Abstract
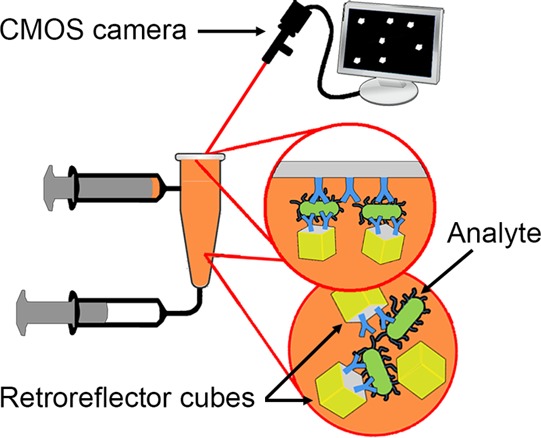
Point-of-care detection of pathogens is medically valuable but poses challenging trade-offs between instrument complexity and clinical and analytical sensitivity. Here we introduce a diagnostic platform utilizing lithographically fabricated micron-scale forms of cubic retroreflectors, arguably one of the most optically detectable human artifacts, as reporter labels for use in sensitive immunoassays. We demonstrate the applicability of this novel optical label in a simple assay format in which retroreflector cubes are first mixed with the sample. The cubes are then allowed to settle onto an immuno-capture surface, followed by inversion for gravity-driven removal of nonspecifically bound cubes. Cubes bridged to the capture surface by the analyte are detected using inexpensive, low-numerical aperture optics. For model bacterial and viral pathogens, sensitivity in 10% human serum was found to be 104 bacterial cells/mL and 104 virus particles/mL, consistent with clinical utility.
Detection of pathogenic organisms traditionally has relied on culture and microscopic observation.1,2 Techniques, such as polymerase chain reaction (PCR), immunofluorescence (IF) assay, and enzyme-linked immunosorbent assay (ELISA), have more recently emerged as important methods for detection of pathogens. While highly effective, these methods are largely confined to laboratory settings, require trained personnel and can involve considerable time and expense.1−8
With the improvement of microfabrication technology, many advances have been made in the development of point of care (POC) diagnostics, particularly in microfluidic devices integrating sample processing and detection into a single device.9−11 Sophisticated methods have been developed for DNA amplification and analysis,9,12−14 cytometry,15−18 whole-organism assays,19−21 and protein detection.22−25 These methods generally rely upon labels such as colored nano- and microparticles, magnetic particles, fluorescent molecules, and liposomes, which can be affected by issues such as photobleaching of fluors and particle stability in the complex sample matrices often used in a POC environment.26−28
Here, we demonstrate the detection of model bacterial and viral pathogens, Escherichia coli and MS2 virus at 104 bacterial cells/mL and 104 virions/mL, respectively, using suspended, microfabricated retroreflector cubes as optical labels conjugated to antibodies. Retroreflectors are widely used on bicycles, traffic signs,29 and safety vests30 because of their high detectability. They return incident light in a narrow beam directly back to the illumination source and, therefore, are significantly brighter than surfaces that scatter light. Our group has previously reported the initial fabrication of retroreflector cubes;31 this paper, however, reports a significantly improved fabrication process and a novel assay concept, demonstrating for the first time, the use of micron-scale retroreflectors in immunoassays. The assay is otherwise based on simple, commercially available materials and requires little or no sample preparation and no exposure to the potentially infectious sample after sample collection. The retroreflector cubes, 5 μm on a side, possess a transparent epoxy core and three mutually perpendicular reflective gold-coated surfaces. Individual cubes are easily detected using a low-cost, low-numerical aperture camera objective, suggesting the possibility of using the cameras of mobile phones with only the addition of a plastic lens and an inline light source. Their highly reflective nature greatly facilitates detection, and photobleaching and storage stability are not concerns with these labels.
In the present assay (detailed in Assay Procedure), a premixed sample is introduced into an in-house-modified qPCR tube, the cap of which is conjugated on its (optical-quality) inner surface with antibodies to the target. The tubes are inverted to allow the dense cubes to settle; any analyte present is captured and bridges the antibody-modified cube onto the antibody-coated cap inner surface. The test vessel is then turned upright after applying a fluidic force discrimination step by pulsing on a vortex mixer to remove weakly bound cubes, and bound retroreflectors are counted using a low numerical aperture camera with inline illumination.
Materials and Methods
Reagents
Optical qPCR tubes (8× strip, catalog no. 401428) and separate caps (8× strip, catalog no. 401425) were obtained from Agilent Technologies (Santa Clara, CA). Rabbit polyclonal anti-E. coli antibodies were obtained from Fitzgerald (Acton, MA). Rabbit polyclonal anti-MS2 antibodies were obtained from Tetracore (Rockville, MD). E. coli (strain MB1457), and bacteriophage MS2 (strain 15597-B1) were obtained from the American Type Culture Collection (Manassas, VA). Phosphate buffered saline (PBS) tablets (10 mM Phosphate, 2.7 mM potassium chloride, 140 mM sodium chloride, pH 7.4) were purchased from Bioline (Taunton, MA). Anonymized human serum was obtained from the Gulf Coast Regional Blood Center (Houston, TX). Fluorescein isothiocyanate (FITC) was obtained from Pierce (Rockford, IL). 1-Ethyl-3-(3-(dimethylamino)propyl) carbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), bovine serum albumin (BSA), Tween-20, hydroxylamine, 6-mercapto-1-hexanol, dimethyl sulfoxide (DMSO), anhydrous chromium trioxide, 96.7% sulfuric acid, dithiobis(succinimidyl propionate) (DSP), and sodium cyanoborohydride were purchased from Sigma-Aldrich (St. Louis, MO).
