Abstract
The knob domains from the fiber proteins of adenovirus serotypes 2 and 12 were labeled with radioiodine and then injected into the bloodstreams of mice. Knob proteins with functional binding sites for the coxsackie and adenovirus receptor (CAR) were cleared rapidly from the circulation, with radioactivity appearing predominantly in the stomach, while knob mutants unable to bind to CAR remained in the blood circulation for a prolonged period. The clearance of radiolabeled wild-type knob from the blood was slowed by coinjecting an excess of unlabeled wild-type knob protein. An earlier study showed that 99mTc-labeled knob protein with intact CAR-binding activity also cleared rapidly from the blood circulation of mice, with radioactivity accumulating predominantly in the liver (K. R. Zinn et al., Gene Ther. 5:798-808, 1998). Together these results suggest that rapid clearance of knob protein from the blood results from specific binding to CAR in the liver and that the bound knob then enters a degradative pathway. The elevated levels of radioiodine in the stomach observed in our experiments are consistent with deiodination of labeled knob by dehalogenases in hepatocyte microsomes and uptake of the resultant free radioiodine by Na/I symporters in the gastric mucosa. Although CAR has been shown to localize in tight junctions of polarized epithelial cells, where it functions in intercellular adhesion, the results of our study suggest that a subset of CAR molecules in the liver is highly accessible to ligands in the blood and able to rapidly deliver bound ligand to an intracellular degradative compartment.
Specific binding to cell surface receptors is necessary for initial infection of host cells by animal viruses. Many human adenoviruses and the group B coxsackieviruses bind to the coxsackie and adenovirus receptor (CAR), a type I membrane glycoprotein with broad tissue distribution (4, 35, 39). The efficiency of virus infection depends on the accessibility and concentration of receptors on host cells and also on the affinity of virus particles for the receptor. For example, the observed poor efficiency of adenovirus-based gene delivery vectors in transducing lung tissue (15, 16, 25, 49) led to the unexpected finding that CAR molecules are localized predominantly on the basal-lateral surface of the respiratory epithelium, where they are inaccessible to virus particles administered via the airway (45). Further investigation showed that CAR is localized in the junctional complex of polarized respiratory epithelial cells, where it is relatively inaccessible to virus particles applied to either the apical or the basal surface of the epithelial sheet (7, 46). CAR expression levels also determine the susceptibilities of various cell lines to adenovirus infection (11, 27, 33, 44, 50).
Adenovirus infections in humans are in most cases restricted to the upper respiratory and gastrointestinal tracts, although rare disseminated infections resulting in liver necrosis have been observed (10). When purposely injected into the mouse bloodstream, adenovirus particles also are taken up preferentially by the liver (29, 31). This marked hepatotropism implies that CAR molecules in the liver may be readily accessible to adenovirus particles in the circulation. Adenoviruses interact with CAR through the knob or head domain of the viral fiber protein (19). Characterization of virus-CAR interaction has been greatly facilitated by the ability of the knob domain to fold correctly during overexpression in Escherichia coli. Knob polypeptides assemble into ∼60-kDa trimers that have three equivalent, high-affinity binding sites for CAR (5, 28, 48). An earlier analysis of the biodistribution of the 99mTc-labeled recombinant knob domain following intravenous administration in mice also suggests that CAR may be highly accessible in the liver relative to other tissues (51). Mouse liver was shown to have about 1013 saturable binding sites for 99mTc-labeled knob (capacity for ∼3 μg of knob protein) and to clear essentially 100% of subsaturating doses of knob from the blood within the first minute of circulation (51). The localization of CAR molecules in the liver has not been examined in detail, but it seems unlikely that CAR sequestered in tight junctions could clear knob from the blood with such rapid kinetics.
Here we analyzed the biodistribution of radioactivity in mice following injection of radioiodinated knob proteins into the bloodstream. Knob proteins with intact CAR-binding activity were cleared rapidly from the blood, but in contrast to the findings with 99mTc-labeled knob, radioactivity did not accumulate in the liver but instead was found at elevated levels in the stomach. Stomach activity typically results from transport of free iodine from the blood into the gut lumen by Na/I symporters in the gastric mucosa and thus indicates that free iodine must be released from the labeled knob protein. Our results suggest that CAR molecules in the liver not only remove knob protein from the blood with high efficiency but also deliver bound knob to a cellular compartment where it is metabolized to release free iodine. Thus, in addition to their role as a homotypic adhesion molecule in epithelial-cell tight junctions, CAR molecules in the liver may also function in ligand uptake and metabolism.
MATERIALS AND METHODS
Expression and purification of recombinant knob domains.
Wild-type and mutant knob domains derived from adenovirus serotypes 2 and 12 (Ad2 and Ad12) were expressed in E. coli with N-terminal His tags by using the pET15b expression vector (Novagen, Inc., Madison, Wis.) as previously described (22). Knob mutants were constructed by PCR mutagenesis using the following mutagenic oligonucleotide primers: 5′-GTACTTGACTCCCGTTCTTTGTAAGAACCA for Ad2 knob C428N, 5′-AATTCTGCAGTTTGCCTCTGGGTCTGGG for Ad2 knob S408E P409A (SP/EA), 5′-GCAGTTTGGTGATGGGTCAGGAG for Ad12 knob P417S, and 5′-AGGCTGCAGTTTGCCTCTGGGTCAGGAGTT for Ad12 knob P408E P409A (PP/EA). Knob proteins were purified from cell lysates by immobilized metal affinity chromatography on nickel-nitrilotriacetic acid coupled agarose beads (Qiagen, Inc., Valencia, Calif.). Bound knob proteins were eluted with imidazole, dialyzed against phosphate-buffered saline (pH 7.2), and stored at −80°C. Proteins were electrophoresed in denaturing polyacrylamide gels containing sodium dodecyl sulfate (SDS) according to the method of Laemmli (26). Conditions for nondenaturing polyacrylamide gel electrophoresis (42) and the use of this method to detect the interaction of wild-type and mutant Ad12 knob proteins with CAR D1 (5) have been described previously. The fluorescence anisotropy-based assay for knob-CAR binding also has been detailed elsewhere (22).
Radioiodination of knob proteins.
