Abstract
Hepatitis C virus (HCV) core protein is suggested to localize to the endoplasmic reticulum (ER) through a C-terminal hydrophobic region that acts as a membrane anchor for core protein and as a signal sequence for E1 protein. The signal sequence of core protein is further processed by signal peptide peptidase (SPP). We examined the regions of core protein responsible for ER retention and processing by SPP. Analysis of the intracellular localization of deletion mutants of HCV core protein revealed that not only the C-terminal signal-anchor sequence but also an upstream hydrophobic region from amino acid 128 to 151 is required for ER retention of core protein. Precise mutation analyses indicated that replacement of Leu139, Val140, and Leu144 of core protein by Ala inhibited processing by SPP, but cleavage at the core-E1 junction by signal peptidase was maintained. Additionally, the processed E1 protein was translocated into the ER and glycosylated with high-mannose oligosaccharides. Core protein derived from the mutants was translocated into the nucleus in spite of the presence of the unprocessed C-terminal signal-anchor sequence. Although the direct association of core protein with a wild-type SPP was not observed, expression of a loss-of-function SPP mutant inhibited cleavage of the signal sequence by SPP and coimmunoprecipitation with unprocessed core protein. These results indicate that Leu139, Val140, and Leu144 in core protein play crucial roles in the ER retention and SPP cleavage of HCV core protein.
Hepatitis C virus (HCV) is a major cause of chronic liver disease (5, 19) and has been estimated to infect more than 170 million people throughout the world (15). Symptoms of persistent HCV infection extend from chronic hepatitis to cirrhosis and finally to hepatocellular carcinoma (18, 42). HCV belongs to the genus Hepacivirus in the family Flaviviridae and possesses a viral genome consisting of a single, positive-strand RNA with a nucleotide length of about 9.4 kb (6, 48). The genome encodes a large precursor polyprotein of approximately 3,000 amino acids (6, 17). The polyprotein is processed co- and posttranslationally into at least 10 viral proteins by host and viral proteases (2, 6, 10, 45). The structural proteins of HCV are located in the N-terminal one-fourth of the polyprotein and are cleaved by host membrane proteases (10, 44). Comparison with other flaviviruses suggests that HCV core protein forms the nucleocapsid, which is surrounded by the envelope containing glycoproteins E1 and E2 (6, 48). Functional analyses suggest that HCV core protein has regulatory roles in host cellular functions. In tissue culture systems, HCV core protein regulates signaling pathways and modulates apoptosis (4, 29, 40, 41, 46, 54, 55). Moreover, transgenic mice expressing HCV core protein developed liver steatosis and thereafter hepatocellular carcinoma (34, 36). Thus, it has been suggested that HCV core protein is a multifunctional molecule that acts as a structural protein but is also involved in the pathogenesis of hepatitis C. HCV core protein has two major forms, p23 and p21 (16, 25, 31, 43, 53). HCV core protein p23 represents a 191-amino-acid product in which the C-terminal hydrophobic region also acts as a signal sequence for E1. HCV polyprotein is cleaved between residues 191 and 192 by host signal peptidase to generate C-terminal and N-terminal polypeptides encompassing the core and E1 proteins, respectively. For the full maturation of HCV core protein, the C-terminal signal-anchor sequence was thought to be further processed by an unidentified microsomal protease (25, 30, 31, 43, 53), and the 21-kDa isoform of core protein is predominantly detected both in cultured cells by transfection with expression plasmid and in viral particles obtained from sera of patients with hepatitis C (53). These results suggest that p21 is the mature form of HCV core protein (53). Immunostaining revealed that most HCV core protein is distributed diffusely throughout the cell, probably in the endoplasmic reticulum (ER) (31, 53). However, a minor population was observed in the nucleus (53).
Recently, a presenilin-related aspartic protease, signal peptide peptidase (SPP), was identified (50). SPP is located in the ER membrane and promotes intramembrane proteolysis of signal peptides. The chemical compound (Z-LL)2-keton inhibits processing of signal peptides by SPP, and it was shown to suppress intramembrane proteolysis of major histocompatibility complex class I molecules, preprolactin, HCV core protein, and others (21, 30, 51). Replacement of Asp265 with Ala in SPP resulted in a loss of catalytic function, although this mutant could bind to TBL4K, a derivative of (Z-LL)2-keton (50). HLA-A was processed into yeast microsomes following the addition of wild-type SPP but not mutant SPP, suggesting that SPP interacts with HLA-A (50). Processing of the signal sequence of HCV core protein by SPP was inhibited by the addition of (Z-LL)2-keton, and Ser183 and Cys184 in the signal sequence of core protein were demonstrated to be important for flexibility and intramembrane proteolysis by SPP (23). Signal sequences generally have a tripartite structure, including a central hydrophobic H region and hydrophilic N- and C-terminal flanking regions (28). SPP recognizes the N- and C-terminal regions and cleaves in the middle of the H region (28). Mutational analyses suggested that the flexibility of signal peptides is generally required for substrate recognition of SPP (23). SPP contains the aspartic protease motifs YD and LGLGD, which are located in the predicted transmembrane region, and it is thought to cleave type II (N terminus in the cytosol and C terminus in the lumen)-oriented substrates (50). However, the effect of the cytoplasmic region of type II membrane substrates on intramembrane proteolysis by SPP is not known. In this study, we examined the regions of HCV core protein that are essential for ER retention and intramembrane cleavage by SPP.
MATERIALS AND METHODS
Plasmids.
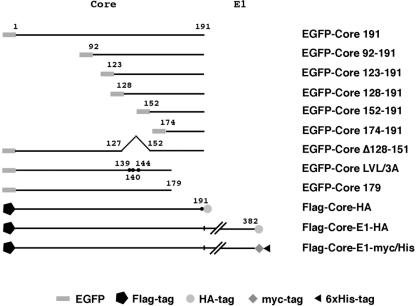
For expression of enhanced green fluorescence protein (EGFP)-fused HCV core proteins in culture cells, the core protein-coding region was amplified by PCR from cDNA encoding full-length HCV polyprotein type 1b (1). The PCR products were subcloned into SalI and BamHI sites 3′ of the EGFP-coding region of pEGFP-C3 (Clontech, Palo Alto, Calif.). The cDNA fragments encoding amino acids 1 to 191, 1 to 179, 92 to 191, 123 to 191, 128 to 191, 152 to 191, and 174 to 191 of HCV core proteins were amplified by PCR and then introduced into pEGFP-C3; these constructs are designated EGFP-Core 191, EGFP-Core 179, EGFP-Core 123-191, EGFP-Core 128-191, EGFP-Core 152-191, and EGFP-Core 174-191, respectively. The genes encoding core proteins with the region between amino acids 128 and 151 deleted and replacement of Leu139, Val140, and Leu144 with Ala were generated by the method of splicing by overlap extension (11, 14, 49) and introduced into pEGFP-C3; these constructs are designated EGFP-Core Δ128-151 and EGFP-Core LVL/3A, respectively (Fig. 1).
FIG. 1.
Expression plasmids used in this study. The genes encoding HCV proteins and their mutants were cloned into pcDNA3.1FlagHA, pcDNA3.1/myc-His C, or pEGFP-C3 as described in Materials and Methods. Other plasmids are described in the text or in the other figure legends.
