Abstract
Epstein-Barr virus (EBV) episomes are stably maintained in permissive proliferating cell lines due to EBV nuclear antigen 1 (EBNA-1) protein-mediated replication and segregation. Previous studies showed the ability of EBV episomes to confer long-term transgene expression and correct genetic defects in deficient cells. To achieve quantitative delivery of EBV episomes in vitro and in vivo, we developed a binary helper-dependent adenovirus (HDA)-EBV hybrid system that consists of one HDA vector for the expression of Cre recombinase and a second HDA vector that contains all of the sequences for the EBV episome flanked by loxP sites. Upon coinfection of cells, Cre expressed from the first vector recombined loxP sites on the second vector. The resulting circular EBV episomes expressed a transgene and contained the EBV-derived family of repeats, an EBNA-1 expression cassette, and 19 kb of human DNA that functions as a replication origin in mammalian cells. This HDA-EBV hybrid system transformed 40% of cultured cells. Transgene expression in proliferating cells was observed for over 20 weeks under conditions that selected for the expression of the transgene. In the absence of selection, EBV episomes were lost at a rate of 8 to 10% per cell division. Successful delivery of EBV episomes in vivo was demonstrated in the liver of transgenic mice expressing Cre from the albumin promoter. This novel gene transfer system has the potential to confer long-term episomal transgene expression and therefore to correct genetic defects with reduced vector-related toxicity and without insertional mutagenesis.
Epstein-Barr virus (EBV) is a gammaherpesvirus that preferentially infects human B lymphocytes (16, 17, 26). The latent form of EBV is capable of lifelong persistence as a circular episome in human B cells. Stable maintenance of the 165-kb EBV episome is conferred mainly by the abilities of EBV episomes to replicate once per S phase and to segregate efficiently into daughter cells during cell division (44). Both mechanisms require the interaction of EBV nuclear antigen 1 (EBNA-1) protein with the EBV origin of replication (oriP) (23). oriP consists of two groups of EBNA-1 binding sites, the family of repeats (FR) and the dyad symmetry element (DS) (31, 43). The FR is composed of 20 imperfect copies of a 30-bp repeat, each one able to bind an EBNA-1 dimer. The binding of EBNA-1 to the FR forms a bridge between chromosomes and oriP-containing plasmids (25). During mitosis, EBV episomes are tethered to metaphase chromosomes through EBNA-1, allowing the efficient segregation of episomes to daughter cells. The association of EBNA-1 with the FR also increases the nuclear import and retention of EBV plasmids and may target the plasmids to nuclear regions conducive to the efficient expression of EBV episome sequences (20).
The DS consists of four copies of the EBNA-1 binding site (43). Upon binding of EBNA-1 through its C-terminal domain, the DS confers the initiation of DNA replication once per S phase (17). In the absence of the DS, EBV episomes fail to undergo replication unless they contain a different source of a replication origin, such as human genomic DNA. Krysan et al. (19) showed that replacement of the DS in EBV plasmids with large human chromosomal fragments resulted in efficient replication and segregation. Further studies demonstrated that the size of the genomic insert affected replication efficiency, favoring inserts of about 20 kb in length (12). EBV plasmids containing human genomic DNA were shown to replicate efficiently in rodent cells, in contrast to plasmids containing EBV-derived oriP (18). The transformation of various cell lines with EBV episomes in vitro has revealed the potential to confer long-term transgene expression and correct genetic defects in deficient cell lines (3, 15, 18, 21, 44).
The delivery of EBV episomes is commonly performed by transfection. However, for quantitative delivery of EBV episomes, particularly in vivo, different techniques are required. Adenovirus recombinants have been shown to efficiently infect a wide variety of mammalian cells and tissues in vitro and in vivo (1). The expression of transgenes from adenovirus vectors is usually temporary, since the adenovirus DNA remains episomal in the majority of infected cells and is only integrated and therefore stably maintained at a frequency of 0.01 to 1% (11). In vivo, immune responses against viral antigens can cause the elimination of infected cells (8, 30, 41). Compared to first-generation adenovirus recombinants, helper-dependent adenovirus (HDA) vectors have no viral coding sequences and have provided more prolonged transgene expression mainly due to reduced toxicity in vitro and in vivo (45, 46). HDA-mediated transgene expression after mouse liver transduction with the α1-antitrypsin gene was shown to occur at a higher level than and to be significantly prolonged compared with first-generation adenovirus transduction (32). In contrast, other investigators described similar levels and duration of transgene expression in rat brain with either a first-generation adenovirus or HDA vector in the absence of preexisting antiadenovirus immunity (36). In another study, the levels of blood clotting factor IX in serum were compared after in vivo transduction of mouse liver with either a recombinant first-generation adenovirus or HDA vector (7). Transgene expression in that study was highest in the first 2 months, with a slow decline to 5% the original level in serum after 1 year for either vector. The amount and duration of factor IX expression were similar for the first-generation adenovirus and the HDA vector.
To combine the superior infectivity of adenovirus vectors with the ability of EBV episomes to confer long-term transgene expression, we have developed an HDA-based strategy to deliver EBV episomes to mammalian cells. This HDA-EBV hybrid system consists of two different HDA vectors. Upon coinfection of cells, Cre recombinase expression from one HDA vector induces the recombination of loxP sites on a second, linear HDA vector that contains all of the sequences necessary to generate an EBV episome. The main component of the resulting circular construct contains the EBV-derived FR, a bicistronic expression cassette for various transgenes and the EBNA-1 protein, and a 19-kb fragment of human DNA capable of initiating replication of the episome once per cell cycle (10). Using different reporter gene assays, we demonstrate that this vector system can efficiently deliver EBV episomes to infected cells and stably transform various mammalian cells with a high efficiency. Furthermore, compared to the previously reported first-generation E1-substituted adenovirus-EBV hybrid system (34), the HDA-based strategy exhibits significantly less toxicity in vitro.
MATERIALS AND METHODS
Cell culture.
The canine osteosarcoma cell line D17 and the human cervical carcinoma cell line HeLa were obtained from the American Type Culture Collection. 293 cells were obtained from Microbix (Toronto, Ontario, Canada) (9), and 293Cre4 cells were obtained from Merck (5). 293Flpe cells have been described elsewhere (37). Cells were grown in high-glucose Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The medium for 293Cre4 cells was supplemented with 400 μg of G418 (Geneticin)/ml, and the medium for 293Flpe cells was supplemented with 1 μg of puromycin (Sigma)/ml.
HDA plasmid design and construction.
All vectors were constructed in bacterial plasmids that contained full-length HDA genomes. The backbones of both the Cre recombinase and the target vector plasmids consisted of a bacterial replication origin, a cos site, kanamycin and ampicillin resistance expression cassettes, and four repeats of the hygromycin resistance expression cassette. These repeats were required to make the plasmid DNA long enough for cosmid cloning. Digestion with the 8-bp cutter PmeI excised the full-length HDA genome with the inverted terminal repeats 3 bp from the DNA ends. All HDA vectors were designed to contain a yellow fluorescent protein (YFP) expression cassette for the titration of vector stocks. The plasmid for the HDA target vectors was constructed to contain all of the sequences necessary to generate a functional EBV episome, including a 19-kb human chromosome 10-derived sequence for replication (LlB41; human origin of replication [hORI]) (19), an expression cassette for the transgene and EBNA-1, and the FR (Fig. 1). Three sets of HDA target vectors were cloned and contained transgenes for puromycin acetyltransferase (PAC; 630 bp), cyan fluorescent protein (CFP; 762 bp), and Renilla luciferase (RenLUC; 933 bp). Each set of vectors consisted of one vector with the FR (HDA.PAC+FR, HDA.CFP+FR, and HDA.RenLUC+FR) and a control vector without the FR (HDA.PAC−FR, HDA.CFP−FR, and HDA.RenLUC−FR). A 2.9-kb stuffer from human chromosome X was inserted between the FR and the simian virus 40 polyadenylation site of the transgene and EBNA-1 expression cassette. The EBV episome sequence was flanked by two parallel loxP sites. Like the HDA target vectors, the Cre recombinase vectors (HDA.Cre) contained a YFP expression cassette for titration and the same 19-kb human chromosome 10-derived sequence as that used in the target vectors. The Cre recombinase was expressed under the control of a human cytomegalovirus (CMV) promoter toward the left end of the vector. The sequences of the cosmids containing the Cre vectors and the target vectors are available upon request.
