these cell types with similar kinetics and (by extrapolation, also assuming similar kinetics of RNA replication) in HEp-2 cells, with only 3 to 6% of the total signal contributed by the input RNA 24 h after transfection. In each of these cell types, the kinetics of RNA replication approximated those described previously for NoV RNAs in transfected BHK21 (6) and BSR T7/5 (23) cells. In contrast, we saw no increase in RNA1 signal for mutant 1 or 7 in these cells over the entire time course (data not shown). These results suggested either (i) that these mutant RNAs failed to replicate in DL-1, HEp-2, and HeLa cells in the absence of B2 or (ii) that the replication and degradation of RNA1 in these cells had reached a steady state in which no net increase could be detected.
As in the earlier experiments (Fig. 1), no reduction in RNA1 accumulation was detected in target BSR T7/5 cells transfected with either mutant RNA (Fig. 2A and B). In other cell types, namely hamster BHK21 cells (Fig. 2A, lanes 8 and 9), monkey Vero-76 cells (lanes 11 and 12), and TRA-171 cells (lanes 14 and 15), the phenotypes were less severe (Fig. 2B). In target BSR T7/5, BHK21, and TRA-171 cells (Fig. 2A, lanes 6, 9, and 15, respectively, and 2B), more RNA1 was detected for mutant 7 than for even the WT, again perhaps due to a lack of competition for RdRp. However, in TRA-171 cells, the ability of mutant 7 to synthesize RNA1 varied from experiment to experiment (compare Fig. 1C, lane 6, with 2A, lane 15). As described above, the signals we detected in BSR T7/5, BHK21, Vero-76, and TRA-171 cells represented an amplification of the input RNA for the WT and both mutants (data not shown). While we cannot rule out the possibility that different RNA transfection efficiencies partially account for the differences in the levels of RNA1 detected in each cell type, such differences are unlikely to contribute much given the exponential nature of RNA replication.
Because BSR T7/5 cells are derived from BHK21 cells, we were surprised to see that the mutant phenotypes were different in these two cell types. However, the stock of BHK21 cells from which BSR T7/5 cells were derived is different from the one we used. The difference between the two stocks is unknown, although they have different passage histories, both before and after transfection with the T7 RNA polymerase expression plasmid.
The severity of the mutant phenotype correlated inversely with RNA replication levels.
The level of WT RNA1 accumulation for each cell type was determined from Northern blots as for the experiment whose results are shown in Fig. 2B. In each case, individual WT RNA1 accumulation values were expressed as percentages of the mean level of WT RNA1 accumulation in BSR T7/5 target cells; the relative RNA1 levels are shown as mean values ± standard deviations (Fig. 2C).
We found that the cell types in which the NoV B2−− and RNA3− mutants had the most severe phenotypes were also the least able to support the replication of WT NoV RNA1. This correlation was strongest for Vero-76 TRA-171, DL-1, and HEp-2 cells (Fig. 2B and C). The correlation was slightly weaker for HeLa cells, in which the RNA accumulation phenotype was the most severe (Fig. 2B). In these cells, WT RNA1 accumulated to a level approximately threefold higher than that seen in HEp-2 cells, yet only half as much RNA1 accumulated for the B2−− mutant in HeLa cells as in HEp-2 cells.
Possible functions of nodavirus B2 proteins.
The results described above showed that the cellular environment into which replicable NoV RNAs were introduced profoundly affected the extent of RNA replication and that the differences were magnified in the absence of protein B2. Our experimental procedure, i.e., the transfection of total intracellular RNAs from cells supporting NoV RNA replication, would be expected to induce cellular defense mechanisms triggered by double-stranded RNA (dsRNA), including the interferon response and RNAi. The observation that small interfering RNAs (siRNAs) can activate the interferon system in a human glioblastoma cell line (39) suggests that, while RNAi and the interferon system are distinct mechanisms, the two responses may overlap in vertebrate cells under certain conditions. Nodavirus B2 proteins may affect both pathways.
However, similar phenotypes were observed whether the replicative cycle was initiated in BSR T7/5, BHK21, or TRA-171 cells by RNA transfection, as shown in Fig. 1 and 2, or by infection with B2−− mutant viruses (data not shown), suggesting that the mutant phenotype was independent of the presence of dsRNA at the time of transfection. Since viral infection would be expected to result in delayed kinetics of interferon induction relative to the RNA transfection protocol, it is unlikely that the mutant phenotype is directly due to an interferon response. This suggestion is strengthened by the observation that the mutants exhibit phenotypic defects, albeit of intermediate magnitude, in Vero-76 cells, which are partially deficient in the interferon pathway (12, 16).