Fabrication of Polypropylene Test Vessel and Preparation of Polypropylene Observation Window
A detailed description of the modification of the qPCR tube test vessels can be found in the Supporting Information (section S1). The final design was optimized to prevent the presence of air bubbles, which can sweep away bound particles by surface tension.23,32
The as-purchased, optical-quality qPCR tube caps, with 3 mm diameter, transparent observation windows, are not optimized for protein adsorption; antibodies, therefore, were covalently attached by the following method. The caps were first cleaned by bath sonication in anhydrous ethanol for 15 min, and then immersed for 1 min in 29:42:29 (w/w/w) concentrated sulfuric acid, distilled water and chromium trioxide at 70 °C.33,34 After oxidation, the carboxylated caps were washed extensively with deionized water purified with a Milli-Q system (Millipore, Billerica, MA), and anhydrous ethanol, and then dried under a stream of nitrogen.
Activation of Carboxylated Observation Window and Immobilization of Antibodies
The carboxylated qPCR tube caps were activated by incubation with 3 mM EDC and 5 mM NHS in 0.1 M MES buffer, pH 6.0 for 15 min at room temperature, then washed with PBS. Rabbit anti-E. coli antibodies (1 mg/mL in 10 mM PBS, pH 7.4) or rabbit anti-MS2 antibodies (1 mg/mL in 10 mM PBS, pH 7.4) were covalently attached to the caps by spotting 10 μL of antibody solution on the activated observation window and incubating at 4 °C, overnight. The tube caps were then washed with PBST (10 mM PBS, pH 7.4, 0.1% Tween-20), and passivated by incubation with a 2% BSA, 100 mM hydroxylamine solution for 3 h. The conjugated caps were then washed with PBS and passivated by adsorption of 2% BSA for 3 h at room temperature.
Fabrication of Retroreflector Cubes and Immobilization of Antibodies
Retroreflector cubes were fabricated using a modification of a process previously reported31 (detailed process and schematic shown in section S2, Figure S2, Supporting Information). Briefly, a 5 μm thick layer of SU-8 5 photoresist was deposited onto a copper-coated silicon wafer by spin-coating at 2000 rpm for 1 min. The coated wafer was baked on a hot plate at 95 °C for 3 min, cross-linked via exposure to 254 nm UV irradiation, then baked once more to cure the epoxy-based resist. A 200 nm layer of copper was then deposited onto the wafer by thermal evaporation followed by spin-coating a 70 nm layer of polystyrene.
An array of squares defining individual cubic retroreflectors was next printed using an in-house ion beam proximity lithography tool, and the polystyrene developed in a toluene bath. The copper layer under the polystyrene was isotropically etched using a copper etchant and the pattern then transferred into the SU-8 layer using an oxygen reactive ion etching process. Citric acid solution was used to etch the final copper layer, leaving an undercut. A 10 nm titanium layer and a 100 nm gold layer were then evaporated at an angle relative the surface normal to coat only three of the optically transparent SU-8 surfaces. The cubes, possessing three gold sides and three SU-8 sides, were released by immersion in citric acid solution to remove the remaining copper and subsequently recovered by centrifugation at 5000 × g for 5 min. Following wash and resuspension steps, the cubes were diluted to 1.5 × 107/mL in PBS and stored at 4 °C for subsequent experiments. It should be noted that water is a weak nucleophile whose reaction with epoxides proceeds at appreciable rates under highly acidic conditions (<pH 4) or at elevated temperatures (>70 °C).35 It was, therefore, imperative either to functionalize the cubes immediately, or to store the particles at low temperatures and neutral pH to retard hydrolysis.
To preferentially functionalize the free SU-8 sides of the cubes, the particles were incubated with 10 mM glycine in 100 mM PBS at pH 9.0 for 1 h to allow the covalent attachment of primary amines to the exposed epoxy groups on the cubes,36 leaving behind carboxyl moieties. The glycine-functionalized cubes were then incubated for 1 h with 100 mM 6-mercapto-1-hexanol in 20:80 (v/v) DMSO in PBS to form a hydroxyl-terminated self-assembled monolayer on the exposed gold surfaces (Figure 1). The self-assembled monolayer provides a degree of passivation by reducing protein adsorption to the gold surfaces. The derivatized cubes were washed with PBS, pelleted, and then resuspended in 1 mL PBS.
Figure 1.
