Abstract
Purpose
Prognostic biomarkers are needed to optimize treatment decisions in prostate cancer. Single nucleotide polymorphisms (SNPs) participate in the individual genetic background modulating risk and clinical outcomes for cancer. In the present study, we tested the hypothesis of whether EGFR polymorphisms are associated with clinical outcomes in prostate cancer.
Materials and Methods
The study population consisted of 212 patients with clinically localized prostate cancer treated with radical prostatectomy from 1997 to 1999. The resected prostatic tissues were genotyped with allele-specific probes for nine haplotype-tagging SNPs. The SNPs were located in intronic, exonic, and flanking regions of linkage disequilibrium in the EGFR gene. Correlations between alleles, recurrence, and survival data were investigated using univariate and multivariate genetic analysis models.
Results
There was a statistically significant association between the SNP rs884419 and prostate cancer recurrence, defined in the study by at least a PSA biochemical recurrence (P < 0.001, log-rank test). The frequency of the recurrence risk-enhancing genotype A/A was 3.1%, compared to 17.4% (A/G) and 80% (G/G) for the risk-reducing genotypes. Based on Cox proportional hazard regression modeling, patients carrying G/G and A/G genotypes were associated with a reduced risk for developing prostate cancer recurrence with hazard ratios of 0.10 (95% CI, 0.02 to 0.41) and 0.13 (95% CI, 0.04 to 0.46), respectively, compared to the A/A genotype (P < 0.002).
Conclusions
These data suggest that a polymorphism flanking the EGFR gene is an independent prognostic genetic biomarker that predicts biochemical recurrence of prostate cancer after radical prostatectomy.
Keywords: EGFR, polymorphism, biomarker, prostate, cancer
INTRODUCTION
The ability to discriminate between aggressive and indolent disease in prostate cancer remains a critical public health issue, considering only 12% of prostate cancer patients will die of their disease.1 Optimal treatment decisions are guided by nomograms that incorporate clinical variables such as clinical stage, Gleason score, and prostate-specific antigen (PSA) levels to calculate the risk of disease recurrence after definitive treatments. With the advent of high-throughput genetic assays, there is rising interest in evaluating genomic biomarkers as variables to aid risk stratification in prostate cancer.
The most common genetic variants in the human genome are single-nucleotide polymorphisms (SNPs). Recent reports indicate associations between polymorphisms in particular genes and clinical outcomes of overall survival, disease-free survival or objective response rates in non-small-cell lung cancer, breast cancer, and leukemia. The majority of studies involving SNPs and prostate cancer have focused on associations between susceptibility loci and androgen biosynthesis and metabolism genes.2 There is rising interest in investigating the potential utility of SNPs as prognostic biomarkers for prostate cancer outcomes.
Epidermal growth factor receptor (EGFR) is a candidate biomarker in prostate cancer. Approximately 39–47% of prostate cancers express EGFR,3 and increased expression has been observed during progression to advanced, androgen-independent stages.4 EGFR expression in preclinical cancer models, including prostate tumors, appears to correlate directly with adverse phenotypes of proliferation, angiogenesis, and migration,5–7 and inversely with tumor radiocurability.8, 9 The mechanisms regulating EGFR gene expression are complex and known to be affected by polymorphic variation.10
EGFR spans a genomic area containing over 1,500 annotated SNPs. Several EGFR polymorphisms can contribute functional variability on gene transcription or in vitro response to drugs in cancer models.10, 11 Few studies have looked for associations between EGFR polymorphisms and clinical presentation or prognosis in prostate cancer.12,13 Therefore, we analyzed the association of EGFR SNPs with prognostic outcomes in clinically localized prostate cancer treated with radical prostatectomy as definitive treatment.
PATIENTS AND METHODS
Study Population and Clinical Data
The study subjects were 212 predominantly Caucasian prostate cancer patients who underwent radical prostatectomy as primary treatment at Vanderbilt University Hospital between 1997 and 1999. The institutional review board at Vanderbilt University School of Medicine approved the study. All patients had adenocarcinoma confirmed histologically. Clinical data from patient follow-up at Vanderbilt Hospital were retrospectively collected using electronic medical records. The mean follow-up for overall survival and assessment of prostate cancer progression were 8.3 ± 2.4 y and 4.4 ± 3.9 y, respectively. The endpoints analyzed were freedom from recurrence (FFR) and overall survival (OS). Recurrence following prostatectomy was classified as biochemical, local, or distant, and the most advanced recurrence type documented was assigned to each patient. Biochemical recurrence was defined as a prostate-specific antigen (PSA) detection of > 0.1 ng/ml in at least two consecutive tests. Salvage treatment modality following recurrence varied among patients and included observation (32%), radiation therapy (21%), hormonal therapy (16%), combined hormonal/radiation therapy (14%), combined hormonal/radiation/chemotherapy (7%), combined hormonal/chemotherapy (4.5%), and unknown salvage treatment.
