Abstract
The phosphoprotein (P) of vesicular stomatitis virus (VSV) is an essential subunit of the viral RNA-dependent RNA polymerase (RdRp) complex. It is phosphorylated at two different domains. Using defective interfering (DI) RNA or minigenomic RNA templates, we previously demonstrated that phosphorylation within the amino-terminal domain I is essential for transcription, whereas phosphorylation within the carboxy-terminal domain II is necessary for replication. For the present study, we examined the role of the phosphorylation of residues in these domains in the life cycle of VSV. Various mutant P coding sequences were inserted into a full-length cDNA clone of VSV, and the virus recovery, kinetics of growth, and mRNA and protein synthesis were examined. We observed that virus recovery was completely abolished when all three phosphate acceptor sites in domain I or both sites in domain II were replaced with alanine. Single or double mutations in domain I (with the exception of P60/64) or single mutations in domain II had no adverse effect on virus recovery. VSVP227, carrying alanine at position 227, showed reduced kinetics of virus growth but increased kinetics of viral mRNA synthesis in infected cells. More interestingly, this particular virus exhibited a significantly reduced cytopathic effects and apoptosis in infected cells, implying that P may be involved in these processes. Furthermore, we found that DI RNAs of different sizes were generated by high-multiplicity passaging of various mutant VSVs, indicating that the viral RdRp may play a significant role in the process of DI particle generation. Taken together, our results suggest that the phosphorylation of residues in domains I and II of VSV P is indispensable for virus growth.
Vesicular stomatitis virus (VSV) is the prototypic rhabdovirus and has a negative-stranded RNA genome of 11,161 nucleotides (50). Within the virus and in infected cells, the genomic RNA is tightly encapsidated by the nucleocapsid (N) protein, forming the viral nucleocapsid, which serves as the template for transcription and replication by the associated viral RNA-dependent RNA polymerase (RdRp) (50). The RdRp is a complex that contains the virally encoded large (L) protein and the phosphoprotein (P protein). While the L protein contains the catalytic center for the polymerization of nucleotides, the P protein serves as an essential subunit of the RdRp. The P protein is multifunctional: in addition to playing a major role in polymerase functions, it binds to the L protein and stabilizes it from proteolytic degradation (5, 12), it complexes with the newly synthesized N protein for the efficient encapsidation of nascent RNA (9, 39, 48), and it interacts with terminal sequences of the viral genome for viral RNA synthesis (27, 29).
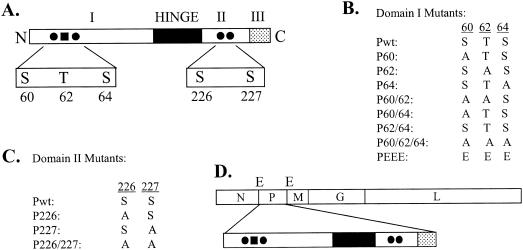
Through mutational and biochemical studies, three functionally homologous domains have been identified in the P proteins (Fig. 1A) of both the Indiana (PI) and New Jersey (PNJ) serotypes of VSV (20, 47). The amino-terminal domain I (residues 1 to approximately 150) is highly acidic and phosphorylated. Domain II, in the carboxy-terminal region spanning residues 210 to about 244, is also phosphorylated. The extreme carboxy-terminal region, domain III, spans 21 to 25 residues and contains a number of basic residues. A hypervariable region (called a hinge) of approximately 50 to 60 amino acids is located between domains I and II, but its function is unknown.
FIG. 1.
(A) Domain structure of P protein. The P protein, with three functionally defined domains (I, II, and III) and a hinge region, is shown. The phosphate acceptor sites Ser-60, Ser-64, Ser-226, and Ser-227 are indicated by solid circles and Thr-62 is indicated by a solid square. (B) Various P protein domain I mutants used for the present study. Substitutions of alanine (A) and glutamic acid (E) residues for serine (S) and threonine (T) residues in domain I are shown. (C) Domain II P protein mutants. (D) Genome organization of VSV. The protein coding regions of VSV genes N, P, M, G, and L are shown in rectangular boxes. E, EcoRV sites flanking the P gene.
The importance of phosphorylation in domain I of the P protein for transcription was elucidated by several in vitro transcription reconstitution studies (1, 3). Purified P protein expressed in Escherichia coli is neither phosphorylated nor active in transcription. In the presence of cellular casein kinase II, it becomes phosphorylated and functions in transcription in vitro (2, 22). By mutational and biochemical studies, the phosphate acceptor sites were mapped to Ser-59 and Ser-61 in PNJ and Ser-60, Thr-62, and Ser-64 in PI (6, 53, 54). Further studies suggested that the phosphorylation of these residues results in multimerization of the P protein, which in turn facilitates its binding to the L protein for transcription (7, 16, 17). Using a plasmid-based reverse genetics system (46), we demonstrated that the phosphorylation of these residues is critical for the function of the P protein in transcription but not in replication (44). In that study, we showed that a P protein in which all of the phosphate acceptor sites were altered to alanine was only marginally active in transcription. Alteration of these residues to glutamic acid or aspartic acid led to proteins that were as active as the wild-type (wt) protein in transcription, suggesting that a net negative charge in the region as a result of phosphorylation is crucial for transcriptional activation of the protein (44). A separate study also reported low levels of transcription under in vitro conditions with the same phosphorylation-defective P mutant (53).
The phosphorylation sites in domain II have been mapped to Ser-236 and Ser-242 in PNJ and Ser-226 and Ser-227 in PI (6). For PNJ, domain I residues must be phosphorylated prior to the phosphorylation of domain II residues (3), but for PI, domain II residues can be phosphorylated without the prior phosphorylation of domain I residues (6). Since phosphorylation in domain I is essential and sufficient for transcription, it seems that the phosphorylation of domain II may not be involved in transcription (16, 38). This contention is further supported by the demonstration that mutations at the domain II phosphate acceptor sites do not affect the transcription functions of the P protein, although its function in replication is severely compromised (26). Most recently, Gupta et al. (23) observed that a transcriptionally inactive P mutant can still form a tripartite complex with the N and L proteins and can function as a replicase for the synthesis of full-length genomic RNA. This study, along with other studies (8, 26, 44), suggests that the transcriptase and replicase complexes of VSV are two distinct entities.
Phosphorylation of the P protein may also play a role in the virus life cycle, during which an infectious unit is assembled in an orchestrated manner involving the transcription, replication, and assembly of functional nucleocapsid templates. Since the P protein interacts with the terminal sequences of the viral genome (27, 29), mutations at the phosphorylation sites might alter such interactions. In the context of the full-length viral genome, such altered polymerase interactions might result in reduced viral growth and potentially lead to the generation of different types of defective interfering (DI) particles. In the case of respiratory syncytial virus (RSV), mutations at the P phosphorylation sites led to highly impaired growth of the virus in animals, although such mutations had mixed effects on the in vitro replication of RSV in different cell types (36). On the other hand, phosphorylation-defective mutants of Sendai virus (SeV) and the wt virus showed no significant differences in growth in cell culture and in mice (25).
For the present study, we examined whether infectious VSV particles containing mutations in the phosphate acceptor sites could be recovered. Our results show that VSV particles containing mutations in phosphate acceptor sites Ser-60, Thr-62, and Ser-64 or in Ser-60 and Ser-64 in domain I of the P protein could not be recovered, although VSV particles containing all other combinations of mutations were readily recovered. Single mutations at residue Ser-226 or Ser-227 in domain II also led to the generation of infectious viruses. However, mutations at both of these residues were detrimental. Furthermore, we observed that VSV particles containing a mutation at residue 227 (VSVP227) were less cytopathic and induced an increased synthesis of mRNAs in infected cells compared to wt VSV.
MATERIALS AND METHODS
Cells and viruses.