Fragments encoding Flag and hemagglutinin (HA) tags were inserted at both ends of the multicloning site of pcDNA3.1 (pcDNA3.1FlagHA). PCR products encoding either HCV core protein alone, core protein followed by E1 (Core-E1), or their mutants were cloned into pcDNA3.1FlagHA, resulting in plasmids encoding recombinant proteins sharing Flag and HA tags at the N and C termini, respectively (Fig. 1). In Flag-Core-HA and its derived mutants, Ala191 was replaced by Arg to avoid processing by signal peptidase for determination of cleavage by SPP, as previously shown for the processing of the E1-E2 junction (7). In addition, the region encoding Flag-Core-E1 or its mutants was cleaved from pcDNA3.1FlagHA constructs and then introduced between the SacI and XhoI sites of pcDNA3.1/myc-His C (Invitrogen Corp., Carlsbad, Calif.). The resulting plasmids encode HCV proteins sharing Flag and myc/His epitopes at the N and C termini, respectively (Fig. 1). Genes encoding core protein with a single amino acid (Leu139, Val140, or Leu144), double amino acids (Leu139 and Val140, Leu139 and Leu144, or Val140 and Leu144), or triple amino acids (Leu139, Val140, and Leu144) replaced with Ala were generated by splicing by overlap extension and introduced into pcDNA3.1FlagHA and pcDNA3.1/myc-His C (Fig. 1; see Fig. 4).
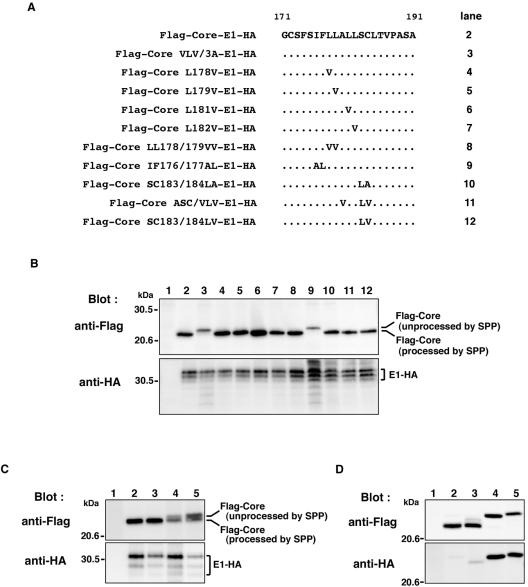
FIG. 4.
Amino acid residues essential for SPP cleavage of the HCV core protein signal sequence of genotype 1a and 1b strains. (A) Mutations in the amino acid residues in the signal sequence of Flag-Core-E1-HA are indicated. Dots indicate unchanged amino acids. (B) Flag-Core-E1-HA (lane 2), Flag-Core LVL/3A-E1-HA (lane 3), Flag-Core L178V-E1-HA (lane 4), Flag-Core L179V-E1-HA (lane 5), Flag-Core L181V-E1-HA (lane 6), Flag-Core L182V-E1-HA (lane 7), Flag-Core LL178/179VV-E1-HA (lane 8), Flag-Core IF176/177AL-E1-HA (lane 9), Flag-Core SC183/184LA-E1-HA (lane 10), Flag-Core ASC/VLV-E1-HA (lane 11), or Flag-Core SC183/184LV-E1-HA (lane 12) was expressed in 293T cells. (C) The gene encoding core and E1 polyprotein of the genotype 1a H77c strain of HCV was introduced into pcDNA3.1FlagHA. Flag-H77c Core-E1-HA (lane 2), Flag-H77c Core ASC/VLV-E1-HA (lane 3), Flag-H77c Core LVL/3A-E1-HA (lane 4), or Flag-H77c Core IF176/177AL-E1-HA (lane 5) was expressed in BHK cells. Cell lysates were analyzed by immunoblotting with anti-Flag (upper panel) and anti-HA (lower panel) antibodies. (D) Expression of Flag-Core 191-HA mutants in 293T cells. The gene encoding core protein with a change of Ala191 to Arg was introduced into pcDNA3.1FlagHA. Flag-Core-HA (lane 2), Flag-Core ASC/VLV-HA (lane 3), Flag-Core LVL/3A-HA (lane 4), and Flag-Core IF176/177AL-HA (lane 5) were analyzed by immunoblotting with anti-Flag (upper panel) and anti-HA (lower panel) antibodies. Cells transfected with an empty plasmid were used as a negative control (lanes 1 in panels B, C, and D).
The genes encoding the ER-targeting and ER retrieval sequences of calreticulin fused with DsRed at the N and C termini, respectively (8, 37, 39), were inserted between the EcoRV and XbaI sites of pcDNA3.1 (pcDNA ER-DsRed) to visualize the ER in culture cells. This recombinant protein is designated ER-DsRed in this study.
Cloning of SPP.
The cDNA encoding SPP was amplified from human liver mRNA (Clontech) by reverse transcription-PCR and cloned into T-vector prepared from pBluescript II SK(−) (27). The gene encoding SPP with an attached HA tag and ER retrieval signal, KEKK, at the C terminus (SPP-HAER) was cloned into pcDNA3.1 to eliminate the possibility that the HA tag suppresses the endogenous ER retrieval signal of SPP. SPP-HAER was colocalized with ER-DsRed on the ER membrane and glycosylated upon transfection into cells (data not shown).
Subcellular localization of wild-type and mutant HCV core proteins.
HeLa cells were maintained in the Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. HeLa cells were seeded on an eight-well chamber slide at 2 × 104 cells per well 24 h before transfection. The cells were transfected with the various plasmids by lipofection with Lipofectamine 2000 (Invitrogen). To determine protein subcellular localizations, transfected cells were fixed with phosphate-buffered saline (PBS) containing 3% paraformaldehyde at 18 h posttransfection and then observed with a confocal laser-scanning microscope (Bio-Rad, Tokyo, Japan). To confirm subcellular localization of the core proteins, transfected cells were fractionated with a subcellular proteome extraction kit (Calbiochem, Darmstadt, Germany). Stepwise extraction resulted in four distinct fractions, which contain mainly cytosolic, membrane-organelle, nuclear, and cytoskelton proteins, respectively. Each fraction was precipitated with trichloroacetic acid and analyzed by immunoblotting, and the densities of the bands were measured with Multi Gauge version 2.2 (Fujifilm, Tokyo, Japan).
Immunoblotting.
After transfection, 293T cells were harvested, washed twice with PBS, and lysed in 20 mM Tris-HCl (pH 7.4) containing 135 mM NaCl, 1% Triton X-100, and 10% glycerol (lysis buffer) supplemented with 1 mM phenylmethylsulfonyl fluoride, 1 mM NaF, and 1 mM Na2VO3. The lysate was centrifuged at 6,500 × g for 5 min at 4°C. The resulting supernatants were subjected to sodium dodecyl sulfate (SDS)-13.5% polyacrylamide gel electrophoresis. The separated proteins were electroblotted onto a Hybond-P polyvinylidene difluoride membrane (Amersham Bioscience, Piscataway, N.J.). These membranes were blocked with PBS containing 5% skim milk and 0.05% Tween 20 (Sigma, St. Louis, Mo.) and incubated with mouse monoclonal anti-Flag M2 (Sigma), anti-HA 16B12 (HA.11; BabCO, Richmond, Calif.), or monoclonal mouse anti-His6-AD1.1.10 (Genzyme/Techne, Tokyo, Japan) immunoglobulin G (IgG) at room temperature for 30 min and then with horseradish peroxidase-conjugated anti-mouse IgG antibody at room temperature for 30 min. Immunoreactive bands were visualized by using the enhanced chemiluminescence Super Signal West Femto substrate (Pierce, Rockford, Ill.).
