Abstract
Noroviruses (NVs) are the most important pathogen of epidemic nonbacterial gastroenteritis. The recent finding that NVs recognize human histo-blood group antigens (HBGAs) as receptors provided a new approach to study the pathogenesis of NVs. Using computational and site-directed mutagenesis approaches, our investigators previously identified a plausible binding pocket in the P domain of the NV capsids. In this study, we further characterize the role of the P domain in the interaction with human HBGA receptors using three NV strains representing three binding patterns. Our results show that the isolated P domain, although it did not form virus-like particles (VLPs), formed dimers, and the dimers bound HBGAs with the same patterns as those of the intact viral capsids. In contrast, the S domain, which formed small, thin-layer VLPs, did not bind A, B, or H HBGAs. A chimera containing the S domain of VA387 and the P domain of MOH revealed a binding pattern of the P donor strain (MOH). Deletion experiments revealed that an intact P domain is necessary for receptor binding. The P domain dimers are stable over a broad range of pH (2 to 11) or under strong denaturing conditions. Taken together, our results suggest that the P domain of NV contains essential elements for strain-specific binding to receptors. Further study of the P domain will provide useful information about the virus-receptor interaction. The high yield and easy production of the recombinant P protein in the Escherichia coli expression system will provide a simple approach to this goal.
Noroviruses (NVs), formally called Norwalk-like viruses, belong to one of two genera of human caliciviruses, the Norovirus and Sapovirus genera, within the Caliciviridae. NVs contain a single-stranded, positive-sense RNA genome of about 7.7 kb (12, 15). The viral genome encodes one major structural protein of ∼60 kDa that is responsible for the building of the viral capsid (15). In addition, a minor structural protein of ∼20 kDa was identified in the recombinant Norwalk virus capsid (7), which is encoded by the third open reading frame of the genome (15). The function and location of the protein in the capsid remain unclear, although a recent study suggested that this basic protein stabilizes the capsid protein and protects the virus-like particles (VLPs) from disassembly (1). NVs are difficult to study due to the lack of a cell culture and an animal model. The successful expression of NV capsid proteins (8, 13, 14, 16, 17) and the fact that the capsid proteins spontaneously form empty VLPs provided a valuable way for development of diagnostic assays for studying the immunology, epidemiology, and pathogenesis of NVs.
The X-ray crystal structure of the prototype Norwalk virus VLPs showed that the Norwalk virus capsid is composed of 180 capsid protein monomers that form a T=3 icosahedral capsid (20). Each of the capsid proteins has two major domains, the S and P domains, linked by a hinge of 8 amino acids (aa). The S domain (residues 1 to 217) is responsible for the formation of the interior shell of the capsid, while the C-terminal P domain (residues 226 to 530) forms the arch-like structures extending from the shell. Morphogenesis studies have shown that the S domain is required for the assembly of the capsid and participates in multiple intermolecular interactions of dimers, trimers, and pentamers, whereas the P domain is mainly involved in dimeric interaction (20). The P domain can be further divided into P1 and P2 subdomains. Since the P2 domain (aa 275 to 405) contains the most variable sequence and is located on the surface of the capsid, it is believed that the P2 domain plays an important role in immune recognition and receptor interaction. Using computational analysis and site-directed mutagenesis approaches, we have recently identified a plausible receptor binding pocket in the P2 domain that is responsible for binding of the capsid with human histo-blood group antigens (HBGAs) (21). This binding pocket is composed of an RGD-like motif at the bottom of the pocket and three strain-specific binding sites surrounding the RGD-like motif (21).
Increasing data have shown that NVs recognize HBGAs as receptors (9-11, 18, 19). Human HBGAs are complex carbohydrates linked to glycoproteins or glycolipids that are present on red blood cells and mucosal epithelial cells or as free antigens in biological fluids, such as blood or saliva. These antigens are produced by sequential addition of monosaccharides to the antigen precursors by several glycosyltransferases that are genetically controlled and defined as the ABO, Lewis, and secretor gene families. The recognition of HBGAs by NVs is strain specific. Six distinct binding patterns have been described (reference 9 and unpublished data). The prototype Norwalk virus represents one of the six binding patterns that binds to HBGAs of type A and O secretors but not of nonsecretors. The other five binding patterns recognize A, B, and O secretors (VA387), A and B secretors (MOH), A secretors (BUDS), and two Lewis-epitope binders (VA207 and Boxer) with slightly different binding affinity to other side chains of oligosaccharides in the antigenic determinants of the blood group antigens. Consistent with these binding patterns, human volunteer studies revealed that saliva from volunteers with nonsecretor status did not bind to Norwalk virus and nonsecretors were resistant to experimental Norwalk virus infection following challenge (18). A retrospective study of volunteers challenged with Norwalk virus also showed that type O individuals had the highest and type B individuals had the lowest infection rates for Norwalk virus compared with that in other blood types following the challenge (10). By analogy, each of the other five binding patterns may have its own host ranges defined by blood types. For example, individuals with blood types of A, B, or O secretors are susceptible to VA387, while MOH may only infect type A and B secretors, although direct evidence for this hypothesis remains to be established.