FIG. 1.
Schematic diagram of the binary HDA-EBV vector system. Delivery of a circular EBV episome to mammalian cells is achieved by coinfection with two HDA vectors. Cre recombinase expressed from HDA.Cre catalyzes recombination between two loxP sites on the HDA target vector, generating a 28-kb EBV episome. The episome contains all sequences necessary for stable maintenance in proliferating cells, including hORI (not drawn to scale), FR, and a bicistronic expression cassette for the transgene and EBNA-1. ITR, adenovirus type 5 inverted terminal repeat; Ψ, adenovirus packaging signal; pCMV, human CMV enhancer-promoter; p(A), simian virus 40 or bovine growth hormone polyadenylation site; IRES, internal ribosome entry site; B, BamHI site; N, NotI site.
HDA propagation.
HDA.Cre was propagated in 293Cre4 cells by using helper virus AdLC8cluc as described by Parks et al. (27). HDA.Cre stocks had titers of 1 × 1010 to 5 × 1010 yellow transducing units (YTU)/ml on 293 cells and an AdLC8cluc helper virus titer of ∼0.1% the YTU titer (determined by comparing PFU on 293 cells to YTU).
The propagation scheme for HDA loxP target vectors was developed by Umana et al. (37) and uses FL helper virus in 293Flpe cells. The HDA plasmid was digested with PmeI to release the full-length HDA genome from the bacterial plasmid backbone. On day 0, the PmeI-digested HDA plasmid was transfected into 293Cre (HDA.Cre) or 293Flpe (HDA loxP target vector) cells at 50% confluence in a 60-mm cell culture dish by calcium phosphate transfection (Profection; Promega). After 18 to 24 h, cells were infected with the respective helper virus at a concentration previously determined to cause complete cytopathic effects (CPE) within 48 h (termed the practical multiplicity of infection [MOI]). When extensive CPE had developed, cells were harvested and frozen-thawed three times in 1 ml of medium, and the viral lysate was passaged into a new 60-mm dish and coinfected with the practical MOI of helper virus. At each successive passage, cells were coinfected with the practical MOI of helper virus and harvested when extensive CPE had developed. At this and subsequent passages, the amount of HDA vector amplification was assayed by infecting 293 cells with serial dilutions of viral lysates and counting the percentage of yellow fluorescent cells at 72 h postinfection (p.i.). Cells were infected at a vector MOI of 3 YTU. Amplification was generally taken to the fifth or sixth passage, with the final passage taking place in 5 to 20 500-cm2 dishes.
Vectors were purified with a CsCl2 step gradient (1.25 to 1.35 g/ml) in 10 mM Tris (pH 8.0) and centrifuged at 36,000 rpm for 90 min in an SW40 rotor (Beckman). This step was followed by centrifugation with a linear CsCl2 equilibrium buoyant density gradient with a hinge density of 1.33 g of CsCl2/ml in 10 mM Tris (pH 8.0) at 20,000 rpm for 24 h in a VTi50 rotor (Beckman), resulting in separation of the HDA vector in the upper band and helper virus that was not restricted by the Cre- or Flp-expressing 293 cells in the lower band. The upper band was dialyzed in 10 mM Tris-HCl (pH 8.0). HDA loxP target vector stocks had titers of 1 × 1010 to 2 × 1010 YTU/ml and contamination of ∼1% FL helper virus, as determined by measuring PFU on 293 cells. For injection into tail veins of mice, the HDA vector stocks were concentrated 10-fold by using an Amicon Ultra-4 100.000 MWCO centrifugal filter device, with close to 100% recovery of YTU.
Clonogenic assay.
A total of 2 × 104 D17 cells were used to seed wells of a 48-well plate 2 days prior to infection. On the day of infection (day 0), the number of cells per well was determined by hemacytometer counting. The MOI was defined as the YTU for the vector divided by the number of cells infected. Infection was performed by using 100 μl of serum-free DMEM at 37°C for 1 h followed by the addition of 1 ml of DMEM with 10% FBS. At 24 h p.i., cells were trypsinized and counted, and 5 × 103 cells were plated on each of four 10-cm dishes. At 7 days p.i., two plates were placed in DMEM with 10% FBS and 1 μg of puromycin/ml, and the remaining two plates were maintained in DMEM with 10% FBS. Colonies were counted after 1 to 2 weeks, when colony numbers had reached a plateau. The transformation efficiency was calculated as the number of colonies generated by infected cells in medium with puromycin divided by the number of colonies generated by mock-infected cells in medium without puromycin times 100. The plating efficiency of infected cells was determined by dividing the number of colonies generated by infected cells in medium without puromycin by the number of colonies generated by mock-infected cells in medium without puromycin.
To assay the persistence of puromycin resistance in stably transformed cells propagated in the absence of selection, plates of puromycin-resistant colonies generated as described above were trypsinized after 14 days of growth in medium with puromycin. Cells were divided and placed on two 10-cm dishes (defined as week 0) for each experimental condition (MOIs for HDA.PAC+FR, 1, 3, 10, 30, and 100). One plate was propagated in medium with puromycin, and one plate was propagated without puromycin. Cells were passaged 1:3 when they reached confluence. At weeks 0, 1, 2, 3, 4, 6, and 11, cells maintained in medium with or without puromycin were assayed for the ability to form colonies in medium with puromycin as described above. The percentage of puromycin-resistant cells was calculated as the number of colonies in medium with puromycin divided by the number of colonies at week 0 times 100. Cells were counted once or twice per week to assess growth rate and doubling time.
Fluorescence assays.
A total of 5 × 104 D17 cells were used to seed each well of a 24-well cell culture plate. At 24 h after plating, cells were coinfected with HDA.Cre (MOI, 30) and HDA.CFP+FR (MOI, 10) in triplicate wells. Cells were trypsinized at 3 and 7 days p.i., placed on a glass slide, and assayed for the expression of YFP and CFP by fluorescence microscopy (Leica fluorescence microscope). For long-term expression experiments, cells were replated at a 1:3 split ratio. The relative percentage of CFP-positive cells was calculated by dividing the number of CFP-positive cells by the total number of cells.
Isolation of DNA.
A modified Hirt DNA (13) isolation procedure was used to isolate low-molecular-weight DNA. Cells were harvested from 60-mm dishes and resuspended in 1 ml of 10 mM Tris-HCl (pH 8.0)-10 mM EDTA-1 mg of pronase/ml. After 2 h of incubation at 37°C, 0.25 ml of 5 M NaCl was added, followed by overnight incubation at 4°C. The precipitate was removed by centrifugation at 15,000 × g for 60 min. The supernatant was precipitated with 0.7 volume of isopropanol, dissolved in 10 mM Tris-HCl (pH 8.0)-1 mM EDTA, and phenol-chloroform extracted. The aqueous supernatant was brought to 0.3 M sodium acetate and precipitated with 2.5 volumes of ethanol. The resulting Hirt DNA was redissolved in 1 ml of 10 mM Tris-HCl (pH 8.0)-1 mM EDTA-25 μg of DNase-free RNase (Boehringer Mannheim) and incubated at 37°C for 30 min.