Knob proteins were labeled on tyrosine with 131I by using Iodogen beads (Pierce Biotechnology, Inc., Rockford, Ill.) according to the manufacturer's instructions. Briefly, a bead was washed with Tris-iodination buffer (TIB) (125 mM Tris-HCl [pH 6.8], 150 mM NaCl) and dropped into a reaction tube containing 100 μl of radioactivity (50 μCi of Na131I). After 5 min was allowed for oxidation of iodide species, knob protein (80 μl; 1 mg/ml) was added to the tubes and the reaction was allowed to proceed for five more minutes. The reaction was stopped by addition of 25 μl of l-tyrosine solution (10 mg/ml in TIB). Unincorporated iodine was removed by ultrafiltration in a 10-kDa-cutoff centrifuge cartridge (Pall Corp., Ann Arbor, Mich.). Protein in the retentate was diluted to 500 μl with saline and filtered through a 300-kDa-cutoff ultrafiltration cartridge to eliminate protein aggregates. Labeled proteins in the filtrate were characterized by electrophoresis in polyacrylamide gels under denaturing and nondenaturing conditions before injection into mice. Proteins in gels were visualized by Coomassie blue staining.
Knob proteins also were labeled with 125I on lysine residues by using Bolton-Hunter reagent to control for possible effects of labeling on protein stability in vivo. For radioiodination using Bolton-Hunter reagent, 125I-labeled Bolton-Hunter reagent (100 μCi) was dried in a stream of nitrogen and kept on ice. Knob protein (25 μg at 1 mg/ml in phosphate-buffered saline) was added to the dried reagent, and the reaction was allowed to proceed for 3 h on ice. The reaction was terminated by addition of 0.25 ml of 0.2 M glycine dissolved in phosphate-buffered saline (pH 8). After incubation at room temperature for 5 min, the radiolabeled knob protein was separated from unincorporated Bolton-Hunter reagent by ultrafiltration as described above.
Animal biodistribution experiments.
Animal experiments were approved by the Institutional Animal Care and Use Committee at Brookhaven National Laboratory (Upton, N.Y.) and performed according to its guidelines. Animals were housed in an isolated animal facility under controlled conditions. Male Swiss Webster mice were placed in a mouse restrainer and injected in the tail vein with radiolabeled proteins (200 μl; ∼2.64 μg; 1 μCi). After specific times (1.5, 6, or 24 h), the animals (n = 3 per group) were sacrificed, and various organs were weighed and counted for radioactivity. Radioactive accumulation was calculated on a per-gram-of-tissue basis. Blood volume, muscle mass, and bone mass were estimated at 7, 43, and 10% of body weight, respectively. Analysis of variance (ANOVA) or the Student t test was performed on the biodistribution data by using GraphPad PRISM software (version 4.0; GraphPad Software, Inc., San Diego, Calif.). The ANOVA was followed by Bonferroni's post hoc test, and a P value of <0.05 was taken as significant for rejection of the null hypothesis. For competition experiments, mice were coinjected with radioiodinated knob and a 10-fold molar excess of unlabeled knob protein.
RESULTS
Activity and stability of knob domains used for biodistribution analysis.
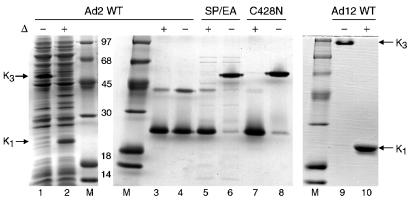
Wild-type knob domains from the fiber proteins of Ad2 and Ad12, and mutant Ad2 and Ad12 knobs with decreased or enhanced affinity for CAR, were expressed in E. coli with N-terminal His tags and purified by metal affinity chromatography. Knob trimers are resistant to denaturation by SDS at moderate temperatures (21), reflecting a highly stable trimer interface (42, 48). We observed that while freshly isolated wild-type Ad2 knob trimers were stable in SDS (Fig. 1, lanes 1 and 2), purified Ad2 knob trimers that had been stored at 4°C for 1 to 2 days were dissociated to polypeptide monomers and an apparent dimer species after incubation at room temperature in gel sample buffer containing 2% SDS and 5% β-mercaptoethanol (Fig. 1, lanes 3 and 4). By contrast, wild-type Ad12 knob trimers were resistant to denaturation by SDS after prolonged storage at 4°C (Fig. 1, lanes 9 and 10).
FIG. 1.
Stability and activity of knob trimers. Cleared lysates of E. coli cells expressing wild-type (WT) Ad2 knob were mixed with Laemmli sample buffer and incubated for 5 min at room temperature (−) or in a boiling water bath (+ and Δ) before electrophoresis in an SDS-polyacrylamide gel. Ad2 knob protein in the unheated sample migrated to a position indicated by K3, corresponding to the intact knob trimer (lane 1). After boiling, Ad2 knob protein migrated to a position indicated by K1, corresponding to the dissociated monomeric knob polypeptide (lane 2). Samples of knob domains from wild-type Ad2 (lanes 3 and 4), Ad2 mutants SP/EA (lanes 5 and 6) and C428N (lanes 7 and 8), and wild-type Ad12 (lanes 9 and 10) were analyzed for trimer stability, as described above, after storage of the purified proteins for several days at 4°C. Wild-type Ad2 knob trimers were dissociated to polypeptide monomers and dimers by incubation in Laemmli buffer at room temperature (lane 4), whereas Ad2 SP/EA, Ad2 C428N, and wild-type Ad12 trimers were stable under these conditions (lanes 6, 8, and 9, respectively). Intact knob trimers do not uniformly bind SDS; therefore, the different electrophoretic mobilities of Ad2 and Ad12 knob trimers result from differences in surface charge rather than in molecular size. Proteins were visualized by staining the gels with Coomassie blue. The molecular sizes (in kilodaltons) of protein standards loaded in lanes M are indicated.