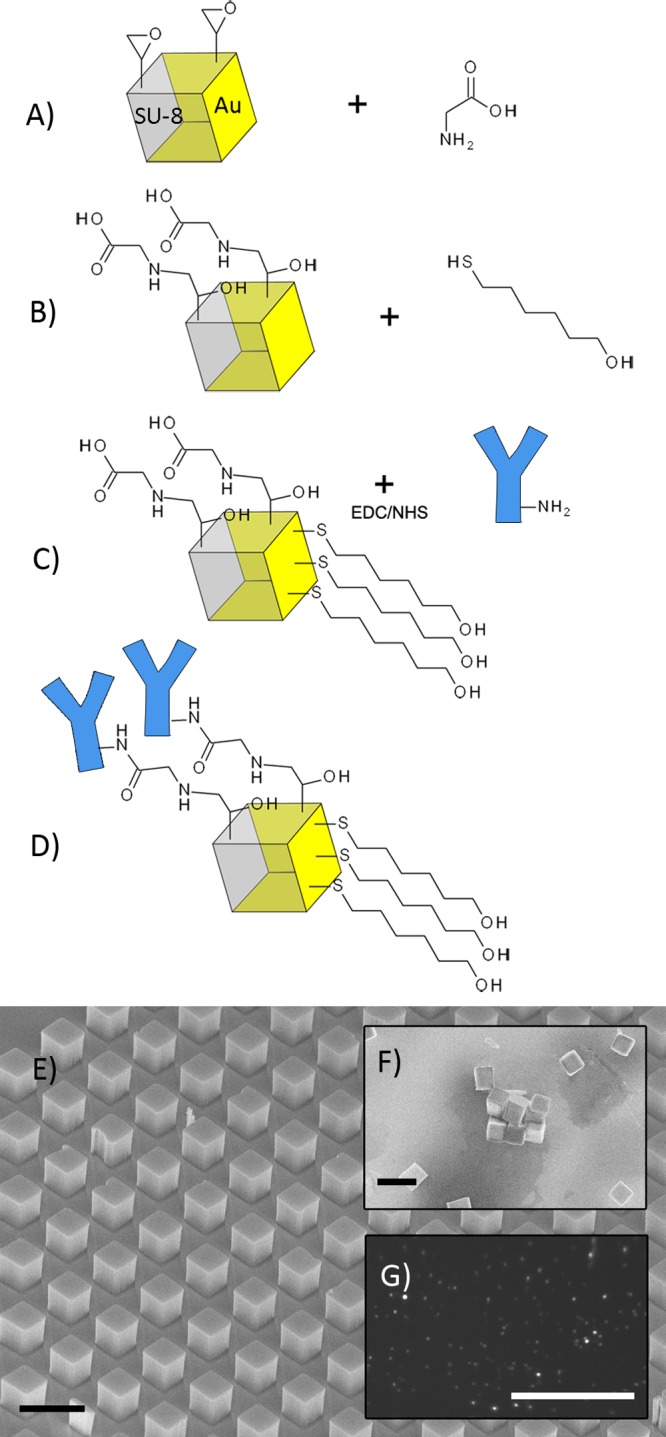
Functionalization of retroreflector cubes. (A) Reaction of glycine with epoxy groups on exposed SU-8 surfaces. (B) Addition of 6-mercapto-1-hexanol to gold surfaces. (C) and (D) EDC/NHS addition of antibodies. (E) Wafer of cubes before release. (F) SEM image of cubes spotted onto a wafer after release and functionalization. Aggregation shown here is an artifact of drying; aggregation of cubes in normally negligible. Black scale bars (E, F) are 10 μm. (G) Image of retroreflector cubes spotted onto a wafer, captured with a 0.085 numerical aperture CMOS camera with inline light source; white scale bar 100 μm.
Carboxyl-functionalized cubes were incubated with EDC/NHS as described in Activation of Carboxylated Observation Window and Immobilization of Antibodies section. The activated cubes were pelleted and resuspended in 400 μL of 0.25 mg/mL antibody solution for overnight incubation at 4 °C with constant mixing by inversion. The antibody concentration used was chosen to avoid cube aggregation during conjugation since conjugated cubes were observed to aggregate at offered antibody concentrations of 25 μg/mL, but showed minimal aggregation at concentrations of greater than 125 μg/mL (section S2, Figure S3, Supporting Information). The cubes were then pelleted and resuspended in 2% BSA, 100 mM hydroxylamine for passivation at room temperature for 3 h. The cubes were then washed with PBST, resuspended in 1 mL PBS and stored at 4 °C.
Assay Procedure
qPCR tube reaction vessels were fabricated by adding inlet and outlet tubes as detailed in the Supporting Information, and fitted with antibody-modified tube caps prepared as described above. To perform the assay (schematic shown in Figure 2), pathogen-spiked samples (340 μL) were mixed with 1.5 × 105 antibody-modified retroreflector cubes in 1 mL Luer-Lok syringes (Becton-Dickinson, Franklin Lakes, NJ) mixed for 1 h at room temperature on a rotator at 12 rpm. The premixed samples were then drawn into reaction vessels through the sample inlet ports by suction from syringes attached to the sample outlets. Once filled, the vessels were allowed to stand inverted (window side down) for 15 min to allow cubes to settle to the observation surface. Tubes were turned upright for 2 min, and then the 15 min sedimentation was repeated to allow cubes initially settled in the wrong orientation a chance to reorient and bind.
Figure 2.
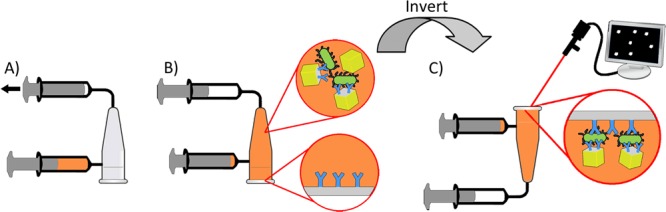
Schematic of assay. (A) Premixed sample is introduced by suction and (B) allowed to settle to the transparent observation window. (C) Tube is inverted, the observation window is imaged and retroreflecting cubes are counted.
Specificity was enhanced by the net downward force exerted by the buoyant weight of the cubes when the tube is in the upright orientation, and additional fluidic force discrimination was applied by way of a 0.5 s pulse on a Vortex Genie 2 (Fisher Scientific, Waltham, MA) at setting 6 to disrupt any remaining nonspecific interactions. The tubes were then held upright for at least 2 min to ensure cubes falling from the surface would be well beyond the depth of field of the CMOS camera and would, therefore, not be imaged.
Retroreflector Cube Imaging and Counting
Images of the observation window were captured using a 0.085 numerical aperture lens and a CMOS camera (EO-5012M, Edmund Optics, Barrington, NJ) with inline illumination (halogen incandescent light source) controlled by a custom LabVIEW-based software application. The camera was angled at 35° from the vertical for selective imaging of retroreflected light in preference to specular reflection, and a low NA was used to discriminate against scattered light.