The baby hamster kidney (BHK-21) cells used for this study were maintained in Eagle's minimal essential medium (MEM) containing 5% fetal bovine serum (FBS). VSV (Indiana serotype, San Juan strain) was propagated in BHK-21 cells for the preparation of virus stock. A recombinant vaccinia virus (vTF7-3) expressing bacteriophage T7 RNA polymerase (15) was grown and titrated by plaque assay in BHK-21 cells. The production and purification of DI T particles (33) were described previously (46).
Plasmids.
The plasmids pN, pP, and pL, carrying the coding sequences of the N, P, and L proteins of VSV (Indiana serotype, San Juan strain) under the control of the T7 RNA polymerase promoter, as well as plasmid pVSVFL(+), containing an antigenomic-sense full-length VSV sequence (a gift of J. K. Rose, Yale University, New Haven, Conn.), have been described previously (32, 46). The plasmids encoding the mutant P proteins of VSV (Indiana serotype, Mudd-Summers strain) have been described in previous studies (26, 44). Additional P protein mutants were generated by PCR-based mutagenesis performed by the megaprimer method (52). The mutant P genes were amplified from their respective plasmids by using Pfu polymerase (Stratagene). The primers used for amplification were as follows: P(+), 5′ATATATGGTACCGATATCATGGATAATCTCACAAAAGTTCGTGAG3′(forward), and P(−), 5′ATATATCTGCAGATATCTGTTACTTTTTTTCATAGTCTACAGAGAATATTTG3′ (reverse),both of which contain EcoRV sites (underlined). The PCR product was purified, digested with EcoRV, and cloned into pVSVFL(+) that had been digested with the same enzyme. Clones were sequenced by cycle sequencing for the presence of the designed mutations in the P gene.
Transfection, recovery, and identification of mutant viruses.
BHK-21 cells were grown in 60-mm-diameter tissue culture dishes to 80 to 90% confluency. The cells were infected with vTF7-3 at a multiplicity of infection (MOI) of 10 PFU/cell. After adsorption for 45 min at 37°C, the cells were washed and transfected with 4 μg of pVSVFL(+), 6 μg of pN, 2 μg of pL, and 4 μg of a wt or mutant pP plasmid by using Lipofectamine 2000 (Invitrogen). At 4 h posttransfection, the cells were washed twice and incubated with 2 ml of fresh medium containing 5% FBS for 48 h. Culture supernatants were harvested, clarified at 16,000 × g for 20 min, and used to infect fresh BHK-21 cells for 14 to 18 h to generate stock viruses. Rescued viruses in the culture supernatants from transfected cells and the amplification passage (P1) were titrated by plaque assays. Stocks of mutant viruses were prepared by infecting BHK-21 cells with P1 viruses at an MOI of 0.1. For confirmation of the identities of the mutant viruses, BHK-21 cells were infected with each virus at an MOI of 10. At 6 h postinfection, total RNAs from the cells were isolated by using TRIzol (Invitrogen). Reverse transcription-PCR (RT-PCR) was performed with Moloney murine leukemia virus reverse transcriptase and Taq DNA polymerase (Invitrogen). PCR products were analyzed in agarose gels and sequenced directly for the presence of the mutations. For sequencing of the P gene of the mutant viruses in its entirety, the RT-PCR products were cloned into pBR322 and five clones of each mutant were sequenced.
Determination of single-step growth kinetics.
BHK-21 cells were grown in 60-mm-diameter tissue culture dishes to 90% confluency. Cells were infected with either wt or mutant virus at an MOI of 20. After adsorption for 45 min, the cells were washed twice with phosphate-buffered saline, and 2.5 ml of fresh medium containing 2% FBS was added. Aliquots of 120 μl of culture supernatant were collected at various times postinfection, clarified at 16,000 × g for 5 min, and frozen at −85°C. The amount of virus present in the culture supernatant was determined by plaque assay.
High-multiplicity passaging of mutant viruses.
BHK-21 cells were grown in 60-mm-diameter tissue culture dishes to 90% confluency. The cells were infected with wt and mutant viruses at an MOI of 10. The cells were incubated at 37°C, and the development of a cytopathic effect (CPE) was monitored for a period of 12 to 24 h. When complete CPE developed, the culture supernatant was harvested and clarified. One hundred microliters of the clarified supernatant from this passage (P1) was used to infect fresh BHK-21 cells as described above. At 16 to 20 h postinfection, 100 μl of clarified supernatant from the next passage (P2) was used to infect fresh BHK-21 cells. The process of high-multiplicity passaging was repeated multiple times to generate P3 to P10 supernatants. Supernatants from each passage were clarified and stored in small aliquots at −85°C until further use.
Metabolic labeling of viral RNAs and electrophoretic analysis.
BHK-21 cells were grown in 35-mm-diameter tissue culture dishes to 90% confluency. Cells were infected with a wt or mutant virus at an MOI of 10 or 100. For an examination of mRNA synthesis at early times after infection, cells infected with the higher MOI were treated with actinomycin D at a concentration of 20 μg per ml for 1 h immediately after infection and the RNAs were radiolabeled with 25 μCi each of [3H]uridine and [3H]adenosine per ml of Dulbecco's modified Eagle medium for 1 h in the presence of actinomycin D. For an examination of mRNA synthesis at late times after infection, cells infected with the lower MOI were treated and labeled similarly at 7 to 8 h postinfection. After labeling, total RNAs from the cells were extracted by the use of TRIzol, analyzed by electrophoresis in acid agarose-urea gels, and detected by fluorography as described earlier (46). The fluorograms were scanned and RNA bands were quantitated with the VersaDoc imaging system and Quantity One software (Bio-Rad).
To analyze viral RNA synthesis in cells infected with culture supernatants from high-multiplicity passages, we infected BHK-21 cells grown as described above with an equal volume of cell culture supernatant collected from each passage. After 45 min of adsorption, the cell monolayers were washed and incubated in medium. The infected cells were labeled for 4 h with [3H]uridine and [3H]adenosine in the presence of actinomycin D as described above. Cytoplasmic extracts were prepared as described previously (46) and total RNAs recovered from one-third of the extracts were analyzed as described above. For analysis of the replication products, the viral nucleocapsids were immunoprecipitated from the remaining cytoplasmic extracts, and RNAs recovered from the immunoprecipitated nucleocapsids were analyzed as described above.
Analysis of viral proteins.
BHK-21 cells grown in 35-mm-diameter tissue culture dishes to 90% confluency were infected with a wt or mutant virus at an MOI of 10 or 100. For an examination of protein synthesis at early times after infection, cells infected with the higher MOI were incubated with methionine- and cysteine-free Dulbecco's modified Eagle medium for 1 h immediately after infection and the proteins were radiolabeled with 30 μCi of Expre35S35S label (Perkin-Elmer) per ml of medium. For an examination of protein synthesis at late times after infection, cells infected with the lower MOI were labeled similarly at 7 to 8 h postinfection. After labeling, cell lysates were prepared, and the viral proteins were analyzed by electrophoresis in 10% polyacrylamide gels and detected by fluorography as described earlier (46).
To examine the phosphorylation status of P proteins, we infected parallel cultures of BHK-21 cells grown as described above with a wt or mutant virus at an MOI of 10. For orthophosphate labeling, infected cells were incubated with phosphate-free medium for 1 h prior to labeling with 100 μCi of [32P]orthophosphate (Perkin-Elmer) per ml of medium. Equal amounts of cell lysates from labeled and unlabeled cells were analyzed by electrophoresis as described above. Gels containing radiolabeled proteins were subjected to autoradiography for the detection of 32P-labeled P proteins. Equal quantities of cytoplasmic extracts from labeled and unlabeled cells were electrophoresed in parallel 10% polyacrylamide gels. The gel containing labeled samples was fixed, dried, and exposed for autoradiography. The gel containing unlabeled samples was subjected to Western blotting. The blots were incubated with rabbit anti-P polyclonal antisera and the protein bands were detected by chemiluminescence by use of an ECL kit (Amersham). The protein or 32P content was quantitated with Quantity One software as described above.