Immunoprecipitation.
Immunoprecipitation analysis was carried out as described previously (32). Plasmids were transfected into 293T cells by lipofection. Transfected cells were harvested at 18 h posttransfection and lysed in lysis buffer with 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxypropanesulfonic acid (CHAPSO) (Dojindo, Kumamoto, Japan). Cell lysates were incubated with monoclonal anti-HA, anti-Glu-Glu (anti-EE) (BabCO), or anti-Flag antibody at 4°C for 1.5 h and then with protein G-Sepharose CL-4B (Amersham Bioscience) at 4°C for 1.5 h. After centrifugation at 6,500 × g for 3 min at 4°C, the pellets were washed five times with lysis buffer. Immunoprecipitates were subjected to immunoblotting.
Deglycosylation.
Plasmids encoding core and E1 proteins were transfected into 293T cells by lipofection, and cell lysates were immunoprecipitated with anti-HA antibody at 18 h posttransfection. Immunoprecipitates were eluted from protein G-Sepharose CL-4B in 0.5% SDS and 1% 2-mercaptoethanol and digested with endo-β-N-acetylglucosaminidase H (Endo H) or peptide-N-glycosidase F (PNGase F) according to the protocol of the manufacturer (Roche, Mannheim, Germany). The resulting mixtures were subjected to immunoblotting.
RESULTS
Region required for ER retention of HCV core protein.
To determine the regions within HCV core protein that are responsible for ER retention, EGFP-fused, N-terminally truncated HCV core protein (Fig. 1) was coexpressed with the ER marker ER-DsRed. EGFP-Core 191 colocalized with ER-DsRed to the ER (Fig. 2), whereas EGFP-Core 179 was localized primarily to the nucleus as reported previously (3, 33, 43, 47), suggesting that the C-terminal signal sequence is essential for anchoring HCV core protein to the ER membrane. However, EGFP-Core 174-191 exhibited diffuse staining similar to that of EGFP, suggesting that the signal sequence alone is not sufficient for ER localization. EGFP-Core 92-191, EGFP-Core 123-191, and EGFP-Core 128-191 were colocalized with ER-DsRed in the ER, but EGFP-Core 152-191 stained similarly to EGFP-Core 174-191 and EGFP. These data suggest that not only the C-terminal signal sequence but also the region from amino acids 128 to 151 is required for ER retention of HCV core protein.
FIG. 2.
Intracellular localization of EGFP-Core mutants. EGFP-Core and its deletion mutants were coexpressed with ER-DsRed in HeLa cells, and the localization of core proteins was examined by confocal microscopy.
Region essential for processing of the signal sequence of HCV core protein by SPP and signal peptidase.
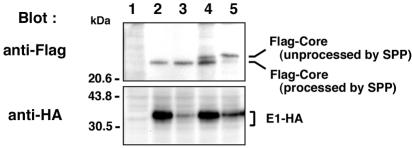
Based on hydrophobicity and a cluster of basic amino acids, HCV core protein was proposed to possess three regions (domains 1 to 3) (Fig. 3A, upper panel) by Hope and McLauchlan (12). To assess the involvement of the region encompassing amino acids 128 to 151 in proteolysis of the signal sequence of HCV core protein by signal peptidase and SPP, three hydrophobic amino acids, Leu139, Val140, and Leu144, in the most hydrophobic peak in domain 2 were replaced with Ala to reduce hydrophobicity, and Ala191 was replaced with Arg to eliminate processing by signal peptidase (Fig. 3A, lower panel). When a wild-type Flag-Core-HA construct was expressed in 293T cells, a single band of 23 kDa was detected by blotting with anti-Flag, but not with anti-HA, suggesting that the HA-fused signal sequence was properly processed by SPP and that Flag-core protein of 23 kDa was generated (Fig. 3B, lanes 2 and 11). In cells expressing the substitution mutants, 25- and 23-kDa bands were detected by the anti-Flag antibody (Fig. 3B, upper panel, lanes 3 to 9) and 25-kDa bands were detected by the anti-HA antibody (Fig. 3B, lower panel, lanes 3 to 9), indicating that the 25- and 23-kDa bands correspond to core proteins that are unprocessed and processed by SPP, respectively. Cleavability of the signal sequence of mutant core proteins by SPP was suppressed in accordance with the number of substitutions, and almost no processing of the signal sequence was observed in cells expressing Flag-Core LVL/3A-HA, which has three amino acid substitutions (Fig. 3B, lane 9). These results indicate that Leu139, Val140, and Leu144 play crucial roles in the processing of the signal-anchor of HCV core protein by SPP. Furthermore, deletion of the hydrophobic region including amino acids 128 to 151 from HCV core protein completely eliminated processing by SPP, and this species was seen only as a single band of 23.5 kDa which was detected by both the anti-Flag and anti-HA antibodies (Fig. 3B, lane 10). Taken together with the observation that Ala180, Ser183, and Cys184 in the signal sequence of HCV core protein of the type 1a Glasgow strain were demonstrated to be essential for SPP proteolysis (13, 23), these results indicate that the hydrophobic region from amino acid 139 to 144 in domain 2 of HCV core protein also participates in the processing of the signal sequence by SPP.
FIG. 3.
Identification of the region responsible for processing of the signal sequence of HCV core protein by SPP and signal peptidase. (A) The hydrophobicity profile of HCV core protein was predicted by the method of Kyte and Doolittle (20). Hope and McLauchlan separated the HCV core protein into three regions, domains 1 to 3 (12). Two hydrophobic regions are predicted in the regions from amino acid 128 to 151 and from amino acid 164 to 186 in the C terminus of the HCV core protein. Mutations and deletions in the region from amino acid 128 to 151 of Flag-Core-HA and Flag-Core-E1-HA constructs are indicated. Dots indicate unchanged amino acids. (B) Expression of Flag-Core-HA polyproteins with changes of Ala191 to Arg in 293T cells. Flag-Core-HA (lanes 2 and 11), Flag-Core L139A-HA (lane 3), Flag-Core V140A-HA (lane 4), Flag-Core L144A-HA (lane 5), Flag-Core LV139/140AA-HA (lane 6), Flag-Core LL139/144AA-HA (lane 7), Flag-Core VL140/144AA-HA (lane 8), Flag-Core LVL/3A-HA (lane 9), and Flag-Core Δ128-151-HA (lane 10) were analyzed by immunoblotting with anti-Flag (upper panel) or anti-HA (lower panel) antibody. Cells transfected with an empty plasmid were used as a negative control (lane 1). (C) Expression of Flag-Core-E1-HA mutants in 293T cells. Flag-Core-E1-HA (lanes 2 and 11), Flag-Core L139A-E1-HA (lane 3), Flag-Core V140A-E1-HA (lane 4), Flag-Core L144A-E1-HA (lane 5), Flag-Core LV139/140AA-E1-HA (lane 6), Flag-Core LL139/144AA-E1-HA (lane 7), Flag-Core VL140/144AA-E1-HA (lane 8), Flag-Core LVL/3A-E1-HA (lane 9), and Flag-Core Δ128-151-E1-HA (lane 10) were analyzed by immunoblotting with anti-Flag (upper panel) or anti-HA (lower panel) antibody. The asterisk indicates unprocessed Flag-Core Δ128-151. Cells transfected with an empty plasmid were used as a negative control (lane 1). (D) The deglycosylation procedure is described in Materials and Methods. After transfection, cell lysates were immunoprecipitated with anti-HA antibody and immunoprecipitates were digested with Endo H (upper panel) or PNGase F (lower panel). Following digestion, proteins were separated by SDS-polyacrylamide gel electrophoresis, and material from cells transfected with vector (lane 1), Flag-Core-E1-HA (lane 2), Flag-Core LVL/3A-E1-HA (lane 3), and Flag-Core Δ128-151-E1-HA (lane 4) was detected by blotting with anti-HA. Nontreated and Endo H- or PNGase F-treated samples are indicated by − and +, respectively. Asterisks indicate mouse IgG heavy chains.
