Abstract
In the F family of conjugative plasmids, TraJ is an essential transcriptional activator of the tra operon that encodes most of the proteins required for conjugation. Here we report for the first time the X-ray crystal structures of the TraJ N-terminal domains from the prototypic F plasmid (TraJF11–130) and from the Salmonella virulence plasmid pSLT (TraJpSLT1–128). Both structures contain similar Per-ARNT-Sim (PAS) folds, which further homodimerize through the N-terminal helix and the structurally conserved β-sheet of the PAS fold from each protomer. Mutational analysis reveals that the observed dimeric interface is critical for TraJF transcriptional activation, indicating that dimerization of TraJ is required for its in vivo function. TraJ is specific in activating its cognate tra operon promoter; however, heterologous PAS domains from pSLT and R100 TraJ can functionally replace the TraJF PAS domain, suggesting that the allelic specificity of TraJ is solely mediated by the region C-terminal to the PAS domain.
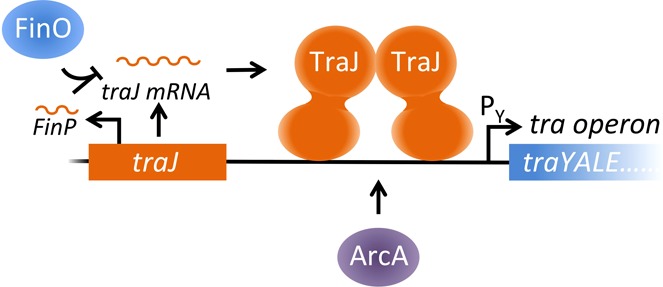
Conjugative DNA transfer is the major cause of rapid dissemination of antibiotic resistance and virulence factors in bacterial pathogens. During bacterial conjugation, a donor bacterium intimately contacts a recipient to form a conjugative pore (a bacterial type IV secretion system) through which a single-stranded DNA is transferred from the donor to the recipient bacterium.1,2 In the F and F-like plasmid-mediated bacterial conjugation systems, more than 20 different proteins are involved in DNA processing and conjugative pore formation, most of which are expressed from the polycistronic tra operon in the plasmid.1 Transcription of the tra operon requires TraJ, an essential activator of the tra operon promoter, PY.
The traJ gene is located immediately upstream of the tra operon in the F and F-like plasmids, encoding a cytoplasmic protein that binds DNA upstream of PY presumably through its putative C-terminal helix–turn–helix (HTH) DNA-binding motif (Figure S1 of the Supporting Information).3 TraJ is thought to function to relieve PY from H-NS (histone-like nucleoid structuring protein) repression as well as to activate PY in an independent mechanism together with a host factor, ArcA.4−6 As a central regulator of bacterial conjugation, the intracellular level of TraJ is stringently regulated at multiple levels by different host and plasmid-encoded factors. CRP (catabolite repressor protein) and Lrp (leucine-responsive protein) regulate traJ transcription,7−9 whereas Hfq (host factor for Qβ replicase) and the plasmid-encoded FinOP (fertility inhibition) system affect TraJ translation.10−12 Heat shock chaperonin protein GroEL and the Escherichia coli CpxAR two-component system have been found to mediate proteolytic degradation of TraJ.13−15
Despite limited pairwise amino acid sequence identity, all F family TraJ homologues are predicted to have a PAS domain in the N-terminal region (Figure S1 of the Supporting Information).16,17 PAS domains commonly function as sensors in signaling proteins to regulate diverse physiological processes in all three kingdoms of life.17,18 PAS domains are characterized by a five-stranded antiparallel β-sheet, flanked by varied numbers of α-helices. Some of the well-characterized PAS proteins include FixL, which is involved in nitrogen fixation in bacteria;19 PER, which controls circadian behavior in insects;20 and HIF proteins, which regulate the response to hypoxia in mammals.21 Typically, PAS domains are covalently linked to and regulate the activities of various effector domains, including enzymes, transcription factors/DNA-binding domains, ion channels, and chemotaxis proteins. PAS domains usually facilitate dimerization or higher-order oligomerization. Some PAS folds bind small-molecule ligands such as metabolites, heme, and flavin nucleotides to exert their physiological activities.18,22
Our recent mutational analysis indicates that the putative PAS domain of TraJF is essential for its homodimerization, intracellular protein stability, and in vivo function.23 Mutation of multiple cysteine residues (Cys30, Cys41, and Cys67) within the TraJF PAS domain significantly inhibits F conjugation, which leads to the hypothesis that the TraJ cysteines form a redox center for sensing oxidative stress.16 As most of the characterized single missense mutations can significantly elevate the susceptibility of TraJF to proteolytic degradation by the protease–chaperone pair HslVU, it was further hypothesized that oxidation of the TraJ PAS domain might trigger HslVU-catalyzed degradation of TraJ, resulting in repression of F plasmid-mediated bacterial conjugation.23
To obtain structural insights into the putative PAS domain of TraJ, we subcloned and crystallized the N-terminal regions of TraJ from the prototypic F plasmid and Salmonella enteric Serovar Typhimurium virulence plasmid pSLT. The two structures, F plasmid TraJ11–130 and pSLT TraJ1–128, were determined at 1.55 and 1.67 Å resolution, respectively. Both structures exhibit characteristic features of a PAS fold and form homodimers through an extensive dimeric interface. Mutational analysis revealed that residues forming the dimeric interface of TraJF11–130 are important for TraJ in activating the traJ operon promoter PY, indicating that homodimerization is required for TraJ function. Both the N-terminal PAS domains of pSLT and R100 TraJ are fully active in replacing the corresponding PAS domain of F plasmid TraJ, despite the fact that each of these TraJ homologues is specific in activating its own cognate PY, indicating that the allelic specificity of TraJ is likely governed by its C-terminal domain. On the basis of these results, we further discuss the role of TraJ dimerization in regulating the in vivo function of TraJ.