Total cellular DNA was isolated by using a blood and tissue DNA extraction kit (Qiagen). The final DNA concentration was determined by measuring the absorbance at 260 nm.
RESULTS
Design of the HDA-EBV hybrid system.
We used two different HDA vectors to deliver functional EBV episomes into the nuclei of mammalian cells (Fig. 1). The first HDA vector (HDA.Cre) expressed the bacteriophage P1-derived Cre recombinase fused to the simian virus 40 T-antigen nuclear localization signal under the control of a CMV promoter. The 19-kb hORI sequence (19) comprised the largest part of the vector and functioned as a stuffer to guarantee that the size of the final HDA genome met the requirements for the efficient packaging of HDA genomes. As shown in an earlier study, HDAs sized at <27 kb propagate inefficiently and show a greater tendency to rearrange their genomes (28). HDA.Cre also contained a CMV promoter-driven YFP expression cassette for titration.
The second HDA vector was designated the target vector and contained the full-length EBV episome sequence. The sequences of the functional EBV episome included a 19-kb human chromosome 10-derived sequence for replication (hORI) (19), an expression cassette for the transgene and EBNA-1, and the EBV-derived FR. The EBV episome sequence was flanked by two parallel loxP sites so that Cre-induced recombination resulted in circularization of the intervening sequence (14). The resulting circular EBV episome, but not the linear target vector DNA, expressed the transgene and EBNA-1. This property of the target vector design was achieved through placement of the CMV promoter upstream of the bicistronic transgene-EBNA-1 expression cassette only after Cre-mediated circularization. Before recombination, the CMV promoter drives the expression of YFP used for the titration of vectors. Based on experience with previous target vector designs, the expression of EBNA-1 during vector propagation can interfere significantly with vector replication, probably due to binding of EBNA-1 to the FR (34). In the EBV episome, EBNA-1 is translated from the same RNA that encodes the transgene by translation initiation from an internal ribosome entry site derived from encephalomyocarditis virus (40).
Generation of EBV episomes in mammalian cells.
We confirmed the delivery and in vivo circularization of the EBV episome following coinfection of cells with the HDA target vector and HDA.Cre. We performed these studies with D17 cells to compare them to earlier studies in which these cells were used with first-generation adenovirus vectors to deliver EBV episomes (34). D17 cells were coinfected with HDA.PAC+FR at an MOI of 30 and HDA.Cre at an MOI of 10. All HDA titers were based on YTU on 293 cells. At 24, 48, and 72 h p.i., cells were harvested, and low-molecular-weight Hirt DNA was isolated from 106 cells, digested with BamHI, and analyzed by Southern blot with the PAC sequence as a probe (Fig. 2). With this strategy, a 3.3-kb fragment was generated from unrecombined DNA; in contrast, a 5.7-kb fragment was generated from the recombined circular episome.
FIG. 2.
Generation of the EBV episome by the HDA-EBV hybrid system. D17 cells were infected with HDA.PAC+FR (MOI, 30) either alone (lane 2) or with HDA.Cre (MOI, 10) (lanes 3 to 5). Hirt DNA from infected cells was prepared at 24, 48, and 72 h p.i. as indicated, digested with BamHI, and analyzed by Southern blotting with the PAC fragment as a probe. This strategy allows distinction of the EBV episome (5.7-kb band) from the linear, unrecombined HDA.PAC+FR DNA (3.3-kb band). Lane 1 contains a marker of the plasmid DNA used to prepare HDA.PAC+FR cut with BamHI and PmeI.
Vector DNA isolated from D17 cells at 72 h after infection with HDA.PAC+FR alone yielded only the 3.3-kb fragment indicative of unrecombined DNA, as expected in the absence of Cre (Fig. 2, lane 2). In contrast, Hirt DNA isolated from D17 cells that were coinfected with HDA.PAC+FR and HDA.Cre generated the EBV episome-specific 5.7-kb band. Based on phosphorimager analysis, the relative amount of episomal DNA compared to total vector DNA increased from 9 to 33% between 24 and 72 h after coinfection. The total amount of linear vector DNA decreased by about 4-fold between 24 and 72 h, whereas the amount of episomal DNA increased by about 1.5-fold. These data suggest that EBV episomes were rapidly generated within the first 24 h p.i. Furthermore, while the amount of EBV episomal DNA increased, linear vector DNA was lost at a rapid rate. We assume that the loss of the linear vector signal resulted from the continuing excision of PAC DNA through the action of Cre and dilution of the linear vector in the replicating cells. We assume again that the increase in the amount of episomal DNA resulted from the continued formation of episomes by the action of Cre on linear target vector DNA and the maintenance of the generated EBV episomes by one replication per S phase in the replicating cells.
Optimization of EBV episome delivery.
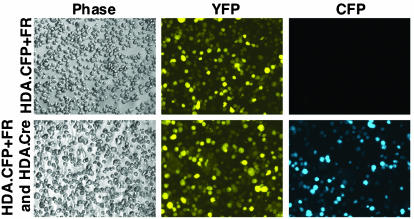
The binary design of our HDA-EBV vector system allowed us to vary the total and relative amounts of each vector used for coinfection. To quantitate the efficiency of episome delivery, we took advantage of the different excitation and emission spectra for YFP and CFP, allowing a clear distinction between the proteins by fluorescence microscopy (6). The target vector HDA.CFP+FR was designed to express only YFP in the linear form, allowing the quantitation of transduction efficiency. After Cre recombination, YFP expression was replaced by CFP expression from the EBV episome. This strategy allowed the simultaneous quantitation of the linear vector and the recombined product. As demonstrated in Fig. 3, D17 cells infected with HDA.CFP+FR alone showed only YFP expression. In contrast, coinfection of D17 cells with HDA.Cre and HDA.CFP+FR generated signals for both YFP- and CFP-derived emissions. The YFP signal originated from both HDA.Cre and unrecombined HDA.CFP+FR. This assay clearly distinguished YFP from CFP and confirmed the requirement for Cre for generation of the CFP-expressing episome.
FIG. 3.
Generation of the EBV episome expressing CFP. D17 cells were infected with HDA.CFP+FR (MOI, 30) either alone or with HDA.Cre (MOI, 10). Fluorescence microscopy performed 3 days p.i. allowed distinction of CFP expression and YFP expression due to different excitation and emission spectra. Cells infected with HDA.CFP+FR alone (upper panels) showed only YFP expression from the linear vector and no CFP expression. Coinfection of cells with both vectors (lower panels) resulted in YFP expression and CFP expression due to generation of the EBV episome. The cells in each column were the same in every field (phase, YFP, and CFP).
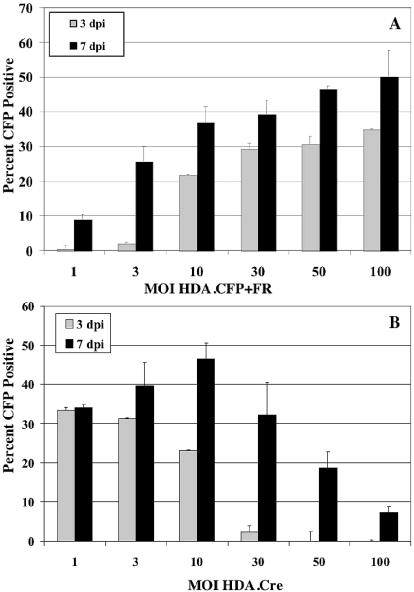
To evaluate the efficiency of EBV episome delivery as a function of vector dose, we initially infected D17 cells with a range of MOIs (1 through 100) for the CFP target vector and a constant MOI of 10 for the Cre-expressing vector. The number of cells showing YFP or CFP expression was assayed at days 3 and 7 after coinfection. The number of CFP-positive cells was compared to the number of total cells (Fig. 4A). The percentage of cells showing EBV episome-mediated CFP transgene expression was dependent on the MOI for HDA.CFP+FR and increased from day 3 to day 7. At an MOI of 1 for HDA.CFP+FR, only 9% of all cells showed CFP expression after 7 days. However, the percentage of CFP-positive cells increased steadily with increasing MOIs. At an MOI of 100 for HDA.CFP+FR, 50% of all cells were positive for CFP 7 days after coinfection. A similar trend was observed 3 days after coinfection, ranging from 1% CFP-positive cells at an MOI of 1 to 35% CFP-positive cells at an MOI of 100. These results indicate that the efficiency of EBV episome delivery is a function of the target vector dose and increases at higher MOIs.