Based on our group's recent finding of a potential receptor binding pocket in the P domain (21), we characterize further in this study the host-pathogen interaction of NVs, focusing on the structure-function relationships of the P domain in receptor binding. We demonstrate that the P domain is responsible for receptor binding and that it contains essential elements for strain binding specificity. The Escherichia coli-expressed P proteins form stable dimers that bind to receptors similar to their parental intact capsids. The biochemical properties and binding profiles of the P proteins with their receptors under different denaturing conditions are also characterized. Finally, the significance of using the P domain as a model to study the structure-function relationship and receptor-ligand interaction is discussed.
MATERIALS AND METHODS
Construction of NV mutants.
The S and the P domain mutants were made by cloning the S or the P domain-encoding sequences into the pFastBac vector (Invitrogen, Carlsbad, Calif.) at the BamHI and NotI sites. Both mutants contained the hinge region. The S domain mutant also contained the first 10 aa of the P domain. In the P domain mutant, a start codon was added to the beginning of the coding sequence. For construction of a VA387/MOH chimeric mutant, an EcoRI recognition site (GAATTC) was first introduced at the beginning of the hinge region of the VA387 capsid gene in pFastBac by mutagenesis using the QuikChange mutation kit (Stratagene, La Jolla, Calif.). The P domain-encoding sequence of MOH was then PCR amplified and replaced the P domain of VA387 between the EcoRI and NotI sites. The constructs for expressing the four C-terminal deleted capsids (CD1 to -4) were made by cloning the corresponding regions (see Fig. 4A) of the VA387 capsid gene into pFastBac. In comparison with wild-type capsid sequence, mutant CD1 lost the last 54 aa of the P1-2 domain; CD2 did not contain the whole P1-2 domain (the last 122 aa); CD3 had a deletion of the last 189 aa (the C-terminal half of the P2 domain and P1-2 domain); CD4 contained the S and the P1-1 domain only (with the last 242 aa deleted). The constructs for expressing the three internal deleted capsids (ID1 to -3) were prepared by cloning the capsid-encoding gene of VA387 with corresponding deletions into pFastBac vector. Mutant ID1 contained a deletion of 16 aa in the vicinity of the RGD motif (aa 281 to 296). ID2 and ID3 both contained an 11-aa deletion at positions between aa 327 and 337 or aa 370 and 380, respectively. The genes with deletions were made by overlapping PCRs as described previously (21). The primers used in this study are listed in Table 1. All constructs were confirmed by sequencing.
FIG. 4.
Expression of capsid proteins with deletions in the P domain. (A) Schematic representation of mutants with C-terminal deletions (CD1 to -4) or internal deletions (ID1 to -3) at or near the beginning of the P domains. (B) Western analysis of the mutated capsid proteins with C-terminal deletions (top panel) or with internal deletions (bottom panel) using antibody against VA387 capsid. Samples were pellet resuspensions after centrifugation of the clarified cell lysates at 100,000 × g for 2 h. Arrows on the top panel indicate signals of the truncated proteins. The weak bands at ∼65 kDa on the bottom panel are unspecific signals from baculovirus. r387 is recombinant wild-type capsid protein of VA387 that served as a positive control (600 ng). Bac and Sf9 are wild-type baculovirus and uninfected insect cells, respectively, which served as negative controls. (C) Schematic representation of the constructs for P2 domain mutants (P2-A to P2-C). Rectangles indicate the locations of the dimeric interaction sites around the P2 subdomain predicted by a previous study (20). NGR and RGD indicate the positions of NGR and RGD motifs, respectively. (D) Western analysis of the truncated P proteins.
TABLE 1.
Primers used to generate constructs for expression of recombinant capsid proteins in insect cells or E. coli
| Name | Sequence (5′ to 3′)a | Sense | Enzyme | Construct(s) generated |
|---|---|---|---|---|
| P502 | GCACGGATCCATGAAGATGGCGTCGAATGAC | + | BamHI | S domain, CD1 to CD4 |
| P404 | AGTCAGCGGCCGCTTATAAGATCGGGACGCTGAATGG | − | NotI | S domain |
| P422 | GCACGGATCCTTCTTGGTGCCACCCACAGTT | + | BamHI | P domain |
| P409 | AGTCAGCGGCCGCTTATAATGCACGTCTGCGCCC | − | NotI | P domain |
| P400 | AGTCAGCGGCCGCTTACAGAACCCTACCTGTGTCTGG | − | NotI | CD1 |
| P401 | AGTCAGCGGCCGCTTAAGCTAGGTGCACATTATGACC | − | NotI | CD2 |
| P402 | AGTCAGCGGCCGCTTATGGAGTGAAGTGGACAGTTCC | − | NotI | CD3 |
| P403 | AGTCAGCGGCCGCTTAATGACTGCCTGCAATGTGGGT | − | NotI | CD4 |
| P405 | GGCACTACCCAGCTGCTGTCTGCTCATGACTATATAATGAATTTGGCA | + | None | ID1 |
| P406 | TGCCAAATTCATTATATAGTCATGAGCAGACAGCTGGGTAGTGCC | − | None | ID1 |
| P410 | CCTCTGGGAACTCCAGATTTCACCACAAGAGAGGATGGCTCG | + | None | ID2 |
| P411 | CGAGCCATCCACTCTTGTGGTGAAATCTGGAGTTCCCAGAGG | − | None | ID2 |
| P407 | GGCAGTGTTCAATCAACCACTACGAAATTCACCCCAGTCGGC | + | None | ID3 |
| P408 | GCCGACTGGGGTGAATTTCGTAGTGGTGTATTGAACACTGCC | − | None | ID3 |
| P471 | CCACCCACAGTTGAATTCAGAACTAAACCATTCACC | + | EcoRI | Chimera |