Apoptosis assay.
BHK-21 cells grown in 35-mm-diameter tissue culture dishes to 90% confluency were infected with a wt or mutant virus at an MOI of 0.5. After adsorption for 45 min, the cells were washed, fresh medium was added, and the cells were incubated at 37°C for 12 to 14 h. The medium was aspirated and the cells were trypsinized. The cells were washed twice in cold phosphate-buffered saline and adjusted to a density of 106 per ml in 1× annexin-binding buffer (Molecular Probes). A 100-μl cell suspension was mixed with 5 μl of annexin V-fluorescein isothiocyanate and 1 μl of propidium iodide (100 μg/ml) (Molecular Probes) and incubated at room temperature for 15 min. Finally, the cells were diluted with 400 μl of 1× annexin-binding buffer and analyzed by flow cytometry for live, apoptotic, and necrotic cells.
Indirect immunofluorescence.
BHK-21 cells were infected with an MOI of 0.5 of wt VSV or VSVP227. At 12 to 14 h postinfection, the cells were fixed with acetone-methanol (1:1) at −20°C and incubated with a polyclonal mouse anti-VSV antibody. The cells were incubated with the secondary antibody, anti-mouse Alexa-488 (Molecular Probes), and analyzed for immunofluorescence under a Nikon fluorescence microscope attached to a camera (Optronics).
RESULTS
To determine the role of the phosphorylation of residues in domains I and II of the P protein (Fig. 1A) of VSV in the virus replication cycle, we wanted to recover VSVs encoding single and multiple substitutions in the phosphate acceptor sites (Fig. 1B and C) of the P protein. We replaced the P protein coding region in pVSVFL(+) (32) with various mutant P protein coding regions. This was facilitated by the use of EcoRV sites that flank the coding region of the P protein in pVSVFL(+) (Fig. 1D). Mutations within the P coding region in various pVSVFL(+) plasmids were confirmed by sequencing. Plasmids encoding various mutant P proteins in pVSVFL(+) were then used along with three support plasmids (pN, pP, and pL) to recover various mutant VSVs by established procedures (32, 43, 45). Viruses encoding wt or mutant P proteins were designated VSVPwt, VSVP60, VSVP62, VSVP64, etc.
Recovery of VSVs containing mutant P proteins with single or double amino acid substitutions at the phosphorylation sites in domain I.
For initial transfection experiments, we used various mutant P plasmids as a component of a support plasmid mix to recover VSV; however, we were unable to recover VSV from the transfected cells. This was most likely due to the intrinsic low transcriptional activities of these mutant P proteins (44). We rationalized that the use of the wt P plasmid in the support plasmid mix would likely drive the transcription and replication of VSVFL templates at levels that would allow us to initially recover viruses from the transfected cells. Subsequent infection and replication of the virus produced from transfected cells would be dependent upon the activity of the encoded mutant P proteins. Therefore, to recover VSV containing a wt or mutant P gene, we transfected BHK-21 cells infected with vTF7-3 with various pVSVFL templates along with the support plasmids pN, pP, and pL. The supernatants from transfected cells were collected at 48 h posttransfection. To determine if infectious VSV was present in the supernatant, we used an aliquot of clarified supernatant to infect fresh BHK-21 cells. The CPE that is characteristic of VSV infections (the rounding of cells and subsequent detachment) was observed within 12 to 24 h postinfection, indicating that infectious VSV was present in the supernatant of transfected cells. In control experiments in which the pL plasmid was omitted from the support plasmid mix, no such CPE was detected. The results from several independent transfection experiments indicated that VSVs encoding all of the single and two double mutant P proteins (VSVP60, VSVP62, VSVP64, VSVP60/62, and VSVP62/64) could be rescued. VSVP60/64, carrying mutations for positions Ser-60 and Ser-64 of the P protein, could not be rescued even after 10 independent rescue attempts and also varying the amounts of support plasmid mix used during transfection. In addition, we attempted to detect VSVP60/64 by passaging the transfected cell supernatant into cells expressing the wt P protein in trans, but this attempt failed to detect any rescued VSVP60/64.
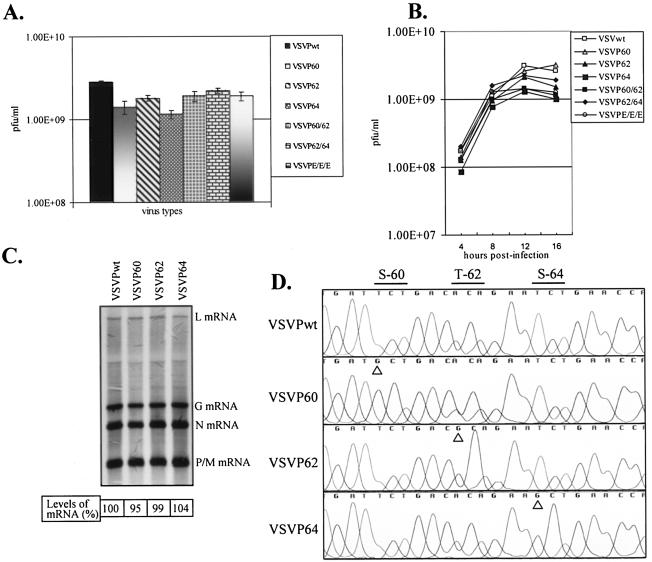
Since the maximal CPE was observed at different times postinfection with supernatants from transfected cells, we suspected that the supernatants contained various amounts of infectious VSV. The supernatants from transfected cells were amplified once in cells to generate a P1 stock virus. To examine the growth potential of various mutant viruses, we infected cells with each of the mutant viruses and determined the overall yield of virus by a plaque assay. The results (Fig. 2A) show that the recovered mutant viruses grew to titers that were similar to that of wt VSV, indicating that the overall growth potential of the mutant viruses was not adversely affected by the phosphorylation status of the encoded P protein. Even after multiple passages of the transfected cell supernatant, we failed to recover virus from cells transfected with pVSVFLP60/64. VSVPE/E/E, which contained glutamic acid residues in place of the phosphate acceptor sites in domain I, readily grew to titers similar to those of other viruses. Taken together, these results indicate that although the mutant P proteins possessed different levels of transcriptional activity in minigenome transcription experiments (44, 53), viruses encoding the mutant proteins, with the exception of VSVP60/64, could be rescued readily from transfected cells and grew to similar titers.
FIG. 2.
(A) Yield of various mutant viruses. BHK-21 cells were infected with mutant viruses at an MOI of 1, and the yield of virus in clarified supernatants from infected cells at 16 h postinfection was determined by plaque assay. Error bars showing standard deviations for five independent experiments are shown. (B) Single-cycle growth kinetics of viruses with substitutions in domain I residues. BHK-21 cells were infected with wt or mutant viruses at an MOI of 20, and culture supernatants were collected at the indicated time points. The viruses in the supernatants were quantitated by plaque assay. The averages of three independent experiments are presented. (C) Analysis of VSV mRNAs in cells infected with domain I mutants. BHK-21 cells were infected with VSVPwt, VSVP60, VSVP62, and VSVP64 at an MOI of 10. RNAs were radiolabeled at 7 to 8 h postinfection in the presence of actinomycin D, analyzed by electrophoresis, and detected by fluorography as described in Materials and Methods. The positions of the VSV mRNAs are indicated on the right. Numbers at the bottom show the average levels of mRNA from three independent experiments. (D) Confirmation of identity of mutant viruses. Total RNAs isolated from wt or mutant virus-infected cells were subjected to RT-PCR amplification of the P coding region. The products were directly sequenced by cycle sequencing. A chromatogram showing the presence of the desired mutations is presented. Specific nucleotide changes corresponding to the mutant proteins are marked with arrowheads.
P protein without phosphorylation in domain I residues cannot support the rescue of infectious VSV.