To examine the role of the region from amino acid 139 to 144 in the cleavage of the HCV core protein signal sequence by signal peptidase and SPP in more detail, substitutions of Leu139, Val140, and/or Leu144 with Ala were introduced into the Flag-Core-E1-HA polyprotein (Fig. 1). Flag-Core-E1-HA protein was cleaved to the expected molecular mass of 23 kDa of Flag-Core protein by signal peptidase and SPP (Fig. 3C, lanes 2 and 11), whereas slightly larger bands corresponding to a core protein unprocessed by SPP were detected in cells expressing polyproteins possessing mutations within amino acids 139 to 144 (Fig. 3C, lanes 3 to 9). A lack of processing by SPP was detected mainly in core proteins containing double amino acid changes of Leu139, Val140, and/or Leu144 to Ala (Fig. 3C, lanes 6 to 8), and only an unprocessed band was detected in a triple amino acid substitution mutant (Fig. 3C, lane 9) and a deletion mutant lacking amino acids 128 to 151 (Fig. 3C, lane 10). In contrast to the processing of core protein, E1 protein processed from the mutant polyproteins exhibited the same molecular mass of 32 to 35 kDa and the same deglycosylation patterns following digestion with Endo H or PNGase F (Fig. 3D). These results indicate that the internal hydrophobic region from amino acid 139 to 144 of HCV core protein is essential for processing by SPP but not for cleavage of the core-E1 junction by signal peptidase and the subsequent translocation of E1 protein into the ER. It was suggested that signal peptides must be liberated from the precursor protein by cleavage with signal peptidase in order for them to become substrates for SPP (23). Our data indicate that processing by SPP is not a prerequisite for cleavage of the core-E1 junction by signal peptidase.
Amino acid sequence essential for SPP cleavage of the signal sequences of HCV core proteins of genotypes 1a and 1b.
Martoglio and colleagues reported that HCV core protein is processed by SPP after cleavage by host signal peptidase and that Ala180, Ser183, and Cys184 residues in the signal sequence of HCV core protein of type 1a Glasgow strain are essential for SPP proteolysis, as they maintain the structure of the breaking α-helix (23, 30). To determine the amino acids essential for SPP cleavage of the signal sequence of type 1b HCV core protein, Flag-Core-E1-HA and its substitution mutants were expressed in 293T cells (Fig. 4). Mutation of one, two, or three amino acids, except for Flag-Core IF176/177AL-E1-HA (Fig. 4B, lane 9), did not affect the processing of the core protein signal sequence. Flag-Core IF176/177AL-E1-HA exhibited the same molecular size as Flag-Core LVL/3A-E1-HA (Fig. 4B, lane 2), suggesting that Ile176 and Phe177 in the signal sequence of core protein are essential for cleavage by SPP in our system. However, the triple amino acid substitution (Ala180, Ser183, and Cys184) in the type 1b J1strain (Flag-Core ASC/LVL-E1-HA) (Fig. 4B, lane 11), which is the same as the spmt mutant of the type 1a Glasgow strain (23, 30), did not affect the processing of the signal sequence of HCV core protein by SPP. All derived E1 proteins exhibited a molecular mass of 32 to 35 kDa irrespective of the presence of mutations, and deglycosylation by digestion with endoglycosidases generated uniform 22-kDa bands of E1 proteins (data not shown). These results indicate that Ile176 and Phe177, but not Ala180, Ser183, and Cys184, in the signal sequence of type 1b HCV core protein are essential for processing by SPP and confirm that processing of signal sequence by SPP is not required for cleavage by signal peptidase and translocation of E1 protein into the ER. To determine whether the difference in cleavage of signal sequence depends on the genotype of HCV, Ala180, Ser183, and Cys184 in the HCV core protein of the genotype 1a H77c strain were replaced with Val, Leu, and Val, respectively. The spmt construct of the type 1a H77c strain did not affect the processing of core and E1 proteins in BHK cells (Fig. 4C, lane 3) and 293T cells (data not shown). In contrast, replacement of Leu139, Val140, and Leu144 by Ala and of Ile176 and Phe177 by Ala and Leu suppressed the processing of the core protein signal sequence of the type 1a H77c strain in BHK cells (Fig. 4C, lanes 4 and 5). These results indicate that three hydrophobic amino acids Leu139, Val140, and Leu144 in the hydrophobic peak in domain 2 and the two amino acids Ile176 and Phe177 in the transmembrane domain play important roles in the intramembrane proteolysis of HCV core protein signal sequence of genotypes 1a and 1b by SPP.
To further examine the cleavage of the signal sequence of HCV core proteins by SPP, we prepared IF176/177AL and the spmt mutant core proteins carrying a substitution of Ala191 to Arg to avoid processing by signal peptidase as described above. In cells expressing a wild-type or LVL/3A mutant core protein, a 23-kDa processed or a 25-kDa unprocessed core protein was detected, as seen in Fig. 3B (Fig. 4D, lanes 2 and 4). The IF176/177AL mutant exhibited a 26-kDa unprocessed band which was detected by anti-HA antibody (Fig. 4D, lane 5). In contrast, the spmt core protein exhibited a major band at 23 kDa and a faint 24-kDa band after blotting with the anti-Flag antibody (Fig. 4D, lane 3). Detection of a small amount of the 24-kDa unprocessed band by the anti-HA antibody indicates that most of the spmt mutant core protein was processed by SPP. The unprocessed core proteins of spmt, LVL/3A and IF176/177AL exhibited different electrophoretic mobilities, estimated to be 24, 25, and 26 kDa, respectively (Fig. 4D, lower panel, lanes 3 to 5). Lemberg and Martoglio pointed out that the mobility of a protein does not necessarily correlate with its molecular mass when analyzed in a Tris-glycine gel system due to the unexpected electrophoretic mobility of the proteins (22). However, detection of HA-tagged unprocessed signal sequence in the core mutants clearly demonstrated that LVL/3A and IF176/177AL mutants substituted with Leu139, Val140, and Leu144 in domain 2 and with Ile176 and Phe177 in the transmembrane domain, respectively, have lost the ability to be cleaved by SPP.
Effect of a loss-of-function mutant of SPP on the processing of the signal sequence of HCV core protein.