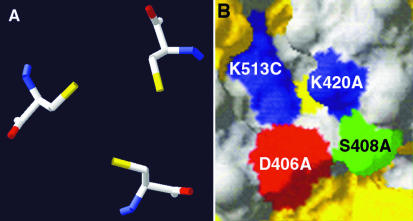
The relatively abundant polypeptide dimer generated by incubation of Ad2 knob in standard gel sample buffer (Fig. 1, lane 4) could be converted to polypeptide monomers by treatment with the strong reducing agent Tris(2-carboxyethyl)-phosphine hydrochloride (TCEP) (data not shown), suggesting that this species might result from disulfide bond formation between polypeptide subunits in knob trimers. The crystal structure of the Ad2 knob trimer (42) shows that the sulfhydryl groups of cysteine 428 of each polypeptide subunit are oriented toward the threefold axis of symmetry at the base of the knob, with a distance of 4.2 Å between sulfur atoms (Fig. 2A). Cysteine is not conserved at this position in knob domains from other Ad serotypes, and the structurally equivalent residue in Ad12 knob is asparagine. When cysteine 428 of Ad2 knob was converted to asparagine (C428N), the mutant trimers were stable in SDS after prolonged storage (Fig. 1, lanes 7 and 8). These results suggest that a disulfide bond can form between the cysteine 428 residues of two subunits of wild-type Ad2 knob trimers upon air oxidation and that formation of the disulfide bond may lock the knob trimer into an asymmetric conformation that is more easily invaded by SDS. As expected, the CAR-binding activity of the C428N mutant was intact, as indicated by a quantitative shift in its electrophoretic mobility after brief incubation with CAR D1 prior to electrophoresis under nondenaturing conditions (Fig. 3A).
FIG. 2.
Structural features that impact Ad2 knob trimer stability. (A) Spatial orientation of the cysteine 428 of each polypeptide subunit of the wild-type Ad2 knob trimer. Disulfide bond formation upon air oxidation may produce the Ad2 knob polypeptide dimer species resolved by SDS-polyacrylamide gel electrophoresis (Fig. 1) and render the knob trimer sensitive to denaturation by SDS. Yellow, sulfur atoms. (B) Portion of the surface of the Ad2 knob trimer, showing the juxtaposition of lysine 420 from polypeptide chain A and lysine 513 from chain C. Electrostatic repulsion between these lysines may destabilize the trimer interface and promote denaturation by SDS. Also shown are aspartate 406 and serine 408 from polypeptide chain A. Mutation of serine 408 to glutamic acid renders the knob trimer resistant to denaturation by SDS. The introduced negative charge of Glu408 therefore may balance the positive charge on lysine 420, reducing the electrostatic repulsion across the trimer interface. Both panels were produced with the SwissPDB Viewer program (17).
FIG. 3.
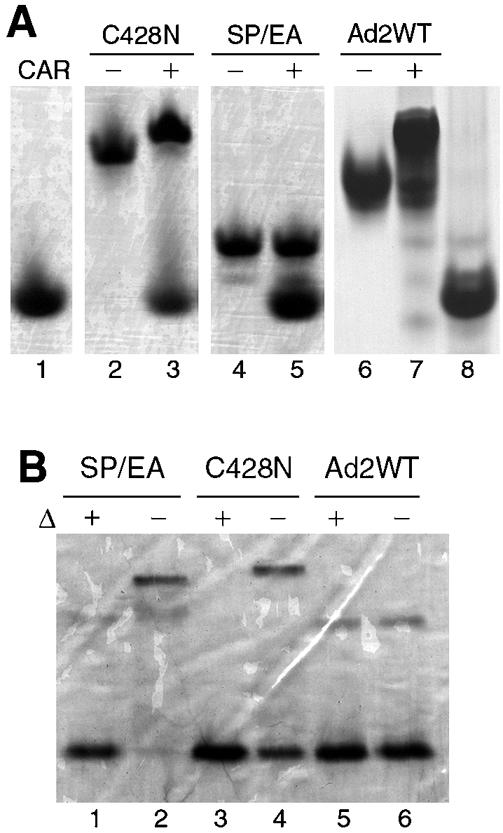
(A) 131I-labeled Ad2 knob mutants (C428N and SP/EA) and unlabeled wild-type (WT) Ad2 knob protein were incubated for 5 min at room temperature in buffer alone (lanes 2, 4, and 6, respectively) or in buffer containing the recombinant immunoglobulin variable-type domain of CAR (lanes 3, 5, and 7, respectively) prior to electrophoresis in polyacrylamide gels under nondenaturing conditions to detect the formation of knob-CAR complexes. Samples of CAR D1 alone were loaded in lanes 1 and 8. Proteins were visualized by staining the gel with Coomassie blue. (B) SDS-polyacrylamide gel analysis, as described for Fig. 1, of wild-type and mutant Ad2 knob proteins after labeling with 131I by the iodobead method. Iodination partially destabilized the C428N (lane 4) but not the SP/EA (lane 2) mutant. Proteins were visualized by staining the gel with Coomassie blue.
The knob AB loop contributes about half of the amino acids in knob that directly contact CAR D1, and it was reported previously that the binding affinity of Ad12 knob for CAR was severely reduced by alteration of two contact residues (P417E P418A) in the AB loop by site-directed mutagenesis (5). To create a non-CAR-binding derivative of Ad2 knob for use as a control in biodistribution experiments, Ad2 knob residues S408 and P409, which correspond to P417 and P418 of Ad12 knob, were similarly altered by mutagenesis (S408E P409A). As expected, the resulting Ad2 knob mutant (SP/EA) was unable to form stable complexes with CAR D1, as shown by the absence of a mobility shift when the mutant protein was mixed with CAR D1 prior to electrophoresis in a nondenaturing polyacrylamide gel (Fig. 3A).
SP/EA mutant trimers unexpectedly were resistant to denaturation by SDS after prolonged storage at 4°C (Fig. 1, lanes 5 and 6), and the Ad2 knob crystal structure again suggested a possible mechanism for stabilization of the mutant trimers. Charge repulsion between the closely juxtaposed lysine 420 and lysine 513 side chains on adjacent subunits of wild-type Ad2 knob trimers might provide an opening into the trimer interface that can be invaded by SDS. The extra negative charge introduced by substitution of glutamic acid for serine at residue 408 is located close to the positively charged side chain of lysine 420 (Fig. 2B) and therefore might weaken electrostatic repulsion across the trimer interface between lysines 420 and 513.
Biodistribution analysis.