Genotyping of EGFR Polymorphisms in Prostate Cancer Samples
Processing of prostatectomy specimens and genomic DNA extraction from deparaffinized specimens were performed as described previously.14 Purified genomic DNA was genotyped for the following haplotype-tagging SNPs in the EGFR gene: rs3735064, rs7780270, rs11543848, rs11976696, rs9642391, rs845560, rs845562, rs7808697, and rs884419. SNP selection was influenced by their possible association with increased cancer risk, based on differential allele representation in a large-scale, population-based, case-control study involving breast cancer patients and healthy controls.15 Allelic discrimination of these EGFR polymorphisms was performed using Taqman® SNP genotyping assays (Applied Biosystems, assay IDs: C_335819_10 [rs3735064], C_2678606_10 [rs7780270], C_16170352_20 [rs11543848], C_321872_10 [rs11976696], C_2678667_10 [rs9642391], C_7610424_10 [rs845560], C_7610434_10 [rs845562], and C_8304143_10 [rs884419]), and the following reagents for rs7808697: Forward primer, 5’-CTC CAT CCA TGT TCT TGC AAA GTA C-3’; Reverse primer, 5’-GAC AGA CTG GAT AAA GAA AAT TGT GGT ACA-3’; and the allele-specific probes 5’-VIC-CTT TTG TGG CTA CCT AGT G-3’ and 5’-FAM- TTG TGG CTG CCT AGT G-3’. The final volume for each reaction was 5 µl, consisting of 2.5 µl TaqMan Universal PCR Master Mix (Applied Biosystems), 900 nM of each primer, 200 nM of each TaqMan probe, and 10 ng genomic DNA. The PCR profile consisted of an initial denaturation step at 95°C for 10 min and 50 cycles with 95°C for 15 sec and 60°C for 1 min. The fluorescence level was measured with the ABI PRISM 7900HT sequence detector (ABI). Genotypes were determined by ABI SDS software. Quality control samples were included in genotyping assays. Concordance for blinded samples was 100% for most SNPs, with the exception of rs11543848 (75%) and rs9642391 (67%).
Statistical Analysis
The primary analysis focused on detecting associations between nine htSNPs in the EGFR gene and FFR/OS. In the univariate analysis, SNPs associated with survival outcomes were selected based on a log-rank or exact log-rank test examining one SNP at a time. In order to control the inflation of false positive rate for the multiple comparisons, we used false discovery rate (FDR) < 0.116 as cutoff points to detect significant associations for any of the nine htSNPs. For the multivariable analysis, the Cox proportional hazards model was used to determine whether the SNP associated with survival could serve as an independent predictor, adjusting for important prognostic factors such as surgical margin, Gleason grade and pre-prostatectomy PSA. OS was calculated from the day of surgery to the day of death or last follow-up. We calculated FFR from the day of surgery to the day of recurrence or last follow-up. Data were censored for live (or recurrence-free) patients as of their last follow-up visits. Kaplan-Meier survival curves were calculated for the subgroups of potential risk factors and were compared using the log rank test. Descriptive statistics, including mean and standard deviation for continuous variables, as well as percentages and frequencies for categorical variables were calculated (Table 1). All P values are based on two-sided tests and differences were considered statistically significant when p-value < 0.05, except in the selection of SNPs associated with survival outcome. Analyses were performed using SAS system version 9.1 and R version 2.1.1.
Table 1.
Patient Demographics
| Characteristic | Number (N=212) |
|
|---|---|---|
| Race, n(%) | ||
| White | 208 (98.11) | |
| Black | 4 (1.89) | |
| Age at diagnosis, n(%), years | ||
| Mean (SD) | 61.2 (7.08) | |
| <60 | 86 (40.57) | |
| 60–70 | 104 (49.06) | |
| >70 | 22 (10.38) | |
| Pre-prostatectomy PSA, n(%), ng/ml | ||
| Mean (SD) | 8.4 (6.21) | |
| <4 | 22 (10.38) | |
| 4–10 | 113 (53.30) | |
| >10 | 43 (20.28) | |
| Missing | 34 (16.04) | |
| Gleason score, n(%) | ||
| 2–6 | 130 (61.32) | |
| 7 | 70 (33.02) | |
| 8–10 | 12 (5.66) | |
| Surgical margin, n(%) | ||
| Negative | 133 (62.74) | |
| Positive | 78 (36.79) | |
| Missing | 1 (0.47) | |
| Extracapsular extension, n(%) | ||
| Negative | 125 (58.96) | |
| Positive | 67 (31.60) | |
| Missing | 20 (9.43) | |
| Disease classification 1, n(%) | ||
| Clinically inapparent (T1) | 8 (3.77) | |
| Confined within prostate (T2) | 134 (63.21) | |
| Regional (T3) | 57 (26.89) | |
| Missing | 13 (6.13) | |
| Recurrence, n(%) | ||
| No | 168 (79.25) | |
| Yes | 44 (20.75) | |
| Overall survival, n(%) | ||
| Alive | 175 (82.55) | |
| Dead | 37 (17.45) | |
Abbreviations: PSA, prostate-specific antigen
Based on the American Joint Committee on Cancer classification
Bioinformatics Tools
Web-based databases used in the present study include NCBI Database of Single Nucleotide Polymorphisms (dbSNP) [Build 128, http://www.ncbi.nlm.nih.gov/SNP] and the International HapMap Project,17 for its allele frequency data in the CEU population (i.e., Utah residents with ancestry from Northern and Western Europe) and for its genome browser and Haploview software18 to measure the extent of linkage disequilibrium (LD) between SNPs and generate an LOD score plot [http://www.hapmap.org]. The HapMap datasource used was HapMap Data Rel 23a/Phase II Mar 08, on NCBI B36 Assembly, dbSNPb126.