| P472 | GGTCAATGGTTTAGTTCTGAATTCAACTGTGGGTGG | − | EcoRI | Chimera |
| P475 | ACAGCCGAATTCTCAAAGACTAAGCCATTTACA | + | EcoRI | MOH P domain, chimera |
| P476 | AGTCAGCGGCCGCTTACTGAAACCTTCTGCGCCG | − | NotI | MOH P domain, chimera |
| P492 | GCACGGATCCTACTTGGTGCCACCAACAGTG | + | BamHI | MOH P domain |
| P493 | GCACGGATCCTTTTTAGTCCCTCCTACGGTG | + | BamHI | Norwalk virus P domain |
| P494 | GGACGCGGCCGCTTATCGGCGCAGACCAAG | − | NotI | Norwalk virus P domain |
| P503 | GCACGGATCCACTACCCAGCTGTCTGCTGTC | + | BamHI | P2-A |
| P504 | GCACGGATCCATTCCTTTGGAAAAGTTGTAC | + | BamHI | P2-B, P2-C |
| P505 | GCGAGCGGCCGCAGCTAGGTGCACATTATGACC | − | NotI | P2-A, P2-C |
| P506 | GCGTGCGGCCGCTGCTGGAGCTGCTTCTTGGTA | − | NotI | P2-B |
Sequences are from strain VA387, except those indicated (MOH or Norwalk virus).
Expression and purification of mutated capsid proteins in insect cells.
Mutated capsid proteins were expressed in insect cells, Spodoptera frugiperda (Sf9), using the Bac-to-Bac baculovirus expression system (Invitrogen) according to the manufacturer's manual as described previously (21). Infected cells were harvested 4 to 5 days postinfection. The cell lysates were centrifuged at 5,000 × g for 15 min to separate the cell debris. VLPs in the supernatant were purified by centrifugation at 100,000 × g for 150 min. For further purification of the VLPs, the resuspended pellets were separated using a sucrose step-gradient (10 to 50%) centrifugation, as described previously (13, 14). The purified mutant or wild-type VLPs were used to perform saliva binding assays and/or electron microscopic observation. Since the P domain mutant does not form VLPs, the supernatant from cell lysate was used to perform saliva binding assays. Capsid protein concentration was determined by quantitative Western analysis as described before (21).
Expression and purification of recombinant P domain in E. coli.
The DNA fragments encoding the P domains plus the hinge regions of strains VA387 (16), MOH (5, 6), and Norwalk virus (14, 15) were PCR amplified and cloned into the vector pGEX-4T-1 (Amersham Bioscience, Piscataway, N.J.) at the BamHI and NotI sites. After sequence confirmation by sequencing, the constructs were expressed in E. coli strain BL21 at room temperature overnight and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside. Purification of the recombinant proteins from bacteria was performed using glutathione-Sepharose 4 Fast Flow (Amersham Bioscience) according to the manufacturer's instructions. The glutathione S-transferase (GST)-P domain fusion protein was eluted by using glutathione (Amersham Bioscience), and the P proteins were cleaved directly from the beads by thrombin (Amersham Bioscience) at room temperature for 16 h. The P proteins were further purified to homogeneity by gel filtration using the size-exclusion column Superdex 75 (Amersham Bioscience). Alternatively, the eluted proteins were purified by fractional precipitation. Protein concentrations were measured spectroscopically or by quantitative Western analysis as described previously (21). The constructs for expressing three truncated P domains in E. coli were made by cloning the corresponding coding sequences of VA387 (aa 262 to 409 for P2-A, aa 245 to 460 for P2-B, and aa 245 to 409 for P2-C; see Fig. 4C, below) into the vector pGEX-4T-1 (Amersham Bioscience). The proteins were expressed and purified using a similar procedure as described for the full P domain.
Assay of NV capsid binding to HBGAs.
The binding of wild-type as well as mutated capsids to HBGAs was measured by saliva binding enzyme immune assays as described previously (9). The saliva samples used in these studies were selected from previous studies, which were determined by enzyme immune assays with monoclonal antibodies specific to human HBGAs (9). The genotypes of these saliva donors were not determined. In order to determine the binding affinity of the mutated capsids relative to that of the wild type, all capsids were assayed under the same dynamic conditions for their binding to HBGAs of A, B, and O type saliva within a comparable range of protein concentrations (0.01 to 5 μM). To confirm the binding properties of the isolated P domains, HBGA binding assays were also performed using synthetic A- and B-trisaccharide-bovine serum albumin conjugates (Glycorex AB, Lund, Sweden), which were used at a concentration of 5 μg/ml.
Detection of P domain dimers by BS3 cross-linker.
BS3 (bis [sulfosuccinimidyl] substrate; Pierce, Rockford, Ill.) is a chemical that can link two molecules of a dimer covalently together so that the dimer will show up as a higher-order signal on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. BS3 solutions with different concentrations (in 1× phosphate-buffered saline) were freshly prepared and were mixed with recombinant P protein (0.5 μg/μl). The mixtures were incubated at room temperature for 1 h. BS3 was then inactivated by 100 mM Tris (pH 7.4). The samples were analyzed on an SDS-PAGE gel and by Western blot analysis. Carbonic anhydrate and GST were used as negative and positive controls, respectively.