We compared the biodistribution in mice of knob domains from two different adenovirus serotypes in order to examine the possible effects of variation in binding affinity for the CAR. The knob domain from Ad2 binds CAR with about eightfold greater affinity than does the knob domain from Ad12 (22). After being labeled with 131I by the iodobead method, C428N trimers were partially dissociated by SDS (Fig. 3B) but still retained their ability to interact with CAR D1 in vitro (Fig. 3A). Labeling with 131I did not measurably destabilize the SP/EA mutant trimers (Fig. 3B) or trimers of Ad12 knob (data not shown).
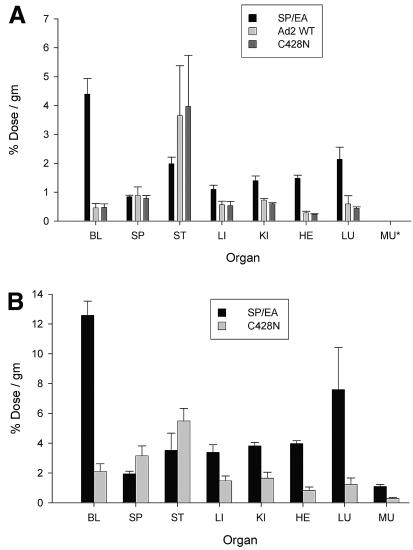
Mice were intravenously injected (via the tail vein) with 131I-labeled knob proteins and were then sacrificed at different times to determine the biodistribution of radioactivity in various organs. Typical biodistribution results with Ad2 knob proteins at 6 h postinjection are shown in Fig. 4A. Knob proteins with intact CAR-binding activity (wild-type and C428N mutant knob proteins) were cleared more rapidly from the blood than SP/EA mutant knob protein, which is unable to bind to CAR and was maintained in the blood circulation for a prolonged period. Radioactivity levels in all organs from mice injected with wild-type or C428N knob were similar to background levels in the blood, except for elevated radioactivity levels in the stomachs of some animals. The absolute amounts of radioactivity in the kidney, heart, and lung tissues were greater in mice injected with the non-CAR-binding SP/EA knob than in mice injected with wild-type or C428N knob. However, it should be noted that blood could not be completely removed from dissected organs before counting; therefore, the higher levels of radioactivity in the kidneys, hearts, and lungs of mice injected with SP/EA knob likely resulted from the elevated levels of the labeled SP/EA protein in the residual blood contained within these organs rather than from binding of the SP/EA knob protein to specific receptor sites within these organs.
FIG. 4.
Biodistribution of radioiodinated Ad2 knob proteins. (A) Wild-type (WT) and mutant Ad2 knob proteins were directly labeled with 131I by the iodobead method (Pierce Biotechnology, Inc.), and then injected intravenously into mice (via the tail vein). After 6 h of circulation, mice were sacrificed, and the radioactivity in dissected organs was measured. BL, blood; SP, spleen; ST, stomach; LI, liver; KI, kidney; HE, heart; LU, lung; MU, muscle (data for muscles were not collected in the experiment shown). Wild-type and C428N mutant Ad2 knobs have equivalent CAR-binding activities, whereas the SP/EA mutant is unable to bind to CAR. Differences between radioactivity levels in organ samples from mice injected with wild-type versus C428N knob were not significant (P > 0.05). Significant differences (P < 0.05) in radioactivity levels in the blood, liver, kidneys, heart, and lungs were observed between mice injected with the non-CAR-binding SP/EA mutant and the CAR-binding wild-type or C428N knob. (B) The same experiment as that for which results are shown in panel A was performed, except that knob proteins were indirectly labeled with 125I by using the Bolton-Hunter reagent (6). Differences in radioactivity levels in organs from mice injected with SP/EA versus C428N knob were all significant (P < 0.05) except for levels in the stomach.
The experiments were repeated with knob proteins that were labeled with 125I-labeled Bolton-Hunter reagent in order to control for possible effects of the different labeling strategies on knob protein stability in vivo. As was observed with the 131I-labeled knob proteins, 125I-labeled C428N mutant knob cleared more rapidly from the blood than 125I-labeled SP/EA mutant knob, which persisted in the blood for a prolonged period (Fig. 4B). In mice injected with the CAR-binding wild-type or C428N knob, radioactivity levels in most organs were at or below the background level in the blood, except for levels significantly elevated over background in the stomachs of all animals. Labeled SP/EA protein in the residual blood contained within the liver, kidney, heart, and lungs probably accounted for the greater absolute levels of radioactivity in these organs than in the corresponding organs from mice injected with the C428N protein. The similar overall results observed in these two experiments suggest that knob biodistribution in mice is not substantially altered by modification of surface-exposed tyrosine or lysine residues with radioiodine.
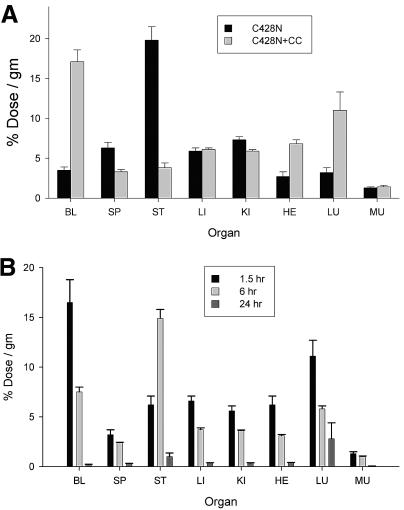
Mice were then coinjected with 131I-labeled C428N protein alone or with a 10-fold molar excess of unlabeled C428N protein, and radioactivity levels in the blood and various organs were determined 6 h later (Fig. 5A). In agreement with the results described above, by 6 h postinjection, C428N protein was mostly cleared from the blood, and most organs showed little accumulation of radioactivity above blood background levels except for the stomach, which had large accumulations above background. Coinjection of an excess of unlabeled C428N protein markedly slowed the clearance of 131I-labeled C428N knob from the blood, suggesting that knob clears from the blood through a specific pathway that requires the binding of knob to a saturable number of receptor sites. A more detailed time course study was then performed, where organs were harvested from mice at 1.5, 6, and 24 h after coinjection with 131I-labeled C428N and a 10-fold excess of unlabeled C428N knob (Fig. 5B). The elevated levels of radioactivity present in the circulation at 6 h postinjection were reduced to near-background levels by 24 h postinjection (Fig. 5B). Decreasing levels of radioactivity were detected at each time point in all organs except the stomach, which had a maximal level at 6 h. By 24 h, radioactivity levels in all organs, including the stomach, were reduced to near the blood background level.