Experimental Procedures
Growth Media and Bacterial Strains
Cells were grown in LB (Luria-Bertani) broth or on LB solid medium unless otherwise specified. Antibiotics were used at the following final concentrations: 100 μg/mL ampicillin and 30 μg/mL kanamycin. The following E. coli strains were used: DH5α [F–ΔlacU169 (Φ80lacZΔM15) supE44 hsdR17 recA1 endA1 gyrA96(Nalr) thi-1 relA1],24 BL21-DE3 [F–ompT hsdSB (rB–mB–) gal dcm] (Invitrogen), BW25113 [Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ–rph-1 Δ(rhaD-rhaB)568 hsdR514],25 JW3686 (ΔtnaA739::kan mutant of BW25113),25 and BE280 [lacZ13(Oc) lacI22 trp-37 rpsL106(strR) xylA13 tnaA4 phoS-3 ilv-280].26
Plasmids, Oligonucleotides, and Plasmid Construction
All plasmids and oligonucleotides used in this work are listed in Table S1 of the Supporting Information. pJLJ2123 was constructed by ligating the 2.5 kb EcoRI–BamHI fragment of pT7-727 to the 0.45 kb EcoRI–BamHI fragment of DNA amplified from pJLJ001 using JLU321 and JLU323 as primers. pJLJ2829 was constructed by ligating the 2.5 kb EcoRI–BamHI fragment of pT7-727 to the 0.4 kb EcoRI–BamHI fragment of DNA amplified from pJLJ001 using JLU328 and JLU329 as primers. pJLJ5629 was constructed by ligating the 2.4 kb EcoRI–BamHI fragment of pK18428 to the 0.4 kb EcoRI–BamHI fragment of DNA amplified from pJLJ001 using JLU356 and JLU329 as primers. pJLJ002 was constructed by ligating the 2.4 kb EcoRI–BamHI fragment of pK18428 to the 0.7 kb EcoRI–BamHI fragment of DNA amplified from pIZ2023 (a gift from J. Casadesus, Universidad de Sevilla, Sevilla, Spain) using JLU390 and JLU394 as primers. pJLJ003 was constructed by ligating the 2.4 kb EcoRI–BamHI fragment of pK18428 to the 0.8 kb EcoRI–BamHI fragment of DNA amplified from plasmid R100 using JLU330B and JLU331B as primers. Overlap extension29 was used to construct pJLJ004 and pJLJ005, expressing hybrid TraJ proteins TraJpSLT1–125:TraJF121–226 and TraJR1001–120:TraJF121–226, respectively. The PCR primer pair of JLU390 and JLU391was used to amplify a fragment encoding the plasmid pSLT TraJ N-terminal region (TraJpSLT1–125) from pJLJ002. The PCR primer pair of JLU330B and JLU347 was used to amplify a fragment encoding the plasmid R100 TraJ N-terminal region TraJR1001–120 from pJLJ003. The primer pair of JLU346 and JLU308 was used to amplify a fragment containing the F plasmid TraJ C-terminal region (TraJF121–226) from pJLJ001.23 The primer pair of JLU390 and JLU308 was used to amplify a pSLT-F hybrid traJ (encoding TraJpSLT1–125:TraJF121–226) fragment, which was further digested by EcoRI and BamHI and cloned into the EcoRI and BamHI sites of pK184, resulting in pJLJ004. The primer pair of JLU330B and JLU308 was used to amplify an R100-F hybrid traJ (encoding TraJR1001–120:TraJF121–226) fragment, which was further digested by EcoRI and BamHI and cloned into the EcoRI and BamHI sites of pK184, resulting in pJLJ005. The gene fragment of the pSLT TraJ PAS-like domain (TraJpSLT1–128) was PCR-amplified using genomic DNA from Salmonella enterica serovar Typhimurium str. 14028S (gi: 267990107) as a template and two primers: forward TraJA and backward TraJB. The PCR product was cloned into vector pMCSG730 according to the ligation-independent cloning procedure,31,32 resulting in pJSLT128. This vector introduces an N-terminal His6 tag followed by a TEV protease recognition site (Table S1 of the Supporting Information).
Protein Expression and Purification
To overexpress His6-tagged TraJF11–130, BL21-DE3 cells containing pJLJ2829 were grown in 1 L of LB broth at 37 °C while being vigorously shaken. After 3 h, IPTG was added to a final concentration of 0.05 mM, and the culture was grown for an additional 16 h at 20 °C before cells were harvested by centrifugation. The cell pellet was suspended in 80 mL of lysis buffer [50 mM Tris-HCl, 10 mM imidazole, 250 mM NaCl, and 10% glycerol (pH 7.0)] with one tablet of Complete, EDTA-free, protease inhibitor cocktail (Roche Applied Science). The suspension was lysed by sonication on ice for 3 min (30 s with a 30 s break, repeated six times) at maximal output. After centrifugation at 27000g for 60 min, the supernatant was loaded on a column with 2 mL of Ni-NTA agarose (Qiagen) pre-equilibrated with 20 mL of lysis buffer. After the sample had been washed with 30 mL of buffer [50 mM Tris-HCl, 20 mM imidazole, 250 mM NaCl, and 10% glycerol (pH 7.0)], the protein bound to the Ni-NTA agarose was eluted with 10 mL of elution buffer [50 mM Tris-HCl, 250 mM imidazole, 250 mM NaCl, and 10% glycerol (pH 7.0)] in 2 mL fractions. The fractions containing His-tagged TraJF11–130 were pooled and mixed with 300 units of AcTEV protease (Invitrogen) at room temperature for 24 h to cleave the His6 tag. The digested mixture was separated by size exclusion chromatography (Hiload 26/60 Superdex 75 prep grade column, Amersham Biosciences), and proteins were eluted with SEC buffer [50 mM Tris-HCl, 250 mM NaCl, and 1 mM dithiothreitol (pH 7.2)]. The peak containing pure TraJF11–130 was concentrated and buffer-exchanged to 0.5 M ammonium acetate, 10% glycerol, and 5 mM dithiothreitol (DTT) by using an Amicon ultracentrifuge filter (Millipore). Selenomethionyl TraJF1–140 was overexpressed from plasmid pJLJ2123 as previously described33 and purified in the same manner as TraJF11–130. The protein concentration was determined by using BCA protein assays (Pierce) following the manufacturer’s instructions.