We had previously observed that a mutant P protein in which the phosphate acceptor sites Ser-60, Thr-62, and Ser-64 were altered to alanine was significantly defective in supporting transcription of a minigenome (44). In a separate study (53), it was shown that this mutant P protein was completely defective in phosphorylation in domain I and possessed low levels of transcriptional activity compared to the wt P protein. Therefore, we examined whether this mutant P protein could rescue infectious VSV from transfected cells. We found that infectious VSV encoding the mutant P60/62/64 protein could not be rescued from transfected cells. This was observed repeatedly in at least 10 independent transfection experiments under various transfection conditions. We also attempted to detect the mutant virus by passaging the transfected cell supernatant into cells expressing the wt P protein in trans, but such attempts failed to detect VSVP60/62/64. These results suggest that VSV encoding a P protein in which all of the phosphate acceptor sites in domain I are altered to alanine cannot be rescued.
Viruses with mutations in phosphorylation sites in domain I exhibit similar kinetics of growth and mRNA synthesis.
Since our previous studies demonstrated that mutations within domain I residues result in a significant downregulation of transcription in a minigenome system (44), we were surprised to observe that VSVs containing these mutant P proteins grew to similar titers during passaging in cell culture. We examined whether these mutant viruses had altered growth kinetics under single-cycle growth conditions. The results (Fig. 2B) show that all of the mutant viruses possessed growth kinetics similar to those of wt VSV. No significant differences in virus titers were observed at any particular time postinfection.
To examine mRNA synthesis, we analyzed metabolically radiolabeled mRNAs in cells infected with various mutant viruses. Our results showed that the levels of mRNA synthesized in cells infected with VSVP60, VSVP62, and VSVP64 were similar to that synthesized in wt VSV-infected cells (Fig. 2C). Similar results were also obtained with the VSVP60/62, VSVP62/64, and VSVPE/E/E viruses (not shown). A quantitative analysis indicated that the levels of mRNA synthesis in cells infected with the mutant viruses were not significantly different from that in wt VSV-infected cells.
In the studies reported above for the recovery of mutant viruses, our approach was based on using the plasmid encoding the wt P protein in the support plasmid mix. It is possible that vaccinia virus-mediated homologous recombination between the plasmid containing the wt P coding region used in the support plasmid mix and the mutant P coding region in the pVSVFL(+) template may have resulted in the generation of a VSVPwt genome. In such a scenario, viruses recovered from the above experiments may predominantly contain wt genomes and may possess similar growth kinetics and mRNA profiles, as shown above (Fig. 2B and C). Although our inability to recover viruses with pVSVFLP60/64 and pVSVFLP60/62/64 suggests otherwise, nevertheless we examined the sequence of the P coding region in various mutant viruses. The results in Fig. 2D show that the introduced mutations were maintained in the mutant virus genomes. We examined the sequences of all of the mutant viruses, but only the results for VSVP60, VSVP62, and VSVP64, along with VSVPwt, are shown. Furthermore, the RT-PCR-amplified products were cloned, and five clones of each mutant virus P coding region were sequenced in their entirety. No other mutations within the P coding region of any of the mutant viruses were observed. Thus, our results suggest that single or double mutations in the phosphate acceptor sites in domain I (with the exception of P60/64) can support replication and growth of VSV.
Phosphorylation status of P protein in mutant virus-infected cells.
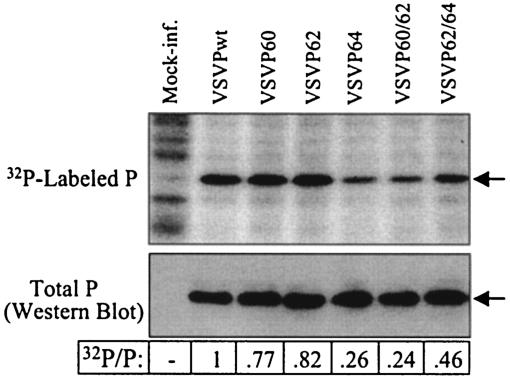
Previous studies have demonstrated that mutations in phosphate acceptor sites in the P protein alter the extent of phosphorylation. However, these studies were performed in vitro or in transfected cells ectopically expressing the mutant P proteins (6, 28). To examine the extent of phosphorylation of P proteins in mutant virus-infected cells, we infected two parallel sets of cells with various mutant viruses. One set of infected cells was radiolabeled with [32P]orthophosphate while the other set was left untreated. Cell lysates were prepared from both cultures and equal amounts of cell lysates were analyzed by electrophoresis. The 32P-labeled samples were examined directly by autoradiography to determine the incorporation of phosphates into the P protein, whereas the unlabeled samples were examined by Western blotting to determine the total amount of P protein. Our results (Fig. 3) show that the extents of phosphorylation of the P protein in various mutant virus-infected cells are different. Interestingly, we observed in independent repeat experiments that the amount of phosphate in the P protein of VSVP64 was significantly lower than that for the other single substitution mutant viruses. It is possible that the mutation in Ser-64 may affect phosphorylation at other sites.
FIG. 3.
Examination of phosphorylation status of P proteins in infected cells. Parallel cultures of BHK-21 cells were infected with wt or mutant viruses. Cells were labeled with [32P]orthophosphate or were left unlabeled. Equal proportions of cytoplasmic extracts were analyzed by electrophoresis in parallel gels. 32P incorporation into the P protein (top) was detected by autoradiography. The total P proteins were examined by Western blotting (bottom). Arrows in the panels identify P proteins. Numbers at the bottom represent average ratios of 32P/P from three independent experiments.
VSV with a mutation in the domain II phosphate acceptor site exhibits reduced kinetics of growth but increased kinetics of mRNA and protein synthesis.
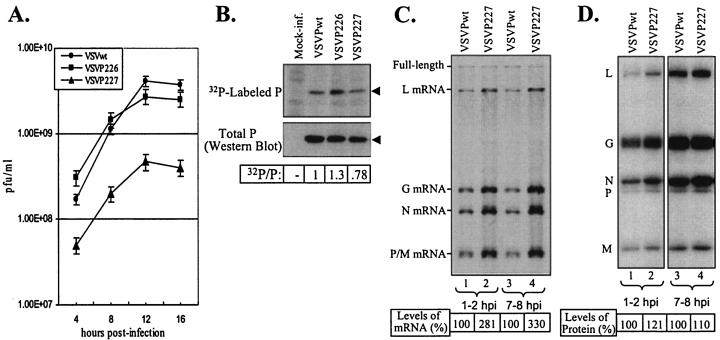
Our previous studies demonstrated that individual mutations in the phosphate acceptor sites (Ser-226 and Ser-227) did not have any adverse effects on transcription or replication but that mutations in both of these sites rendered the protein significantly defective in replication without affecting its transcription functions (26). Therefore, we were interested in examining whether VSVs encoding single or double amino acid substitutions at these sites in the P protein could be rescued. pVSVFL(+) plasmids encoding single or double amino acid substitutions in Ser-226 and Ser-227 of the P protein were used to rescue VSV from transfected cells as described above. We observed that VSVP226 and VSVP227, containing individual amino acid substitutions, could be rescued readily. Upon passaging, VSVP226 grew to titers similar to those of VSVPwt, whereas VSVP227 consistently grew to titers that were about 10-fold lower than those of VSVPwt. The virus encoding a double mutation (VSVP226/227) could not be rescued after multiple attempts under various transfection conditions. An analysis of single-cycle growth kinetics suggested that the VSVP227 virus grows slowly and to lower final titers than the VSVPwt or VSVP226 virus (Fig. 4A).
FIG. 4.