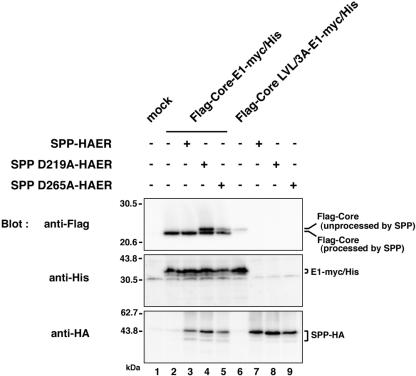
Although there are two reports suggesting that SPP is involved in the processing of the signal sequence of HCV core protein by using the SPP inhibitor (Z-LL)2-keton (23, 30), a direct interaction of HCV core protein with SPP has not been demonstrated. To determine the direct involvement of SPP in the processing of HCV core protein signal sequence, the C-terminal HA tag in the Flag-Core-E1-HA constructs used in the experiments described above was replaced with a myc/His tag and coexpressed with wild-type SPP (SPP-HAER) or with a mutant SPP with amino acid substitutions in the putative protease active sites, i.e., Asp219 (SPP D219A-HAER) or Asp265 (SPP D265A-HAER) to Ala. The signal sequence of HCV core protein was processed in cells coexpressing Flag-Core-E1-myc/His and SPP-HAER (Fig. 5, anti-Flag, lane 3), whereas two bands corresponding to processed and unprocessed (the same size as Flag-Core LVL/3A-E1-myc/His [lane 6]) core proteins were detected in cells coexpressing Flag-Core-E1-myc/His and the mutant SPP constructs (Fig. 5, anti-Flag, lanes 4 and 5). Proper cleavage and glycosylation of E1 proteins in cells coexpressing Flag-Core-E1-myc/His and the SPP mutants (Fig. 5, anti-His, lanes 4 and 5) and those expressing Flag-Core LVL/3A-E1-myc/His (Fig. 5, anti-His, lane 6) indicates that processing of signal sequence by SPP is not required for the cleavage of the core-E1 junction by signal peptidase and translocation of E1 protein into the ER. These results indicate that loss-of-function mutants of SPP inhibit the intramembrane proteolysis of HCV core protein signal sequence and further confirm that the slightly larger bands detected in cells expressing Flag-Core LVL/3A-E1-HA or Flag-Core IF176/177AL-E1-HA are immature core proteins unprocessed by SPP (Fig. 4B, lanes 3 and 9).
FIG. 5.
Effect of loss-of-function mutants of SPP on the processing of the signal sequence of HCV core protein. SPP-HAER, SPPD219A-HAER, or SPPD265A-HAER was coexpressed with Flag-Core-E1-myc/His or Flag-Core LVL/3A-E1-myc/His in 293T cells. Cell lysates were analyzed by immunoblotting with anti-Flag (upper panel), anti-His6 (middle panel), or anti-HA (lower panel) antibody. + and −, presence or absence of each plasmid, respectively. Lane 1, mock; lanes 2, 6, 7, 8, and 9, single expression of Flag-Core-E1-myc/His, Flag-Core LVL/3A-E1-myc/His, SPP-HAER, SPPD219A-HAER, and SPPD265A-HAER, respectively; lanes 3 to 5, coexpression of Flag-Core-E1-myc/His with SPP-HAER, SPPD219A-HAER, and SPPD265A-HAER, respectively.
Interaction of HCV core protein with SPP.
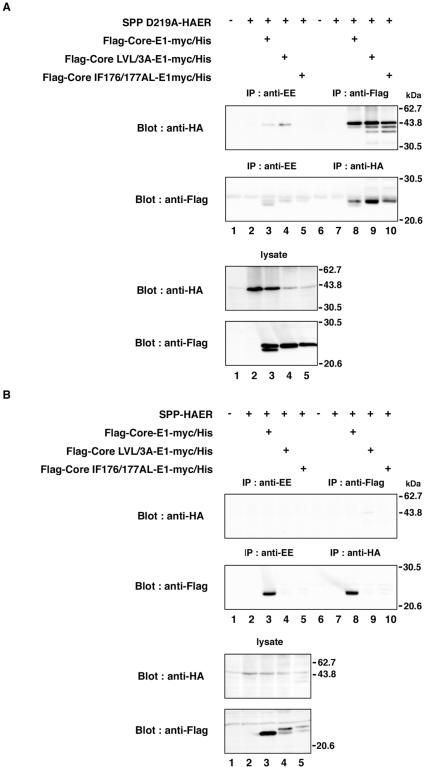
To examine the specific interaction of HCV core protein with SPP, Flag-Core-E1-myc/His, Flag-Core LVL/3A-E1-myc/His, or Flag-Core IF176/177AL-E1-myc/His was coexpressed with SPP-HAER or SPP D219A-HAER in 293T cells and immunoprecipitated with anti-Flag or anti-HA antibody. In cells coexpressing the loss-of-function mutant, SPP D219A-HAER, and one of the three HCV polyprotein substrates, nonspecific bands were detected by immunoblotting with the anti-HA and anti-Flag antibodies in the immunoprecipitates (Fig. 6A, upper and second panels, lanes 3 to 5). Therefore, lysates immunoprecipitated with anti-Flag and anti-HA antibodies were evaluated by comparison with those precipitated with anti-EE. Three bands corresponding to SPP D219A-HAER were coimmunoprecipitated with core proteins by anti-Flag immunoprecipitation (Fig. 6A, upper panel, lanes 8 to 10). SPP has two glycosylation sites (50), and therefore the upper, middle, and lower bands seem to correspond to SPP possessing two glycans, one glycan, and no glycan, respectively. Deglycosylation by PNGase F treatment reduced the molecular sizes of all bands to that of the lowest band (data not shown). Only unprocessed core protein was coimmunoprecipitated with SPP D219A-HAER by anti-HA (Fig. 6A, second panel, lanes 8 to 10). Coexpression of Flag-Core LVL/3A-E1-myc/His or Flag-Core-IF176/177AL-E1-myc/His reduced the expression of SPP D219A-HAER (Fig. 6A, third panel, lanes 4 and 5), suggesting that the core mutants suppress the expression of the SPP mutant. Clear reduction or elimination of the processing of the HCV core protein signal sequence was observed in cells coexpressing SPP D219A-HAER in comparison with those coexpressing wild-type SPP (Fig. 6A and B, bottom panels, lanes 3 to 5). Conversely, no interaction of HCV core protein with wild-type SPP was observed in cells coexpressing SPP-HAER and the HCV polyprotein substrates (Fig. 6B, upper panels, lanes 8 to 10). Broad bands were detected in immunoprecipitates with anti-Flag or anti-EE antibody by immunoblotting with the anti-Flag antibody, probably due to nonspecific binding of the processed core protein to protein G-Sepharose (Fig. 6B, second panel, lanes 3 and 8). These results indicate that a direct interaction of SPP with HCV core protein only between the unprocessed core protein and the loss-of-function mutant of SPP is verifiable. SPP should bind to the signal sequence of HCV core protein and release it after proteolysis, whereas SPP D219A cannot liberate the substrate after binding due to lack of the catalytic activity, suggesting that the SPP mutant may possess dominant negative effects.
FIG. 6.
Interaction of HCV core protein with SPP. SPP D219A-HAER (A) or SPP-HAER (B) was coexpressed with Flag-Core-E1-myc/His, Flag-Core LVL/3A-E1-myc/His, or Flag-Core IF176/177AL-E1-myc/His in 293T cells and immunoprecipitated (IP) with anti-Flag or anti-HA antibody. The immunoprecipitates were analyzed by immunoblotting with anti-HA or anti-Flag antibody. As a control, immunoprecipitation was carried out with anti-EE antibody. + and −, presence or absence of each plasmid, respectively.
Processing of the signal sequence of HCV polyprotein in a human hepatoma cell line.