E. coli BL21 cells containing pJSLT128 were grown with ampicillin and kanamycin. A selenomethionine (SeMet) derivative of the expressed protein was prepared and purified using Ni affinity chromatography as described previously.34,35 Briefly, the harvested cells, containing SeMet-labeled protein, were resuspended in lysis buffer [500 mM NaCl, 5% (v/v) glycerol, 50 mM HEPES (pH 8.0), 10 mM imidazole, and 10 mM 2-mercaptoethanol], and the lysate was clarified by centrifugation, filtered through a 0.44 μm membrane, and applied to a 5 mL HiTrap Ni-NTA column (GE Health Systems) on an AKTAxpress system (GE Health Systems). The His6-tagged protein was removed by treatment with recombinant His7-tagged TEV protease (a gift from D. Waugh, National Cancer Institute, Bethesda, MD). Subtractive Ni-NTA affinity chromatography was used to remove the His6 tag, uncut protein, and His7-tagged TEV protease. The protein of TraJ PAS-like domain was concentrated and exchanged into crystallization buffer containing 250 mM NaCl, 20 mM HEPES (pH 8.0), and 2 mM DTT through an Amicon Ultra centrifugal filter device. The protein concentration was determined by using UV spectroscopy at 280 nm.
Crystallization and Data Collection
TraJF11–130 or selenomethionyl TraJF1–140 was concentrated to ∼3 mg/mL in 0.5 M ammonium acetate, 10% glycerol, and 5 mM DTT. Crystals were obtained using the hanging drop vapor diffusion technique with 3 μL of protein mixed with 3 μL of the well solution at 4 °C for 3–6 weeks. The well solution containing 2.0 M (NH4)2SO4 and 100 mM sodium citrate (pH 5.5) was used for growing both crystals, which were subsequently soaked in the well solution with 25% glycerol for 20 min prior to being flash-frozen in liquid nitrogen. Data were collected from exposure of single crystals at SIBYLS Beamline of the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA). A three-wavelength MAD data set was collected in inverse-beam mode to 2.3 Å resolution for selenomethionyl TraJF1–140 (SeMet-TraJF1–140), and a native data set was collected to 1.55 Å for TraJF11–130. Data were processed and scaled using the HKL2000 package, and the statistics are listed in Table S2 of the Supporting Information.36
TraJpSLT1–128 at 30 mg/mL was crystallized using sitting drop vapor diffusion at room temperature in a Crystal Quick VR 96-well round-bottom plate (Greiner Bio-One North America). The protein solution (0.4 μL) was mixed with 0.4 μL of crystallization reagent using the Mosquito VR nanoliter liquid workstation (TTP LabTech) and allowed to equilibrate against 135 μL of crystallization reagent. Four different crystallization MCSG (the Midwest Center for Structural Genomics) screens (Microlytic) were used: MCSG-1, MCSG-2, MCSG-3, and MCSG-4. The best crystals were obtained under the 67th condition of MCSG-2 that contains 0.1 M dl-malic acid (pH 7.0) and 20% PEG 3500. Prior to data collection, the crystals were cryoprotected using the solution prepared by adding 15% (v/v) glycerol to the crystallization condition and flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K at beamline 19-ID of the Structural Biology Center at the Advanced Photon Source (Argonne National Laboratory). Single-wavelength anomalous dispersion (SAD) data at 0.9792 Å were collected from a single SeMet-labeled protein crystal. The data were collected using SBCCOLLECT and processed and scaled by the HKL3000 suite.37
Determination and Analysis of Structure
SOLVE38 located four expected selenium atoms in the asymmetric unit using the 2.3 Å selenomethionyl TraJF1–140 MAD data. The best solution from SOLVE had a Z score of 57.2 and a mean figure of merit of 0.3. The maximum likelihood density modification in RESOLVE39 was used to improve initial phases, yielding an overall figure of merit of phasing of 0.58. Automatic building by RESOLVE40 using the protein sequence generated a model with ∼200 residues built in an asymmetric unit, which contain a homodimer of SeMet-TraJF1–140. The model was used for molecular replacement with MOLREP41 to generate a TraJF11–130 model from the TraJF11–130 native data set at 3 Å, which was used as a starting model against the 1.55 Å TraJF11–130 native data set for automated model building in ARP/wARP.42 Iterative runs of ARP/wARP combined with manual model building and iterative cycles of refinement in REFMAC43 were used to complete and refine the model at 1.55 Å resolution. The final model of the crystallographic asymmetric unit contains two TraJF11–130 dimers. There was no interpretable electron density for residues 10, 11, and 128–130. Refinement statistics are summarized in Table 1.
Table 1. Data Collection and Refinement Statisticsa.
| SeMet-TraJF1–140 |
|||||
|---|---|---|---|---|---|
| peak | remote | inflection | native TraJF11–130 | SeMet-TraJpSLT1–128 | |
| Data Collection | |||||
| space group | P62 | P61 | P3221 | ||
| cell dimensions [a, b, c (Å)] | 94.54, 94.54, 52.73 | 95.48, 95.48, 102.3 | 57.08, 57.08, 67.64 | ||
| wavelength (Å) | 0.9796 | 0.9537 | 0.9797 | 1.1158 | 0.9792 |
| resolution (Å) | 50–2.30 (2.30–2.38) | 50–2.30 (2.30–2.38) | 50–2.30 (2.30–2.38) | 50–1.55 (1.58–1.55) | 1.67–50 (1.67–1.70) |
| Rmerge (%) | 10.5 (69.7) | 11.2 (76.0) | 10.4 (70.9) | 5.8 (70.8) | 4.9 (45.2) |
| I/σI | 19.4 (2.7) | 18.7 (2.4) | 19.8 (2.7) | 38.6 (2.3) | 14.9 (3.4) |
| completeness (%) | 100 (100) | 100 (99.6) | 100 (99.8) | 99.8 (97.2) | 99.4 (98.7) |
| redundancy | 7.0 (6.6) | 6.9 (6.5) | 7.0 (6.6) | 10.5 (6.1) | 6.0 (5.0) |
| Refinement | |||||
| resolution (Å) | 50–1.55 | 1.67–27.9 | |||
| no. of reflections | 75754 | 15152 | |||
| Rwork/Rfree | 16.0/17.8 | 19.5/22.8 | |||
| no. of atoms | |||||
| protein | 3911 | 1008 | |||
| ligand/ion | 30 | 26 | |||
| water | 333 | 42 | |||
| B factor | |||||
| protein | 27.0 | 35.3 | |||
| ligand/ion | 30.5 | 43.0 | |||
| water | 37.4 | 40.7 | |||
| root-mean-square deviation | |||||
| bond lengths (Å) | 0.007 | 0.006 | |||
| bond angles (deg) | 1.27 | 0.976 | |||
| Ramachandran (%) | |||||
| favored | 97.3 | 100 | |||
| allowed | 2.7 | 0 | |||
| outlier | 0 | 0 | |||
Numbers in parentheses are values for the highest-resolution bin.