(A) Single-cycle growth kinetics of viruses with substitutions in domain II phosphate acceptor sites. The experiment was performed as described in the legend for Fig. 2B. Error bars represent standard deviations. (B) Phosphorylation status of P proteins of domain II mutant viruses. Data were obtained as described in the legend for Fig. 3. Arrowheads in the panels identify P proteins. (C) Kinetics of RNA synthesis in cells infected with VSVPwt and VSVP227. BHK-21 cells infected with VSVPwt (lanes 1 and 3) or VSVP227 (lanes 2 and 4) at an MOI of 100 (lanes 1 and 2) or 10 (lanes 3 and 4) were radiolabeled with [3H]uridine and [3H]adenosine in the presence of actinomycin D for 1 h at 1 or 7 h postinfection as described in Materials and Methods. Total RNAs were isolated and analyzed by electrophoresis. The positions of full-length as well as the five VSV mRNAs are indicated on the left. Numbers at the bottom show the average levels of total viral mRNAs from three experiments. (D) Analysis of viral proteins in cells infected with VSVPwt and VSVP227. BHK-21 cells were infected with VSVPwt (lanes 1 and 3) or VSVP227 (lanes 2 and 4) at an MOI of 100 (lanes 1 and 2) or 10 (lanes 3 and 4). The proteins were radiolabeled with 35S, and equal proportions of cell lysates were analyzed by electrophoresis as described in Materials and Methods. The positions of the viral proteins are shown on the left. The levels of viral protein represent average values from three independent experiments.
An examination of the phosphate incorporation into P proteins (Fig. 4B) showed that the extents of phosphorylation of the P protein in mutant virus-infected cells were different from that seen with wt VSV-infected cells. Surprisingly, the P protein of VSVP226 was found to be hyperphosphorylated. The reason(s) for this is unclear, but it is possible that the mutation in Ser-226 may have activated other cryptic phosphate acceptor sites in the P protein. A similar hyperphosphorylation of this particular mutant P protein has been observed under in vitro conditions (6).
Since VSVP227 consistently grew to titers about 10-fold lower than that of VSVPwt, we suspected that VSVP227 had altered kinetics of mRNA and protein synthesis. To examine the kinetics of mRNA synthesis, we infected parallel cultures of cells with an MOI of 100 or 10 and radiolabeled mRNAs at 1 to 2 h or 7 to 8 h postinfection. The rationale for using such a high MOI was to provide enough templates at the beginning so that mRNAs could be readily detected at early times after infection. The results show that all VSV mRNAs as well as the genomic and antigenomic full-length RNAs could readily be detected (Fig. 4C). Surprisingly, we observed that the levels of mRNAs from VSVP227 virus-infected cells were significantly higher than those produced from VSVPwt virus-infected cells. This observation was reproducible in multiple independent experiments. In addition, the synthesis of increased levels of mRNA in VSVP227 virus-infected cells compared to VSVPwt virus-infected cells was seen at early (1 to 2 h) or late (7 to 8 h) times after infection. It should be noted that the total amounts of genomic and antigenomic RNAs were similar for both VSVP227 and VSVPwt. A quantitative analysis of the levels of mRNAs indicated that VSVP227 synthesizes about a threefold excess of mRNAs per template compared to VSVPwt.
The above results led us to examine whether increased levels of VSV proteins are synthesized in cells infected with VSVP227 compared to those infected with VSVPwt. In a similar experiment, we analyzed the levels of VSV proteins in VSVP227 and VSVPwt virus-infected cells by radiolabeling them at early and late times after infection. Slightly increased levels of viral proteins were detected in VSVP227 virus-infected cells compared to those in VSVPwt virus-infected cells (Fig. 4D).
Taken together, our results suggest that VSVP227, encoding a mutant P protein, grows to lower titers but produces significantly higher levels of mRNAs in infected cells than does the wt.
Reduced cytopathogenicity of VSVP227 virus-infected cells.
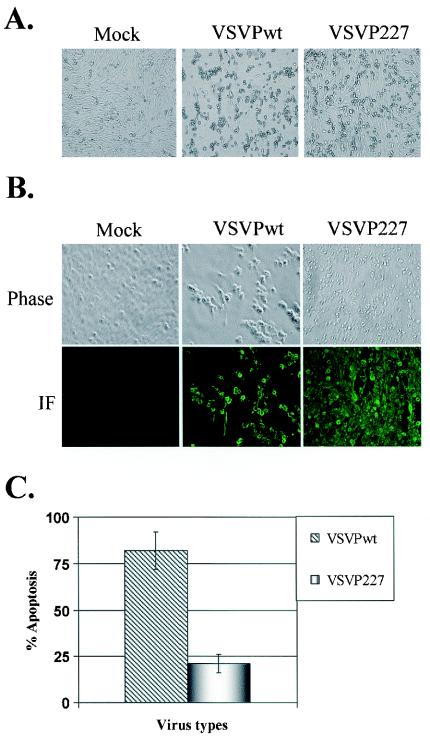
During our studies, we made an interesting observation with the VSVP227 virus. Although the formation of distinct plaques was seen with VSVP227 on monolayers of BHK-21 cells, only about 25% of the cells in the plaque appeared to have rounded off and detached, resulting in a partial clearing of the cell monolayer at the site of plaque formation. This contrasted with the normal plaque morphology of VSV on BHK-21 cells, in which >90% of the cells within the plaque show the CPE that is characteristic of a VSV infection and which normally results in the almost complete clearance of dead cells at the site of the plaque.
This observation suggested that VSVP227 is perhaps less cytopathic in infected cells than is the wt or other mutant viruses. To examine this, we monitored the development of CPE over time in BHK-21 cells infected with VSVPwt or VSVP227. We found that by 12 to 14 h postinfection, infection with VSVwt resulted in the rounding and detachment of >90% of the cells in the monolayer (Fig. 5A). However, infection with VSVP227 resulted in the rounding and detachment of only about 20 to 30% of the cells. Furthermore, at a low MOI (0.5), the cell monolayer of wt VSV-infected cells was completely lost due to a severe CPE by 24 h postinfection, whereas about 50% of the cells in the monolayer of VSVP227-infected cells still did not show CPE even at 48 h postinfection. Indirect immunofluorescence studies using anti-VSV antibodies showed that the majority of cells in the monolayer expressed VSV proteins (Fig. 5B), indicating that the cells in the monolayer that appeared normal were supporting replication of VSVP227.
FIG. 5.
(A) Phase-contrast microscopic images of BHK-21 cells infected with VSVPwt or VSVP227. BHK-21 cells were infected with each virus at an MOI of 0.5. At 12 h postinfection, live cells were observed under a Nikon microscope at a magnification of ×20, and images were processed through a camera (Optronics) attached to the microscope. (B) Expression of VSV proteins in VSVPwt- or VSVP227-infected cells. BHK-21 cells were infected with each virus at an MOI of 0.5 in 12-well plates. At 12 h postinfection, the cells were fixed and incubated with a mouse anti-VSV polyclonal antibody followed by a secondary antibody (anti-mouse Alexa-488). The fluorescence was observed at a magnification of ×40. The top panel shows cells under phase-contrast microscopy and the bottom panel shows the same field under fluorescence microscopy. (C) Quantitative analysis of apoptosis. BHK-21 cells infected with each virus at an MOI of 0.5 were trypsinized at 12 h postinfection and stained for apoptotic and necrotic cells with annexin V-fluorescein isothiocyanate and propidium iodide. The fluorescing cells were analyzed by flow cytometry, and the means of three independent experiments are presented, with error bars representing standard deviations.
Since one of the hallmark features of CPE caused by VSV is the induction of apoptosis (31, 37, 51), we examined the level of apoptosis in cells infected with VSVPwt or VSVP227. The results (Fig. 5C) show that about 22% of cells infected with VSVP227 were undergoing apoptosis compared to about 80% of cells infected with VSVwt. Taken together, these studies indicate that VSVP227 exhibits reduced cytopathogenicity in infected cells.
VSVs encoding various mutant P proteins show various patterns of RNA synthesis upon high-multiplicity passaging.