FIG. 4.
(A) Optimization of the HDA loxP target vector dose. D17 cells were coinfected with HDA.Cre at a constant MOI (10) and with HDA.CFP+FR at various MOIs. The expression of CFP and YFP was evaluated by fluorescence microscopy after 3 and 7 days (dpi, days p.i.). The percentage of cells with EBV episome-mediated expression of CFP was calculated by dividing the total number of CFP-positive cells by the total number of cells. (B) Optimization of the HDA.Cre recombinase vector dose. D17 cells were coinfected with HDA.CFP+FR at a constant MOI (30) and with HDA.Cre at various MOIs. The expression of CFP and YFP was evaluated by fluorescence microscopy after 3 and 7 days. The percentage of cells with EBV episome-mediated expression of CFP was calculated by dividing the total number of CFP-positive cells by the total number of cells. Error bars indicate standard deviations.
Similarly, we studied the effect of varying the MOI for HDA.Cre on the efficiency of EBV episome delivery. For these experiments, D17 cells were coinfected with a range of MOIs (1 to 100) for HDA.Cre and at a constant MOI of 30 for HDA.CFP+FR (Fig. 4B). At a low MOI (1) for HDA.Cre, 34% of all cells showed EBV episome-mediated CFP expression after 7 days. This percentage increased to 46% at an MOI of 10 for HDA.Cre. Surprisingly, at a higher MOI (100) for HDA.Cre, the percentage of CFP-positive cells decreased significantly to 7% after 7 days. This unexpected observation suggests that increasing amounts of HDA.Cre may compete with the target vector during the coinfection process and therefore cause a decrease in EBV episome delivery.
Our binary HDA-EBV system allowed the use not only of various MOIs for each vector but also modifications of the timing of infection. The experiments described above all used infection of both vectors at the same time. We investigated the possibility that a sequential rather than a simultaneous infection of cells with these vectors might further improve the efficiency of transduction. We infected D17 cells with either HDA.CFP+FR at an MOI of 30 or HDA.Cre at an MOI of 10 on day 1. On the following day, cells that had previously been infected with HDA.CFP+FR were infected with HDA.Cre, and cells with prior HDA.Cre transduction were infected with HDA.CFP+FR. Cells that were infected with the CFP target vector on day 1 and the HDA.Cre vector on day 2 showed less CFP expression on days 3 and 7 (20 and 30%, respectively) than cells that were initially infected with the HDA.Cre vector and infected with the CFP target vector a day later (33 and 43% on days 3 and 7, respectively). Coinfection with both vectors yielded 39% CFP-positive cells on day 7 in this experiment. Therefore, sequential infection with HDA.Cre on day 1 and HDA.CFP+FR on day 2 was only marginally better than coinfection.
In summary, we performed a series of experiments to optimize the efficiency of EBV episome delivery. Based on the described data, we determined that coinfection of cells with an HDA target vector MOI of 30 and an HDA.Cre vector MOI of 10 yielded the highest percentage of acutely transformed D17 cells without significant cytotoxicity.
Stable transformation of D17 and HeLa cells by means of the HDA-EBV hybrid system.
We assayed the ability of the HDA-EBV hybrid system to stably transform mammalian cells in cultures to puromycin resistance. For these experiments, D17 and HeLa cells were chosen because both cell lines support EBV episome function and are nonpermissive for the production of ΔE1 adenovirus recombinants that could result from coinfection with a low level of helper virus. HeLa cells were analyzed to provide evidence that this HDA-EBV hybrid system functions in human cells.
To assess stable transformation, we performed a clonogenic assay similar to the previously described assay for the first-generation adenovirus system (34). D17 or HeLa cells were coinfected with HDA.Cre and HDA.PAC+FR on day 0. On day 1, 5 × 103 cells were divided, placed on duplicate 10-cm dishes, and allowed to grow without selection until day 7. At that time, 1 μg of puromycin/ml was added to one set of plates, and the appearance of drug-resistant colonies was monitored over 2 to 4 weeks. The growth of colonies under selective conditions requires recombination of the linear vector into an EBV episome, followed by efficient replication and segregation of the PAC-expressing episome into daughter cells. The number of puromycin-resistant colonies is a measure of stable transformation efficiency. Infected cells were also grown under nonselective conditions on the second set of plates to determine the plating efficiency of infected cells compared to the plating efficiency of uninfected cells. The plating efficiency was used to estimate the degree of cell toxicity induced by infection with the HDA vectors.
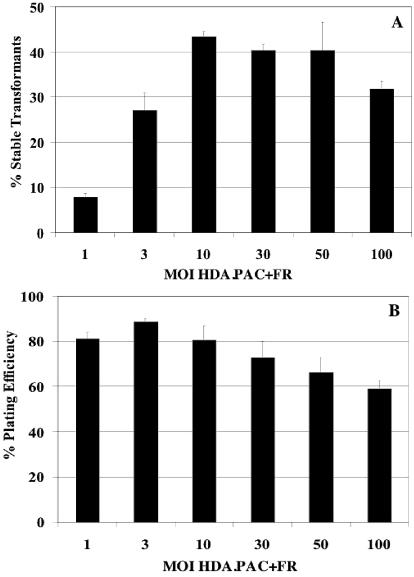
D17 cells infected with HDA.PAC+FR alone did not yield any puromycin-resistant transformants, demonstrating the requirement for Cre expression and subsequent recombination. Similarly, cells coinfected with HDA.Cre and the control vector HDA.PAC−FR, which lacks the FR, did not yield any puromycin-resistant colonies. In contrast, a significant number of D17 cells coinfected with HDA.Cre and HDA.PAC+FR were able to grow into colonies in puromycin-containing selection medium (Fig. 5A). The transformation efficiency, calculated by dividing the number of puromycin-resistant colonies in infected cells grown in medium with puromycin by the number of colonies generated by mock-infected cells grown in medium without puromycin, was found to be dependent on the MOI for HDA.PAC+FR, with a maximum of 43% at an MOI of 10. At higher MOIs for HDA.PAC+FR, the percentages of cells capable of forming puromycin-resistant colonies were similar (39% at an MOI of 30) or slightly decreased (32% at an MOI of 100). The transformation efficiencies at lower MOIs for HDA.PAC+FR were reduced to 27% for an MOI of 3 and to 8% for an MOI of 1.
FIG. 5.
(A) Generation of stable puromycin-resistant D17 transformants by the HDA-EBV hybrid system. D17 cells were coinfected with a constant amount of HDA.Cre (MOI, 10) and a range of MOIs for HDA.PAC+FR. Cells were selected in medium containing 1 μg of puromycin/ml starting at 7 days after the coinfection, and the appearance of colonies was monitored. The transformation efficiency was calculated as the number of colonies generated by infected cells in medium with puromycin divided by the number of colonies generated by mock-infected cells in medium without puromycin. (B) The cell plating efficiency as an estimate of cell viability was calculated by dividing the number of colonies generated by infected cells in medium without puromycin by the number of colonies formed by mock-infected cells in medium without puromycin. Error bars indicate standard deviations.