To count the cubes, images were taken of 3 separate regions within the observation areas of the caps. The three images were then cropped along their focal planes to dimensions 1.4 mm by 0.35 mm and stitched together using Adobe Photoshop (Adobe Systems, San Jose, CA). The number of cubes within each imaged region was then determined using ImageJ 1.46r (National Institutes of Health, Bethesda, MA) by setting a binary threshold to differentiate the cubes from background illumination followed by automated counting (section S3, Figure S4–S9, Supporting Information).
Results and Discussion
Polypropylene Activation and Antibody Immobilization
We characterized the functionalization of the polypropylene observation window of the qPCR tube caps at different stages by XPS and ATR-FTIR analysis.
FTIR spectra (Figure S10, Supporting Information) show that after oxidation with chromic acid, peaks at 3710–3100 and 1760–1550 cm–1 appear and a broadening at 1320–1210 cm–1, occurs, indicating the presence of hydrogen bonded –OH stretch, carboxyl C=O stretch, and carboxylic acid C–OH stretch, respectively. After conjugation to antibodies, the broad –OH stretch peak at 3600–3000 cm–1 remains, and a shifted peak at 1670 cm–1 indicates the presence of amide C=O.34,37−39 These chemical functionalities were not seen on the untreated material.
XPS survey scans (Figure S11, Supporting Information) show that atomic oxygen on the surface of the polypropylene material increased from 0.67% to 14.9% after reaction with the chromic acid solution. High resolution C1s scans of the oxidized sample were fitted with four Gaussian peaks representing the bonds C–C/C–H at 284.6 eV, C–CO2 at 286.0 eV, C–O at 287.2 eV, and CO2 at 288.6 eV indicating the presence of carboxyl and other oxygenated carbon moieties on the surface after treatment.34,39,40
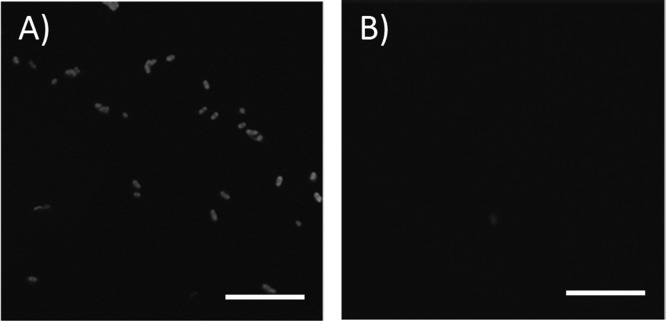
The presence of functional antibodies on the polypropylene cap surface after conjugation was confirmed by the direct capture of fluorescein-conjugated E. coli bacteria on caps functionalized with anti-E. coli antibodies. Caps covalently modified with BSA were used as a negative control. Figure 3 shows fluorescence microscopy images demonstrating the successful capture of FITC-labeled E. coli onto the covalently coupled caps, and the absence of binding on the BSA surfaces.
Figure 3.
Demonstration of functional anti-E. coli antibodies on polypropylene observation window by specific capture of FITC-labeled E. coli. ( A) Fluorescence image of covalently coupled anti-E. coli surface and of (B) covalently coupled BSA surface after incubation with the bacteria. Scale bars are 10 μm; images were equally and uniformly postprocessed for brightness.
Though the images demonstrate the successful capture of fluorescently labeled bacteria onto the antibody coated surface, as opposed to the BSA surface, there is some concern that electrostatic interactions might play a role in this outcome. The isoelectric point (pI) of BSA is estimated to be 4.8,41,42 and the pI of various subclasses of IgG, present in polyclonal sera, can vary from below 6 to 9.5.43,44 This experiment was conducted at physiological pH (100 mM PBS, pH 7.4), at which BSA is generally anionic and a subpopulation of the anti-E. coli antibodies can be cationic. Unmodified E. coli has been reported to possess a net negative surface charge and could therefore be attracted to positively charged surface-bound groups and be repelled by negatively charged surfaces.45−47 However, it is well-known that high ionic strength media (such as ∼0.1 M NaCl) suppress nonspecific binding due to compression of the electrostatic double layer, therefore binding is believed to be the result of specific antibody–antigen interactions.48
Fabrication of Retroreflector Cubes and Immobilization of Antibodies
The previously reported method of fabricating the micron-scale retroreflector cubes resulted in low (32%) and variable release of the cubes from the silicon substrate, as well as delamination of the reflective gold layers, over time, from the sides of the released cubes. The current process, detailed in the Materials and Methods section, is improved by the addition of a titanium adhesion layer on top of the cubes, prior to deposition of gold and a sacrificial copper layer below the cubes. Gold has poor adhesion to SU-8 and to silicon;49 the addition of a titanium layer ensures proper adhesion and limits delamination of the gold layers from the released cubes. The sacrificial copper layer, once etched away, allows more uniform release of the cubes into solution with average recovery of 85%.
The cube functionalization protocol was optimized to avoid aggregation of cubes; as with commercially available particles. We have characterized the homogeneity of the conjugated cube population by optical microscopy of wet preparations; aggregation of the conjugated cubes appears to be minimal.
Specificity of immobilized antibodies was verified by conjugating anti-rabbit IgG antibodies, anti-mouse IgG antibodies and D1.3 anti-hen egg lysozyme (HEL) antibodies to thermally evaporated gold spots on silicon via well-established DSP thiol coupling chemistry. Anti-E. coli and anti-MS2 cubes were spotted on the antibody coated gold areas and then washed gently, by aspiration, with PBST. The resulting substrates were then observed and the bound cube density determined.