The software MatIspector19 was used to identify potential transcription factor binding sites in the 3’ flanking region of EGFR containing the rs884419 SNP [http://www.genomatix.de/cgi-bin/matinspector_prof]. Binding site sequences are presented with capital letters denoting the core sequence (i.e., the highest conserved, consecutive positions of the transcription factor matrix).
RESULTS
Patient Characteristics
Demographic, clinicopathologic, and outcomes information is summarized in Table 1. The mean age of study subjects at the time of prostate cancer diagnosis and definitive treatment with prostatectomy was 61.2 ± 7 y. After a median follow-up time of 9.1 y, 83% of the patients were alive. The median follow-up time for prostate cancer progression was 3.4 y. Recurrence rate was 21%, with the majority classified as biochemical and a lower incidence for local or distant recurrence (80%, 5% and 16% of recurrences, respectively). Based on the risk stratification scheme suggested by the National Comprehensive Cancer Network® guidelines,20 which incorporates Gleason grade, PSA levels and staging parameters to predict probability of biochemical failure after definitive local therapy, the majority of our patient population (64%) could be classified as having an intermediate risk for biochemical failure.
Impact of Genotype for EGFR SNPs on Recurrence and Survival Outcomes in Prostate Cancer
To determine whether polymorphic variants in EGFR are associated with prognostic outcomes in clinically localized prostate cancer treated with surgery, we genotyped genomic DNA from prostatectomy specimens for nine SNPs within the EGFR gene (Table 2). For the majority of SNPs, allelic discrimination assays were informative for at least 86% of genomic DNA samples. The genotype frequency distribution for the SNPs in our study population closely matched the one previously reported in the HapMap database for the CEU population,17 adding confidence to the accuracy of our genotyping reaction.
Table 2.
EGFR SNP Frequencies Among Prostate Cancer Patients
| dbSNP ID 1 | Chromosomal Position 2 | Location (relative to EGFR 3) | Alleles 1 | Aminoacid Change |
Population Diversity | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype Frequency in Study Population |
HapMap-CEU Gen Frequency |
|||||||||
| Patient Number 4 |
Genotypes | Genotypes | ||||||||
| rs3735064 | chr7:55,112,327 | intron 1 | C>T | N/A5 | 137 | C/C 0.628 |
C/T 0.270 |
T/T 0.102 |
C/C 0.596 |
C/T 0.246 |
| rs7780270 | chr7:55,119,380 | intron 1 | G>T | N/A5 | 42 | G/G 0.452 |
G/T 0.405 |
T/T 0.143 |
G/G 0.340 |
G/T 0.468 |
| rs11543848 | chr7:55,196,749 | exon 13 (nonsynonymous) | G>A | K521R | 162 | A/A 0.062 |
A/G 0.259 |
G/G 0.679 |
A/A 0.064 |
A/G 0.404 |
| rs11976696 | chr7:55,199,827 | intron 14 | A>G | N/A5 | 198 | A/A 0.652 |
A/G 0.298 |
G/G 0.051 |
A/A 0.700 |
A/G 0.283 |
| rs9642391 | chr7:55,212,858 | intron 19 | G>C | N/A5 | 194 | G/G 0.593 |
C/G 0.304 |
C/C 0.103 |
G/G 0.491 |
C/G 0.404 |
| rs845560 | chr7:55,218,288 | intron 20 | C>T | N/A5 | 158 | C/C 0.557 |
C/T 0.317 |
T/T 0.127 |
C/C 0.567 |
C/T 0.350 |
| rs845562 | chr7:55,222,299 | intron 20 | G>A | N/A5 | 183 | A/A 0.038 |
A/G 0.240 |
G/G 0.721 |
A/A 0.033 |
A/G 0.233 |
| rs7808697 | chr7:55,231,056 | intron 22 | G>A | N/A5 | 184 | A/A 0.027 |
A/G 0.212 |
G/G 0.761 |
A/A 0.038 |
A/G 0.226 |
| rs884419 | chr7:55,243,774 | flanking (1.249 Kbp downstream) | G>A | N/A5 | 196 | A/A 0.031 |
A/G 0.174 |
G/G 0.796 |
A/A 0.017 |
A/G 0.183 |
Abbreviations: EGFR, Epidermal growth factor receptor; dbSNP, database single nucleotide polymorphism; chr, chromosome; HapMap, International HapMap project; CEU, population consisting of Utah residents with ancestry from Northern and Western Europe
Data obtained from NCBI Database of Single Nucleotide Polymorphisms (dbSNP) Build ID:128, http://www.ncbi.nlm.nih.gov/SNP/, accessed April 2008
Based on alignment with the NCBI Build 36.1 human reference sequence using the BLAT software, http://genome.ucsc.edu/, accessed April 2008
Relative to annotated genomic sequence for human EGFR isoform a, chr7:55,054,219–55,242,525
Indicates number of patients (from n=212) for which the genotyping reaction yielded informative results
A log-rank univariate analysis was performed to identify significant associations between probability for FFR or OS and patient-specific factors, including the genotypes for EGFR SNPs rs3735064, rs7780270, rs11543848, rs11976696, rs9642391, rs845560, rs845562, rs884419 and rs7808697. Results are shown on Table 3. Known prognostic factors including pre-prostatectomy PSA levels, Gleason score, surgical margin, and AJCC tumor category21 were associated with statistically significant differences in probability for FFR. A statistically significant association was also found for EGFR rs884419 genotype and probability of prostate cancer recurrence within 3y, with the G allele having an apparent protective effect. No statistically significant associations were found for the other SNPs analyzed. In the present study, only Gleason score was significantly associated with differential probability of OS. Salvage treatment following recurrence was not strongly associated with OS (data not shown).