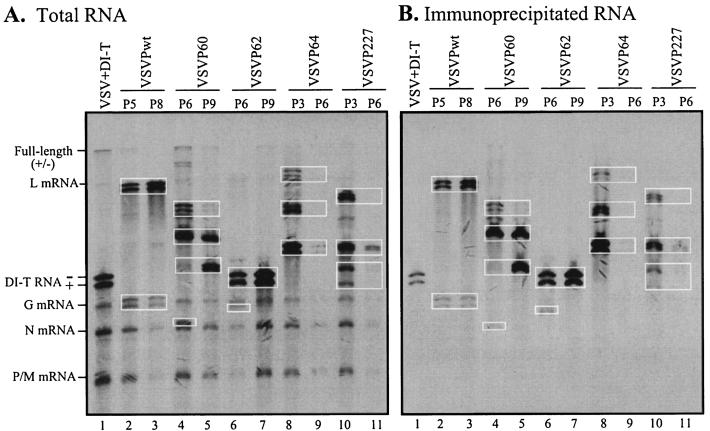
Serial undiluted passaging of VSV in cultured cells generates DI particles containing truncated genomes. The mechanisms of the generation of DI particles are not known, but host cell factors as well as viral factors, such as polymerase and the template, and their interactions are presumed to play a significant role in the process. Using the panel of VSV mutants with altered polymerase functions, we were interested to examine whether the polymerase per se plays a role in the process. Individual mutant viruses (VSVP60, VSVP62, VSVP64, and VSVP227) were used to infect BHK-21 cells at an initial MOI of 10, and equal aliquots of undiluted culture supernatants were passaged nine successive times in BHK-21 cells. We observed severe CPE in early passages followed by drastically reduced CPE in later passages. The pattern of increased and decreased CPE followed the cyclic nature of a prey-predator relationship (14, 42), indicating the production of DI particles in various passages. In initial experiments, culture supernatants from all passages were used to infect naïve cells and the synthesis of RNAs was examined by metabolic labeling. Several unique species of DI RNAs were detected in infected cells. Some DI RNAs appeared early and were lost in late passages, some remained stable over passages, and others appeared in late passages and remained stable (data not shown). Figure 6A shows the pattern of DI RNAs generated by various mutant VSVs in representative passages. The pattern of DI RNAs for each of the VSV mutants was unique. Only VSVP62 produced a DI RNA (lanes 6 and 7) that comigrated with the DI T genome (lane 1). This particular DI RNA appeared early (passage 3) and was amplified with passaging. The pattern of RNA species synthesized in cells infected with various passages appeared to be specific for specific mutant viruses since a similar independent multiple-passage experiment with the mutant viruses resulted in the generation of RNA patterns similar to those shown in Fig. 6A.
FIG. 6.
Analysis of DI particles generated by high-multiplicity passages of wt or mutant viruses. BHK-21 cells in 60-mm-diameter dishes were infected with 50 μl of undiluted cell culture supernatants from the indicated high-multiplicity passages of wt or mutant viruses. At 2 h postinfection, the cells were labeled for 4 h with [3H]uridine and [3H]adenosine in the presence of actinomycin D. Cytoplasmic extracts were prepared as described in Materials and Methods, and one-third of the extracts was used for the isolation of total RNAs. Two-thirds of the extract was incubated with an anti-VSV antibody to immunoprecipitate the nucleocapsids. Total RNAs (A) or RNAs recovered from immunoprecipitated nucleocapsids (B) were analyzed by electrophoresis as described above. DI RNAs seen as closely migrating doublets are identified in rectangular boxes. The positions of full-length genomic and antigenomic (+/−) RNAs as well as the five VSV mRNAs are indicated on the left.
Since these viruses contained mutations in the P protein, which is involved in both transcription and replication of the genome, it is possible that some of these truncated RNAs may have been generated during transcription. The RNA products generated during transcription do not associate with the viral N protein, whereas the products of replication are encapsidated by the N protein and can be specifically immunoprecipitated by an anti-N protein or anti-VSV antibody. To determine if any of the truncated RNAs were generated as a result of defects during transcription of the viral genome, we immunoprecipitated lysates of cells infected with supernatants of various passages and labeled with [3H]uridine and [3H]adenosine with an anti-VSV antibody. An analysis of the RNAs (Fig. 6B) recovered from immunoprecipitated complexes showed that almost all of the truncated RNA species were immunoprecipitable with the anti-VSV antibody, indicating that they are the products of replication. It should be noted that under the gel conditions used here, which separate RNAs based on their size and adenosine residue content, most RNA species were seen as closely migrating doublets representing the genomic and antigenomic RNAs. However, some of the immunoprecipitated RNAs appeared as single species. It is possible that both genomic and antigenomic RNAs of these DI particles have similar adenosine residue contents and therefore could not be resolved from each other.
DISCUSSION
The phosphorylation of viral proteins by cellular or virus-associated kinases is one of the posttranslational modifications essential for their functioning. The phosphorylation of the nucleoprotein (N) of rabies virus (55), the VP30 protein of Marburg virus (41), and the P proteins of RSV (36) and SeV (25) plays a role in the transcription and replication of these viruses. The P protein of VSV has a modular structure. In vitro studies as well as studies with transfected cells using DI RNAs or minigenomic RNAs strongly suggest that the phosphorylation of different structural domains of the P protein regulates its multiple functions in viral genome transcription and replication (2, 3, 7, 8, 16, 17, 26, 44, 53). In the present study, we addressed the role of P protein phosphorylation in the life cycle of VSV. Our results suggest that phosphorylation of the viral P protein is indispensable for the growth of VSV.
Using P proteins with mutations in the phosphate acceptor sites in domain I, we and others have demonstrated previously that phosphorylation in this domain is required for the transcriptional activity of the P protein (44, 53). Our inability to recover infectious virus with a P60/62/64 mutant even after multiple independent attempts and under various transfection conditions suggests that the phosphorylation of domain I residues is absolutely critical for virus growth and multiplication. Our attempts to detect the virus in undiluted supernatants from transfected cells by plaque assay yielded negative results. The P60/64 mutant could not rescue infectious VSV, although it possessed low levels of transcriptional activity in a minigenome assay (44). We suspect that P60/64 is not phosphorylated at all since previous studies have suggested that phosphorylation of Thr-62 requires the prior phosphorylation of either Ser-60 or Ser-64 (28) and that the phosphate at Thr-62 is prone to dephosphorylation by cellular phosphatases (6). The lack of rescue of VSV with P60/64 further strengthens our conclusion that phosphorylation within domain I residues is absolutely required for the recovery of VSV. These results differ from those obtained with other negative-strand RNA viruses, such as RSV and SeV, in that unphosphorylated forms of the P protein can rescue infectious viruses from cells (25, 36). Our present results, as well as those from previous studies, suggest that the requirement of phosphorylation of the P protein of VSV for viral genome replication, transcription, and the generation of infectious viruses is different from that of the paramyxoviruses studied so far. It would be interesting to examine if the mutant VSVs have altered pathogenic characteristics.
We had previously shown that P proteins with single and multiple amino acid substitutions (with the exception of P60) in domain I were <10% active in transcription compared to the wt P protein (44). Interestingly, all of these mutants (with the exception of P60/64 and P60/62/64) could support the rescue of VSVs with kinetics of growth and mRNA synthesis that were similar to those of wt VSV. This observation suggests that the low levels of transcription supported by these mutant proteins were sufficient to allow the production of infectious VSVs. It is possible that in these viruses, compensatory mutations in other regions of the P protein may have resulted in a fully functional protein that could support the growth of VSV. Sequencing of the P protein coding region from various mutants did not reveal any mutations that could account for the observed phenotype, although compensatory mutations in the L and/or N protein have not been ruled out. It is generally believed that viral proteins are synthesized in a large excess in virus-infected cells compared to what may be necessary for growth of the virus. The results presented here suggest that the low levels of transcriptional activity of the mutant P proteins were adequate to support the growth of VSV. At least one of the phosphate acceptor sites must be phosphorylated for the protein to be functional in the full-length viral genome. Alternatively, it is possible that the activity of the mutant proteins in the minigenome transcription and replication system is different from that seen in full-length genomes encoding the mutant proteins. It will be interesting to determine how the mutant P proteins become functionally active in the context of a full-length viral genome.