We hypothesize that the cell-specific differences we observed were the result of differential activation of one or both of these cellular defense mechanisms and their suppression by the NoV B2 protein(s). Human HeLa cells, one of the cell lines in which we found NoV RNA replication to be the most inhibited, have been shown to support RNAi responses (14, 18, 26, 40) and are approximately twofold more responsive to siRNAs than mouse SW3T3 cells are (20), which is consistent with the idea of differential activation of RNAi.
We therefore tested whether a subset of the cell types we used (BSR T7/5, BHK21, and HeLa cells) differed in their abilities to support an RNAi response. Two fluorescent marker proteins (enhanced green fluorescent protein [EGFP] and HcRed1) were expressed in these target cells from constitutive RNA polymerase II promoters, and the effect of EGFP-specific siRNAs on EGFP expression was tested, as described previously (9), except that we used flow cytometry to measure the expression of EGFP and HcRed1 within each cell population.
BSR T7/5, BHK21, and HeLa cells were cotransfected with plasmids pEGFP-N1 and pHcRed1-C1 (BD Biosciences Clontech), either with or without the addition of EGFP-specific siRNAs (Dharmacon RNA Technologies, Lafayette, Colo.), as described previously (9). The total amount of nucleic acid transfected was adjusted with yeast tRNA to 6 μg per 10-cm2 well; BSR T7/5 and BHK21 cells were transfected with Lipofectamine 2000, and HeLa cells were transfected with Lipofectamine. Forty-eight hours after transfection, the cells were fixed in 4% paraformaldehyde, as described previously (31), and subjected to flow cytometry. Data were collected on a Becton-Dickinson FACStarPLUS sorter-cytometer by using BD CELLQUEST software. HcRed1 was excited at 570 nm with a Coherent I90-krypton laser and read at 610 nm by using a 610/20-nm bandpass filter. EGFP was excited at 488 nm with a Coherent I70-argon ion laser and read at 510 nm by using a 510/20-nm bandpass filter.
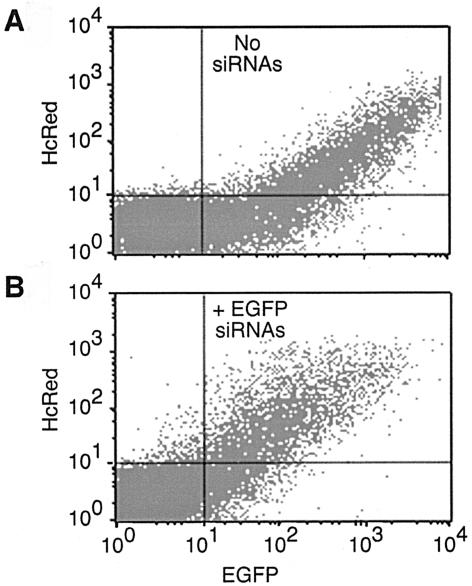
In BHK21 cells, the cotransfection of the EGFP and HcRed1 plasmids resulted in the expression of both fluorescent proteins in 14% of the cells (Fig. 3A, upper right quadrant, and data not shown). The cotransfection of EGFP-specific siRNAs with both EGFP and HcRed1 plasmids resulted in the reduction of EGFP expression in the same population of cells (Fig. 3B, upper right quadrant), evident as a shift of the expression profile to the left (compare Fig. 3A and B). These graphs are representative of those obtained on transfection of BSR T7/5 and HeLa cells, where similar expression profiles were seen (data not shown). Cell populations corresponding to 40% of BSR T7/5 cells and 15% of HeLa cells expressed both fluorescent proteins (data not shown). The relative mean fluorescence intensity of EGFP expression observed in each cell type is summarized in Table 2. In each case, the addition of EGFP-specific siRNAs resulted in a reduction of EGFP in the population of cells expressing both proteins. By this assay, BHK21 cells were 2.5-fold more responsive to siRNAs than BSR T7/5 cells, and HeLa cells (Table 2) and HEp-2 cells (data not shown) were 4.5-fold more responsive than BSR T7/5 cells. This pattern exactly parallels the cell specificity of the mutant phenotype, which was most severe in HeLa and HEp-2 cells, intermediate in BHK21 cells, and WT-like in BSR T7/5 cells (Fig. 2). Whether the magnitudes of these differences are sufficient to account for the phenotypic differences remains to be determined. However, the severity of the NoV mutant phenotype in a given cell type correlated with the ability of that cell type to respond to the induction of an RNAi response by siRNA.
FIG. 3.
Induction of RNAi by siRNAs in BHK21 cells. BHK21 cells were cotransfected with plasmids pEGFP-N1 and pHcRed1-C1, either without (A) or with (B) the addition of EGFP-specific siRNAs, as described previously (9). Forty-eight hours after transfection, the cells were fixed in 4% paraformaldehyde and subjected to flow cytometry as described in the text. EGFP and HcRed1 fluorescence were plotted on the x and y axes, respectively, with the cell population that was positive for the expression of both proteins appearing in the upper right quadrant. Representative data are shown; the same pattern was obtained for BSR T7/5 and HeLa cells (data not shown).