Table 3.
Effect of Patient Factors on Post-prostatectomy Outcomes, Univariate Analysis
| Factor | Number of patients1 (N=212) |
Probability of 3-y recurrence-free surviva |
Probability of 10-y survival |
|||
|---|---|---|---|---|---|---|
| %(95% CI) | P2 | %(95% CI) | P2 | |||
| Race, n(%) | 0.7785 | 0.2738 | ||||
| White | 208 | 88 (83–92) | 76 (68–85) | |||
| Black | 4 | N/A3 | N/A3 | |||
| Age at diagnosis (years) | 0.8123 | 0.3466 | ||||
| <60 | 86 | 89 (83–96) | N/A3 | |||
| 60–70 | 104 | 85 (78–92) | 77 (63–90) | |||
| >70 | 22 | 81 (65–98) | 36 (0–86) | |||
| Pre-prostatectomy PSA (ng/ml) | 0.0430 | 0.5031 | ||||
| <4 | 22 | N/A3 | 0 (0-0) | |||
| 4–10 | 113 | 88 (82–94) | 77 (64–90) | |||
| >10 | 43 | 84 (72–95) | N/A3 | |||
| Gleason score | <0.001 | 0.0050 | ||||
| 2–6 | 130 | 94 (89–98) | 78 (66–90) | |||
| 7 | 70 | 77 (67–87) | 67 (42–93) | |||
| 8–10 | 12 | 67 (40–93) | N/A3 | |||
| Surgical margin | <0.001 | 0.3790 | ||||
| Negative | 133 | 95 (91–98) | 73 (62–85) | |||
| Positive | 78 | 74 (64–84) | N/A3 | |||
| Extracapsular extension | <0.001 | 0.4393 | ||||
| Negative | 125 | 93 (88–97) | N/A3 | |||
| Positive | 67 | 76 (65–86) | 72 (55–88) | |||
| Disease classification, n(%) | <0.001 | 0.0645 | ||||
| Clinically inapparent (T1) | 8 | 71 (38–100) | N/A3 | |||
| Confined within prostate (T2) | 134 | 94 (90–98) | N/A3 | |||
| Regional (T3) | 57 | 71 (59–83) | 64 (42–86) | |||
| EGFR rs884419 Genotype | 0.0096 | 0.9665 | ||||
| G/G | 156 | 89 (85–94) | 74 (64–84) | |||
| A/G | 34 | 88 (76–99) | 0 (0-0) | |||
| A/A | 6 | N/A3 | N/A3 | |||
| EGFR rs7808697 Genotype | 0.2161 | 0.2451 | ||||
| G/G | 140 | 87 (81–92) | N/A3 | |||
| A/G | 39 | 87 (76–98) | 71 (51–91) | |||
| A/A | 5 | N/A3 | N/A3 | |||
| EGFR rs845562 Genotype | 0.7954 | 0.4686 | ||||
| G/G | 132 | 89 (83–94) | N/A3 | |||
| A/G | 44 | 79 (67–91) | 69 (48–89) | |||
| A/A | 7 | N/A3 | N/A3 | |||
| EGFR rs845560 Genotype | 0.2611 | 0.0148 | ||||
| C/C | 88 | 84 (76–92) | 78 (65–91) | |||
| C/T | 50 | 94 (87–100) | N/A3 | |||
| T/T | 20 | 89 (74–100) | N/A3 | |||
| EGFR rs9642391 Genotype | 0.7870 | 0.4411 | ||||
| C/C | 20 | N/A3 | N/A3 | |||
| C/G | 59 | 82 (72–92) | 72 (58–87) | |||
| G/G | 115 | 89 (84–95) | N/A3 | |||
| EGFR rs11976696 Genotype | 0.1277 | 0.1195 | ||||
| G/G | 10 | 60 (30–90) | N/A3 | |||
| A/G | 59 | 79 (69–90) | 54 (10–98) | |||
| A/A | 129 | 90 (85–96) | 67 (51–84) | |||
| EGFR rs11543848 Genotype | 0.5300 | 0.2001 | ||||
| G/G | 110 | 88 (82–94) | 69 (43–94) | |||
| A/G | 42 | 85 (74–96) | 64 (44–84) | |||
| A/A | 10 | N/A3 | N/A3 | |||
| EGFR rs7780270 Genotype | 0.4841 | 0.9378 | ||||
| G/G | 19 | 82 (64–100) | N/A3 | |||
| G/T | 17 | N/A3 | N/A3 | |||