Denaturation and renaturation of P domain dimers.
To test the stability of P domain dimers and their ability to renature, samples were maintained at different pHs or high salt concentration for 1 h and then detected dimer by BS3. To remove the extreme pHs or high salt concentration, the treated samples were repeatedly diluted with 1× PBS (pH 7.4) and concentrated using Amicon or Microcon (Millipore, Bedford, Mass.). Samples were kept at room temperature for 1 h before cross-linking by BS3.
RESULTS
NV capsid P domain contains elements required for binding to viral receptor.
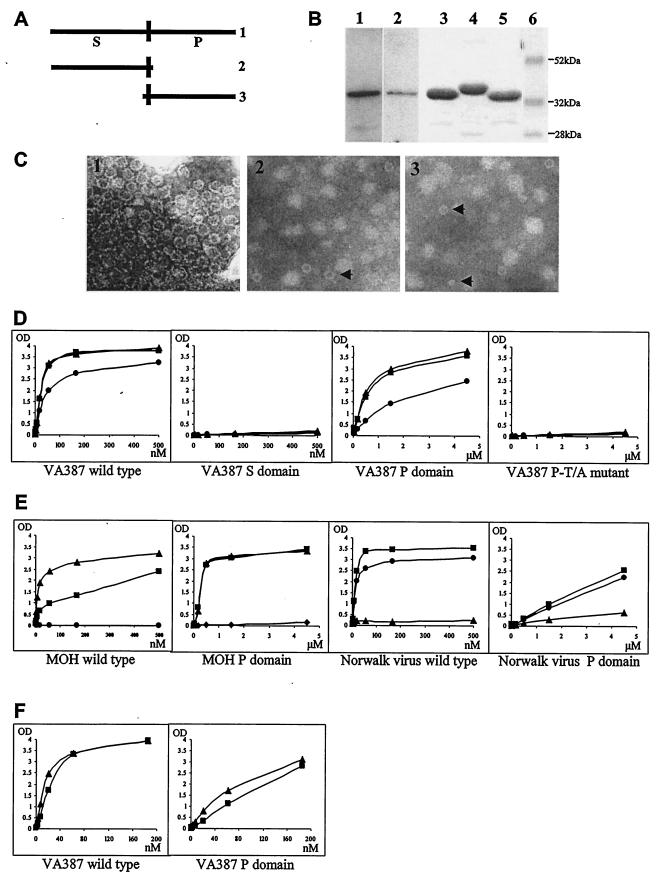
Our previous computational analysis and site-directed mutagenesis data showed that there is a binding pocket in the P domain of NV that is responsible for binding to human HBGAs (21). To determine whether the P domain alone is responsible for the binding, two truncated capsid proteins containing the S or P domains of VA387, respectively, were constructed (Fig. 1A). When these truncated proteins were expressed in the insect cells, the S protein formed small, thin-layer VLPs (Fig. 1C) similar to those observed for the Norwalk virus recombinant S protein (2), but these VLPs did not bind human HBGAs. In contrast, the P domain did not form VLPs, but it bound to the viral receptors with the same binding pattern as that of the intact viral capsid (Fig. 1D).
FIG. 1.
Expression of the S and the P domains of NV capsid proteins, as well as their binding affinities to HBGAs. (A) Schematic representation of the S and the P domain mutants, showing wild-type capsid protein of VA387 (1), S domain mutant (2), and P domain mutant (3). (B) Expression of the truncated capsid proteins in insect cells (lanes 1 and 2) or in E. coli (lanes 3 to 5). Lanes 1 and 2, immunodetection of the S domain (lane 1) and the P domain (lane 2) using antibody against VA387 capsid. Lanes 3 to 5, the P domains of VA387 (lane 3), MOH (lane 4), or Norwalk virus (lane 5) after purification and separation on SDS-PAGE gels stained with Brilliant Blue (G-250). Lane 6, prestained protein marker (low range; Bio-Rad). (C) Wild-type (1) and the smooth, thin-layer VLPs (indicated by arrows) from the S domain mutant (2 and 3) of VA387. Magnification, ×31,500. (D and E) Binding curves of wild-type and truncated capsid proteins to A, B, and H antigens. The nonsecretor saliva was used as a negative control, and no binding was observed in any of the mutants tested (data not shown). (F) Binding curves of VA387 wild-type capsid and P domain to synthetic A- or B-trisaccharides. Data were averaged from at least three independent experiments. ▪, A antigen; ▴, B antigen; •, H antigen (type O saliva).
Due to the low yield of the recombinant P protein in the insect cell culture (Fig. 1B), we also expressed the protein in E. coli. The expressed GST-fusion proteins were insoluble when the culture was induced at 37°C. However, the majority of the P proteins became soluble when the expression was carried out at room temperature. High yields of the proteins were obtained after cleavage of the P protein from the GST tag using thrombin proteinase (Fig. 1B). Both the fused and the released P proteins revealed a typical binding pattern with the A, B, and H antigens as the intact viral capsid (Fig. 1D; see also Fig. 3B, below), providing useful materials for further study of the mechanism of virus-receptor interaction. To confirm the above observation, binding assays were also performed using synthetic A- and B-trisaccharide-bovine serum albumin conjugates (Glycorex AB), which revealed a similar result (Fig. 1F). These data indicated that the P domain is responsible and contains essential components for binding to HBGA receptors.