FIG. 5.
Biodistribution of radioiodinated Ad2 C428N protein in the presence of excess unlabeled C428N protein. (A) Ad2 C428N mutant protein was directly iodinated with 131I and injected intravenously into mice (via the tail vein) in the presence (+CC) or absence of a 10-fold excess of coinjected unlabeled C428N protein. After 6 h of circulation, mice were sacrificed, and radioactivity in dissected organs was measured. Organ abbreviations are explained in the legend to Fig. 4. All differences in radioactivity levels in organs between mice injected with 131I-labeled C428N alone and mice coinjected with excess unlabeled C428N were significant (P < 0.05) except for levels in the liver and muscle. (B) Mice were coinjected intravenously with 131I-labeled Ad2 C428N protein and a 10-fold molar excess of unlabeled C428N protein. Mice were sacrificed after 1.5, 6, or 24 h of circulation, and radioactivity in dissected organs was measured. Differences in organ radioactivity levels between the 1.5- and 24-h samples were all significant (P < 0.05). Differences between samples taken at 1.5 versus 6 h were significant (P < 0.05) for all organs except the spleen, lungs, and muscle. Differences between samples taken at 6 versus 24 h were significant for all organs except the lungs.
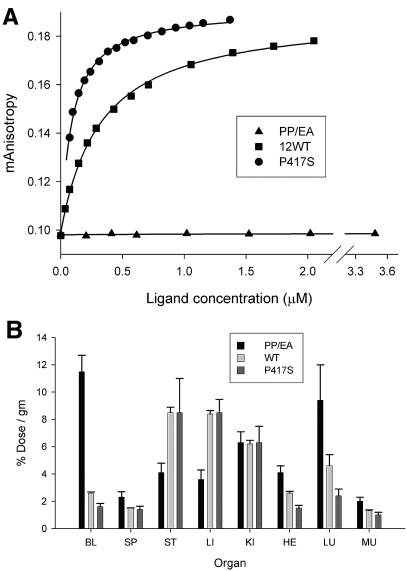
A parallel set of experiments was conducted with the recombinant knob domain from Ad12, which binds CAR with ∼8-fold-weaker affinity than Ad2 knob (22). Two Ad12 knob mutants were included in the analysis: the PP/EA mutant, which cannot bind to CAR, and the P417S mutant, which has a binding affinity for CAR approximately equivalent to that of Ad2 knob (Fig. 6A). As was observed with Ad2 knob, Ad12 knob proteins that can interact with CAR (wild-type and P417S mutant) were mostly cleared from the blood by 6 h postinjection, whereas the nonbinding PP/EA mutant was retained in the blood for a prolonged period (Fig. 6B). At 6 h postinjection, differences in blood radioactivity levels between mice injected with wild-type versus P417S knob were not significant. The overall biodistribution of radioactivity in major organs was similar in mice injected with 131I-labeled wild-type versus P417S Ad12 knob. Relative to blood background levels, mice injected with 131I-labeled wild-type or P417S Ad12 knob showed elevated levels of radioactivity in the stomach, as observed for CAR-binding Ad2 knob proteins. However, in contrast to the results with Ad2 knob, mice injected with 131I-labeled wild-type or P417S Ad12 knob also had substantial accumulations of radioactivity (relative to blood background) in the liver and kidneys.
FIG. 6.
Biodistribution of wild-type and mutant Ad12 knob proteins. (A) CAR-binding activities of wild-type (WT) Ad12 knob and the Ad12 knob P417S and PP/EA mutants were measured by fluorescent anisotropy using a fluorescein-labeled CAR probe as described previously (22). (B) Wild-type and mutant Ad12 knob proteins were directly labeled with 131I and injected intravenously into mice (via the tail vein). After 6 h of circulation, mice were sacrificed, and radioactivity in dissected organs was measured. Organ abbreviations are explained in the legend to Fig. 4. Differences in radioactivity levels in organs from mice injected with the CAR-binding wild-type or P417S knob were not significant except for levels in the heart (P < 0.05). Differences between samples from mice injected with the nonbinding PP/EA mutant versus wild-type knob were significant (P < 0.05) except for the spleen, kidneys, and lungs. Differences between PP/EA and P417S mutant samples were all significant (P < 0.05) except for kidney samples.
DISCUSSION
Here we investigated the biodistribution in mice of radioiodinated knob domains derived from the fiber proteins of Ad2 and Ad12. The biodistribution in mice of the 99mTc-labeled knob domain derived from the Ad5 fiber protein was described in an earlier report (51). The knob domains from these three serotypes bind specifically to CAR with high affinity (22, 24, 28). The results of the two biodistribution studies are in good agreement with regard to the rapid clearance of knob proteins from the blood. Clearance of 99mTc-labeled Ad5 knob from the blood was inhibited by coinjection of an excess of unlabeled Ad5 knob but not by coinjection of an excess of unlabeled knob derived from Ad3, which is unable to bind to CAR (8, 35). In the present study, clearance of radioiodinated Ad2 knob was inhibited by coinjection of unlabeled Ad2 knob. Futhermore, Ad2 and Ad12 knob mutants that cannot bind to CAR were cleared from the blood more slowly than the corresponding wild-type, CAR-binding forms of these proteins. Together these results support the conclusion that rapid clearance of knob proteins from the blood results from interaction of knob proteins with CAR.