To confirm the data obtained for 293T cells with human liver cells, processing of core-E1 polyprotein in FLC4 cells, a human hepatoma cell line, was examined (Fig. 7). Processing by signal peptidase and SPP was evident in cells expressing Flag-Core-E1-HA or Flag-Core ASC/VLV-E1-HA (lanes 2 and 3), whereas clear processing by signal peptidase, but not complete cleavage by SPP, was observed in FLC-4 cells expressing Flag-Core LVL/3A-E1-HA or Flag-Core IF176/177AL-E1-HA (lanes 4 and 5). These results are consistent with data obtained with 293T cells, suggesting that the processing of the signal sequence of HCV core protein is not cell type dependent or an artifact of the techniques used in this study.
FIG. 7.
Processing of HCV core-E1 polyprotein in the human hepatoma cell line FLC-4. Flag-Core-E1-HA (lane 2), Flag-Core ASC/VLV-E1-HA (lane 3), Flag-Core LVL/3A-E1-HA (lane 4), or Flag-Core IF176/177AL-E1-HA (lane 5) was expressed in FLC-4 cells and analyzed by immunoblotting with anti-Flag (upper panel) or anti-HA (lower panel) antibody. Cells transfected with an empty plasmid were used as a negative control (lane 1).
Localization of mutant HCV core proteins.
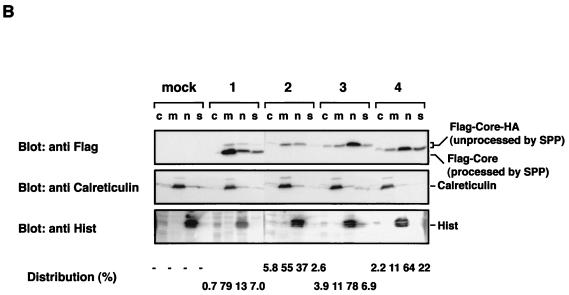
To determine the effect of mutations on the localization of HCV core protein, EGFP-Core 191and its mutants that are defective in cleavage by SPP were expressed in HeLa cells (Fig. 8A). EGFP-Core 191 was processed by SPP and colocalized with an ER marker. EGFP-Core IF176/177AL, which bears a mutation that confers α-helix structure to the signal sequences, was diffusely distributed but did not completely colocalize with ER-DsRed as seen with EGFP-Core 191. EGFP-Core LVL/3A was localized mainly to the nucleus and, to a lesser extent, the cytoplasm, and EGFP-Core Δ128-151 exhibited complete nuclear localization. To confirm the subcellular localization of mutant HCV core proteins, cells were transfected with expression plasmids encoding N-terminally Flag-tagged and C-terminally HA-tagged core proteins to minimize the effect of fusion protein and fractionated, as described in Materials and Methods (Fig. 8B). Consistent with the subcellular localization of EGFP-Core proteins, Flag-Core 191-HA was detected mainly in the membrane-organelle fraction and Flag-Core LVL/3A-HA and Flag-Core Δ128-151-HA were localized mainly in the nuclear fraction. Although EGFP-Core IF176/177AL did not completely colocalize with the ER marker, 55% of Flag-Core IF176/177AL-HA was detected in membrane-organelle fraction. Since we could not separate ER and Golgi fractions by the fractionation method used, it is possible that Flag-Core IF176/177AL-HA localizes mainly in the Golgi rather than the ER. These results indicate that not only the C-terminal signal sequence but also the hydrophobic region from amino acid 139 to 144 in domain 2 and proper processing by SPP are involved in the ER retention of HCV core protein.
FIG. 8.
Localization of mutant HCV core proteins. (A) Putative processing mechanisms of wild-type and mutant core proteins is illustrated at the top. EGFP-Core-191 (column 1), EGFP-Core IF176/177AL (column 2), EGFP-Core LVL/3A (column 3), and EGFP-Core Δ128-151 (column 4) were coexpressed with ER-DsRed in HeLa cells, and subcellular localization of core proteins was examined by confocal microscopy. (B) Subcellular fractionation of HeLa cells transfected with plasmids encoding Flag-Core-HA polyproteins. Cells transfected with an empty plasmid (lane M) or plasmid encoding Flag-Core 191-HA (lanes 1), Flag-Core IF176/177AL-HA (lanes 2), Flag-Core LVL/3A-HA (lanes 3), or Flag-Core Δ128-151-HA (lanes 4) were extracted into four fractions, as described in Materials and Methods. Each fraction was concentrated and subjected to immunoblotting with anti-Flag antibody (upper panel). Lanes c, m, n and s, cytosol, membrane-organelle, nuclear, and cytoskeleton fractions, respectively. Calreticulin and histone (His) were used as markers for membrane-organelle and nuclear fractions, respectively. To determine the distributed ratio of processed and unprocessed core proteins in each fraction, the density of core protein in each fraction was measured and is indicated as a percentage at each bottom of lane.
DISCUSSION
Hope and McLauchlan identified three regions in the HCV core protein, including two hydrophobic regions in the C-terminal one-third of the protein and the region from amino acid 119 to 174, which was designated domain 2 (12). Domain 2 was hypothesized to interact with lipid droplets and confer stability to the HCV core protein (12). Although a mutation in the signal sequence of core protein that renders it resistant to SPP proteolysis restored retention on lipid droplets and overall stability (30), deletion of domain 2 from HCV core protein leads to diffusion in the cytoplasm and degradation after processing by signal peptidase (30). Indeed, these data suggest that mature HCV core protein is retained on lipid droplets via domain 2, but they do not necessarily indicate that the region is required for the ER retention of HCV core protein. The intramembrane proteolysis of the signal sequence of HCV core protein by SPP is abolished when helix-breaking and -bending residues in the C-terminal signal-anchor sequence are replaced by basic amino acids (30). However, the involvement of other regions of HCV core protein in processing by SPP is not known. In this study, we could demonstrate that not only the C-terminal signal-anchor domain but also three hydrophobic amino acids Leu139, Val140, and Leu144 in domain 2 are required for intramembrane proteolysis by SPP. However, this domain is not essential for the cleavage of the core-E1 junction by signal peptidase or for translocation of E1 into the ER. Furthermore, the nuclear localization of domain 2 core mutants possessing an unprocessed C-terminal signal-anchor sequence indicates that association with the ER membrane through domain 2 is required for ER retention of HCV core protein.
Martoglio and colleagues demonstrated, by using a Semliki Forest virus expression system, that Ala180, Ser183, and Cys184 break the α-helical structure within the signal sequence and are essential for the intramembrane proteolysis of HCV core protein of the type 1a Glasgow strain by SPP in BHK and Huh7 cell lines (23, 30). However, mutation of Ala180, Ser183, and Cys184 in core proteins of the type 1b J1 and type 1a H77 strains could not inhibit signal sequence processing by SPP in the BHK and 293T cell lines by expression with plasmid. HCV core protein of the type 1a Glasgow strain shares 96.3 and 95.8% amino acid homology to those of the H77c and J1 strains, respectively. Furthermore, the signal sequences of the core proteins of these three strains are almost identical, and therefore the observed differences in cleavability by SPP might be attributable to sequence other than the signal sequence or expression system used. We reexamined the HCV core protein signal sequence and, using the method of Garnier et al. (9), chose to further examine Ile176 and Phe177 as residues that may interfere with the assumption of a compact α-helix structure and allow for intramembrane proteolysis by SPP. Mutation of Ile176 and Phe177 (Core IF176/177AL) of genotype 1a and 1b strains, which is predicted to confer α-helical structure to the signal sequences, inhibited processing by SPP. EGFP-Core IF176/177AL exhibited no colocalization with an ER marker; this differs from the case for the wild-type core protein. These data suggest that the processing of the signal sequence by SPP may play a role in the ER retention of HCV core protein.