Figure 4 and scanning electron microscope images (Figure S12, Supporting Information) reveal that cubes conjugated with rabbit antibodies were captured by anti-rabbit antibody spots at much higher densities than the anti-mouse and (murine) D1.3 anti-lysozyme antibody spots, providing an indication of the specificity of the cubes.
Figure 4.
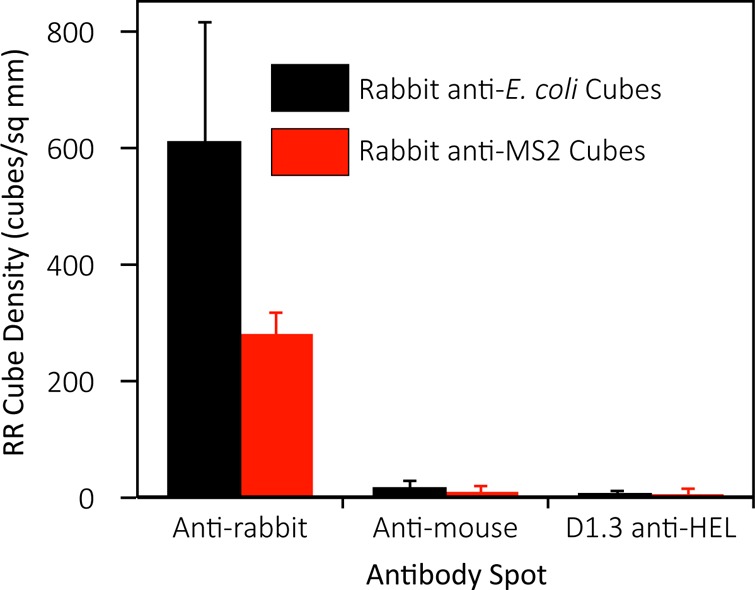
Specific capture of microretroreflector cubes bearing rabbit anti-E. coli and anti-MS2 polyclonal antibodies. Error bars are 1 SD from at least 4 independent antibody spots.
Assay Performance
Detection of E. coli was performed in PBS and in 10% human serum diluted in PBS at concentrations of 103, 104, and 106 cells/mL. Detection of MS2 bacteriophage was performed in the same media at concentrations of 104, 105, 106, and 108 virus particles/mL. The assay system design was optimized to prevent the presence of air bubbles, which may sweep away bound particles by surface tension.32 To achieve specificity, during upright orientation of the assay tubes, the buoyant weight of the cubes applies a net downward force of 1.8 pN, as estimated by simple force balance, enough to break weak nonspecific interactions which are on the order of 0.1–1.0 pN. The optional radial fluidic force discrimination step by pulse vortexing applied a lateral acceleration of approximately 4.3 × g, which was enough to disrupt any remaining nonspecific interactions.23,24
As shown in Figure 5, as the concentration of bacteria increases, the number of retroreflecting cubes detected also increases; however, binding of cubes in 10% human serum is somewhat lower than observed in buffer. This is likely because of serum proteins directly interacting with the analyte or blocking binding sites on the assay antibodies.50−52
Figure 5.
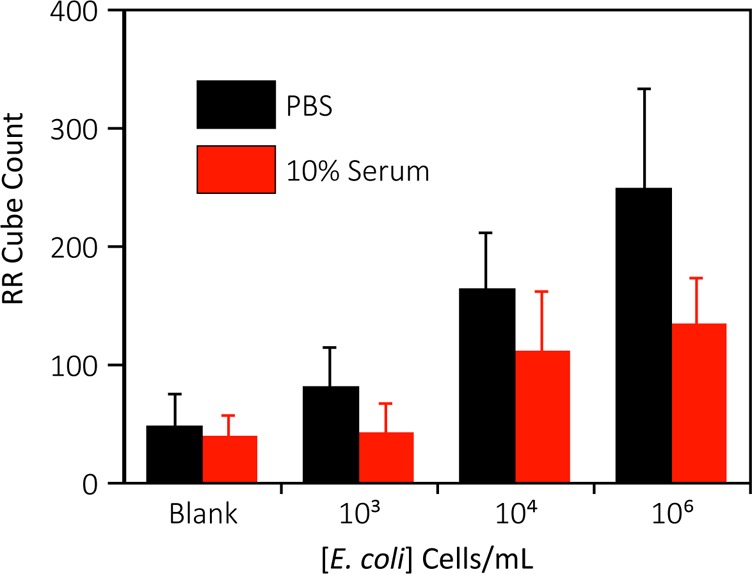
Dose–response for detection of E. coli via sedimentation of retroreflector cubes. Error bars are 1 SD from at least 3 independent experiments.
The detection of MS2 bacteriophage in both buffer and 10% human serum is shown in Figure 6. As expected, an increase in virus concentration produces an increase in the retroreflector cube count. At very high virus concentrations the cube density decreases, however, probably due to a hook effect as excess virus coats both cubes and the observation surface.52−54
Figure 6.