A key difference between the results of these two studies was the site of accumulation of radioactivity in mice after injection of radiolabeled CAR-binding knob proteins. While the bulk of radioactivity accumulated in the livers of mice injected with 99mTc-labeled Ad5 knob, only minor accumulations of radioactivity were detected in the livers of mice injected with 131I-labeled Ad12 knob, and no accumulation over background was detected in mice injected with iodinated wild-type or C428N Ad2 knob. Since knob proteins from all three serotypes bind to CAR with similarly high affinities and have significant structural and sequence homologies, the different biodistributions observed in the two studies likely result from different properties of the radionuclides used for protein labeling. Technetium-99m is a radiometal that can bind either directly to protein sulfhydryl groups or indirectly to chelating groups that are incorporated into the protein by chemical or genetic modification (9, 14). Ad5 knob was labeled with 99mTc by an indirect method using the chelator succinimidyl 6-hydrazinonicotinate (HYNIC), which was incorporated into knob protein by chemical modification (51). Catabolism of 99mTc-HYNIC-labeled proteins in hepatocyte microsomes has been reported in other studies (34) to generate a stable radiometabolite, 99mTc-(HYNIC-lysine)(tricine)(tricine), which is eliminated from the liver only very slowly. Because these stable radiometabolites are retained in the liver, it is not possible to determine from tissue counting or in vivo imaging whether radioactivity corresponds to intact radiolabeled knob protein or to products of knob degradation.
Proteins can be labeled with radioiodine either directly on tyrosine residues, as in the Iodobead method, or by attachment of preformed iodotyrosine to lysine epsilon amino groups, as in the Bolton-Hunter method (6). However, the iodine-tyrosine bond is labile to hydrolysis by deiodinase enzymes, which are particularly abundant in hepatocyte microsomes (30, 38). In contrast to the intracellular sequestration of 99mTc-labeled metabolites, free iodine released from labeled proteins by deiodinase activity readily diffuses out of the liver and enters the bloodstream, where it is taken up by Na/I symporters in the gastric mucosa and transported into the gut lumen (23). In control experiments, we injected mice with free 131I and observed rapid uptake of radioactivity in the gut (data not shown), in good agreement with published results (23). The limited accumulation of radioactivity in the livers of mice injected with 131I-labeled knob proteins therefore likely results from efficient deiodination of knob protein in the liver and diffusion of the resultant free iodine into the bloodstream. Consistent with this model, an earlier study showed that 99mTc-labeled low-density lipoprotein particles were trapped in the liver whereas 125I-labeled low-density lipoprotein particles were deiodinated in the liver, with subsequent accumulation of free iodine in the intestine (41). In mice injected with 131I-labeled Ad2 or Ad12 knob mutants that were unable to interact with CAR, radioactivity was retained in the blood circulation for a prolonged period and did not accumulate in the stomach, indicating that knob protein is not exposed to significant levels of deiodinase activity in the circulation.
While the fate of the knob protein itself cannot be directly determined from our results, it is likely that proteolytic degradation of knob protein would accompany deiodination in hepatocyte microsomes. The slight accumulation of radioactivity in the livers and kidneys of mice injected with 131I-labeled Ad12 knob (Fig. 6B) that was not seen for Ad2 knob suggests that intracellular uptake and/or deiodination may occur at a lower rate for Ad12 knob than for Ad2 knob. Radioactivity in the kidneys of these mice (Fig. 6B) might correspond to peptide degradation products of knob generated in the liver that still retain the radioiodine label.
A role for CAR in protein uptake and metabolism in the liver is unexpected, based on the current understanding of CAR physiology as a tight junction membrane protein (3). While CAR mediates high-affinity attachment of adenovirus particles to the cell plasma membrane, rapid endocytosis of virus-CAR complexes by epithelial cells typically is promoted by integrin coreceptors that interact with an Arg-Gly-Asp (RGD) motif on the viral penton base subunits (2, 47). Ad2 mutants lacking functional penton base RGD motifs are infectious, but entry of these mutant virus particles into adherent cultured cells is delayed by several hours relative to that of wild-type virus (2). The knob domain does not have an RGD motif and is not known to interact with integrins; therefore, CAR alone may be sufficient for endocytosis of knob-CAR complexes from the surfaces of mouse hepatocytes in vivo. The absence of significant accumulation of radioactivity in the liver suggests that once bound to CAR molecules on the hepatocyte surface, knob is rapidly delivered to an intracellular compartment where it is exposed to deiodinase activity. Consistent with this model of integrin-independent uptake of knob, an earlier study showed that adenovirus-CAR complexes on rat hepatocytes can be internalized by an integrin-independent pathway (18).
In polarized epithelial cells, CAR is localized to tight junctions, where it functions as a homotypic cell adhesion molecule (7, 20, 45). CAR-mediated homotypic cell adhesion is likely to result from dimerization of the distal extracellular domain (D1) of opposing CAR molecules on adjacent cells (43). The surface of the CAR D1 domain that forms the homodimer interface is the same surface that is recognized by the knob domain (5, 43). CAR molecules in the junctional complex of polarized epithelial cells are relatively inaccessible to ligands applied to either the apical or the basal surface of the epithelial-cell sheet (7). If CAR is also sequestered in the junctional complex of hepatocytes, then binding of knob to these receptors would likely be inefficient. However, the mouse liver was estimated to have 17,000 binding sites per hepatocyte for 99mTc-labeled Ad5 knob, and these sites were fully saturated with 99mTc-labeled Ad5 knob after circulation of knob in the blood for only 1 min (51). CAR molecules on hepatocytes therefore extract knob from the blood with high efficiency, suggesting that at least a subset of CAR molecules may be localized on the hepatocyte sinusoidal (basal) surface with their D1 domains unoccupied and available for binding to knob. CAR protein and mRNA levels are particularly abundant in the rodent liver relative to levels in other rodent tissues (39, 40), possibly accounting for localization of a fraction of CAR to the hepatocyte sinusoidal surface. Alternatively, localization of CAR to distinct membrane subdomains could result from modifications to the CAR cytoplasmic tail, as suggested by a recent study (1).