TABLE 2.
RNA interference in mammalian cell lines is differentially induced by siRNAsa
| Cell type | Relative mean fluorescence intensityb
|
Fold Decrease in EGFP expressionc | |
|---|---|---|---|
| No siRNAs | With siRNAs | ||
| BSR T7/5 | 3,813 | 1,593 | 2 |
| BHK21 | 1,030 | 224 | 5 |
| HeLa | 1,164 | 131 | 9 |
BSR T7/5, BHK21, and HeLa cells were cotransfected with plasmids pEGFP-N1 and pHcRed1-C1, either in the absence (no siRNAs) or the presence (with siRNAs) of EGFP-specific siRNAs, as described in the legend to Fig. 3. Forty-eight hours later, cells were fixed in 4% paraformaldehyde and subjected to flow cytometry as described in the text.
The relative mean fluorescence intensity for EGFP for each cell line, in the absence or presence of siRNAs, was measured in the population of cells expressing both EGFP and HcRed1 fluorescent proteins.
Relative effect of siRNAs on EGFP expression.
The proposal of differential activation and suppression of cellular responses to dsRNA could also account for the observation that B2− mutants of FHV failed to maintain their original WT levels of RNA replication when they were transfected serially into BHK21 cells (5). It could also explain why FHV RNA1 ceased to replicate at late times after transfection into BHK21 or CHO cells, despite the continued presence of replication-competent RNA (22). A more severe replication defect was observed in Drosophila cells, where eliminating the FHV B2 protein greatly decreased RNA replication and prevented the recovery of viable virus (21, 27). In these cells, infection with the WT FHV resulted in the accumulation of FHV-specific siRNAs, suggesting that an RNAi response against FHV had been established (27). The depletion of Argonaute2, an essential component of the Drosophila RNAi machinery (19), led to a two- to threefold increase in the accumulation of FHV RNA replication products, suggesting that the observed decrease was indeed due to RNAi. Li et al. further showed that FHV B2 suppressed RNAi in both Drosophila cells and transgenic plants (27).
It is possible that the NoV B2 proteins also suppress RNAi, leading to the cell-specific phenotypes we describe here. While FHV and NoV B2 proteins share only 29% sequence identity, they have other properties in common, including similar sizes (FHV, 106 amino acids; NoV, 134 and 137 amino acids), similar enrichment levels in charged (FHV, 28%; NoV, 25%) and hydrophilic (FHV, 26%; NoV, 26%) amino acids, and similar isoelectric points (FHV, 7.46; NoV, 8.66). Moreover, the recent observation that two heterologous dsRNA-binding proteins, Escherichia coli RNase III and reovirus σ3, could suppress RNA silencing in plants, presumably by sequestering dsRNA (29), raises the intriguing possibility that suppression may be a consequence of dsRNA binding. It remains to be determined whether the FHV or NoV B2 proteins can bind dsRNA. However, recent work from Li et al. has shown that the NoV B2 protein suppresses RNAi in cultured cells from D. melanogaster and Anopheles gambiae mosquitoes (28), lending further support to the suggestion that the cell-specific phenotype described above may be due to differential activation and suppression of RNAi. If so, the NoV B2 protein would be the first example of a suppressor of RNAi shown to function in human cells.
Acknowledgments
We thank Marion L. Spell of the UAB Center for AIDS Research (CFAR) Flow Cytometry Core Facility for flow cytometry and the UAB CFAR DNA sequencing facility for the determination of DNA sequences. We thank César Albariño for many helpful discussions and César Albariño, Tokinori Iwamoto, and Cindy Luongo (UAB) for critical reviews of the manuscript.
This work was supported by NIH grant RO1 AI18270 to L.A.B., NIH grant P30 DK54781 to E. J. Sorscher (K.L.J., Pilot Project 1), and Cystic Fibrosis Foundation grant JOHNSO03I0 to K.L.J. This work has been facilitated by the infrastructure and resources provided by the NIH CFAR core grant P30 AI27767.