FIG. 3.
GST tag affects binding affinities of the fused P domain to HBGAs. (A) Western blot analysis of the GST-P fusion protein before (lanes 1 to 3) and after (lanes 5 to 7) thrombin cleavage using pooled antibodies against capsids of VA387, MOH, and Norwalk virus. The proteins before and after cleavage are at the same molar concentration. Lanes 1 and 5, P protein of VA387; lanes 2 and 6, P protein of MOH; lanes 3 and 7, P protein of Norwalk virus. Lane 4 is protein standard. (B to D) Binding curves of GST-P fusion protein before and after thrombin digestion. Data were averaged from at least three independent experiments. ▪, A antigen; ▴, B antigen; •, H antigen (type O saliva). (E) Binding curves of GST alone served as a negative control.
In a previous study, our investigators observed that mutation of a single amino acid, threonine (T302) at the binding pocket of VA387, to an alanine resulted in a total loss of binding to A, B, and H antigens (21). To prove the requirement of this amino acid in receptor binding, a P domain mutant with the same mutation was constructed. As expected, this mutant completely lost its ability to bind to A, B, and H antigens (Fig. 1D), confirming the importance of T302 for binding to the viral receptors.
To verify that the results described above are common to other NV strains, the P domains of strains MOH and Norwalk virus, each representing a different receptor binding pattern (9), were also expressed in E. coli. High yields of soluble proteins were obtained for both strains using the same expression conditions as those for strain VA387 (Fig. 1B). The recombinant P proteins of these two strains also revealed typical binding patterns as their parental capsid: the MOH P protein bound to A and B but not H antigens, and the Norwalk virus P protein bound to A and H, but not B antigens (Fig. 1E). These data further support our conclusion that the P domain contains essential elements required for NV capsid binding to viral receptors.
A further question is whether the S domain of one strain is compatible structurally and functionally with the P domain of another strain representing a different receptor binding pattern. To answer this question, we constructed a chimeric capsid with the S domain of VA387 and the P domain of MOH using the baculovirus expression system (Fig. 2A). Among five trials of expression of this protein in insect cells, two trials resulted in VLP-like structures as the expressed chimeric protein migrated into the sucrose gradient, with the peaks containing 30 and 40% sucrose (Fig. 2B, left panel). Examination of this peak fraction by electron microscopy revealed scattered VLP-like structures (Fig. 2C). However, in the other three trials, the protein only migrated to fractions containing 10 to 20% sucrose (Fig. 2B, right panel), suggesting that while the protein did not form typical VLPs it formed some types of aggregates, because otherwise it would not enter the gradient. Saliva binding assays of this protein revealed receptor binding patterns identical to those of MOH, which bind to A and B but not H antigens (Fig. 2D). Based on these data, we conclude that heterologous S and P domains may not be fully compatible for VLP formation, but the interaction between the S and the P domains does not affect receptor binding, and that the P domain is sufficient for strain specificity.
FIG. 2.
Chimeric capsid and its binding pattern to HBGAs. (A) Schematic representation of chimeric capsid protein containing the S domain of VA387 and the P domain of MOH. (B) Immunodetection of the chimeric protein in fractions from sucrose gradient after separation on SDS-PAGE gels using antibody against VA387 capsid. (Left) The signals occurred in fractions containing 20 to 40% sucrose, with peak intensity between 30 and 40% sucrose. (Right) Signals occurred in fractions containing 10 to 20% sucrose. Arrow shows the direction of sedimentation. (C) Electron microscopic pictures showing the VLP-like structures formed by the chimeric capsid protein. Magnification, ×31,500. (D) Binding curves of the chimera and its parental capsids to A, B, and H antigens. Data were averaged from at least three independent experiments. ▪, A antigen; ▴, B antigen; •, H antigen (type O saliva).
In the studies of receptor binding of the P proteins from the three NV strains, we noticed that the binding affinities of the P domain were affected by the GST tag, and such effects were not the same for individual strains. For MOH, the GST-fusion protein had significantly reduced binding activities to A and B antigens compared with the released P protein (Fig. 3C). For Norwalk virus and 387, however, the GST tag had a positive effect, although mild for 387 (Fig. 3B and D). These effects clearly are not expected to occur under the natural conditions of infected cells; however, this observation is important, because a similar effect could occur from the S domain, since it is linked with the P domain in forming the viral capsid.
An intact P domain may be necessary for binding to HBGAs.