| T/T | 6 | N/A3 | 0 (0-0) | |||
| EGFR rs3735064 Genotype | 0.8846 | 0.7085 | ||||
| C/C | 86 | 88 (81–95) | 70 (56–84) | |||
| C/T | 37 | 84 (72–96) | N/A3 | |||
| T/T | 14 | N/A3 | 0 (0-0) | |||
Analyses were limited to patients for whom data were available
Raw P values for each factor were calculated by the log-rank test or from exact log-rank test (Race and EGFR rs884419)
Unable to calculate because all recurrence or death events occurred before 3-y or 10-y, respective
EGFR SNP rs88419 A/A Genotype is Risk-Enhancing for Recurrence in Prostate Cancer Patients Treated with Prostatectomy
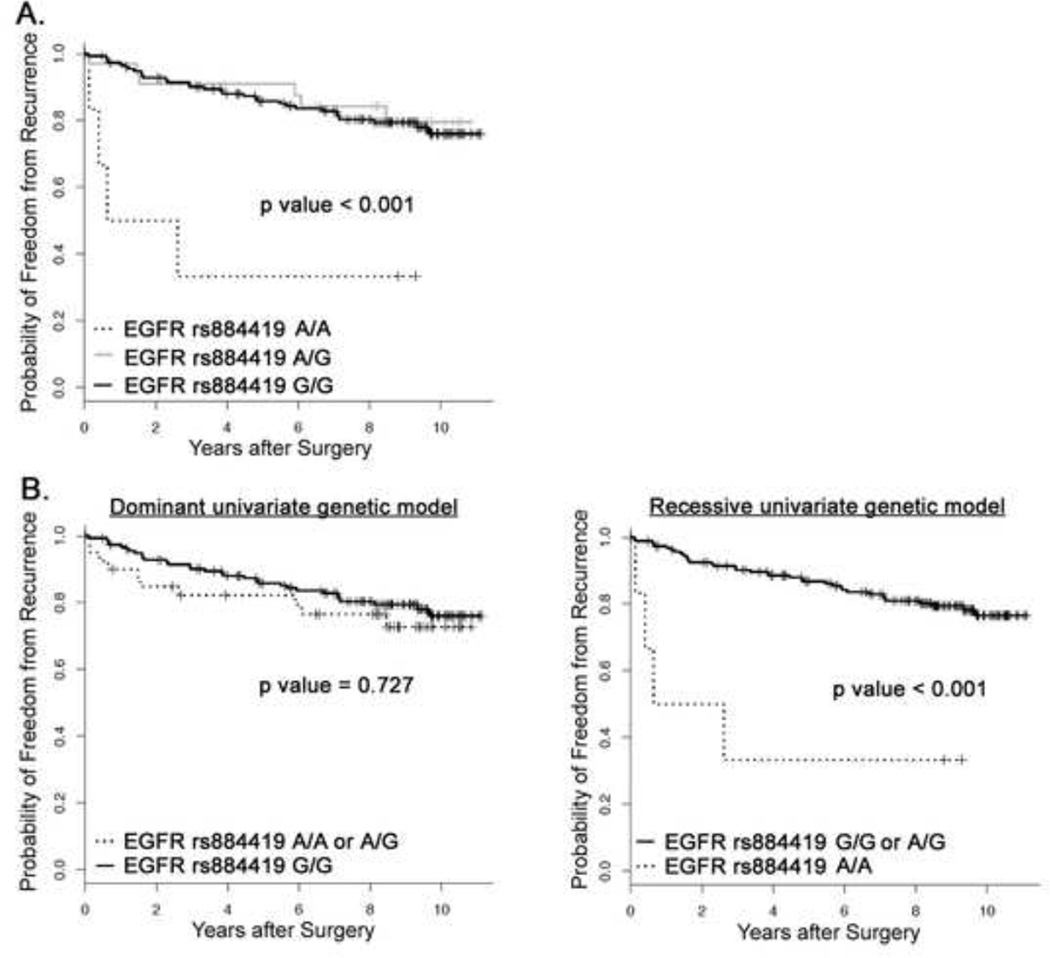
The Kaplan-Meier method was used to estimate the probability distribution for FFR as a function of rs884419 genotype, and the log-rank test was used to determine the significance of survival differences across the different genotypes. The rs884419 A/A genotype enhanced the risk for prostate cancer recurrence within 3 y following prostatectomy (Figure 1A). Dominant and recessive Kaplan-Meier models were generated to investigate the genetic effect pattern of the A allele. In the dominant model, we hypothesized that the genotypes A/A or A/G contributed equally to the risk of recurrence; in the recessive model, we hypothesized that only the A/A genotype contributed to the risk. As presented in Figure 1B, the rs88419 A allele displayed a recessive effect on reducing FFR in our prostate cancer patient population.
Fig 1.
Kaplan-Meier estimates of freedom from recurrence (FFR) following radical prostatectomy as treatment for localized prostate cancer. (A) FFR curves were plotted for the individual EGFR rs884419 genotypes. (B) FFR curves were plotted to test whether allele A of EGFR rs884419 displayed a dominant or recessive genetic effect.
EGFR rs884419 SNP Genotype Predicts FFR Independently of Tumor Grade, PSA Levels, and Surgical Margin Status
To test whether the increased risk of recurrence in patients with rs884419 A/A genotype was linked to an unfavorable risk profile conferred by other clinical risk factors, we performed multivariate analysis using the Cox proportional hazards regression model. The rs884419 genotype was an independent predictor for recurrence (Table 4). This association retained significance when other factors that predicted for FFR in the univariate analysis were included in the model (i.e., Gleason grade, pre-prostatectomy PSA, and surgical margin). Patients carrying G/G and A/G genotypes were associated with a reduced risk for developing prostate cancer recurrence with hazard ratios of 0.10 (95% CI, 0.02 to 0.41) and 0.13 (95% CI, 0.04 to 0.46), respectively, compared to A/A genotype (P < 0.002).