The structure of TraJpSLT1–128 was determined by SAD phasing using the HKL3000 suite.37 SHELXD was used for heavy atom search, and initial phases were obtained from SHELXE.44 The heavy atom sites were refined, and improved phases were calculated by iterations of MLPHARE45 and DM.46 The initial protein models were built in ARP/wARP.47 Manual model rebuilding was conducted in COOT,48 and crystallographic refinement was performed in PHENIX51.49 The final model refined to 1.67 Å was evaluated by MolProbity and a Ramachandran plot with good R and Rfree values and stereochemistry. There was no interpretable electron density for N-terminal residues 1–13 and C-terminal residues 127 and 128. The details of the data collection, structure refinement, and model quality are shown in Table S2 of the Supporting Information. The molecular structure figures were prepared by using PyMOL (http://www.pymol.org) with surface of internal cavities created using HOLLOW (http://hollow.sourceforge.net). CASTp was used for calculating the area and volume of protein internal cavities.50 Areaimol was used for calculating the area of the protein contact surface.51
TraJ in Vivo Activity Assays
Plasmid pJLJ001 (constitutively expressing TraJF at close to physiological levels) or one of its TraJF mutant derivatives was transformed into an E. coli DH5α strain containing plasmid pJLac101-PY (carrying an F plasmid PY-lacZ fusion). A fresh, single transformant was inoculated into LB broth containing appropriate antibiotics and grown at 37 °C while being shaken for 4 h to an OD600 between 0.5 and 1. The function of TraJF (or its mutant) in activating PY is represented by the β-galactosidase activity (LacZ activity) determined as described by Miller52 and reported as Miller units (MU) calculated using the equation 1000[A420/(tvOD600)], where t is the time of reaction (minutes) and v is the volume of culture added (milliliters).
Site-Directed Mutagenesis
All the primers and plasmids used are listed in Table S1 of the Supporting Information. All missense mutations of traJ are generated as a derivative of plasmid pJLJ001 that constitutively expresses wild-type TraJF close to physiological levels.23 L18A was constructed by ligating the 2.4 kb EcoRI–BamHI fragment of pK184 to the 0.7 kb EcoRI–BamHI fragment of DNA amplified from pJLJ001 using JLU349 and JLU308 as primers. Site-directed mutagenesis through overlap extension29 was used to construct plasmids expressing TraJF-F105A and TraJF-Q116A. The primer pairs used for introducing point mutations into TraJF are JLU350 and JLU351 for F105A and JLU352 and JLU353 for Q116A. Primers JLU307 and JLU308 were used with the primers listed above to amplify the mutated traJ DNA fragments, which were digested by EcoRI and BamHI and cloned into pK184, resulting in different pJLJ001 mutant derivatives.
Results
The N-Terminal Region of TraJ Adopts a Conserved PAS Fold Structure
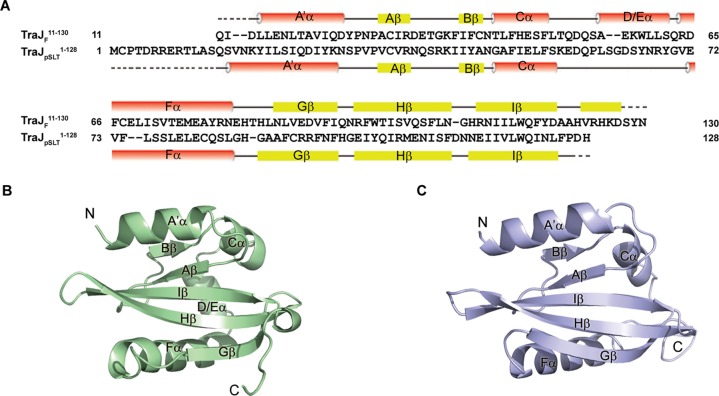
Our previous study indicated that the N-terminal fragment of the F plasmid TraJ, TraJF1–140, can be stably overexpressed in E. coli.23 We crystallized and determined the structure of this domain (Table 1). Better crystals were obtained with TraJF11–130 after deletion of the disordered regions in the N- and C-termini of TraJF1–140, and its structure was further determined as the final model of the TraJF PAS domain (Table 1). We also crystallized a related domain of TraJ from S. enterica Serovar Typhimurium virulence plasmid pSLT, TraJpSLT1–128, which shares ∼20% pairwise amino acid sequence identity with its TraJF counterpart (Figure 1A). Both TraJF and TraJpSLT structures were independently determined using Se-methionine-substituted proteins and anomalous scattering techniques, and each was refined to high resolution [1.55 Å for TraJF and 1.67 Å for TraJpSLT (see Experimental Procedures and Table 1)].
Figure 1.
Overall structures of TraJF11–130 and TraJpSLT1–128. (A) Alignment of amino acid sequences of TraJF11–130 and TraJpSLT1–128. Secondary structure elements of TraJF11–130 and TraJpSLT1–128 are shown above and below the corresponding sequences, respectively. Dotted lines indicate residues not modeled in the crystal structures because of a lack of electron density. (B) Crystal structure of a TraJF11–130 monomer. Secondary structural elements as well as the N- and C-termini are labeled. (C) Crystal structure of a TraJpSLT1–128 monomer.
Both the F and pSLT TraJ N-terminal domains adopt PAS folds, characterized by a highly conserved five-stranded antiparallel β-sheet with a 2-1-5-4-3 strand topology (Figure 1A–C).17,18 An N-terminal helix (A′α) and two or three less structurally conserved helices are packed against the β-sheet on each side of both PAS domains. In spite of the limited sequence similarity, the two structures are nevertheless highly similar with an rmsd (root-mean-square deviation) calculated on Cα atom positions of ∼2 Å. On the basis of an rmsd of superimposed domains, the two TraJ PAS domains are structurally more similar to one another than to any of the other PAS domains deposited in the protein structure database. The major difference between the two structures is that the TraJpSLT PAS lacks a helix corresponding to the D/Eα helix of the TraJF PAS, and this region instead adopts a loop that is more flexible with local B factors approximately twice the overall B factor of the whole protein chain (Figure 1B,C).