We estimated the toxicity induced in D17 cells by infection with the HDA-EBV hybrid system by comparing the plating efficiency of infected cells with the plating efficiency of uninfected cells. As shown in Fig. 5B, the plating efficiency was found to be dependent on the MOI for HDA.PAC+FR. Cells showed greater survival at lower MOIs, with plating efficiencies of 81, 88, and 80% at MOIs of 1, 3, and 10, respectively. At higher MOIs, survival decreased slightly, to 73% at an MOI of 30, to 66% at an MOI of 50, and to 59% at an MOI of 100. The level of cytotoxicity observed in the HDA-EBV system was significantly lower than that previously reported for first-generation vector systems. With the first-generation adenovirus-EBV hybrid system, doses of virus necessary to achieve maximum transformation of surviving cells resulted in about 95% cell death (34).
The ability of the HDA-EBV hybrid system to stably transform cells was also studied with the human cervical cancer cell line HeLa. As in the experiments described above, HeLa cells were coinfected with HDA.Cre and HDA.PAC+FR, followed by selection with puromycin. For HeLa cells, the absolute transformation efficiency increased with increasing MOIs of the target vector up to 16% at an MOI of 10 for HDA.PAC+FR. Higher MOIs for HDA.PAC+FR resulted in lower transformation efficiencies because of increased cytotoxicity, as assayed on the basis of plating efficiency (99% plating efficiency at an MOI of 10 and 61% at an MOI of 100). The results obtained with HeLa cells demonstrate that the function of the HDA-EBV hybrid system is not confined to a specific cell system.
Loss of transgene expression in proliferating D17 cells in the absence of selection for maintenance of the EBV episome.
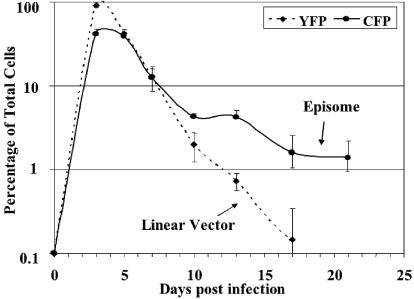
We analyzed the persistence of CFP expression from EBV episomes generated by Cre recombination and of YFP expression from linear vectors in proliferating D17 cells after coinfection with HDA.Cre at an MOI of 10 and HDA.CFP+FR at an MOI of 30. Cells were grown continuously after coinfection, trypsinized upon reaching confluence, and replated at a 1:3 ratio. Cells were assayed for CFP and YFP fluorescence at every passage and counted to determine the number of cell divisions. As shown in Fig. 6, YFP expression from the unrecombined form of the target vector and HDA.Cre reached a peak at 3 days (92%). However, YFP expression was rapidly lost from the proliferating cells. After day 13, the percentage of YFP-expressing cells decreased to <1%.
FIG. 6.
Loss of EBV episome-mediated CFP expression in proliferating D17 cells. D17 cells were infected with HDA.CFP+FR at an MOI of 30 and HDA.Cre at an MOI of 10 on day 0. Cells were divided twice a week at a 1:3 ratio and assessed for YFP and CFP expression at the indicated times. The percentages of fluorescent cells relative to total cells are shown for EBV-mediated CFP expression or YFP expression from linear vectors. Error bars indicate standard deviations.
In contrast, the percentage of cells showing episome-mediated CFP expression increased to 42% at day 3 after coinfection, followed by declines to 12% at day 7 and 4% at day 10. This initial rapid loss of episomal transgene expression was followed by a much slower loss after 10 days. On day 21 p.i., 2 to 4% of all cells showed persistence of CFP expression. These data indicate that the loss of EBV episomes from cells in which there is no selection for maintenance of the episomes follows a biphasic pattern, with an initial rapid loss followed by a subsequent slower loss.
Persistence of the EBV episome in stable D17 transformants.
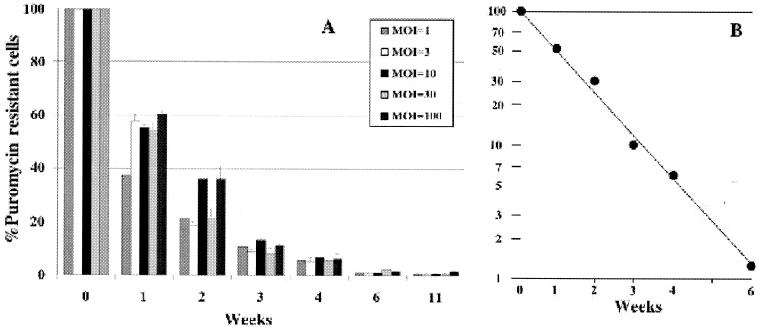
To assess the persistence of transgene expression in a cell population initially selected for the presence of the EBV episome, stable puromycin-resistant D17 transformants were generated by coinfection with HDA.Cre and HDA.PAC+FR. Cells were grown in puromycin-containing selection medium for 14 days and subsequently divided into two sets placed on 10-cm dishes. This time point was designated week 0. One set of plates was cultured with medium containing puromycin, and one set was cultured with medium not containing puromycin. Cells were replated at a ratio of 1:3 with the same medium each time they reached confluence. Colony formation in medium containing puromycin was assayed at weeks 0, 1, 2, 4, 6, and 11 for each set of plates. The number of cells under every experimental condition was counted to calculate cell doubling times.
Cells maintained in medium with puromycin up to 20 weeks continued to form puromycin-resistant colonies at the same high rate as at week 0. In contrast, D17 transformants grown under nonselective conditions demonstrated a slow, first-order kinetic loss in the ability to form puromycin-resistant colonies (Fig. 7). A semilog plot of the data revealed a half-time for puromycin resistance of 1 week, equal to seven doublings of the continuously passaged cells. This value corresponds to a probability of segregation of the EBV episome to daughter cells of 0.91 at each cell doubling. This probability is similar to the probability of EBV episome maintenance under nonselective conditions in several mammalian cell lines initially selected for transformation to drug resistance following plasmid DNA transfection (44).
FIG. 7.
Persistence of puromycin resistance of D17 transformants in nonselective medium. (A) D17 cells stably transformed to puromycin resistance after coinfection with various MOIs for HDA.PAC+FR and an MOI of 10 for HDA.Cre were passaged in medium with puromycin for 2 weeks and then in medium without puromycin (week 0). Colony formation in medium with puromycin was assayed at the indicated times after transfer to medium without puromycin. The number of colonies at each time point was compared to the number of colonies at week 0 for every MOI and is given as a percentage. (B) Semilogarithmic plot of the average percentage of puromycin-resistant colonies for all MOIs for HDA.PAC+FR.
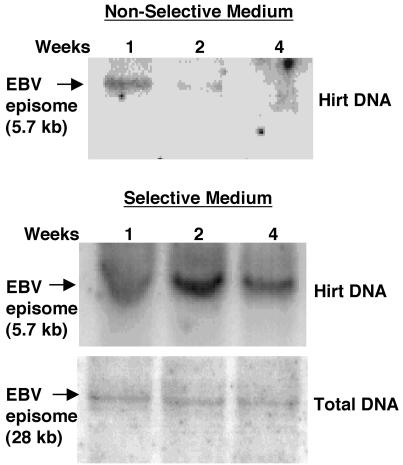
To analyze the maintenance of episomal DNA, low-molecular-weight and total DNAs were extracted from stable puromycin-resistant D17 transformants passaged under selective or nonselective conditions. Southern blot analysis was performed by digesting the DNA with either BamHI or NotI, followed by hybridization with the PAC probe. BamHI digestion releases a 5.7-kb band from the episome and a 3.3-kb band from the unrecombined linear target vector DNA. However, this restriction pattern does not distinguish between the EBV episome and integrated EBV episomal DNAs. In contrast, the NotI restriction site is present only once in the EBV episome and yields a 28-kb band for the extrachromosomal episome but fragments of various lengths for integrated episomal DNA.