Table 4.
Independent Factors Predictive of Recurrence in Prostate Cancer
| Factor | Recurrence | ||
|---|---|---|---|
| Hazard ratio (95%, CI) | P value1 | ||
| EGFR rs884419 genotype | |||
| A/A | 1 | ||
| A/G | 0.094 (0.021–0.413) | 0.0018 | |
| G/G | 0.130 (0.037–0.463) | 0.0016 | |
| Surgical Margin | |||
| Negative | 1 | ||
| Positive | 2.51 (1.22–5.17) | 0.0126 | |
| Gleason Grade2 | 1.87 (1.27–2.77) | 0.0017 | |
| Pre-prostatectomy PSA2 | 1.04 (0.996–1.08) | 0.0769 | |
P values were derived from the Cox proportional hazards regression model, with simultaneous inclusion of all factors shown
For continuous variables, the hazard ratio is applied per increments of one unit of measurement
Potential Influence of rs884419 Genotype on EGFR Genetic Regulation
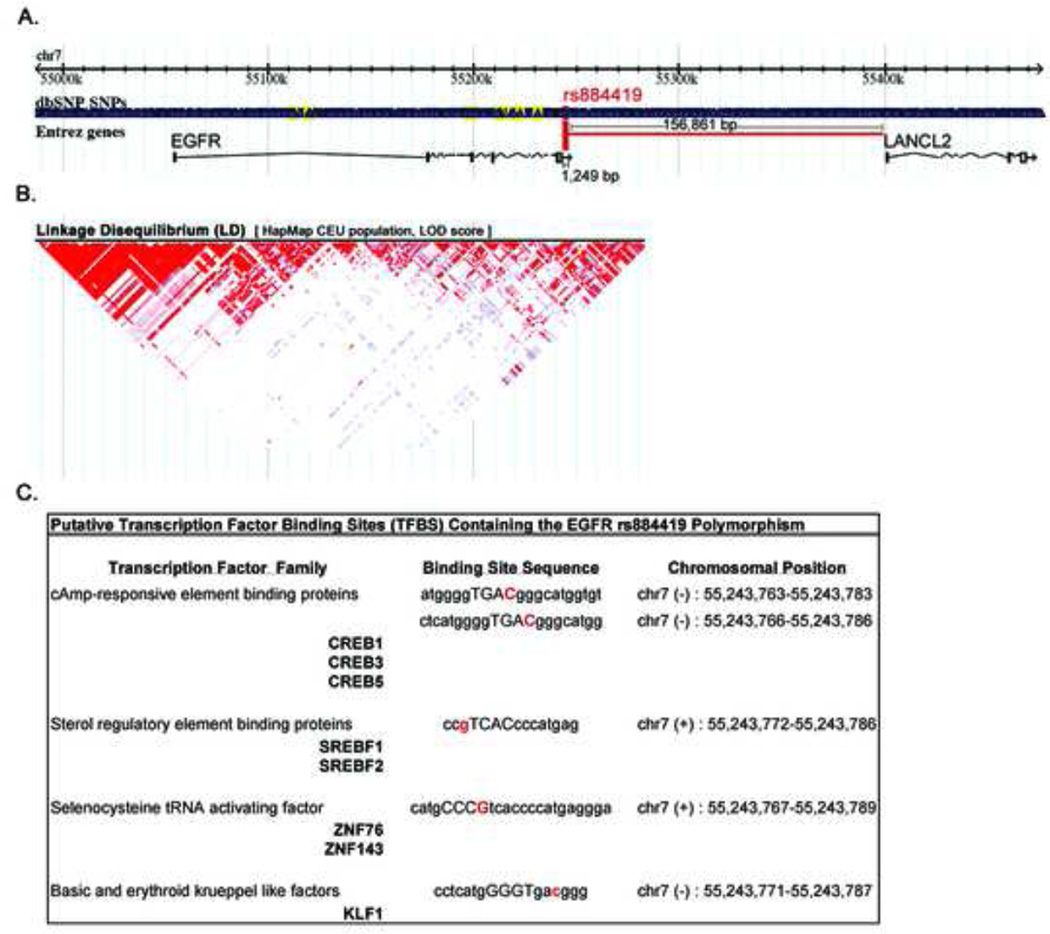
The phenotypic function of rs884419 is currently unknown. As indicated in Figure 2A, its genomic location is nearest to EGFR (1.2 Kbp distance) and significantly upstream from LANCL2 (157 Kbp). Because the association between SNP genotype and recurrence outcomes was only found for rs884419, we hypothesized that this SNP is not in high linkage disequilibrium (LD) with the neighboring EGFR SNPs analyzed in the present study. As indicated by the LD plot in Figure 2B, the LD block encompassing the studied SNPs does not display a tight, high LD-scoring pattern, and supports an independent haplotype-tagging function for rs884419.