To further pinpoint the binding domain, three sets of deletion mutants of 387 capsids were constructed (Fig. 4A and C). The first set included four mutants with C-terminal deletions (CD1 to CD4), while the second set included three mutants with internal deletions (ID1 to ID3) of 11 to 16 aa at or near the N terminus of the P domain. The third set included three mutants of truncated P domains, each containing the P2 subdomain plus the NGR motif (P2-A) or one (P2-B) or both (P2-C) predicted dimeric interaction sites near the two ends of the P2 subdomain, respectively. These mutants were expressed in insect cells (CD and ID mutants) or in bacterial expression system (P2 mutants). They all expressed well except for mutant CD2 (Fig. 4B). All the mutants with the S domain formed VLPs as determined by high-speed centrifugation analysis (data not shown). However, none of these 10 mutants showed detectable binding activities to HBGA receptors (data not shown). In characterizing these mutant proteins, we have noticed that all mutants with a C-terminal deletion were easily degraded, as shown by multiple smaller molecular mass bands on the SDS gels (Fig. 4B and D). Thus, we conclude that an intact P domain is necessary for receptor binding, although we still cannot exclude the possibility of small dispensable sequence or gaps for the binding activity. The C terminus of the P domain seems to be more important for the stability of the capsid protein.
The E. coli-expressed P protein forms dimers.
Based on the crystal structure of the Norwalk virus capsid, it has been suggested that the Norwalk virus capsid protein forms dimers and the P domain facilitates this dimer formation (20). In this study, we tested whether the isolated P domain can form dimers by conducting cross-linking experiments using BS3 substrate (Pierce) as the cross-linker. As show in Fig. 5B, C, and D, a considerable amount of the P proteins of the three strains appeared as dimers in the SDS gels, with a typical dose response with the cross-linker. Under a ratio of 0.45 BS3 to 1 P protein molecule (3 μM BS3), about 40% of the proteins were linked as dimers (see lanes for 3 μM). The GST protein that is known to form dimer showed the same results, while as a negative control, carbonic anhydrate did not show any dimer signal (Fig. 5A, GST and CA lanes). These results indicated that the cross-linking experiment is specific, and the isolated P domain forms dimer in solution. A higher concentration of BS3 resulted in a higher percentage of dimer signal but also lead to higher-order signals, which we believe were artifact. Further studies by analytical gel filtration also confirmed the results. A single peak of the P protein with a retention volume consistent with a molecular mass of 50 to 60 kDa was observed (Fig. 5E). There were no higher- or lower-order peaks in the chromatographic experiments, suggesting that the P domain exists exclusively as a dimer in solution.
FIG. 5.
Isolated P domains of NVs form dimers. (A) Brilliant Blue-stained SDS-PAGE gel showing that carbonic anhydrate (CA) did not show dimer signal, but GST and the P domain formed dimers after cross-linking by BS3 (3 to 30 μM). The GST lane shows that there was no higher-order signal of GST without BS3 cross-linking. (B to D) Western blot analysis of dimers formed by recombinant P domain of VA387 (B), MOH (C), and Norwalk virus (D) after cross-linking by BS3 using antibodies against capsids of VA387, MOH, and Norwalk virus, respectively. Concentrations of BS3 are indicated above each lane. Protein loaded on each lane is ∼800 ng. (E) Chromatograph of gel filtration of the P protein of VA387. The P proteins at ∼4 mg/ml in 700 mM NaCl at pH 7.5 were loaded on a calibrated size-exclusion column (Superdex 75). A single peak of the P protein with a retention volume consistent with a molecular mass of 50 to 60 kDa was observed, as indicted by the molecular mass markers.
The P domain dimer is stable over a wide range of pH values.
NVs are gastrointestinal pathogens and are believed to be able to survive under the acidic conditions of the human gastrointestinal tract. To test whether the P domain dimer can survive under the same conditions, we performed stability experiments by incubating the recombinant P protein of VA387 in solutions at different pH. The experiments showed that the majority of dimers survived incubation at pH 3 to 11 for at least 60 min (Fig. 6A). Immunodetection revealed that at least 10% of the protein remained as dimer after incubation of the protein in a solution at pH 2.0 (Fig. 6A, right panel). These observations were correlated with the receptor binding results of the protein. Considerable binding activities were observed after the incubation of the proteins at pH levels from 2 to 11, with optimum binding at pH 5 to 6 (Fig. 6B), indicating that the P protein remains functional under these conditions.
FIG. 6.
P protein dimer is stable over a wide range pH and under high-salt conditions. (A) Detections of P protein dimer (D) and monomer (M) at different pHs. Recombinant VA387 P proteins were treated with different pH solutions and then were cross-linked by 30 μM BS3. The treated samples were separated on SDS-PAGE gels. (Left) A Brilliant Blue-stained SDS-PAGE gel. (Right) Western analysis of the P protein after treatments at different pHs and BS3 using antibody against VA387 capsid protein. The different pH solutions are indicated for each lane. Stand, prestained protein standard (broad range; Bio-Rad); control, untreated and un-cross-linked P protein. (B) Binding assay of the pH-treated P protein to A-type saliva. (C) Immunodetection of P protein dimers after denaturation and/or renaturation. P proteins were denatured (de) by 8 M urea (U), 6 M guanidine (G), or extreme high or low pHs. Before (de) and after (re) removal of the denaturation factors, samples were cross-linked by 30 μM BS3. The treated samples were separated on an SDS-PAGE gel. Control, untreated and un-cross-linked P protein; U-de, 8 M urea-treated P protein; U-re, renatured P protein after removal of the 8 M urea; G-re, renatured P protein after removal of the 6 M guanidine; pH 1.5-de, protein denatured by pH 1.5 solution; pH 1.5-re, renatured protein after neutralization from pH 1.5; pH 12.5-de, protein treated by pH 12.5 solution; pH 12.5-re, renatured protein after neutralization from pH 12.5. The BS3 concentration was 30 μM. Each lane contained ∼800 ng of P protein.