The present study and the earlier work with 99mTc-labeled knob (51) demonstrate that the recombinant knob domain is a highly effective reagent for targeting CAR molecules in vivo. The knob protein scaffold has several features that might be exploited for general applications in imaging and molecular recognition. Knob trimers are large enough (60 kDa) to avoid rapid renal clearance, as indicated by the prolonged circulation of knob mutants in the mouse bloodstream and the delayed clearance of iodinated wild-type knob by coinjection of excess unlabeled knob, yet they are small relative to most antibody-based targeting reagents and thus should have good penetration into tumors or peripheral tissues. Although the binding specificity of knob domains derived from natural adenovirus serotypes is restricted to CAR and a limited number of alternate receptors (4, 13, 32, 37, 39), the contact residues that determine binding specificity are localized to surface loops that are highly tolerant of sequence variation (5, 36). Therefore, it should be possible to generate knob proteins with novel binding specificities by altering the sequence and length of the specificity-determining loops. Avidity effects resulting from trivalent binding increase the overall affinity of knob for target molecules (28), which could potentially circumvent the need to develop individual binding sites with very high affinities. Recombinant knob domains from several adenovirus serotypes fold correctly and assemble into active trimeric species during overexpression in bacteria (12, 19, 42), providing an abundant and inexpensive source of protein reagents.
Acknowledgments
This work was supported by Public Health Service grant AI-36251 (to P.F.) from the National Institute of Allergy and Infectious Diseases and by a grant from the Medical Sciences Division of the U.S. Department of Energy Office of Biological and Environmental Research.
REFERENCES
- 1.Ashbourne Excoffon, K. J., T. Moninger, and J. Zabner. 2003. The coxsackie B virus and adenovirus receptor resides in a distinct membrane microdomain. J. Virol. 77:2559-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, M., B. Harfe, and P. Freimuth. 1993. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J. Virol. 67:5198-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergelson, J. M. 2003. Virus interactions with mucosal surfaces: alternative receptors, alternative pathways. Curr. Opin. Microbiol. 6:386-391. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 5.Bewley, M. C., K. Springer, Y. B. Zhang, P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 6.Bolton, A. E., and W. M. Hunter. 1973. The labelling of proteins to high specific radioactivities by conjugation to a 125I-containing acylating agent. Biochem. J. 133:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, C. J., J. T. Shieh, R. J. Pickles, T. Okegawa, J. T. Hsieh, and J. M. Bergelson. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 98:15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Defer, C., M. T. Belin, M. L. Caillet-Boudin, and P. Boulanger. 1990. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J. Virol. 64:3661-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckelman., W. C. 1995. Radiolabeling with technetium-99m to study high-capacity and low-capacity biochemical systems. Eur. J. Nucl. Med. 22:249-263. [DOI] [PubMed] [Google Scholar]
- 10.Faden, H., H. Jockin, M. R. Talty, R. Fazilli, and H. Barrett. 1985. Fatal disseminated adenovirus infection featuring liver necrosis and prolonged viremia in an immunosuppressed child with malignant histiocytosis. Am. J. Pediatr. Hematol. Oncol. 7:15-20. [PubMed] [Google Scholar]
- 11.Freimuth, P. 1996. A human cell line selected for resistance to adenovirus infection has reduced levels of the virus receptor. J. Virol. 70:4081-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freimuth, P., K. Springer, C. Berard, J. Hainfeld, M. Bewley, and J. Flanagan. 1999. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J. Virol. 73:1392-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 14.Goel, A., J. Baranowska-Kortylewicz, S. H. Hinrichs, J. Wisecarver, G. Pavlinkova, S. Augustine, D. Colcher, B. J. Booth, and S. K. Batra. 2001. 99mTc-labeled divalent and tetravalent CC49 single-chain Fv's: novel imaging agents for rapid in vivo localization of human colon carcinoma. J. Nucl. Med. 42:1519-1527. [PubMed] [Google Scholar]
- 15.Goldman, M. J., and J. M. Wilson. 1995. Expression of αvβ5 integrin is necessary for efficient adenovirus-mediated gene transfer in the human airway. J. Virol. 69:5951-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grubb, B. R., R. J. Pickles, H. Ye, J. R. Yankaskas, R. N. Vick, J. F. Engelhardt, J. M. Wilson, L. G. Johnson, and R. C. Boucher. 1994. Inefficient gene transfer by adenovirus vector to cystic fibrosis airway epithelia of mice and humans. Nature 371:802-806. [DOI] [PubMed] [Google Scholar]
- 17.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 18.Hautala, T., T. Grunst, A. Fabrega, P. Freimuth, and M. J. Welsh. 1998. An interaction between penton base and αv integrins plays a minimal role in adenovirus-mediated gene transfer to hepatocytes in vitro and in vivo. Gene Ther. 5:1259-1264. [DOI] [PubMed] [Google Scholar]
- 19.Henry, L. J., D. Xia, M. E. Wilke, J. Deisenhofer, and R. D. Gerard. 1994. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J. Virol. 68:5239-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda, T., H. Saitoh, M. Masuko, T. Katagiri-Abe, K. Tominaga, I. Kozakai, K. Kobayashi, T. Kumanishi, Y. G. Watanabe, S. Odani, and R. Kuwano. 2000. The coxsackievirus-adenovirus receptor protein as a cell adhesion molecule in the developing mouse brain. Brain Res. Mol. Brain Res. 77:19-28. [DOI] [PubMed] [Google Scholar]
- 21.Hong, J. S., and J. A. Engler. 1996. Domains required for assembly of adenovirus type 2 fiber trimers. J. Virol. 70:7071-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howitt, J., M. C. Bewley, V. Graziano, J. M. Flanagan, and P. Freimuth. 2003. Structural basis for variation in adenovirus affinity for the cellular coxsackievirus and adenovirus receptor. J. Biol. Chem. 278:26208-26215. [DOI] [PubMed] [Google Scholar]