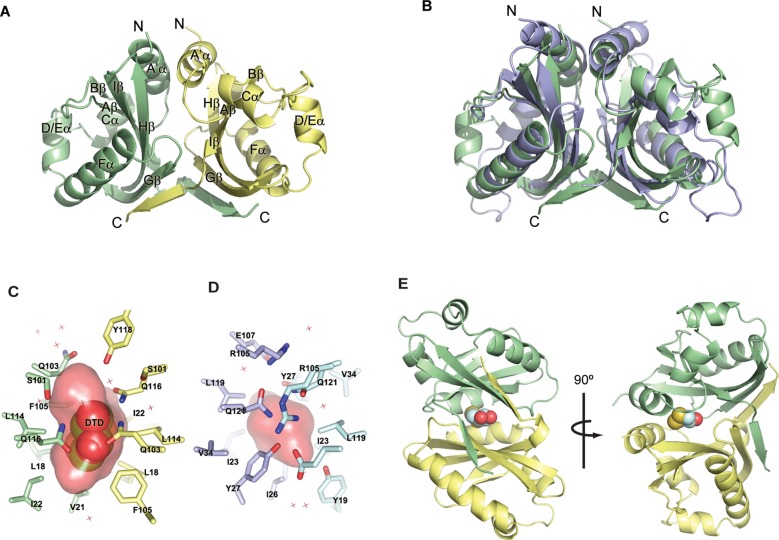
TraJ PAS Domains Form Homodimers
The TraJF PAS domain crystallographic asymmetric unit contains four protomers that are arranged as a pair of nearly identical dimers (Figure 2A). The asymmetric unit of the TraJpSLT PAS domain contains only a single protomer; however, a crystallographic 2-fold structure generates a dimer that can virtually be superimposed on the TraJF PAS dimer (Figure 2B). In the conserved TraJ dimer packing, one protomer is offset from the other by ∼120°. The dimer interface is stabilized by both the coiled coil formed by the N-terminal helices (A′α) and interactions between the conserved β-sheets, which is consistent with other known structures of the PAS domain dimers. Each TraJF11–130 molecule has ∼1600 Å2 of its ∼5600 Å2 total surface area buried at the dimeric interface, whereas ∼1800 Å2 of the ∼6100 Å2 total surface area of TraJpSLT1–128 is involved in dimeric contacts.
Figure 2.
Homodimerization of TraJF11–130 and TraJpSLT1–128. The two protomers of each dimer are colored differently. (A) Crystal structure of a TraJF11–130 dimer. (B) Superposition of the TraJF11–130 (green) and TraJpSLT1–128 (blue) dimers. (C) TraJF11–130 residues surrounding DTD (shown as spheres) at the dimeric interface. The surface of the internal pocket is colored red. (D) Residues surrounding a small cavity at the dimeric interface of TraJpSLT1–128. (E) Position of the DTD (spheres) pocket in the overall structure of a TraJF11–130 dimer.
In both structures, there is an internal cavity enclosed in the dimeric interface of the PAS domain (Figure 2C,D). In the TraJF11–130 structure, the cavity has a total surface area of ∼367 Å2 and a buried volume of ∼370 Å3, and it also contains an artificial ligand, DTD (dithiane diol, oxidized DTT) (Figure 2C,E and Figure S2 and Results of the Supporting Information). The cavity formed by TraJpSLT1–128 dimerization is smaller and contains no density corresponding to DTD in spite of the fact that DTT was used during protein purification (Figure 2D). It appears that Tyr27 of TraJpSLT1–128 (corresponding to Ile22 of TraJF11–130) from both protomers partially occludes the pocket, resulting in a smaller buried volume (∼103 Å3). Despite our efforts (see Results and Figures S3 and S4 of the Supporting Information), we were unable to determine if a specific physiological ligand exists in the pocket to regulate TraJ dimerization or its in vivo function.
Mutations That Affect the PAS Domain and Dimerization of TraJF
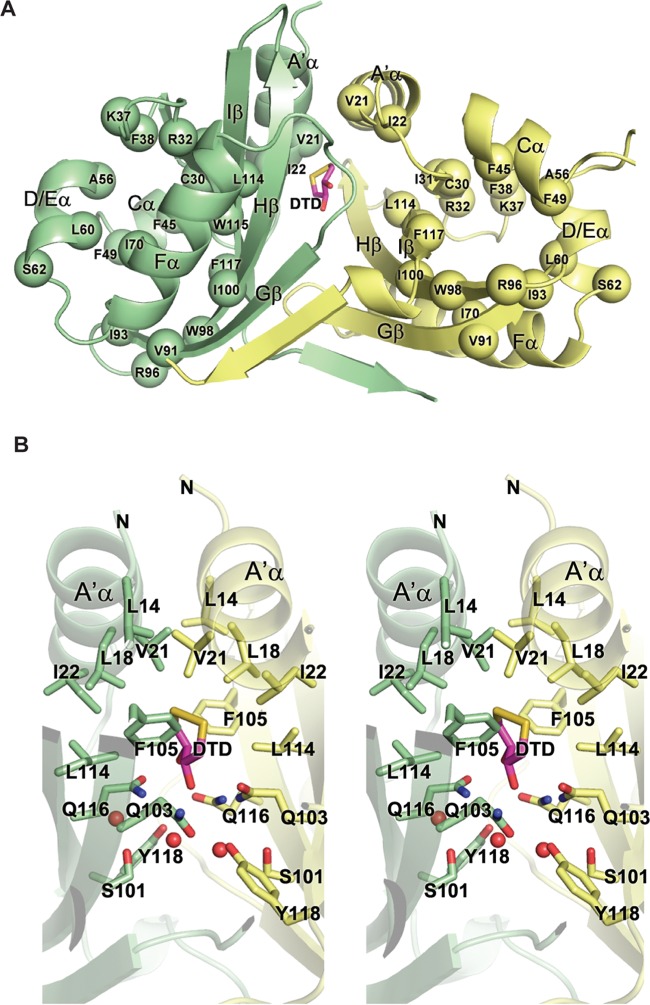
We have previously characterized a large collection of missense mutations in TraJF, which were selected from error-prone PCR mutagenesis based on their deleterious effects on activation of the plasmid PY promoter.23 All of these mutations are clustered in either an N-terminal region (residues 21–117) or a C-terminal region (residues 150–219), roughly overlapping the PAS domain and the putative helix–turn–helix DNA-binding domain, respectively (Figure S1 of the Supporting Information). Each of the N-terminal mutations affects the intracellular stability, dimerization, and in vivo function of TraJF, suggesting that these features of TraJF are intrinsically related.23 The structure of TraJF11–130 revealed that most of those functionally conserved N-terminal residues are involved in proper folding of the domain (Figure 3A). Residues Cys30, Phe38, Phe45, Phe49, Ala56, Leu60, Ile70, Val91, Ile93, Trp98, Ile100, Trp115, and Phe117 pack within the hydrophobic core of an individual PAS fold. Residues Ile22, Ile31, and Leu114 are involved in hydrophobic interactions between the conserved N-terminal helix (A′α) and the β-sheet of the PAS fold. The four functionally conserved hydrophilic residues (Arg32, Lys37, Ser62, and Arg96) are surface-exposed and are involved in hydrogen bonding or electrostatic interactions with nearby residues and also likely stabilize the fold.