The results of the Southern blot analysis are shown in Fig. 8. Under nonselective conditions, the EBV episome band was clearly visible after 1 week but was lost rapidly thereafter, was at the limits of detection by Southern blot and phosphoimager analyses after 2 weeks, and was undetectable at 4 weeks. This loss of EBV episomes in the absence of selection for their maintenance is consistent with the loss of the ability of the cells to form puromycin-resistant colonies. In contrast, when cells were selected for continued puromycin resistance, the full-length 28-kb plasmid was maintained. Of particular interest, most of the EBV episomes were maintained without integration into chromosomal DNA for at least 4 weeks, equivalent to >30 cell doublings. However, we cannot determine from this analysis whether a small fraction of the episomes were integrated, since random integration of a fraction of the episomes in this nonclonal population would produce a low-intensity smear that would not be observable.
FIG. 8.
Analysis of low-molecular-weight and total DNAs from stable D17 transformants. Low-molecular-weight and total DNAs from puromycin-resistant D17 transformants grown in nonselective medium (upper panel) and selective medium (lower panels) were analyzed by Southern blotting with the PAC sequence as a probe at the indicated times (weeks). Low-molecular-weight DNA was digested with BamHI to generate the episome-specific 5.7-kb band. Total DNA was digested with NotI to yield a 28-kb band indicative of the EBV episome.
In vivo transformation with the HDA-EBV hybrid system.
Renilla luciferase expressed after recombination of the HDA.RenLUC target vectors is able to generate light by catalyzing the oxidation of coelenterazine (2). The different substrates for Renilla and firefly luciferases allow these two reporter enzymes to be assayed independently. To assay the ability of target vectors to recombine in vivo, transgenic mice engineered to express the Cre recombinase under the control of the liver-specific albumin promoter (29) were infected with 2 × 109 YTU of HDA.RenLUC−FR by means of tail vein injection. Wild-type control mice without Cre recombinase expression received the same vector dose. Animals were sacrificed 12 days after the infection, and liver tissue was analyzed for the expression of firefly and Renilla luciferases. As shown in Fig. 9, Renilla luciferase expression was 30-fold higher in albumin-Cre-expressing mice than in control mice. Firefly luciferase expression from the contaminating helper virus was low in both groups, as expected based on the low level of helper virus contamination. These data suggest that the HDA.RenLUC−FR target vector can undergo Cre-mediated recombination and generate transgene-expressing episomes in vivo.
FIG. 9.
EBV episome-mediated expression of Renilla luciferase in mouse liver after injection of HDA.RenLUC−FR. Mice transgenic for expression of the Cre recombinase under the control of the liver-specific albumin promoter were injected intravenously with 2 × 109 YTU of HDA.RenLUC−FR. Liver tissue was analyzed for Renilla and firefly luciferases 12 days after the injection. Wild-type C57BL/6 mice were used as controls. Error bars indicate standard deviations.
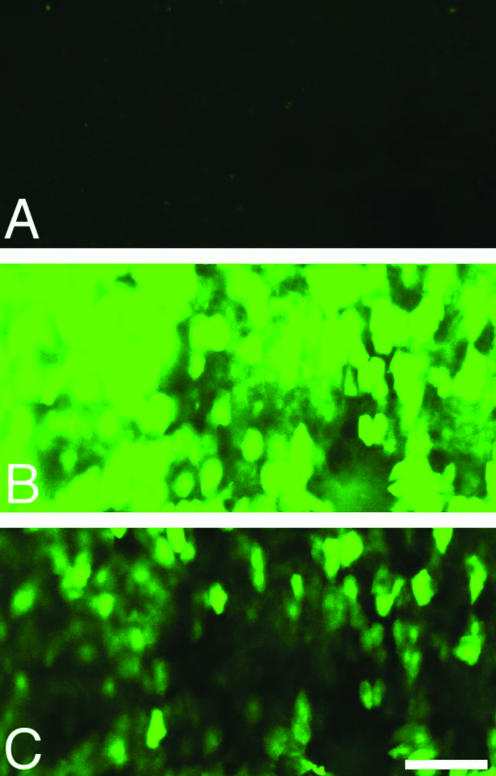
Our strategy of infecting cells with two different HDA vectors in order to generate EBV episomes, a Cre-expressing vector and a loxP target vector, can be successful in vivo only when cells are coinfected with both vectors. To test whether a large fraction of hepatocytes can be infected by our HDA vectors, HDA.Cre at 1.5 × 109 YTU was administered to nude mice by means of tail vein injection 6 h following tail vein injection of 6.4 mg of murine immunoglobulin to inhibit sequestration of the vector in Kupffer cells (35). Transduction of liver tissue was analyzed by confocal fluorescence microscopy of liver sections prepared 19 days p.i. YFP expression from the HDA.Cre vector was apparent in a large fraction of the hepatocytes (Fig. 10). These results suggest that it should be possible to coinfect hepatocytes with two vectors administered by means of tail vein injection at the vector concentrations currently available. Moreover, Yant et al. reported high-efficiency hepatocyte coinfection with two separate HDA vectors in mice (42).
FIG. 10.
Transduction of mouse hepatocytes following tail vein injection. A dose of 6.4 mg of mouse immunoglobulin was administered to an adult female nude mouse by means of tail vein injection to inhibit sequestration of the vector by Kupffer cells (35). After 6 h, HDA.Cre at 1.5 × 109 YTU was administered by means of tail vein injection. The mouse was sacrificed 19 days later, and 500-μm sections of liver embedded in agarose were prepared and examined for YFP fluorescence through a 15-μm z-axis stack under a fluorescein isothiocyanate channel by using a Bio-Rad confocal microscope. (A) Section from a control mouse injected with immunoglobulin only. (B) Section from a mouse injected with HDA.Cre at the same confocal settings as in panel A. (C) Same section as in panel B but at 38% the laser intensity used for panel B. Bar, 100 μm.
DISCUSSION
We have designed a novel HDA-EBV hybrid vector system capable of stably transforming mammalian cells with a high efficiency. Using coinfection of cells with two different HDA vectors, we were able to successfully deliver EBV episomes to different mammalian cell lines, as demonstrated by Southern blot and reporter gene assays. The HDA-EBV strategy resulted in acute transformation of about 40 to 50% of cells after 7 days and was therefore significantly more effective than EBV episome delivery by transfection of HDA plasmid DNA (≈5%). These data demonstrate that HDA-mediated delivery of EBV episomes is highly efficient compared to other methods. Furthermore, the binary nature of our system allowed manipulation of the doses for both the Cre-expressing and the target vectors and therefore optimization of transformation efficiency. As might be expected, the acute transformation efficiency increased significantly at higher target vector doses. However, higher total doses of HDA vectors also resulted in increasing cytopathic effects, possibly due to the exposure of cells to large amounts of adenovirus structural proteins. Interestingly, increasing the ratio of the Cre-expressing vector to the target vector resulted in a decrease in the acute transformation efficiency, likely due to competition between the two vectors during the multistep process of infection and recombination. The dose of vector necessary to generate a maximum number of episomes in a given cell population might depend on the cell type and the infection conditions.