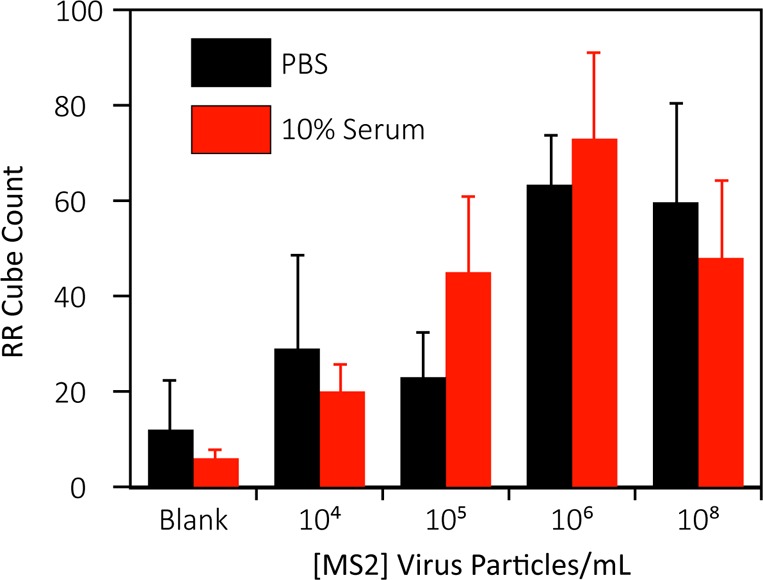
Dose–response for detection of MS2 via sedimentation of retroreflector cubes. Error bars are 1 SD from at least 3 independent experiments.
Statistical analysis was performed using a one-tailed Student’s t test to determine the limit of detection of the assay based on the concentrations tested, with 95% (p < 0.05) confidence level. The LOD for E. coli was found to be 104 cells/mL in both buffer and 10% serum, comparable to more sophisticated rapid diagnostic formats.11,20 Estimated LODs for MS2 were more matrix-sensitive at 106 virus particles/mL in buffer and 104 virus particles/mL in 10% serum. While these values differ due to residual variability in this early stage assay, each is well below the levels of viremia seen in many diseases55−57,46−48 and competitive with many established laboratory methods such as qPCR, ELISA, and microscopy, as well as POC lab-on-chip devices which range, in their respective limits of detection, from 103–108 virus particles/mL.21,58
The relatively small fraction of the 150 000 offered cubes observed bound to the window deserves comment. The observation window covers only 36% of the area of the tube cap, and only those cubes which fall and bind in the proper orientation (i.e., with transparent sides facing the camera) will be detected. Also, since we image the observation window from a single direction, only cubes illuminated from a direction within the 90° arc of retroreflectance would be detected. We therefore estimate the percentage of cubes which we would detect within the observation window to be 36% × 1/2 × 1/4 = 4.5% of the total number bound. While this fraction is small and could be increased by, for example, imaging from multiple directions, it is sufficient for successful assay performance.
During the assay, multiple cubes could potentially bind to a single bacterium, or the same virus particles. Clumps of cubes bridged by analyte potentially could sterically hinder the binding of those analytes to the capture surface and reduce the number of cubes observed during the assay readout, negatively affecting the limit of detection and the linearity of the dynamic range. However, this point-of-care assay is not meant for quantification of target, and we, therefore, do not believe this potential effect greatly affects the outcome of the test results for a yes/no assay.
In MS2 detection, lower numbers of cubes are bound to the observation regions than those seen in E. coli experiments. This may be due to the potential limitations of small virus particles in bridging large cubes to the surface. Inherent heterogeneity of the unpurified polyclonal anti-MS2 antibody stock preparations also may have resulted in lower antibody densities on the anti-MS2 cube surfaces as opposed to the affinity purified (by the vendor) antibodies conjugated to the anti-E. coli cubes. This hypothesis is supported by the lower capture densities of cubes by the anti-rabbit antibody spots shown in Figure 4, suggesting that optimized capture and detection antibody pairs may further improve the detection limit for virus particles.
Sample-to-result time for this experimental assay is currently 95 min and is dominated by the mixing and settling steps, but has the potential to be greatly shortened with further optimization59,60 and centrifugation. Preliminary studies demonstrate that settling time can be reduced to 1 min using mild centrifugation at 50 × g. Preliminary studies also support the possibility of multiplexing the assay by multiwavelength observation of dye-doped colored retroreflecting cubes.
Conclusion
We have developed a novel POC assay utilizing ultrabright, retroreflective, cubic microparticles in an inexpensive, potentially rapid and simple format with sensitivity comparable to conventional laboratory-based methods. An improved fabrication process has resulted in better recovery of cubes which can be functionalized using well-established methods. Detection of model pathogens was demonstrated in both buffer and 10% human serum with similar detection limits of 104 cells/mL and 104 virus particles/mL for E. coli and MS2 bacteriophage, respectively. Future work will involve the development of a multiplexed assay via the fabrication of colored retroreflector cubes doped with photoresist-soluble dyes. Differently colored cubes bearing antibodies against different pathogens may be used simultaneously within the same vessel and detected automatically in the presence of target.
Acknowledgments
The project described was supported by the NIAID/NIH (Grant No. U54 AI057156). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the RCE Programs Office, NIAID, or NIH. Support was also provided by the NSF (Grant No. CMMI-0900743), the Alliance for Nanohealth Competitive Research Program (Grant No. W81XWH-09-2-0139), the Welch Foundation (Grant No. E-1264), and the University of Houston (Project No. EEC-0647775). The authors wish to acknowledge Meenu Adhikari for the gift of MS2 phage. The authors would also like to thank the Center for Integrated Bio and Nanosystems and the University of Houston Nanofabrication Facility for providing excellent facilities and support. The authors also acknowledge Tecnológico de Monterrey Research chair (Grant CAT161) and CONACyT for fellowship no. 295368.