Precursor HCV core protein consists of 191 amino acids and is processed by signal peptidase from a polyprotein after translocation of the C-terminal signal-anchor sequence into the ER. This is then cleaved by SPP into the mature core protein and localizes primarily to the ER. The mature core protein, further processed by an unidentified protease, is composed of amino acids 151 to 153 and is detected in the nucleus (33, 47). Actually, HCV core protein is observed in the cytoplasm, nucleus, and nucleoli in transgenic mice expressing HCV core protein (35). Under normal conditions, the precursor core protein is processed by SPP to 173 to 179 amino acids and localizes to the ER. In contrast, a 179-amino-acid construct containing a limited C-terminal anchor-signal sequence, Core 179, localizes primarily to the nucleus and to the ER to a lesser extent. This striking difference in the subcellular localizations of Core 191 and Core 179, in conjunction with data from the Core IF176/177AL construct, indicates that the presence of the full-length signal-anchor sequence and proper processing by SPP is required for retention of HCV core protein on the ER membrane.
We also demonstrated that the reduction in hydrophobicity in domain 2 affects proteolysis of the signal sequence by SPP and localization of HCV core protein. It was suggested that HCV core protein interacts with lipid droplets containing triacylglycerol and/or ER membrane through domain 2 irrespective of intramembrane proteolysis of the signal sequence (30). A mutant HCV core protein in domain 2, EGFP-Core LVL/3A with Leu139, Val140, and Leu144 replaced by Ala, was processed by signal peptidase but not by SPP and localized to the nucleus in spite of the presence of an unprocessed hydrophobic signal sequence in the C terminus. This result suggests that penetration of the HCV core protein signal sequence into the ER membrane is necessary, but not sufficient, for ER retention of HCV core protein. Insertion of the C-terminal signal-anchor sequence of core protein into the ER may induce conformational changes in domain 2 to render it accessible to the ER membrane and/or lipid droplets by exposure of hydrophobic residues in the domain, residues that are well conserved among various genotypes of HCV. Although it was suggested that processed HCV core protein was retained on the ER membrane via an interaction with unprocessed core protein (25) or with the C-terminal transmembrane region of E1 (26), our data provide a new model of the ER retention of HCV core protein. HCV core protein is a structural protein that forms the nucleocapsid, and virus particles are thought to be released into ER. Therefore, retention of HCV core protein on the ER membrane should be essential for the assembly of HCV.
Intramembrane-cleaving proteases have been shown to play pivotal roles in cell regulation and signaling and are involved in diseases such as Alzheimer's disease (52). SPP belongs to a family of aspartic proteases family and has two aspartic acid residues, Asp219 and Asp265, in the enzyme active site (50). Signal peptidase II also belongs to this aspartic protease family and cleaves the signal sequence by attacking a proton of a water molecule via an aspartic acid of the enzyme (38). Mutant SPP bearing an Asp265-to-Ala substitution was deficient in the processing of HLA-A but retained binding activity to the SPP substrate analogue TBL4K (50). We could demonstrate a direct interaction by immunoprecipitation of unprocessed HCV core proteins with mutant SPP lacking catalytic and substrate-releasing activities by replacement of Asp219 with Ala. Binding of the loss-of-function SPP mutants with unprocessed core proteins irrespective of mutation or deletion in domain 2 indicates that the domain is not directly involved in the interaction.
It has been demonstrated that expression of HCV core protein alone is sufficient for the induction of hepatic steatosis and hepatocellular carcinoma in transgenic mice (24, 34, 35). Furthermore, we demonstrated that nuclear localization and degradation of HCV core protein is regulated by PA28γ-dependent proteolysis (33). These findings suggest that HCV core protein plays a pivotal role in the development of hepatocellular carcinoma and that intramembrane proteolysis may regulate the subcellular localization of HCV core protein. Although the SPP inhibitor (Z-LL)2-keton suppresses cleavage of signal sequence essential for homeostasis, host defense, etc., a specific inhibitor against the intramembrane proteolysis of HCV core protein, such as antagonists for the binding of HCV core protein to ER membrane via domain 2, will be an effective antiviral drug for patients with chronic hepatitis C. Furthermore, involvement of intramembrane proteolysis by SPP in the processing of other HCV proteins and the fates of the peptides cleaved by SPP in the replication and pathogenesis of hepatitis C are subjects of future studies.
Acknowledgments
We gratefully thank T. Shioda for advice on confocal microscopy and J. McLauchlan for valuable discussions.
This work was supported in part by grants-in-aid from the Ministry of Health, Labor and Welfare; the program for Promotion of Fundamental Studies in Health Sciences of the Organization for Drug ADR Relief, R&D Promotion, and Product Review; the Ministry of Education, Culture, Sports, Science and Technology; and the 21st Century Center of Excellence Program of Japan.
REFERENCES
- 1.Aizaki, H., Y. Aoki, T. Harada, K. Ishii, T. Suzuki, S. Nagamori, G. Toda, Y. Matsuura, and T. Miyamura. 1998. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology 27:621-627. [DOI] [PubMed] [Google Scholar]
- 2.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, S. C., J. H. Yen, H. Y. Kang, M. H. Jang, and M. F. Chang. 1994. Nuclear localization signals in the core protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 205:1284-1290. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. M., L. R. You, L. H. Hwang, and Y. H. Lee. 1997. Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-beta receptor modulates the signal pathway of the lymphotoxin-beta receptor. J. Virol. 71:9417-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 6.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubuisson, J., S. Duvet, J. C. Meunier, A. Op De Beeck, R. Cacan, C. Wychowski, and L. Cocquerel. 2000. Glycosylation of the hepatitis C virus envelope protein E1 is dependent on the presence of a downstream sequence on the viral polyprotein. J. Biol. Chem. 275:30605-30609. [DOI] [PubMed] [Google Scholar]
- 8.Fliegel, L., K. Burns, D. H. MacLennan, R. A. Reithmeier, and M. Michalak. 1989. Molecular cloning of the high affinity calcium-binding protein (calreticulin) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 264:21522-21528. [PubMed] [Google Scholar]
- 9.Garnier, J., J. F. Gibrat, and B. Robson. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266:540-553. [DOI] [PubMed] [Google Scholar]
- 10.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 12.Hope, R. G., and J. McLauchlan. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913-1925. [DOI] [PubMed] [Google Scholar]
- 13.Hope, R. G., D. J. Murphy, and J. McLauchlan. 2002. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 277:4261-4270. [DOI] [PubMed] [Google Scholar]
- 14.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 15.Houghton, M., A. Weiner, J. Han, G. Kuo, and Q. L. Choo. 1991. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral. disease. Hepatology 14:381-388. [PubMed] [Google Scholar]