Our studies with mutants in domain II of the P protein also suggest that phosphorylation of either Ser-226 or Ser-227 is essential for the growth of VSV. Our inability to rescue VSV encoding a P protein with mutations at both of these positions was not surprising since this protein was shown to be significantly defective in replication (26). Although VSVs with single amino acid substitutions in this domain of the P protein were rescued easily, the growth kinetics of VSVP227 were slower than those of wt VSV. In several independent experiments, we observed that VSVP227 grew to titers that were about 10-fold lower than those of wt VSV. More interestingly, VSVP227 synthesized mRNAs that were present, on average, in threefold larger amounts than the wt VSV. Although some increase in viral proteins was observed in VSVP227-infected cells, proportionately increased amounts of viral proteins were not seen, indicating that some level of translational control may be operative in VSVP227-infected cells. The reason for the increased synthesis of mRNA is not clear, but it is not due to elevated levels of full-length templates (Fig. 4C). Sequencing of the P gene of this virus indicated that no other mutations were present in the P protein. It is possible that extragenic mutations in another protein may have been responsible for this phenotype. Attempts are being made to address this possibility.
Perhaps one of the most interesting aspects of the present study is the observation that cells infected with VSVP227 exhibited significantly reduced CPE and apoptosis compared to wt VSV-infected cells (Fig. 5). Rounding and subsequent detachment of the cells, which are characteristics of VSV infections, were substantially reduced. The reason(s) for the reduced cytopathogenicity and apoptosis of VSVP227 is not known, but it could be due to several factors. Apoptosis in VSV-infected cells is caused by the direct effects of the M protein and possibly another viral component (30). This other component has been suggested to be either the viral double-stranded RNA replication intermediates (10, 34), the leader RNA (11, 21, 40, 49), or the G protein (13). A role for the P protein in apoptosis could be envisioned from the results reported here. The introduced mutation at P227 may disrupt the apoptotic function of the protein, and therefore VSVP227 containing this mutation may induce significantly reduced apoptosis. The viral M protein is known to induce CPE (4, 30) similar to that seen in VSV-infected cells. Extragenic mutations in the M protein of VSVP227 that disrupt its apoptotic and CPE-inducing functions may be one of the possible reasons. Alternatively, it is likely that an antiviral state is established in infected cells, leading to reduced cytopathogenicity and viral titers. In this context, VSVP227 may have acquired properties to activate NF-κB, which is an important mediator of innate and adaptive host defense mechanisms (18, 35). It will be interesting to examine whether NF-κB is activated in VSVP227-infected cells, and if so, whether this activation occurs through interferon-dependent or interferon-independent pathways. Further work will be needed to determine the mechanism and factors responsible for the conversion of a highly cytopathic VSV to a mildly cytopathic virus.
Multiple factors control the generation of DI particles in high-multiplicity passages of VSV. Host cells appear to play a major role in the process (24). Our observation that mutant VSVs with altered polymerase functions generated different sets of DI particles in the same host environment indicates that the polymerase as well as polymerase-template interactions due to an altered P protein may also play critical roles in the process. It is also possible that the alteration of the P protein affects the host cell environment in a manner that leads to the generation of different types of DI RNAs. Whether certain mutant viruses preferentially generate certain types of DI RNAs (such as the copy-back type or internal deletion type) is not known. A further characterization of DI RNAs may shed light into the role of the viral RNA polymerase in generating specific types of DI RNAs. Previous studies have demonstrated that an altered replicase specificity results in the resistance of a VSV mutant (Sdi−) to interference by a DI particle (19). In this regard, it will be interesting to examine whether DI particles generated by one VSV mutant can interfere with the replication of another VSV mutant. Such studies will provide insights into an understanding of the selection of viral termini that are involved in interactions with altered polymerase molecules for the evolution of DI particles during persistent and acute viral infections.
In conclusion, our studies reported here suggest that the phosphorylation of residues in the P protein is essential for the recovery and propagation of infectious VSV. Our results also suggest that the viral RNA polymerase plays a critical role in the generation of DI particles. Furthermore, we report that a VSV with a mutation at Ser-227 in domain II of the P protein is less cytopathic than the wt VSV, indicating that the P protein may be involved in cytopathology in infected cells. The molecular basis of the reduced cytopathogenicity of VSVP227 is currently under investigation.
Acknowledgments
We thank Debasis Nayak for assistance with the construction of some of the mutants. We also thank You Zhou, Center for Biotechnology, University of Nebraska—Lincoln, for help with fluorescence microscopic studies.
This investigation was supported by Public Health Service grant AI-34956 from the NIH and also in part by grant P20RR15635 from the COBRE program of the National Center for Research Resources, NIH.
Footnotes
Publication 14425 of the Agricultural Research Division of the Institute of Agriculture and Natural Resources of the University of Nebraska—Lincoln.
REFERENCES
- 1.Barik, S., and A. K. Banerjee. 1991. Cloning and expression of the vesicular stomatitis virus phosphoprotein gene in Escherichia coli: analysis of phosphorylation status versus transcriptional activity. J. Virol. 65:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barik, S., and A. K. Banerjee. 1992. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein P. Proc. Natl. Acad. Sci. USA 89:6570-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barik, S., and A. K. Banerjee. 1992. Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcription activation. J. Virol. 66:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blondel, D., G. G. Harmison, and M. Schubert. 1990. Role of matrix protein in cytopathogenesis of vesicular stomatitis virus. J. Virol. 64:1716-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canter, D. M., and J. Perrault. 1996. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology 219:376-386. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. L., T. Das, and A. K. Banerjee. 1997. Phosphorylated states of vesicular stomatitis virus P protein in vitro and in vivo. Virology 228:200-212. [DOI] [PubMed] [Google Scholar]
- 7.Das, T., A. K. Gupta, P. W. Sims, C. A. Gelfand, J. E. Jentoft, and A. K. Banerjee. 1995. Role of cellular casein kinase II in the function of the phosphoprotein (P) subunit of RNA polymerase of vesicular stomatitis virus. J. Biol. Chem. 270:24100-24107. [DOI] [PubMed] [Google Scholar]
- 8.Das, T., A. K. Pattnaik, A. M. Takacs, T. Li, L. N. Hwang, and A. K. Banerjee. 1997. Basic amino acid residues at the carboxy-terminal eleven amino acid region of the phosphoprotein (P) are required for transcription but not for replication of vesicular stomatitis virus genome RNA. Virology 238:103-114. [DOI] [PubMed] [Google Scholar]
- 9.Davis, N. L., H. Arnheiter, and G. W. Wertz. 1986. Vesicular stomatitis virus N and NS proteins form multiple complexes. J. Virol. 59:751-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Der, S. D., Y. L. Yang, C. Weissmann, and B. R. Williams. 1997. A double-stranded RNA-activated protein kinase-dependent pathway mediating stress-induced apoptosis. Proc. Natl. Acad. Sci. USA 94:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunigan, D. D., S. Baird, and J. Lucas-Lenard. 1986. Lack of correlation between the accumulation of plus-strand leader RNA and the inhibition of protein and RNA synthesis in vesicular stomatitis virus infected mouse L cells. Virology 150:231-246. [DOI] [PubMed] [Google Scholar]