REFERENCES
- 1.Ajello, C. A. 1979. Evaluation of Aedes albopictus tissue culture for use in association with arbovirus isolation. J. Med. Virol. 3:301-306. [DOI] [PubMed] [Google Scholar]
- 2.Albariño, C. G., B. D. Price, L. D. Eckerle, and L. A. Ball. 2001. Characterization and template properties of RNA dimers generated during flock house virus RNA replication. Virology 289:269-282. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, L., J. F. E. Newman, and J. S. Porterfield. 1975. The multiplication of Nodamura virus in insect and mammalian cell cultures. J. Gen. Virol. 26:15-20. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, L., and H. A. Scott. 1973. The pathogenicity of Nodamura virus for insects. Nature 241:545. [DOI] [PubMed] [Google Scholar]
- 5.Ball, L. A. 1995. Requirements for the self-directed replication of flock house virus RNA 1. J. Virol. 69:720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball, L. A., J. M. Amann, and B. K. Garrett. 1992. Replication of Nodamura virus after transfection of viral RNA into mammalian cells in culture. J. Virol. 66:2326-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchholz, U. J., S. Finke, and K.-K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, T. R. 1988. Re-evaluation of HeLa, HeLa S3, and HEp-2 karyotypes. Cytogenet. Cell Genet. 48:19-24. [DOI] [PubMed] [Google Scholar]
- 9.Chiu, Y. L., and T. M. Rana. 2002. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell 10:549-561. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Dasmahapatra, B., R. Dasgupta, A. Ghosh, and P. Kaesberg. 1985. Structure of the black beetle virus genome and its functional implications. J. Mol. Biol. 182:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. USA 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckerle, L. D., and L. A. Ball. 2002. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology 296:165-176. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 15.Friesen, P. D., and R. R. Rueckert. 1981. Synthesis of Black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J. Virol. 37:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 17.Garzon, S., and G. Charpentier. 1991. Nodaviridae, p. 351-370. In J. R. Adams and J. R. Bonami (ed.), Atlas of invertebrate viruses. CRC Press, Boca Raton, Fla.
- 18.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 19.Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi, and G. J. Hannon. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146-1150. [DOI] [PubMed] [Google Scholar]
- 20.Harborth, J., S. M. Elbashir, K. Vandenburgh, H. Manninga, S. A. Scaringe, K. Weber, and T. Tuschl. 2003. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 13:83-105. [DOI] [PubMed] [Google Scholar]
- 21.Harper, T. A. 1994. Characterization of the proteins encoded from the nodaviral subgenomic RNA. Ph. D. thesis. University of Wisconsin, Madison.
- 22.Johnson, K. L., and L. A. Ball. 1999. Induction and maintenance of autonomous flock house virus RNA1 replication. J. Virol. 73:7933-7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, K. L., B. D. Price, and L. A. Ball. 2003. Recovery of infectivity from cDNA clones of Nodamura virus and identification of small nonstructural proteins. Virology 305:436-451. [DOI] [PubMed] [Google Scholar]
- 24.Krishna, N. K., and A. Schneemann. 1999. Formation of an RNA heterodimer upon heating of nodavirus particles. J. Virol. 73:1699-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuno, G. 1980. New cell line. A continuous cell line of a nonhematophagous mosquito, Toxorhynchites amboinensis. In Vitro 16:915-917. [Google Scholar]
- 26.Lagos-Quintana, M., R. Rauhut, W. Lendeckel, and T. Tuschl. 2001. Identification of novel genes coding for small expressed RNAs. Science 294:853-858. [DOI] [PubMed] [Google Scholar]
- 27.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 28.Li, W.-X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. A. Brydon, K. L. Johnson, A. García-Sastre, L. A. Ball, P. Palese, and S.-W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichner, Z., D. Silhavy, and J. Burgyan. 2003. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol. 84:975-980. [DOI] [PubMed] [Google Scholar]
- 30.Newman, J. F. E., and F. Brown. 1977. Further physicochemical characterization of Nodamura virus. Evidence that the divided genome occurs in a single component. J. Gen. Virol. 38:83-95. [DOI] [PubMed] [Google Scholar]
- 31.Oomens, A. G. P., A. G. Megaw, and G. W. Wertz. 2003. Infectivity of a human respiratory syncytial virus lacking the SH, G, and F proteins is efficiently mediated by the vesicular stomatitis virus G protein. J. Virol. 77:3785-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Policastro, P. F., M. G. Peacock, and T. Hackstadt. 1996. Improved plaque assays for Rickettsia prowazekii in Vero-76 cells. J. Clin. Microbiol. 34:1944-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Scherer, W. F. 1968. Variable results of sodium deoxycholate tests of Nodamura virus, an ether and chloroform resistant arbovirus. Proc. Soc. Exp. Biol. Med. 129:194-199. [DOI] [PubMed] [Google Scholar]
- 36.Scherer, W. F., and H. S. Hurlbut. 1967. Nodamura virus from Japan: a new and unusual arbovirus resistant to diethyl ether and chloroform. Am. J. Epidemiol. 86:271-285. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353-365. [PubMed] [Google Scholar]
- 38.Selling, B. H., and R. R. Rueckert. 1984. Plaque assay for Black beetle virus. J. Virol. 51:251-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sledz, C. A., M. Holko, M. J. de Veer, R. H. Silverman, and B. R. G. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 40.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tesh, R. B. 1980. Infectivity and pathogenicity of Nodamura virus for mosquitoes. J. Gen. Virol. 48:177-182. [Google Scholar]