- 16.Hussy, P., H. Langen, J. Mous, and H. Jacobsen. 1996. Hepatitis C virus core protein: carboxy-terminal boundaries of two processed species suggest cleavage by a signal peptide peptidase. Virology 224:93-104. [DOI] [PubMed] [Google Scholar]
- 17.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiyosawa, K., T. Sodeyama, E. Tanaka, Y. Gibo, K. Yoshizawa, Y. Nakano, S. Furuta, Y. Akahane, K. Nishioka, R. H. Purcell, and H. J. Alter. 1990. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology 12:671-675. [DOI] [PubMed] [Google Scholar]
- 19.Kuo, G., Q. L. Choo, H. J. Alter, G. L. Gitnick, A. G. Redeker, R. H. Purcell, T. Miyamura, J. L. Dienstag, M. J. Alter, C. E. Stevens, M. J. Alter, C. E. Stevens, G. E. Tegtmeier, F. Bonino, M. Colombo, W. S. Lee, C. Kuo, K. Berger, J. R. Shuster, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science 244:362-364. [DOI] [PubMed] [Google Scholar]
- 20.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 21.Lemberg, M. K., F. A. Bland, A. Weihofen, V. M. Braud, and B. Martoglio. 2001. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol. 167:6441-6446. [DOI] [PubMed] [Google Scholar]
- 22.Lemberg, M. K., and B. Martoglio. 2003. Analysis of polypeptides by sodium dodecyl sulfate-polyacrylamide gel electrophoresis alongside in vitro-generated reference peptides. Anal. Biochem. 319:327-331. [DOI] [PubMed] [Google Scholar]
- 23.Lemberg, M. K., and B. Martoglio. 2002. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10:735-744. [DOI] [PubMed] [Google Scholar]
- 24.Lerat, H., M. Honda, M. R. Beard, K. Loesch, J. Sun, Y. Yang, M. Okuda, R. Gosert, S. Y. Xiao, S. A. Weinman, and S. M. Lemon. 2002. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology 122:352-365. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Q., C. Tackney, R. A. Bhat, A. M. Prince, and P. Zhang. 1997. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol. 71:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo, S. Y., M. J. Selby, and J. H. Ou. 1996. Interaction between hepatitis C virus core protein and E1 envelope protein. J. Virol. 70:5177-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchuk, D., M. Drumm, A. Saulino, and F. S. Collins. 1991. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 19:1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martoglio, B., and B. Dobberstein. 1998. Signal sequences: more than just greasy peptides. Trends Cell. Biol. 8:410-415. [DOI] [PubMed] [Google Scholar]
- 29.Marusawa, H., M. Hijikata, T. Chiba, and K. Shimotohno. 1999. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-kappaB activation. J. Virol. 73:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moradpour, D., T. Wakita, K. Tokushige, R. I. Carlson, K. Krawczynski, and J. R. Wands. 1996. Characterization of three novel monoclonal antibodies against hepatitis C virus core protein. J. Med. Virol. 48:234-241. [DOI] [PubMed] [Google Scholar]
- 32.Moriishi, K., M. Koura, and Y. Matsuura. 2002. Induction of Bad-mediated apoptosis by Sindbis virus infection: involvement of pro-survival members of the Bcl-2 family. Virology 292:258-271. [DOI] [PubMed] [Google Scholar]
- 33.Moriishi, K., T. Okabayashi, K. Nakai, K. Moriya, K. Koike, S. Murata, T. Chiba, K. Tanaka, R. Suzuki, T. Suzuki, T. Miyamura, and Y. Matsuura. 2003. Proteasome activator PA28γ-dependent nuclear retention and degradation of hepatitis C virus core protein. J. Virol. 77:10237-10249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriya, K., H. Fujie, Y. Shintani, H. Yotsuyanagi, T. Tsutsumi, K. Ishibashi, Y. Matsuura, S. Kimura, T. Miyamura, and K. Koike. 1998. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 4:1065-1067. [DOI] [PubMed] [Google Scholar]
- 35.Moriya, K., H. Fujie, H. Yotsuyanagi, Y. Shintani, T. Tsutsumi, Y. Matsuura, T. Miyamura, S. Kimura, and K. Koike. 1997. Subcellular localization of hepatitis C virus structural proteins in the liver of transgenic mice. Jpn. J. Med. Sci. Biol. 50:169-177. [DOI] [PubMed] [Google Scholar]
- 36.Moriya, K., H. Yotsuyanagi, Y. Shintani, H. Fujie, K. Ishibashi, Y. Matsuura, T. Miyamura, and K. Koike. 1997. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol. 78:1527-1531. [DOI] [PubMed] [Google Scholar]
- 37.Munro, S., and H. R. Pelham. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899-907. [DOI] [PubMed] [Google Scholar]
- 38.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 39.Pelham, H. R. 1996. The dynamic organisation of the secretory pathway. Cell Struct. Funct. 21:413-419. [DOI] [PubMed] [Google Scholar]
- 40.Ray, R. B., K. Meyer, R. Steele, A. Shrivastava, B. B. Aggarwal, and R. Ray. 1998. Inhibition of tumor necrosis factor (TNF-alpha)-mediated apoptosis by hepatitis C virus core protein. J. Biol. Chem. 273:2256-2259. [DOI] [PubMed] [Google Scholar]
- 41.Ruggieri, A., T. Harada, Y. Matsuura, and T. Miyamura. 1997. Sensitization to Fas-mediated apoptosis by hepatitis C virus core protein. Virology 229:68-76. [DOI] [PubMed] [Google Scholar]
- 42.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, Q. Choo, M. Houghton, and G. Kuo. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selby, M. J., Q. L. Choo, K. Berger, G. Kuo, E. Glazer, M. Eckart, C. Lee, D. Chien, C. Kuo, and M. Houghton. 1993. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J. Gen. Virol. 74:1103-1113. [DOI] [PubMed] [Google Scholar]
- 45.Shoji, I., T. Suzuki, M. Sato, H. Aizaki, T. Chiba, Y. Matsuura, and T. Miyamura. 1999. Internal processing of hepatitis C virus NS3 protein. Virology 254:315-323. [DOI] [PubMed] [Google Scholar]
- 46.Shrivastava, A., S. K. Manna, R. Ray, and B. B. Aggarwal. 1998. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J. Virol. 72:9722-9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki, R., Y. Matsuura, T. Suzuki, A. Ando, J. Chiba, S. Harada, I. Saito, and T. Miyamura. 1995. Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C terminus deleted. J. Gen. Virol. 76:53-61. [DOI] [PubMed] [Google Scholar]
- 48.Takamizawa, A., C. Mori, I. Fuke, S. Manabe, S. Murakami, J. Fujita, E. Onishi, T. Andoh, I. Yoshida, and H. Okayama. 1991. Structure and organization of the hepatitis C virus genome isolated from human carriers. J. Virol. 65:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallejo, A. N., R. J. Pogulis, and L. R. Pease. 1995. Mutagenesis and synthesis of novel recombinant genes using PCR, p. 603-612. In C. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Weihofen, A., K. Binns, M. K. Lemberg, K. Ashman, and B. Martoglio. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296:2215-2218. [DOI] [PubMed] [Google Scholar]
- 51.Weihofen, A., M. K. Lemberg, H. L. Ploegh, M. Bogyo, and B. Martoglio. 2000. Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J. Biol. Chem. 275:30951-30956. [DOI] [PubMed] [Google Scholar]
- 52.Weihofen, A., and B. Martoglio. 2003. Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell. Biol. 13:71-78. [DOI] [PubMed] [Google Scholar]
- 53.Yasui, K., T. Wakita, K. Tsukiyama-Kohara, S. I. Funahashi, M. Ichikawa, T. Kajita, D. Moradpour, J. R. Wands, and M. Kohara. 1998. The native form and maturation process of hepatitis C virus core protein. J. Virol. 72:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You, L. R., C. M. Chen, and Y. H. Lee. 1999. Hepatitis C virus core protein enhances NF-κB signal pathway triggering by lymphotoxin-beta receptor ligand and tumor necrosis factor alpha. J. Virol. 73:1672-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, N., A. Khoshnan, R. Schneider, M. Matsumoto, G. Dennert, C. Ware, and M. M. Lai. 1998. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J. Virol. 72:3691-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]