- 12.Emerson, S. U., and M. Schubert. 1987. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 84:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florkiewicz, R. Z., A. Smith, J. E. Bergmann, and J. K. Rose. 1983. Isolation of stable mouse cell lines that express cell surface and secreted forms of the vesicular stomatitis virus glycoprotein. J. Cell Biol. 97:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank, S. A. 2000. Within-host spatial dynamics of viruses and defective interfering particles. J. Theor. Biol. 206:279-290. [DOI] [PubMed] [Google Scholar]
- 15.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, Y., and J. Lenard. 1995. Cooperative binding of multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerase (L) and template: pathways of assembly. J. Virol. 69:7718-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, Y., and J. Lenard. 1995. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase-II. EMBO J. 14:1240-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109(Suppl.):S81-S96. [DOI] [PubMed] [Google Scholar]
- 19.Giachetti, C., and J. J. Holland. 1988. Altered replicase specificity is responsible for resistance to defective interfering particle interference of an Sdi− mutant of vesicular stomatitis virus. J. Virol. 62:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill, D. S., D. Chattopadhyay, and A. K. Banerjee. 1986. Identification of a domain within the phosphoprotein of vesicular stomatitis virus that is essential for transcription in vitro. Proc. Natl. Acad. Sci. USA 83:8873-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grinnell, B. W., and R. R. Wagner. 1983. Comparative inhibition of cellular transcription by vesicular stomatitis virus serotypes New Jersey and Indiana: role of each viral leader RNA. J. Virol. 48:88-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta, A. K., T. Das, and A. K. Banerjee. 1995. Casein kinase II is the P protein phosphorylating cellular kinase associated with the ribonucleoprotein complex of purified vesicular stomatitis virus. J. Gen. Virol. 76:365-372. [DOI] [PubMed] [Google Scholar]
- 23.Gupta, A. K., D. Shaji, and A. K. Banerjee. 2003. Identification of a novel tripartite complex involved in replication of vesicular stomatitis virus genome RNA. J. Virol. 77:732-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holland, J. J., L. P. Villarreal, and M. Breindl. 1976. Factors involved in the generation and replication of rhabdovirus defective T particles. J. Virol. 17:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, C. J., A. Kato, M. C. Bowman, K. Kiyotani, T. Yoshida, S. A. Moyer, Y. Nagai, and K. C. Gupta. 1999. Role of primary constitutive phosphorylation of Sendai virus P and V proteins in viral replication and pathogenesis. Virology 263:195-208. [DOI] [PubMed] [Google Scholar]
- 26.Hwang, L. N., N. Englund, T. Das, A. K. Banerjee, and A. K. Pattnaik. 1999. Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J. Virol. 73:5613-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isaac, C. L., and J. D. Keene. 1982. RNA polymerase-associated interactions near template promoter sequences of defective interfering particles of vesicular stomatitis virus. J. Virol. 43:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson, R. L., D. Spadafora, and J. Perrault. 1995. Hierarchical constitutive phosphorylation of the vesicular stomatitis virus P protein and lack of effect on P1 to P2 conversion. Virology 214:189-197. [DOI] [PubMed] [Google Scholar]
- 29.Keene, J. D., B. J. Thornton, and S. U. Emerson. 1981. Sequence-specific contacts between the RNA polymerase of vesicular stomatitis virus and the leader RNA gene. Proc. Natl. Acad. Sci. USA 78:6191-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopecky, S. A., M. C. Willingham, and D. S. Lyles. 2001. Matrix protein and another viral component contribute to induction of apoptosis in cells infected with vesicular stomatitis virus. J. Virol. 75:12169-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyama, A. H. 1995. Induction of apoptotic DNA fragmentation by the infection of vesicular stomatitis virus. Virus Res. 37:285-290. [DOI] [PubMed] [Google Scholar]
- 32.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leamson, R. W., and M. Reichman. 1974. The RNA of defective vesicular stomatitis virus particles in relation to viral cistrons. J. Mol. Biol. 85:551-568. [DOI] [PubMed] [Google Scholar]
- 34.Lee, S. B., and M. Esteban. 1994. The interferon-induced double-stranded RNA-activated protein kinase induces apoptosis. Virology 199:491-496. [DOI] [PubMed] [Google Scholar]
- 35.Li, Q., and I. M. Verma. 2002. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2:725-734. [DOI] [PubMed] [Google Scholar]
- 36.Lu, B., C. H. Ma, R. Brazas, and H. Jin. 2002. The major phosphorylation sites of the respiratory syncytial virus phosphoprotein are dispensable for virus replication in vitro. J. Virol. 76:10776-10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyles, D. S. 2000. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 64:709-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massey, D. M., N. Deans, and J. Lenard. 1990. Phosphorylation of NS protein by vesicular stomatitis virus nucleocapsids: lack of effect during RNA synthesis and separation of kinase from L protein. J. Virol. 64:3259-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masters, P. S., and A. K. Banerjee. 1988. Resolution of multiple complexes of phosphoprotein NS with nucleocapsid protein N of vesicular stomatitis virus. J. Virol. 62:2651-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGowan, J. J., S. U. Emerson, and R. R. Wagner. 1982. The plus-strand leader RNA of VSV inhibits DNA-dependent transcription of adenovirus and SV40 genes in a soluble whole-cell extract. Cell 28:325-333. [DOI] [PubMed] [Google Scholar]
- 41.Modrof, J., C. Moritz, L. Kolesnikova, T. Konakova, B. Hartlieb, A. Randolf, E. Muhlberger, and S. Becker. 2001. Phosphorylation of Marburg virus VP30 at serines 40 and 42 is critical for its interaction with NP inclusions. Virology 287:171-182. [DOI] [PubMed] [Google Scholar]
- 42.Palma, E. L., and A. Huang. 1974. Cyclic production of vesicular stomatitis virus caused by defective interfering particles. J. Infect. Dis. 129:402-410. [DOI] [PubMed] [Google Scholar]
- 43.Pattnaik, A. K., L. A. Ball, A. W. LeGrone, and G. W. Wertz. 1992. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 69:1011-1020. [DOI] [PubMed] [Google Scholar]
- 44.Pattnaik, A. K., L. Hwang, T. Li, N. Englund, M. Mathur, T. Das, and A. K. Banerjee. 1997. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J. Virol. 71:8167-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pattnaik, A. K., and G. W. Wertz. 1991. Cells that express all five proteins of vesicular stomatitis virus from cloned cDNAs support replication, assembly, and budding of defective interfering particles. Proc. Natl. Acad. Sci. USA 88:1379-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pattnaik, A. K., and G. W. Wertz. 1990. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J. Virol. 64:2948-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul, P. R., D. Chattopadhyay, and A. K. Banerjee. 1988. The functional domains of the phosphoprotein (NS) of vesicular stomatitis virus (Indiana serotype). Virology 166:350-357. [DOI] [PubMed] [Google Scholar]
- 48.Peluso, R. W. 1988. Kinetic, quantitative, and functional analysis of multiple forms of the vesicular stomatitis virus nucleocapsid protein in infected cells. J. Virol. 62:2799-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poirot, M. K., W. M. Schnitzlein, and M. E. Reichmann. 1985. The requirement of protein synthesis for VSV inhibition of host cell RNA synthesis. Virology 140:91-101. [DOI] [PubMed] [Google Scholar]
- 50.Rose, J. K., and M. Schubert. 1987. Rhabdovirus genomes and their products, p. 129-166. In R. R. Wagner (ed.), The rhabdoviruses. Plenum Press, New York, N.Y.
- 51.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 52.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 53.Spadafora, D., D. M. Canter, R. L. Jackson, and J. Perrault. 1996. Constitutive phosphorylation of the vesicular stomatitis virus P protein modulates polymerase complex formation but is not essential for transcription or replication. J. Virol. 70:4538-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takacs, A. M., S. Barik, T. Das, and A. K. Banerjee. 1992. Phosphorylation of specific serine residues within the acidic domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. J. Virol. 66:5842-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, X., X. Gong, H. D. Foley, M. J. Schnell, and Z. F. Fu. 2002. Both viral transcription and replication are reduced when the rabies virus nucleoprotein is not phosphorylated. J. Virol. 76:4153-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]