Figure 3.
Functionally deleterious mutations in TraJF11–130 reveal conserved structural components. (A) Mapping of deleterious missense mutations (spheres) onto the TraJF11–130 dimer. The two protomers are colored green and yellow. See Figure S1 of the Supporting Information for the identity of the missense mutations. (B) Stereoview of the dimerization interface of the TraJF PAS domain formed by interactions between the N-terminal helices (A′α) and the β-sheets from both protomers. Water molecules are represented by red spheres.
Among residues that make direct contacts with the partner protomer across the dimerization interface of TraJF11–130 (Figure 3B), only one (Val21) was identified as being functionally important in our previous screen. Val21 not only mediates coiled-coil interactions between A′α helices of the two protomers but also interacts with Phe105 in Hβ of the partner protomer. This explains the fact that substitution of Val21 with a negatively charged aspartic acid leads to a >20-fold decrease in TraJF activity.23 To further probe the physiological relevance of the conserved dimeric interface of TraJ PAS domains, we generated less disruptive alanine mutations at three residues (Leu18, Phe105, and Gln116) and assayed the ability of the TraJF mutants to activate the PY promoter (Table 2 and Figure 3B). Leu18 appears to be involved in coiled-coil interactions between A′α helices across the partner protomers, and the L18A mutation modestly inhibited PY transcription. The benzyl rings of Phe105 from Hβ of each protomer form intimate hydrophobic interactions with A′α helices of the partner protomer, and the F105A mutation reduced the activity of TraJF by >6-fold. The side chains of Gln116 from both protomers are part of the hydrogen bonding network across the dimerization interface also involving Gln103, Tyr118, and two water molecules. The Q116A mutation reduced the activity of TraJF by ∼50%. These results indicate that the dimeric interface observed in the crystal structure of TraJF11–130 is relevant for the TraJ in vivo function.
Table 2. Function of F Plasmid TraJ or Its Mutants in Activating the F Plasmid PYa.
| form of TraJb | codon change | LacZ activity (MU) |
|---|---|---|
| TraJ | none | 3011 ± 239 |
| TraJ– | frameshift | 55 ± 9 |
| V21D | GTT → GAT | 130 ± 10 |
| L18A | CTG → GCG | 2500 ± 110 |
| F105A | TTT → GCG | 458 ± 37 |
| Q116A | CAA → GCG | 1747 ± 131 |
Determined by assaying the β-galactosidase (LacZ) activity of E. coli DH5α cells containing the reporter plasmid pJLAC101-PY (containing an F plasmid PY-lacZ fusion) and the TraJ expression plasmid pJLJ001 (expressing wild-type TraJ), or a pJLJ001 derivative containing a frameshifted traJ (TraJ–) obtained previously,23 or one of the pJLJ001 derivatives expressing TraJ missense mutants.
TraJ mutants are named after their corresponding amino acid residue substitutions.
Functional Interchangeability of the TraJ PAS Domains in the F Family of Conjugative Plasmids
A recent study indicates that F-like plasmid R1 TraJ (TraJR1) shares a certain level of functional interchangeability with TraJpSLT but not with TraJF or another F-like plasmid R100 TraJ (TraJR100).4 To further test the allelic specificity of different TraJ proteins, we assessed the ability of TraJ from F, pSLT, and R100 to drive transcription from an F PY-lacZ fusion reporter plasmid, pJLac101-PY.23 Each of the TraJ proteins was expressed from the pK184 expression plasmid such that each was produced at close to physiological levels.23 The results showed that only TraJF activated F plasmid PY whereas neither pSLT or R100 TraJ was able to activate transcription above the level of the traJ control, suggesting that each TraJ homologue is functionally specific to its own cognate plasmid (Table 3).
Table 3. Ability of TraJ Homologues and Hybrid TraJ To Activate the F Plasmid PYa.
| TraJF | TraJ– | TraJpSLT | TraJR100 | TraJpSLT1–125:TraJF121–226 | TraJR1001–120:TraJF121–226 | |
|---|---|---|---|---|---|---|
| LacZ activity (MU) | 2870 ± 126 | 55 ± 9 | 46 ± 3 | 41 ± 5 | 2873 ± 76 | 2796 ± 310 |
The ability of different TraJ proteins to activate PY was determined in E. coli cells containing a corresponding TraJ construct (from pJLJ001 to pJLJ005) and the F plasmid PY-lacZ fusion reporter plasmid pJLac101-PY as described in Experimental Procedures. The LacZ activity shown is the average of results from two independent samples. As a negative control, pJLJ2729 (a pJLJ001 derivative missing the C-terminal region of the F plasmid traJ) has a LacZ activity of 43 ± 3 MU.
To determine which function domain of TraJ carries this allelic specificity, we performed a domain swapping experiment based on the amino acid sequence and structural alignments of the F family TraJ homologues (Figure 1A and Figure S1 of the Supporting Information). We replaced the PAS domain in the F plasmid TraJ with the PAS domain of pSLT or R100 TraJ to test the ability of the hybrid TraJ to activate F plasmid PY in pJLac101-PY (Table 3). Interestingly, both the hybrid versions of TraJ (TraJpSLT1–125:TraJF121–226 and TraJR1001–120:TraJF121–226) activated the F plasmid PY at levels comparable to that of the F plasmid TraJ, indicating that the allelic specificity of the F family plasmid TraJ lies in its C-terminal functional domain that includes a putative HTH DNA-binding motif whereas the PAS domains of TraJ homologues are functionally interchangeable in spite of their limited amino acid sequence homology.