Supporting Information Available
(1) Fabrication of assay tubes, (2) fabrication and functionalization of retroreflector cubes, (3) image capture and counting of cubes, (4) FTIR and XPS spectra depicting functionalization of polypropylene observation window, and (5) specific capture of antibody-conjugated cubes. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Pitt T. L.; Saunders N. A. J. Clin. Pathol. 2000, 53, 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyburn H.; Mbakilwa H.; Mwangi R.; Mwerinde O.; Olomi R.; Drakeley C.; Whitty C. J. M. BMJ [Br. Med. J.] 2007, 334, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naber S. P. N. Engl. J. Med. 1994, 331, 1212–1215. [DOI] [PubMed] [Google Scholar]
- Engleberg N. C.; Eisenstein B. I. Annu. Rev. Med. 1992, 43, 147–155. [DOI] [PubMed] [Google Scholar]
- Tang Y.-W.; Procop G. W.; Persing D. H. Clin. Chem. 1997, 43, 2021–2038. [PubMed] [Google Scholar]
- Erlich H. A.; Gelfand D.; Sninsky J. J. Science 1991, 252, 1643–1651. [DOI] [PubMed] [Google Scholar]
- Pawlak M.; Schick E.; Bopp M. A.; Schneider M. J.; Oroszlan P.; Ehrat M. Proteomics 2002, 2, 383–393. [DOI] [PubMed] [Google Scholar]
- Kellenberger C. A.; Wilson S. C.; Sales-Lee J.; Hammond M. C. J. Am. Chem. Soc. 2013, 135, 4906–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Cui D.; Liu C.; Li H.; Chen J. Anal. Chim. Acta 2007, 584, 237–243. [DOI] [PubMed] [Google Scholar]
- Chin C. D.; Laksanasopin T.; Cheung Y. K.; Steinmiller D.; Linder V.; Parsa H.; Wang J.; Moore H.; Rouse R.; Umviligihozo G.; Karita E.; Mwambarangwe L.; Braunstein S. L.; van de Wijgert J.; Sahabo R.; Justman J. E.; El-Sadr W.; Sia S. K. Nat. Med. 2011, 17, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Verbarg J.; Plath W. D.; Shriver-Lake L. C.; Howell P. B. Jr.; Erickson J. S.; Golden J. P.; Ligler F. S. Anal. Chem. 2013, 85, 4944–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auroux P. A.; Koc Y.; deMello A.; Manz A.; Day P. J. R. Lab. Chip 2004, 4, 534–546. [DOI] [PubMed] [Google Scholar]
- Kokoris M.; Nabavi M.; Lancaster C.; Clemmens J.; Maloney P.; Capadanno J.; Gerdes J.; Battrell C. F. Methods 2005, 37, 114–119. [DOI] [PubMed] [Google Scholar]
- Zhang C.; Xing D. Nucleic Acids Res. 2007, 35, 4223–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard G. R.; Sanders C. K.; Martin J. C.; Kaduchak G.; Graves S. W. Anal. Chem. 2007, 79, 8740–8746. [DOI] [PubMed] [Google Scholar]
- Bouzid M.; Steverding D.; Tyler K. M. Curr. Opin. Biotechnol. 2008, 19, 302–306. [DOI] [PubMed] [Google Scholar]
- Jani I. V.; Janossy G.; Brown D. W. G.; Mandy F. Lancet Infect. Dis. 2002, 2, 243–250. [DOI] [PubMed] [Google Scholar]
- Simonnet C.; Groisman A. Anal. Chem. 2006, 78, 5653–5663. [DOI] [PubMed] [Google Scholar]
- McClain M. a.; Culbertson C. T.; Jacobson S. C.; Ramsey J. M. Anal. Chem. 2001, 73, 5334–5338. [DOI] [PubMed] [Google Scholar]
- Oh S.; Jadhav M.; Lim J.; Reddy V.; Kim C. Biosens. Bioelectron. 2013, 41, 758–763. [DOI] [PubMed] [Google Scholar]
- Ferris M. M.; Stepp P. C.; Ranno K. A.; Mahmoud W.; Ibbitson E.; Jarvis J.; Cox M. M. J.; Christensen K.; Votaw H.; Edwards D. P.; Rowlen K. L. J. Virol. Methods 2011, 171, 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goluch E. D.; Nam J.-M.; Georganopoulou D. G.; Chiesl T. N.; Shaikh K. a.; Ryu K. S.; Barron A. E.; Mirkin C. a.; Liu C. Lab Chip 2006, 6, 1293–1299. [DOI] [PubMed] [Google Scholar]
- Mulvaney S. P.; Cole C. L.; Kniller M. D.; Malito M.; Tamanaha C. R.; Rife J. C.; Stanton M. W.; Whitman L. J. Biosens. Bioelectron. 2007, 23, 191–200. [DOI] [PubMed] [Google Scholar]
- Mulvaney S. P.; Myers K. M.; Sheehan P. E.; Whitman L. J. Biosens. Bioelectron. 2009, 24, 1109–1115. [DOI] [PubMed] [Google Scholar]
- Connelly J. T.; Kondapalli S.; Skoupi M.; Parker J. S. L.; Kirby B. J.; Baeumner A. J. Anal. Bioanal. Chem. 2012, 402, 315–323. [DOI] [PubMed] [Google Scholar]
- Chiodini P. L.; Bowers K.; Jorgensen P.; Barnwell J. W.; Grady K. K.; Luchavez J.; Moody A. H.; Cenizal A.; Bell D. Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 331–337. [DOI] [PubMed] [Google Scholar]
- Jorgensen P.; Chanthap L.; Rebueno A.; Tsuyuoka R.; Bell D. Am. J. Trop. Med. Hyg. 2006, 74, 750–754. [PubMed] [Google Scholar]