Fig 2.
rs884419 as a potential cis-acting EGFR regulator. (A) Genomic context for SNPs analyzed; red, rs884419; yellow, all others. (B) LD plot for SNPs in/near EGFR gene. Color intensity is proportional to LD strength between SNP pairs. (C) MatInspector-predicted TFBS in the EGFR 3’ flanking region containing the rs884419 polymorphic variant in their binding sequence.
A possible functional role for rs884419 is as a cis-acting genetic regulator of EGFR expression, based on its location at the 3’ flanking region of this gene. We therefore investigated whether the rs884419 polymorphism resides in a consensus sequence recognized by known transcription factors, using an in silico prediction method. As shown in Figure 2C, the MatInspector software tool19 identified a series of putative transcription factor binding sites (TFBSs) mapping to the genomic location of rs884419 (location for the SNP allele is in red font in the binding site sequence).
DISCUSSION
We conducted a retrospective molecular epidemiology study to identify novel markers for risk stratification in prostate cancer. Our data indicates that the EGFR SNP rs884419 is a genetic marker for prostate cancer recurrence risk within 3 y following radical prostatectomy, and this variant genotype remained an independent predictive factor after adjusting for other covariates (Figure 1, Table 3–4). To our knowledge, our study is the first to show a prognostic role for a polymorphism located in the 3’ flanking region of EGFR on the outcome of PSA recurrence after radical prostatectomy.
Although the mechanism underlying any functional effect for rs884419 is currently unknown, the location of this SNP in putative TFBSs for CREB, SREBF, ZNF, and KLF1 (Figure 2) suggests that there may be functional differences between rs884419 alleles in regulating gene expression. The 3’ regulatory region of human genes is rich in regulatory elements that can modulate gene expression over long distances.22, 23 SNPs in the 3’ flanking region of genes such as SERPINA1, HBD, and SLC9A3R1 have been shown to affect the binding of transcription factors regulating their expression.23 Thus, it is possible that the EGFR SNP rs884419 may modulate expression of the EGFR gene. We are currently in the process of determining whether the SNP is functional and results in variation in EGFR transcription and expression.
Current nomogram-based tools use clinical and pathologic parameters to predict either pre- or postoperatively the probability of prostate cancer recurrence after radical prostatectomy. Incorporating biomarker variables into the existing predictive nomograms may further enhance the accurate discrimination of patients with high risk of prostate cancer relapse. In addition, the use of genomic markers such as SNPs avoids the problem of heterogeneity of prostate cancer within the same specimen that contributes to sampling error and subsequently limits pathologic parameters accuracy. The list of biomarkers with possible prognostic roles in for prostate cancer continues growing. In particular, there is increasing support for SNPs having a role as genomic biomarkers for outcomes in prostate cancer. Associations have been reported between polymorphisms in XRCC1 and recurrence risk following radical prostatectomy,25 in IL-6 and prostate cancer progression to bone metastases,26 in Osteoprotegerin and risk of metastatic prostate cancer disease,27 in SOD2, XRCC1, and XRCC3 and development of late injury following radiation treatment,28 in TGFβ1 and adverse quality of life following radiotherapy,29 and in EGF and earlier relapse in patients treated with androgen suppression therapy.28 How these SNPs interact with other genetic factors and clinicopathologic variables to affect outcomes in prostate cancer is currently unknown.
The association found in the present study was between genetic marker rs884419 and biochemical (PSA) progression in prostate cancer. This endpoint does not always translate into clinically significant recurrence following curative prostatectomy. Accordingly, we did not observe a statistically significant correlation between the rs884419 genotype and 10 yr survival (Table 3). Of note, an expected association was found between Gleason score and OS, since this is a parameter known to be associated with prostate cancer-specific mortality following biochemical recurrence.30 Biochemical recurrence is a valid outcome for assessing the efficacy of radical prostatectomy as a treatment choice in our prostate cancer patient population. Therefore, this genetic marker could be informative for treatment selection in early stage disease and for the identification of high-risk patients that may benefit from adjuvant treatment modalities and/or intense follow-up.
This study has several limitations. First, the prognostic genetic marker identified in the study population was not confirmed in an independent validation set. Also, the population size number may have limited the power to detect existing associations between the other SNPs studied and prostate cancer outcomes. Finally, the study population was primarily Caucasian (98%) and therefore the results may have limited application across different races. However, given the limited sample size, having one ethnic group possibly helped avoid extreme bias due to population stratification. Of note, the potential for sampling error that could affect the pathologic or staging values such as the Gleason score was minimized by the use of prostatectomy specimens.24 Despite these limitations, the present study highlights a molecular marker that may enhance the currently available risk stratification schemes for prostatectomy-treated clinically localized prostate cancer.