The observed correlation between dimer formation and receptor binding prompted us to test if the loss of binding of mutant P T/A of VA387 (21) was due to a failure of dimer formation. Cross-linking experiments indicated that the mutant formed dimers (data not shown). Although this result does not exclude the possibility of involvement of dimer formation in receptor binding, it suggests that threonine T320 was not involved in dimer formation, which is a significant addition to our previous observations.
The P domain dimer survives treatment with high concentrations of denaturing reagents.
NV is one of the unique pathogens that particularly cause infection following environmental surface contamination. We thus characterized the stability of the P protein dimer against high salt concentration. As shown in Fig. 6C, at least 30% of the protein was still dimeric after a 60-min 8 M urea treatment (lane U-den). After removing the urea and then allowing the P protein to renature for 60 min at room temperature (see Materials and Methods), more than 70% of the protein appeared as dimers or polymers (lane U-re). The protein treated with 6 M guanidine showed a similar result (lane G-re). In both cases, the dimer seemed to be more stable than the polymer (compare lane U-den to lane U-re), further indicating that the dimer may be a natural form of the P domain. Similar renaturation was also observed after the protein was treated with strong pH, and the renaturation seemed to be more efficient under an acidic rather than a basic condition (compare lane pH 1.5-re to lane pH 12.5-re). In conclusion, our data suggested that the P domain dimer is highly stable even at extremes of pH and in the presence of denaturants.
DISCUSSION
NVs are the most important viral pathogens of nonbacterial epidemics of acute gastroenteritis affecting people in both developed and developing countries. In the United States, NVs account for over 90% of epidemic outbreaks (3, 4). The pathogenesis of NVs remains unclear, mainly because there are no animal or cell culture models available for replicating the viruses. The recent finding that NVs recognize HBGAs as receptors provided a new approach to study the host specificity of NVs. In this study, we have shown that the P domain of the NV capsid protein contains the essential elements for receptor binding and that the bacterially expressed P proteins form dimers that bind to viral receptors. These results suggested that further characterization of the P domain would provide useful information for understanding the interaction between the viral capsids and HBGA receptors and that the bacterially expressed P proteins could serve as a valuable source of materials for this purpose. Further study of the crystal structure of the P proteins could also generate additional data that may lead to a description of the interface between the receptors and the binding pocket, as suggested by our group's previous study (21).
A growing literature has shown that HBGAs play an important role in the host specificity for many viral and bacterial pathogens. The NV-receptor interaction provides a unique system among these pathogens. NVs are genetically and antigenically diverse, and different NVs recognize different receptors. So far, six binding patterns of NVs to HBGAs have been described according to host ABO, secretor, and Lewis blood types (9). The facts that the NV capsid is structurally simple compared with other human viral and bacterial pathogens and that each of the NV binding patterns involves a specific receptor-epitope interaction make it an excellent model system for studying the host-pathogen interaction for NV as well as for other HBGA-related pathogens.
The fact that the P domain may contain elements for strain-specific binding has been suggested by its high sequence variation compared with other regions of the capsid protein and by its surface location on the capsid, as determined by the crystallography study of Norwalk virus capsid (20). In this study, we have provided direct evidence that the P domain participates in receptor binding by mutational analyses, including a series of deletion mutants of the capsids, truncated capsid proteins of the S or P domains, respectively, a chimeric capsid with the S and P domains from two different parent strains, and expression of functional P proteins from three NV strains. Our data clearly demonstrate that the P domain is responsible for NV receptor binding. In addition, we have shown that the recombinant P domain spontaneously forms dimers, which complements previous observations made during the crystallographic analysis of Norwalk virus (20).
This is the first report that a truncated NV capsid protein can bind to viral receptors and that the binding does not require formation of VLPs. In a previous study, our investigators showed that a binding pocket in the P domain is responsible for viral receptor binding and that the formation of this pocket involves only intramolecular interactions (21). We noticed that the binding efficiencies of the P proteins to various HBGAs were 10- to 50-fold lower than those of the intact capsids, based on the comparisons of the concentrations of different proteins needed to reach the maximum reactivities in the enzyme immune assays. One possible reason is that a VLP is a big complex that is composed of 180 capsid protein monomers. As a result, a VLP should be 90-fold more sensitive to detection in the receptor binding assay compared with single P protein dimer. Thus, capsid formation could have an amplification effect in the binding assay. However, it does not exclude the possibility of misfolding of the isolated P domain.
We have also shown that the S domain of VA387 forms smooth, thin-layer particles, similar to those previously reported for the Norwalk virus S domain (2), despite Norwalk virus and VA387 belonging to two different genogroups. Moreover, our demonstration that the NV P protein spontaneously forms dimers confirms the structural independence of the P domain from the S domain, in which the P domain is linked with the S domain by a hinge that allows the P domain to freely form dimers during capsid assembly (20). The high stability of the P dimer also agrees with the prediction of multiple interaction sites between two P protein monomers. The resistance of the dimer to acidic conditions provides insight into why the virus pathogen survives in the human intestinal tract. The high resistance to a strong denaturing reagent is also consistent with the fact that NV commonly causes infection through environmental surface contamination, although the stability of intact viral capsids of NV remains to be determined.