Discussion
Consistent with previous bioinformatics analysis,16,17 the X-ray crystal structures of TraJF11–130 and TraJpSLT1–128 presented here indicate that the F family TraJ proteins contain structurally similar N-terminal PAS domains despite limited amino acid sequence homology (Figure 1 and Figure S1 of the Supporting Information). TraJ proteins also homodimerize similarly, forming an extensive dimeric interface (Figure 2). The observed structural details of the TraJF110–130 PAS fold and its dimeric interface are fully supported by extensive mutational and functional analysis (Figure 3 and Table 2). Although F, R100, and pSLT TraJ appear to activate only their cognate tra operon promoters, the PAS domains of these TraJ homologues are functionally interchangeable (Table 3), indicating that the allelic specificity of F family TraJ is carried by only the region C-terminal to the PAS domain. Because the C-terminal domain is thought to act as a DNA-binding domain,3 our finding suggests that plasmid specificity is governed at the level of protein–DNA interactions, whereas the PAS domain might be dedicated to regulatory functions, possibly in response to cellular signals, as signal sensing is a common feature in many known PAS domains.18
In the crystal structures of both TraJF11–130 and TraJpSLT1–128, there is an internal pocket formed at the dimer interface (Figure 2). To the best of our knowledge, this is the first observation of an internal pocket formed through homodimerization of a PAS domain. While some PAS domains have ligand-binding pockets as part of their signaling function, these ligands commonly bind within a single PAS unit.18,22 We were unable to determine whether there is a specific physiological ligand in the TraJ PAS dimer pocket to regulate its function (see Results and Figures S3 and S4 of the Supporting Information); however, we cannot rule out the possibility that a small-molecule ligand may regulate TraJ PAS dimerization in response to environmental cues.
PAS domains promote oligomerization of many proteins, and it has been hypothesized that signal-induced changes in protein quaternary structure are involved in the signal transduction function of many PAS proteins.18 A previous study suggested that multiple cysteines in TraJ could be part of a metal-containing redox center to regulate transcription of the tra operon in response to oxidative pressure.16 The crystal structures of TraJF11–130 and TraJpSLT1–128 reveal that these cysteines do not interact with one another or metal ligands (Figure 3A and Figure S1 of the Supporting Information), and therefore, it is unlikely that they coordinate with a metal ion to form a redox center. Instead, these cysteines in the observed structures pack separately in the hydrophobic core of the PAS domain, explaining the previous observation that mutation of Cys30 to a serine or a tryptophan severely affects protein stability, dimerization, and in vivo function of TraJF.23
TraJ PAS domains dedicate a large surface area to homodimerization (Figure 1), and all dimerization-defective TraJF missense mutants that we have identified previously are significantly more sensitive than wild-type TraJF to proteolytic degradation by the HslVU protease–chaperone pair in vivo.23We hypothesize that folding of the TraJ PAS domain relies on its dimerization. Cellular signals could potentially modulate TraJ dimerization and therefore influence the susceptibility of TraJ to HslVU, providing a mechanism for regulating cellular TraJ levels and tra operon activity.
Acknowledgments
We thank S. Classen for assistance with crystallographic data collection (Advanced Light Source, SIBYLS beamline 12.3.1). We acknowledge Robert Jedrzejczak for providing a clone of the TraJ from S. enterica TraJpSLT1–128 and Josep Casadesus for providing plasmid pIZ2023.
Glossary
Abbreviations
- BS3
bis(sulfosuccinimidyl) suberate
- DTD
dithiane diol
- CRP
catabolite repressor protein
- DTT
dithiothreitol
- GC/MS-EI
gas chromatography/mass spectrometry-electron ionization
- Hfq
host factor for Qβ replicase
- HTH
helix–turn–helix
- H-NS
histonelike nucleoid structuring protein
- LB
Luria-Bertani
- Lrp
leucine-responsive protein
- MALDI-TOF
matrix-assisted laser desorption ionization time-of-flight
- MCSG
Midwest Center for Structural Genomics
- MU
Miller units
- PAS
Per-ARNT-Sim
- SAD
single-wavelength anomalous dispersion
- SeMet
selenomethionine.
Supporting Information Available
Methods that are used only for work described in the Results in the Supporting Information, our finding that indole or DTD is not required for TraJF to function under optimal growth conditions, a list of all the references cited in the Supporting Information, all the plasmids and oligonucleotides used in this work (Table S1), amino acid sequence alignment of the major TraJ orthologues, ranges of PAS domains and the putative HTH motif, and amino acid substitutions that affect TraJF function (Figure S1), electron density at the DTD pocket of the TraJF11–130 dimer (Figure S2), GC/MS-EI analysis of purified DTT-free TraJF11–130, which indicates the presence of indole present in the protein sample (Figure S3), and evidence that indole or DTD does not affect the protein stability, dimerization, or in vivo function of TraJF (Figure S4). This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
Coordinates and structure factors for TraJpSLT1–128 and TraJF11–130 have been deposited in Protein Data Bank as entries 4EW7 and 4KQD, respectively.