Using the HDA-EBV hybrid system, we were able to generate a high percentage of stable puromycin-resistant transformants. The transformation efficiency of 43% was significantly greater than that (maximum of 10%) of a previously described first-generation adenovirus-EBV hybrid system (34). The greatly enhanced efficiency of the HDA-EBV hybrid system is primarily a function of decreased cytotoxicity compared to that of the ΔE1 system, particularly at higher MOIs. Puromycin-resistant D17 transformants cultured in selective medium were able to maintain EBV episomes over 30 cell doublings, as evidenced by the persistent ability to form colonies in puromycin-containing medium and by detection of the full-length, unintegrated episome by Southern blotting (Fig. 8). In the absence of selective pressure, the ability of D17 transformants to generate colonies in puromycin-containing selection medium as an indicator of the presence of episomal transgene expression decreased over time. The maintenance rate for D17 transformants was found to be 91% per cell division and was consistent with observations from previous studies indicating that the loss rate for EBV episomes was about 2 to 5% per generation in cells selected to be drug resistant following plasmid DNA transfection (25).
It was somewhat surprising that the maintenance of EBV episomes was not dependent on the HDA target vector MOI. Cells initially infected at higher MOIs for HDA.PAC+FR were expected to contain a larger number of EBV episomes per cell and therefore to maintain puromycin resistance longer than cells infected at lower MOIs. It is possible, however, that the number of recombination products per cell reaches a maximum that is dependent on the level of Cre expression or the availability of target vector genomes inside the nucleus. Furthermore, cells may only be able to maintain a certain limited number of functional EBV episomes by means of chromosomal attachment mechanisms. Other EBV episomes that are not tethered to metaphase chromosomes may be lost at much higher rates during cell division.
Transgene expression in proliferating cells cultured under conditions that did not select for the presence of episomes revealed a biphasic loss of EBV episomes. After transformation of D17 cells by coinfection with HDA.CFP+FR and HDA.Cre, an initially rapid decline in the fractions of cells expressing CFP was followed by decline at a much lower rate (Fig. 6). About 2 to 4% of cells continued to express CFP from an EBV episome at the end of 3 weeks. These findings are consistent with observations from previous studies in which plasmid DNA transfection was used to introduce EBV episomes (23). Leight and Sugden (23) estimated that the percentage of initially transfected cells able to maintain EBV episomes in a high percentage of daughter cells is between 1 and 10% under nonselective conditions. They suggested that the ability of cells to retain EBV episomes is dependent on epigenetic factors present in only a small fraction of cells in a given cell population. Even though EBV episomes were initially lost from dividing cells at a rapid rate, at least one daughter cell from every initially transformed cell must have retained the episome. Such cells remained puromycin resistant, explaining why the fraction of cells that were coinfected with HDA.PAC+FR and HDA.Cre and that gave rise to a drug-resistant colony (Fig. 5) were similar to the fraction of cells initially expressing CFP after coinfection with HDA.CFP+FR and HDA.Cre (Fig. 4). These cells remained puromycin resistant and gave rise to cells that segregated the plasmid to daughter cells at a high frequency, resulting in the formation of a drug-resistant colony. Once cells that segregate the episome to daughter cells arise, their descendants continue to segregate the episome at a high frequency, as shown by the low rate of episome loss from cells initially selected for growth in puromycin for 2 weeks (Fig. 7).
The epigenetic mechanism underlying the efficient segregation of EBV episomes to daughter cells (23) remains to be elucidated. However, the interaction of EBNA-1-bound episomes with cellular chromosomes required for episome segregation during mitosis likely involves cellular proteins in addition to EBNA-1. The cellular proteins p32/TAP and EBP2 have been reported to associate with the DNA-linking regions of EBNA-1 and may facilitate the interaction of EBNA-1 with mitotic chromosomes (4, 39). Hung et al. showed that a high-mobility group 1 or histone H1 protein sequence can substitute for EBNA-1 amino acids 1 to 378 and mediate the efficient accumulation of replicated oriP-containing plasmids, the association with mitotic chromosomes, nuclear retention, and long-term episome persistence (15).
Extrachromosomally replicated and maintained EBV episomes have been studied for gene therapeutic applications by a number of investigators (3). In cell cultures, an oriP- and EBNA-1-based construct was used to deliver the cystic fibrosis transmembrane conductance regulator gene to correct the cyclic AMP-dependent chloride transport defect in transformed human airway epithelial cells (22). With an EBV episome, the stable expression of the human hypoxanthine phosphoribosyltransferase gene in a deficient lung fibroblast cell line was observed over 6 months (38). EBV episomes have also shown promising results in animal models. A human factor IX-expressing EBV episome was used to transduce mouse liver and resulted in a 10- to 100-fold increase in transgene expression in the presence of EBNA-1 and the FR (33). Furthermore, high levels of factor IX were found up to 8 months after injection. Lee et al. used an EBV episome for transfer of the human multidrug resistance (MDR-1) gene. In the context of EBNA-1 and oriP, transfected cells showed higher-level and more prolonged expression of MDR-1 in vitro and in vivo than did controls (21).
The ability to deliver functional EBV episomes in vivo with our HDA-EBV system was confirmed after liver transduction of mice transgenic for albumin-Cre with a target vector. Transgene expression from the EBV episome was clearly dependent on the expression of Cre recombinase. In vivo, hepatocytes are mainly postmitotic, and liver tissue is therefore likely to maintain the EBV episome even in the absence of selection. It is also possible that only a small number of transduced cells are sufficient for therapeutic levels of transgene expression. Furthermore, EBV episomes may persist longer than linear HDA DNA because of resistance to exonucleases. An immune response to EBNA-1 is unlikely to present a problem, since the protein is poorly immunogenic (24). However, countering an immune response against the products of potentially therapeutic transgenes may prove to be a formidable challenge. Future studies will assess the long-term persistence of EBV-mediated transgene expression in animals transgenic for albumin-Cre and in mice coinjected with HDA target vectors and HDA.Cre.
In summary, we have demonstrated the feasibility of delivering EBV episomes to mammalian cells in vitro and in vivo by using an HDA vector system. Further studies will evaluate the persistence of EBV episome-mediated transgene expression in vivo. Our system has the potential to significantly improve the duration and increase the level of transgene expression for therapeutic and diagnostic gene therapy applications.
Acknowledgments
This work was supported by a generous grant from the American Association of Obstetricians and Gynecologists Foundation (AAOGF) to O.D., In Vivo Molecular and Cellular Imaging Center (ICMIC) award P50 CA88306 from the National Cancer Institute, and a UCLA Human Gene Therapy seed grant to A.J.B. S.D.G. received partial support from the Predoctoral Training Program in Genetic Mechanisms at UCLA (T32-GM07104), and J.S.G. received partial support from U.S. Public Health Service National Research Service award GM07185 and a Cota Robles Fellowship.
We are particularly grateful for the technical assistance of Carol Eng. Kohnosuke Mitani, Harvey Herschman, and Lily Wu contributed to this work during numerous discussions.