The facts that the NV P domain contains essential elements for receptor binding and that the protein spontaneously forms dimers indicate that a crystallographic analysis of the P proteins would be informative. As the first step toward this goal, the E. coli-expressed recombinant P proteins of the three NOR strains were used in crystallization screens, and we have been successful in obtaining small crystals for the P proteins of all three strains. Although further improvement of the quality and size of the crystals is necessary for a crystallographical analysis, the ready formation of crystals of the P proteins is encouraging. A comparison of the crystal structures of the P proteins representing different binding patterns will provide a detailed three-dimensional view of the binding pocket as well as the structure for the strain specificity. In addition, a crystal structure of the P protein in the presence of receptors will generate a direct image of the interface between the ligand and the receptor. Finally, the crystal structure of the P protein may provide additional new information that may not be possible for the crystals of the intact VLPs, because the crystallography data collected from the crystals of single P proteins or P protein dimers are expected to have a higher resolution than those for the full capsids.
Acknowledgments
We thank Tibor Farkas for helpful discussions, and we thank Irene Hofmann and Dan Song for technical support.
The research described in this article was supported by the National Institute of Allergy and Infectious Diseases (RO1 AI37093-6).
REFERENCES
- 1.Bertolotti-Ciarlet, A., S. E. Crawford, A. M. Hutson, and M. K. Estes. 2003. The 3′ end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein VP1: a novel function for the VP2 protein. J. Virol. 77:11603-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertolotti-Ciarlet, A., L. J. White, R. Chen, B. V. Prasad, and M. K. Estes. 2002. Structural requirements for the assembly of Norwalk virus-like particles. J. Virol. 76:4044-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 4.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 5.Farkas, T., T. Berke, G. Reuter, G. Szucs, D. O. Matson, and X. Jiang. 2002. Molecular detection and sequence analysis of human caliciviruses from acute gastroenteritis outbreaks in Hungary. J. Med. Virol. 67:567-573. [DOI] [PubMed] [Google Scholar]
- 6.Farkas, T., S. A. Thornton, N. Wilton, W. Zhong, M. Altaye, and X. Jiang. 2003. Homologous versus heterologous immune responses to Norwalk-like viruses among crew members after acute gastroenteritis outbreaks on 2 US Navy vessels. J. Infect. Dis. 187:187-193. [DOI] [PubMed] [Google Scholar]
- 7.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale, A. D., S. E. Crawford, M. Ciarlet, J. Green, C. Gallimore, D. W. Brown, X. Jiang, and M. K. Estes. 1999. Expression and self-assembly of Grimsby virus: antigenic distinction from Norwalk and Mexico viruses. Clin. Diagn. Lab. Immunol. 6:142-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, P., T. Farkas, S. Marionneau, W. Zhong, N. Ruvoen-Clouet, A. L. Morrow, M. Altaye, L. K. Pickering, D. S. Newburg, J. LePendu, and X. Jiang. 2003. Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J. Infect. Dis. 188:19-31. [DOI] [PubMed] [Google Scholar]
- 10.Hutson, A. M., R. L. Atmar, D. Y. Graham, and M. K. Estes. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 185:1335-1337. [DOI] [PubMed] [Google Scholar]
- 11.Hutson, A. M., R. L. Atmar, D. M. Marcus, and M. K. Estes. 2003. Norwalk virus-like particle hemagglutination by binding to H histo-blood group antigens. J. Virol. 77:405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang, X., D. Y. Graham, K. N. Wang, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, X., D. O. Matson, G. M. Ruiz-Palacios, J. Hu, J. Treanor, and L. K. Pickering. 1995. Expression, self-assembly, and antigenicity of a Snow Mountain agent-like calicivirus capsid protein. J. Clin. Microbiol. 33:1452-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, X., W. M. Zhong, T. Farkas, P. W. Huang, N. Wilton, E. Barrett, D. Fulton, R. Morrow, and D. O. Matson. 2002. Baculovirus expression and antigenic characterization of the capsid proteins of three Norwalk-like viruses. Arch. Virol. 147:119-130. [DOI] [PubMed] [Google Scholar]
- 17.Lew, J. F., A. Z. Kapikian, X. Jiang, M. K. Estes, and K. Y. Green. 1994. Molecular characterization and expression of the capsid protein of a Norwalk-like virus recovered from a Desert Shield troop with gastroenteritis. Virology 200:319-325. [DOI] [PubMed] [Google Scholar]
- 18.Lindesmith, L., C. Moe, S. Marionneau, N. Ruvoen, X. Jiang, L. Lindblad, P. Stewart, J. LePendu, and R. Baric. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548-553. [DOI] [PubMed] [Google Scholar]
- 19.Marionneau, S., N. Ruvoen, B. Le Moullac-Vaidye, M. Clement, A. Cailleau-Thomas, G. Ruiz-Palacois, P. Huang, X. Jiang, and J. Le Pendu. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 21.Tan, M., P. Huang, J. Meller, W. Zhong, T. Farkas, and X. Jiang. 2003. Mutations within the P2 domain of Norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 77:12562-12571. [DOI] [PMC free article] [PubMed] [Google Scholar]