We acknowledge support via Canadian Institutes of Health Research Grant CIHR 42447 (J.N.M.G.), National Institutes of Health Grants NIGMS PSI-Biology GM094585 (A.J., MCSG) and GM094623 (J.N.A.), and the U.S. Department of Energy, Office of Biological and Environmental Research, via Contract DE-AC02-06CH11357.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Frost L. S.; Ippen-Ihler K.; Skurray R. A. (1994) Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58, 162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanka E.; Wilkins B. M. (1995) DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64, 141–169. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Maillard J. M.; Arutyunov D.; Frost L. S. (2010) The F plasmid transfer activator TraJ is a dimeric helix-turn-helix DNA-binding protein. FEMS Microbiol. Lett. 310, 112–119. [DOI] [PubMed] [Google Scholar]
- Wagner M. A.; Bischof K.; Kati D.; Koraimann G. (2013) Silencing and activating type IV secretion genes of the F-like conjugative resistance plasmid R1. Microbiology 159, 2481–2491. [DOI] [PubMed] [Google Scholar]
- Will W. R.; Lu J.; Frost L. S. (2004) The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol. Microbiol. 54, 769–782. [DOI] [PubMed] [Google Scholar]
- Silverman P. M.; Wickersham E.; Harris R. (1991) Regulation of the F plasmid traY promoter in Escherichia coli by host and plasmid factors. J. Mol. Biol. 218, 119–128. [DOI] [PubMed] [Google Scholar]
- Camacho E. M.; Casadesus J. (2005) Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol. Microbiol. 57, 1700–1718. [DOI] [PubMed] [Google Scholar]
- Starcic M.; Zgur-Bertok D.; Jordi B. J.; Wosten M. M.; Gaastra W.; van Putten J. P. (2003) The cyclic AMP-cyclic AMP receptor protein complex regulates activity of the traJ promoter of the Escherichia coli conjugative plasmid pRK100. J. Bacteriol. 185, 1616–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho E. M.; Casadesus J. (2002) Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44, 1589–1598. [DOI] [PubMed] [Google Scholar]
- Will W. R.; Frost L. S. (2006) Hfq is a regulator of F-plasmid TraJ and TraM synthesis in Escherichia coli. J. Bacteriol. 188, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur D. C.; Ghetu A. F.; Gubbins M. J.; Edwards R. A.; Frost L. S.; Glover J. N. (2003) FinO is an RNA chaperone that facilitates sense-antisense RNA interactions. EMBO J. 22, 6346–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K. N.; Andres I.; Achtman M. (1978) Fertility repression of F-like conjugative plasmids: Physical mapping of the R6–5 finO and finP cistrons and identification of the finO protein. Proc. Natl. Acad. Sci. U.S.A. 75, 5836–5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahrl D.; Wagner A.; Tscherner M.; Koraimann G. (2007) GroEL plays a central role in stress-induced negative regulation of bacterial conjugation by promoting proteolytic degradation of the activator protein TraJ. J. Bacteriol. 189, 5885–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau-Wong I. C.; Locke T.; Ellison M. J.; Raivio T. L.; Frost L. S. (2008) Activation of the Cpx regulon destabilizes the F plasmid transfer activator, TraJ, via the HslVU protease in Escherichia coli. Mol. Microbiol. 67, 516–527. [DOI] [PubMed] [Google Scholar]
- Gubbins M. J.; Lau I.; Will W. R.; Manchak J. M.; Raivio T. L.; Frost L. S. (2002) The positive regulator, TraJ, of the Escherichia coli F plasmid is unstable in a cpxA* background. J. Bacteriol. 184, 5781–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arutyunov D.; Rodriguez-Maillard J. M.; Frost L. S. (2011) A PAS domain within F plasmid TraJ is critical for its function as a transcriptional activator. Biochem. Cell Biol. 89, 396–404. [DOI] [PubMed] [Google Scholar]
- Taylor B. L.; Zhulin I. B. (1999) PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moglich A.; Ayers R. A.; Moffat K. (2009) Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W.; Hao B.; Mansy S. S.; Gonzalez G.; Gilles-Gonzalez M. A.; Chan M. K. (1998) Structure of a biological oxygen sensor: A new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. U.S.A. 95, 15177–15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O.; Doi M.; Yujnovsky I.; Cardone L.; Berndt A.; Hennig S.; Schulze S.; Urbanke C.; Sassone-Corsi P.; Wolf E. (2005) Crystal structure and interactions of the PAS repeat region of the Drosophila clock protein PERIOD. Mol. Cell 17, 69–82. [DOI] [PubMed] [Google Scholar]
- McIntosh B. E.; Hogenesch J. B.; Bradfield C. A. (2010) Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72, 625–645. [DOI] [PubMed] [Google Scholar]
- Henry J. T.; Crosson S. (2011) Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65, 261–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Peng Y.; Arutyunov D.; Frost L. S.; Glover J. N. (2012) Error-prone PCR mutagenesis reveals functional domains of a bacterial transcriptional activator, TraJ. J. Bacteriol. 194, 3670–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Baba T.; Ara T.; Hasegawa M.; Takai Y.; Okumura Y.; Baba M.; Datsenko K. A.; Tomita M.; Wanner B. L.; Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono H.; Otsuji N. (1968) Genetic mapping of regulator gene phoS for alkaline phosphatase in Escherichia coli. J. Bacteriol. 95, 1182–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S.; Richardson C. C. (1985) A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U.S.A. 82, 1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling M. G.; Holmes R. K. (1990) Construction of vectors with the p15a replicon, kanamycin resistance, inducible lacZ α and pUC18 or pUC19 multiple cloning sites. Nucleic Acids Res. 18, 5315–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N.; Hunt H. D.; Horton R. M.; Pullen J. K.; Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Stols L.; Gu M.; Dieckman L.; Raffen R.; Collart F. R.; Donnelly M. I. (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expression Purif. 25, 8–15. [DOI] [PubMed] [Google Scholar]
- Aslanidis C.; de Jong P. J. (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenfeldt W. H.; Stols L.; Millard C. S.; Joachimiak A.; Mark I. D. (2009) A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530. [PubMed] [Google Scholar]
- Kim Y.; Dementieva I.; Zhou M.; Wu R.; Lezondra L.; Quartey P.; Joachimiak G.; Korolev O.; Li H.; Joachimiak A. (2004) Automation of protein purification for structural genomics. J. Struct. Funct. Genomics 5, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.; Babnigg G.; Jedrzejczak R.; Eschenfeldt W. H.; Li H.; Maltseva N.; Hatzos-Skintges C.; Gu M.; Makowska-Grzyska M.; Wu R.; An H.; Chhor G.; Joachimiak A. (2011) High-throughput protein purification and quality assessment for crystallization. Methods 55, 12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski L.; Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Minor W.; Cymborowski M.; Otwinowski Z.; Chruszcz M. (2006) HKL-3000: The integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D62, 859–866. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C.; Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C. (2002) Automated structure solution, density modification and model building. Acta Crystallogr. D58, 1937–1940. [DOI] [PubMed] [Google Scholar]
- Vagin A.; Teplyakov A. (2000) An approach to multi-copy search in molecular replacement. Acta Crystallogr. D56, 1622–1624. [DOI] [PubMed] [Google Scholar]
- Morris R. J.; Perrakis A.; Lamzin V. S. (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244. [DOI] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A64, 112–122. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z.Maximum likelihood refinement of heavy atom parameters. (1991) Proceedings of the CCP4 Study Weekend 25–26 January 1991, pp 80–85, Daresbury Laboratory, Warrington, U.K. [Google Scholar]
- Cowtan K. (1994) DM: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 34–38. [Google Scholar]
- Langer G.; Cohen S. X.; Lamzin V. S.; Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundas J.; Ouyang Z.; Tseng J.; Binkowski A.; Turpaz Y.; Liang J. (2006) CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.; Richards F. M. (1971) The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 55, 379–400. [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.