- Suzuki M.; Husimi Y.; Komatsu H.; Suzuki K.; Douglas K. T.. 2008, 5720–5725. [DOI] [PubMed]
- Hawkins H.; Carlson P. Transp. Res. Rec. 2001, 1754, 11–20. [Google Scholar]
- Luoma J.; Schumann J.; Traube E. C. Accid. Anal. Prev. 1996, 28, 377–383. [DOI] [PubMed] [Google Scholar]
- Sherlock T.; Nasrullah A.; Litvinov J.; Cacao E.; Knoop J.; Kemper S.; Kourentzi K.; Kar A.; Ruchhoeft P.; Willson R. J. Vac. Sci. Technol. B 2011, 29, 06FA01–06FA01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skottrup P. D.; Hansen M. F.; Lange J. M.; Deryabina M.; Svendsen W. E.; Jakobsen M. H.; Dufva M. Acta Biomater. 2010, 6, 3936–3946. [DOI] [PubMed] [Google Scholar]
- Goddard J. M.; Hotchkiss J. H. Prog. Polym. Sci. 2007, 32, 698–725. [Google Scholar]
- Holmes-Farley S. R.; Reamey R. H.; McCarthy T. J.; Deutch J.; Whitesides G. M. Langmuir 1985, 1, 725–740. [Google Scholar]
- Hu Y. L.; Jiang H.; Zhu J.; Lu M. New J. Chem. 2011, 35, 292–298. [Google Scholar]
- Hermanson G. T.Bioconjugate Techniques; Academic Press: Amsterdam, 2008. [Google Scholar]
- Holmes-Farley S. R.; Bain C. D.; Whitesides G. M. Langmuir 1988, 4, 921–937. [Google Scholar]
- Lin J. C.; Tseng S. M. J. Mater. Sci. Mater. Med. 2001, 12, 827–832. [DOI] [PubMed] [Google Scholar]
- Bergbreiter D. E. Prog. Polym. Sci. 1994, 19, 529–560. [Google Scholar]
- Tao G.; Gong A.; Lu J.; Sue H.-J.; Bergbreiter D. E. Macromolecules 2001, 34, 7672–7679. [Google Scholar]
- Shouren G.; Kojio K.; Takahara A.; Kajiyama T. J. Biomater. Sci., Polym. Ed. 1998, 9, 131–150. [DOI] [PubMed] [Google Scholar]
- Wang C.; Wang J.; Deng L. Nanoscale Res. Lett. 2011, 6, 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y.; Miyai K.; Hata N.; Ichihara K. Anal. Biochem. 1984, 143, 249–255. [DOI] [PubMed] [Google Scholar]
- Subramanian G.Antibodies: Novel Technologies and Therapeutic Use, Volume 2; Springer: New York, 2004. [Google Scholar]
- Lytle D. A.; Rice E. W.; Johnson C. H.; Fox K. R. Appl. Environ. Microbiol. 1999, 65, 3222–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottenbos B.; Grijpma D. W.; van der Mei H. C.; Feijen J.; Busscher H. J. J. Antimicrob. Chemother. 2001, 48, 7–13. [DOI] [PubMed] [Google Scholar]
- Lee E.-H.; Yoo G.; Jose J.; Kang M.-J.; Song S.-M.; Pyun J.-C. BioChip J. 2012, 6, 221–228. [Google Scholar]
- Diamandis E. P., Christopoulos T. K., Eds. Immunoassay; Elsevier Science: San Diego, CA, 1996. [Google Scholar]
- Vogt K. W.; Kohl P. A.; Carter W. B.; Bell R. A.; Bottomley L. A. Surf. Sci. 1994, 301, 203–213. [Google Scholar]
- Fountoulakis M.; Juranville J. F.; Jiang L.; Avila D.; Röder D.; Jakob P.; Berndt P.; Evers S.; Langen H. Amino Acids 2004, 27, 249–259. [DOI] [PubMed] [Google Scholar]
- Nemzek J. A.; Newcomb D. E.; Call D. R.; Remick D. G. Immunol. Invest. 1999, 28, 209–221. [DOI] [PubMed] [Google Scholar]
- Selby C. Ann. Clin. Biochem. 1999, 36, 704–721. [DOI] [PubMed] [Google Scholar]
- Rodbard D.; Feldman Y.; Jaffe M. L.; Miles L. E. M. Immunochemistry 1978, 15, 77–82. [DOI] [PubMed] [Google Scholar]
- Ylander P. J.; Bicskei Z.; Hänninen P.; Soini J. T. Biophys. Chem. 2006, 123, 141–145. [DOI] [PubMed] [Google Scholar]
- Gubler D. J.; Suharyono W.; Tan R.; Abidin M.; Sie a. Bull. W. H. O. 1981, 59, 623–630. [PMC free article] [PubMed] [Google Scholar]
- Njenga M. K.; Paweska J.; Wanjala R.; Rao C. Y.; Weiner M.; Omballa V.; Luman E. T.; Mutonga D.; Sharif S.; Panning M.; Drosten C.; Feikin D. R.; Breiman R. F. J. Clin. Microbiol. 2009, 47, 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrell D. A.; Cox T. M.; Firth J. D.; Török E. e.. Oxford Textbook of Medicine: Infection, 5th ed.; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Driskell J. D.; Kwarta K. M.; Lipert R. J.; Porter M. D.; Neill J. D.; Ridpath J. F. Anal. Chem. 2005, 77, 6147–6154. [DOI] [PubMed] [Google Scholar]
- Parsa H.; Chin C. D.; Mongkolwisetwara P.; Lee B. W.; Wang J. J.; Sia S. K. Lab Chip 2008, 8, 2062–2070. [DOI] [PubMed] [Google Scholar]
- Driskell J. D.; Kwarta K. M.; Lipert R. J.; Vorwald A.; Neill J. D.; Ridpath J. F.; Porter M. D. J. Virol. Methods 2006, 138, 160–169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.