Acknowledgments
We thank Judith N. Roberts and Ping Cao for their assistance with collection of clinical data and sample processing. This work was supported by CA093240-06, R01 CA90899, P30 CA68485, and DOD W23RYX-3305-N603.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Ettinger DS, Bepler G, Bueno R, et al. Non-small cell lung cancer clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2006;4:548. doi: 10.6004/jnccn.2006.0046. [DOI] [PubMed] [Google Scholar]
- 3.Rocha-Lima CM, Soares HP, Raez LE, et al. EGFR targeting of solid tumors. Cancer Control. 2007;14:295. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- 4.Di Lorenzo G, Tortora G, D'Armiento FP, et al. Expression of epidermal growth factor receptor correlates with disease relapse and progression to androgen-independence in human prostate cancer. Clin Cancer Res. 2002;8:3438. [PubMed] [Google Scholar]
- 5.Cross M, Dexter TM. Growth factors in development, transformation, and tumorigenesis. Cell. 1991;64:271. doi: 10.1016/0092-8674(91)90638-f. [DOI] [PubMed] [Google Scholar]
- 6.Olayioye MA, Neve RM, Lane HA, et al. The ErbB signaling network: receptor heterodimerization in development and cancer. Embo J. 2000;19:3159. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prenzel N, Fischer OM, Streit S, et al. The epidermal growth factor receptor family as a central element for cellular signal transduction and diversification. Endocr Relat Cancer. 2001;8:11. doi: 10.1677/erc.0.0080011. [DOI] [PubMed] [Google Scholar]
- 8.Milas L, Fan Z, Andratschke NH, et al. Epidermal growth factor receptor and tumor response to radiation: in vivo preclinical studies. Int J Radiat Oncol Biol Phys. 2004;58:966. doi: 10.1016/j.ijrobp.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 9.Nasu S, Ang KK, Fan Z, et al. C225 antiepidermal growth factor receptor antibody enhances tumor radiocurability. Int J Radiat Oncol Biol Phys. 2001;51:474. doi: 10.1016/s0360-3016(01)01671-6. [DOI] [PubMed] [Google Scholar]
- 10.Araujo A, Ribeiro R, Azevedo I, et al. Genetic polymorphisms of the epidermal growth factor and related receptor in non-small cell lung cancer--a review of the literature. Oncologist. 2007;12:201. doi: 10.1634/theoncologist.12-2-201. [DOI] [PubMed] [Google Scholar]
- 11.Qian W, Liu J, Jin J, et al. Arsenic trioxide induces not only apoptosis but also autophagic cell death in leukemia cell lines via up-regulation of Beclin-1. Leuk Res. 2007;31:329. doi: 10.1016/j.leukres.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 12.Wolff H, Saukkonen K, Anttila S, et al. Expression of cyclooxygenase-2 in human lung carcinoma. Cancer Res. 1998;58:4997. [PubMed] [Google Scholar]
- 13.Kumar VL, Majumder PK, Kumar V. Observations on EGFR gene amplification and polymorphism in prostatic diseases. Int Urol Nephrol. 2000;32:73. doi: 10.1023/a:1007160202229. [DOI] [PubMed] [Google Scholar]
- 14.Browning REt, Li H, Shinohara ET, et al. ATM polymorphism IVS62+60G>A is not associated with disease aggressiveness in prostate cancer. Urology. 2006;67:1320. doi: 10.1016/j.urology.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Hallahan D, Geng L, Qu S, et al. Integrin-mediated targeting of drug delivery to irradiated tumor blood vessels. Cancer Cell. 2003;3:63. doi: 10.1016/s1535-6108(02)00238-6. [DOI] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B. 1995:289. [Google Scholar]
- 17.The International HapMap Project. Nature. 2003;426:789. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 18.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 19.Cartharius K, Frech K, Grote K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 20.NCCN Practice Guidelines in Oncology-v.1.2008. 2008 In: http://www.nccn.org/professionals/physician_gls/PDF/prostate.pdf. [Google Scholar]
- 21.AJCC Cancer Staging Manual. 6th ed. New York: Springer; 2002. pp. 309–316. [Google Scholar]
- 22.West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14(Spec No 1):R101. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 23.Batchelor TT, Sorensen AG, di Tomaso E, et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11:83. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muntener M, Epstein JI, Hernandez DJ, et al. Prognostic significance of Gleason score discrepancies between needle biopsy and radical prostatectomy. Eur Urol. 2008;53:767. doi: 10.1016/j.eururo.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Sarkar S, Davies JE, Huang Z, et al. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 26.Kesarwani P, Ahirwar DK, Mandhani A, et al. Association between-174 G/C promoter polymorphism of the interleukin-6 gene and progression of prostate cancer in North Indian population. DNA Cell Biol. 2008;27:505. doi: 10.1089/dna.2008.0742. [DOI] [PubMed] [Google Scholar]
- 27.Narita N, Yuasa T, Tsuchiya N, et al. A genetic polymorphism of the osteoprotegerin gene is associated with an increased risk of advanced prostate cancer. BMC Cancer. 2008;8:224. doi: 10.1186/1471-2407-8-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burri RJ, Stock RG, Cesaretti JA, et al. Association of single nucleotide polymorphisms in SOD2, XRCC1 and XRCC3 with susceptibility for the development of adverse effects resulting from radiotherapy for prostate cancer. Radiat Res. 2008;170:49. doi: 10.1667/RR1219.1. [DOI] [PubMed] [Google Scholar]
- 29.Peters CA, Stock RG, Cesaretti JA, et al. TGFB1 single nucleotide polymorphisms are associated with adverse quality of life in prostate cancer patients treated with radiotherapy. Int J Radiat Oncol Biol Phys. 2008;70:752. doi: 10.1016/j.ijrobp.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Freedland SJ, Humphreys EB, Mangold LA, et al. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. Jama. 2005;294:433. doi: 10.1001/jama.294.4.433. [DOI] [PubMed] [Google Scholar]