- 23.Josefsson, M., T. Grunditz, T. Ohlsson, and E. Ekblad. 2002. Sodium/iodide-symporter: distribution in different mammals and role in entero-thyroid circulation of iodide. Acta Physiol. Scand. 175:129-137. [DOI] [PubMed] [Google Scholar]
- 24.Kirby, I., R. Lord, E. Davison, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 2001. Adenovirus type 9 fiber knob binds to the coxsackie B virus-adenovirus receptor (CAR) with lower affinity than fiber knobs of other CAR-binding adenovirus serotypes. J. Virol. 75:7210-7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles, M. R., K. W. Hohneker, Z. Zhou, J. C. Olsen, T. L. Noah, P. C. Hu, M. W. Leigh, J. F. Engelhardt, L. J. Edwards, K. R. Jones, et al. 1995. A controlled study of adenoviral-vector-mediated gene transfer in the nasal epithelium of patients with cystic fibrosis. N. Engl. J. Med. 333:823-831. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Li, D., L. Duan, P. Freimuth, and B. W. O'Malley, Jr. 1999. Variability of adenovirus receptor density influences gene transfer efficiency and therapeutic response in head and neck cancer. Clin. Cancer Res. 5:4175-4181. [PubMed] [Google Scholar]
- 28.Lortat-Jacob, H., E. Chouin, S. Cusack, and M. J. van Raaij. 2001. Kinetic analysis of adenovirus fiber binding to its receptor reveals an avidity mechanism for trimeric receptor-ligand interactions. J. Biol. Chem. 276:9009-9015. [DOI] [PubMed] [Google Scholar]
- 29.Mizuguchi, H., N. Koizumi, T. Hosono, A. Ishii-Watabe, E. Uchida, N. Utoguchi, Y. Watanabe, and T. Hayakawa. 2002. CAR- or αv integrin-binding ablated adenovirus vectors, but not fiber-modified vectors containing RGD peptide, do not change the systemic gene transfer properties in mice. Gene Ther. 9:769-776. [DOI] [PubMed] [Google Scholar]
- 30.Morrell, S. L., J. A. Fuchs, and J. L. Holtzman. 2000. Effect of methoxychlor administration to male rats on hepatic, microsomal iodothyronine 5′-deiodinase, form I. J. Pharmacol. Exp. Ther. 294:308-312. [PubMed] [Google Scholar]
- 31.Nakamura, T., K. Sato, and H. Hamada. 2003. Reduction of natural adenovirus tropism to the liver by both ablation of fiber-coxsackievirus and adenovirus receptor interaction and use of replaceable short fiber. J. Virol. 77:2512-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemerow, G. R. 2000. Cell receptors involved in adenovirus entry. Virology 274:1-4. [DOI] [PubMed] [Google Scholar]
- 33.Okegawa, T., Y. Li, R. C. Pong, J. M. Bergelson, J. Zhou, and J. T. Hsieh. 2000. The dual impact of coxsackie and adenovirus receptor expression on human prostate cancer gene therapy. Cancer Res. 60:5031-5036. [PubMed] [Google Scholar]
- 34.Ono, M., Y. Arano, T. Uehara, Y. Fujioka, K. Ogawa, S. Namba, T. Mukai, M. Nakayama, and H. Saji. 1999. Intracellular metabolic fate of radioactivity after injection of technetium-99m-labeled hydrazino nicotinamide derivatized proteins. Bioconjug. Chem. 10:386-394. [DOI] [PubMed] [Google Scholar]
- 35.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roelvink, P. W., G. Mi Lee, D. A. Einfeld, I. Kovesdi, and T. J. Wickham. 1999. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science 286:1568-1571. [DOI] [PubMed] [Google Scholar]
- 37.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sorimachi, K., A. Niwa, and Y. Yasumura. 1980. Phenolic ring deiodination in cultured rat hepatoma cells, and subcellular localization of deiodinases in cultured rat hepatoma, monkey hepatocarcinoma cells and normal rat liver homogenates. Biochim. Biophys. Acta 630:469-475. [DOI] [PubMed] [Google Scholar]
- 39.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomko, R. P., C. B. Johansson, M. Totrov, R. Abagyan, J. Frisen, and L. Philipson. 2000. Expression of the adenovirus receptor and its interaction with the fiber knob. Exp. Cell Res. 255:47-55. [DOI] [PubMed] [Google Scholar]
- 41.Vallabhajosula, S., M. Paidi, J. J. Badimon, N. A. Le, S. J. Goldsmith, V. Fuster, and H. N. Ginsberg. 1988. Radiotracers for low density lipoprotein biodistribution studies in vivo: technetium-99m low density lipoprotein versus radioiodinated low density lipoprotein preparations. J. Nucl. Med. 29:1237-1245. [PubMed] [Google Scholar]
- 42.van Raaij, M. J., N. Louis, J. Chroboczek, and S. Cusack. 1999. Structure of the human adenovirus serotype 2 fiber head domain at 1.5 Å resolution. Virology 262:333-343. [DOI] [PubMed] [Google Scholar]
- 43.van Raaij, M. J., E. Chouin, H. van der Zandt, J. M. Bergelson, and S. Cusack. 2000. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 Å resolution. Structure Fold Des. 8:1147-1155. [DOI] [PubMed] [Google Scholar]
- 44.van't Hof, W., and R. G. Crystal. 2002. Fatty acid modification of the coxsackievirus and adenovirus receptor. J. Virol. 76:6382-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walters, R. W., T. Grunst, J. M. Bergelson, R. W. Finberg, M. J. Welsh, and J. Zabner. 1999. Basolateral localization of fiber receptors limits adenovirus infection from the apical surface of airway epithelia. J. Biol. Chem. 274:10219-10226. [DOI] [PubMed] [Google Scholar]
- 46.Walters, R. W., P. Freimuth, T. O. Moninger, I. Ganske, J. Zabner, and M. J. Welsh. 2002. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell 110:789-799. [DOI] [PubMed] [Google Scholar]
- 47.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 48.Xia, D., L. J. Henry, R. D. Gerard, and J. Deisenhofer. 1994. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure 2:1259-1270. [DOI] [PubMed] [Google Scholar]
- 49.Zabner, J., B. G. Zeiher, E. Friedman, and M. J. Welsh. 1996. Adenovirus-mediated gene transfer to ciliated airway epithelia requires prolonged incubation time. J. Virol. 70:6994-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zabner, J., P. Freimuth, A. Puga, A. Fabrega, and M. J. Welsh. 1997. Lack of high affinity fiber receptor activity explains the resistance of ciliated airway epithelia to adenovirus infection. J. Clin. Investig. 100:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zinn, K. R., J. T. Douglas, C. A. Smyth, H. G. Liu, Q. Wu, V. N. Krasnykh, J. D. Mountz, D. T. Curiel, and J. M. Mountz. 1998. Imaging and tissue biodistribution of 99mTc-labeled adenovirus knob (serotype 5). Gene Ther. 5:798-808. [DOI] [PubMed] [Google Scholar]