REFERENCES
- 1.Benihoud, K., P. Yeh, and M. Perricaudet. 1999. Adenovirus vectors for gene delivery. Curr. Opin. Biotechnol. 10:440-447. [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik, S., and S. S. Gambhir. 2002. Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc. Natl. Acad. Sci. USA 99:377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, J., and J. M. Vos. 2002. Establishment of an oriP/EBNA1-based episomal vector transcribing human genomic beta-globin in cultured murine fibroblasts. Gene Ther. 9:1447-1454. [DOI] [PubMed] [Google Scholar]
- 4.Ceccarelli, D. F., and L. Frappier. 2000. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J. Virol. 74:4939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L., M. Anton, and F. L. Graham. 1996. Production and characterization of human 293 cell lines expressing the site-specific recombinase Cre. Somat. Cell Mol. Genet. 22:477-488. [DOI] [PubMed] [Google Scholar]
- 6.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 7.Ehrhardt, A., and M. A. Kay. 2002. A new adenoviral helper-dependent vector results in long-term therapeutic levels of human coagulation factor IX at low doses in vivo. Blood 99:3923-3930. [DOI] [PubMed] [Google Scholar]
- 8.Gahery-Segard, H., F. Farace, D. Godfrin, J. Gaston, R. Lengagne, T. Tursz, P. Boulanger, and J. G. Guillet. 1998. Immune response to recombinant capsid proteins of adenovirus in humans: antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J. Virol. 72:2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 10.Haase, S. B., and M. P. Calos. 1991. Replication control of autonomously replicating human sequences. Nucleic Acids Res. 19:5053-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harui, A., S. Suzuki, S. Kochanek, and K. Mitani. 1999. Frequency and stability of chromosomal integration of adenovirus vectors. J. Virol. 73:6141-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinzel, S. S., P. J. Krysan, C. T. Tran, and M. P. Calos. 1991. Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol. Cell. Biol. 11:2263-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 14.Hoess, R. H., and K. Abremski. 1984. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc. Natl. Acad. Sci. USA 81:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan, G., E. M. Miyashita, B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1996. Is EBV persistence in vivo a model for B-cell homeostasis? Immunity 5:173-179. [DOI] [PubMed] [Google Scholar]
- 17.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2396. In B. N. Fields (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 18.Krysan, P. J., and M. P. Calos. 1993. Epstein-Barr virus-based vectors that replicate in rodent cells. Gene 136:137-143. [DOI] [PubMed] [Google Scholar]
- 19.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langle-Rouault, F., V. Patzel, A. Benavente, M. Taillez, N. Silvestre, A. Bompard, G. Sczakiel, E. Jacobs, and K. Rittner. 1998. Up to 100-fold increase of apparent gene expression in the presence of Epstein-Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J. Virol. 72:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, C. G., W. D. Vieira, I. Pastan, and M. M. Gottesman. 2001. An episomally maintained MDR1 gene for gene therapy. Hum. Gene Ther. 12:945-953. [DOI] [PubMed] [Google Scholar]
- 22.Lei, D. C., K. Kunzelmann, T. Koslowsky, M. J. Yezzi, L. C. Escobar, Z. Xu, A. R. Ellison, J. M. Rommens, L. C. Tsui, M. Tykocinski, and D. C. Gruenert. 1996. Episomal expression of wild-type CFTR corrects cAMP-dependent chloride transport in respiratory epithelial cells. Gene Ther. 3:427-436. [PubMed] [Google Scholar]
- 23.Leight, E. R., and B. Sugden. 2000. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev. Med. Virol. 10:83-100. [DOI] [PubMed] [Google Scholar]
- 24.Levitskaya, J., M. Coram, V. Levitsky, S. Imreh, P. M. Steigerwald-Mullen, G. Klein, M. G. Kurilla, and M. G. Masucci. 1995. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 375:685-688. [DOI] [PubMed] [Google Scholar]
- 25.Middleton, T., and B. Sugden. 1994. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J. Virol. 68:4067-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyashita, E. M., B. Yang, K. M. Lam, D. H. Crawford, and D. A. Thorley-Lawson. 1995. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell 80:593-601. [DOI] [PubMed] [Google Scholar]
- 27.Parks, R. J., L. Chen, M. Anton, U. Sankar, M. A. Rudnicki, and F. L. Graham. 1996. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. USA 93:13565-13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parks, R. J., and F. L. Graham. 1997. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 71:3293-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Postic, C., and M. A. Magnuson. 2000. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis 26:149-150. [DOI] [PubMed] [Google Scholar]
- 30.Reichel, M. B., R. R. Ali, A. J. Thrasher, D. M. Hunt, S. S. Bhattacharya, and D. Baker. 1998. Immune responses limit adenovirally mediated gene expression in the adult mouse eye. Gene Ther. 5:1038-1046. [DOI] [PubMed] [Google Scholar]
- 31.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 5:1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiedner, G., N. Morral, R. J. Parks, Y. Wu, S. C. Koopmans, C. Langston, F. L. Graham, A. L. Beaudet, and S. Kochanek. 1998. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat. Genet. 18:180-183. [DOI] [PubMed] [Google Scholar]
- 33.Sclimenti, C. R., A. S. Neviaser, E. J. Baba, L. Meuse, M. A. Kay, and M. P. Calos. 2003. Epstein-Barr virus vectors provide prolonged robust factor IX expression in mice. Biotechnol. Prog. 19:144-151. [DOI] [PubMed] [Google Scholar]
- 34.Tan, B. T., L. Wu, and A. J. Berk. 1999. An adenovirus-Epstein-Barr virus hybrid vector that stably transforms cultured cells with high efficiency. J. Virol. 73:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao, N., G. P. Gao, M. Parr, J. Johnston, T. Baradet, J. M. Wilson, J. Barsoum, and S. E. Fawell. 2001. Sequestration of adenoviral vector by Kupffer cells leads to a nonlinear dose response of transduction in liver. Mol. Ther. 3:28-35. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, C. E., G. Schiedner, S. Kochanek, M. G. Castro, and P. R. Lowenstein. 2000. Peripheral infection with adenovirus causes unexpected long-term brain inflammation in animals injected intracranially with first-generation, but not with high-capacity, adenovirus vectors: toward realistic long-term neurological gene therapy for chronic diseases. Proc. Natl. Acad. Sci. USA 97:7482-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umana, P., C. A. Gerdes, D. Stone, J. R. Davis, D. Ward, M. G. Castro, and P. R. Lowenstein. 2001. Efficient FLPe recombinase enables scalable production of helper-dependent adenoviral vectors with negligible helper-virus contamination. Nat. Biotechnol. 19:582-585. [DOI] [PubMed] [Google Scholar]
- 38.Wade-Martins, R., R. E. White, H. Kimura, P. R. Cook, and M. R. James. 2000. Stable correction of a genetic deficiency in human cells by an episome carrying a 115 kb genomic transgene. Nat. Biotechnol. 18:1311-1314. [DOI] [PubMed] [Google Scholar]
- 39.Wang, Y., J. E. Finan, J. M. Middeldorp, and S. D. Hayward. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236:18-29. [DOI] [PubMed] [Google Scholar]
- 40.Witherell, G. W., and E. Wimmer. 1994. Encephalomyocarditis virus internal ribosomal entry site RNA-protein interactions. J. Virol. 68:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, Y., F. A. Nunes, K. Berencsi, E. E. Furth, E. Gonczol, and J. M. Wilson. 1994. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc. Natl. Acad. Sci. USA 91:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yant, S. R., A. Ehrhardt, J. G. Mikkelsen, L. Meuse, T. Pham, and M. A. Kay. 2002. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat. Biotechnol. 20:999-1005. [DOI] [PubMed] [Google Scholar]
- 43.Yates, J., N. Warren, D. Reisman, and B. Sugden. 1984. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc. Natl. Acad. Sci. USA 81:3806-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, H., L. Pastore, and A. L. Beaudet. 2002. Helper-dependent adenoviral vectors. Methods Enzymol. 346:177-198. [DOI] [PubMed] [Google Scholar]
- 46.Zou, L., P. Yotnda, T. Zhao, X. Yuan, Y. Long, H. Zhou, and K. Yang. 2002. Reduced inflammatory reactions to the inoculation of helper-dependent adenoviral vectors in traumatically injured rat brain. J. Cereb. Blood Flow Metab. 22:959-970. [DOI] [PubMed] [Google Scholar]