Abstract
Genetic circuits perform computational operations based on interactions between freely diffusing molecules within a cell. When transcription factors are combined to build a circuit, unintended interactions can disrupt its function. Here, we apply “part mining” to build a library of 73 TetR-family repressors gleaned from prokaryotic genomes. The operators of a subset were determined using an in vitro method and this information was used to build synthetic promoters. The promoters and repressors were screened for cross-reactions. Of these, 16 were identified that both strongly repress their cognate promoter (5- to 207-fold) and do not interact with other promoters. Each repressor:promoter pair was converted to a NOT gate and characterized. Used as a set of 16 NOR gates, there are >1054 circuits that could be built by changing the pattern of input and output promoters. This represents a large set of compatible gates that can be used to construct user-defined circuits.
Introduction
Living cells can be programmed by incorporating integrated genetic gates into their DNA1. These gates rely on biochemical interactions to perform computational operations, including switches, logic, and memory2,3. Gates can be connected to each other when they are designed to be extensible, meaning that the form of their input and output signals are the same. For example, if both the inputs and outputs are promoters, then this signal is defined as the flux of RNA polymerase (RNAP) on DNA4. To date, the complexity of circuits has been low, consisting of the few available gates based on the same transcription factors re-used across labs and projects5. Increasing the number of available gates will enable the construction of larger circuits to encode more sophisticated algorithms6. The challenge has been that all of the gates within a circuit need to be orthogonal; in other words, the biochemical interactions on which they are based cannot cross-react7. It becomes increasingly difficult to add gates because the number of potential cross-reactions grows quickly as N2-N.
NOT and NOR gates are simple and broadly useful functions. A transcriptional NOT gate can be implemented by using an input promoter to drive expression of a repressor, which turns off expression of an output promoter (Figure 1a)8. Even these simple gates can perform signal-processing functions, for example to convert a dark sensor into a light sensor9 and a male sensor into a female sensor10. A NOR gate is a logic function where the output is ON only when both inputs are OFF. NOR gates are Boolean complete, meaning that they can be combined to generate any computational operation. A genetic NOR gate can be built by adding a second input promoter in series to the NOT gate, so that both input promoters drive the expression of the repressor11. Two gates are orthogonal if their repressors do not bind to each other's promoters. Obtaining more gates that can be used as part of the same circuit requires having a set of repressors that bind to different operator sequences.
Figure 1. A large repressor library is compiled using genome mining.
(a) A genetic NOT gate (symbol shown) can be built using a repressor (pink arrow) that binds to an operator (pink box) in an output promoter. (b) The pipeline for the discovery and characterization of orthogonal repressors is shown. The second panel depicts a portion of the CSI microarray used to determine the operator sequence. (c) The complete library of 73 synthesized repressors (plus TetR) are organized into a phylogenetic tree diagram, where carets indicate those repressors that appear in the final orthogonality matrix illustrated in Figure 3d. The tree was aligned based on respective repressor protein sequences and branch lengths correspond to relative divergence in amino acid sequence. The sequences and sources of each repressor are provided in Supplementary Data Set 1. The two IcaR orthologs originate from two distinct host organisms where (A) indicates Staphylococcus aureus and (E) indicates Staphylococcus epidermidis.
There are a number of biochemical mechanisms that could be used to produce the repressing function required by a NOT gate. The most common is to use a protein-based repressor, which binds to an operator DNA sequence within its target promoter. Various classes of natural repressors exist, including phage repressors (e.g., cI), LacI, and TetR-family repressors, all of which have been used to build NOT gates8,12,13. Several modular scaffolds – zinc finger proteins (ZFPs)14 and transcription activator-like effectors (TALEs)15 – have a domain architecture that allows proteins to be designed to bind target sequences. ZFPs and TALEs have been used to control expression in eukaryotic cells16-18 and to a lesser degree in prokaryotes19,20. Recently, it has been shown that transcription can be repressed with a CRISPR-Cas system (“CRISPRi”) that uses a nuclease-null Cas9 protein and an RNA guide sequence to block transcription at a specific site21. Because of the programmability of the RNA-DNA interaction, this system holds promise for building orthogonal repressors; use of CRISPRi to build layerable gates has the potential to be a powerful tool in the construction of circuits.
In this paper, we decided to target TetR homologues for several reasons. First, TetR is one of the earliest and most pervasive transcription factors used in biotechnology and has appeared in numerous applications22. As an inducible system, it is part of a classic multi-plasmid system23 and has been used in a broad range of host organisms, including bacteria/archea24, fungi25, insects26, plants27, mammalian cells28, and live animals29. Second, it has been used in many genetic circuits in synthetic biology, including a toggle switch12 and oscillator13 in Escherichia coli. It has also been used to build a time-delay circuit in mice30 and a NOT gate in mosquitoes10. Third, TetR and most homologues have a simple mode of repression where dimers bind to a promoter and physically block RNAP31. Fourth, they are able to achieve specificity with relatively short operator sequences22. Finally, tens of thousands of homologues are available from many host organisms and there is evidence that they exhibit sequence specific binding to disparate operator sequences22. Small differences in the amino acid sequence and operator nucleotides have been shown to yield high-affinity, orthogonal interactions32,33. The potential for orthogonality is also large; coding theory predicts that there is an upper limit of 130 helix-turn-helix repressors that could function in one cell without exhibiting crosstalk34.
To increase the number of available gates, we used DNA synthesis to access repressors selected from the sequence database and screened them to identify an orthogonal subset. Using an in vitro microarray assay, the DNA binding preferences for individual repressors were comprehensively examined, from which well-defined motifs were obtained. This information, together with previously identified operator sequences, was used to construct synthetic promoter libraries to identify those that were highly repressed. The resulting repressor:promoter pairs were systematically converted into NOT gates, their cross reactions measured in all combinations, and then used to construct composite circuits in vivo. Overall, this work represents a large set of compatible, orthogonal components from which user-defined circuits can be constructed by simply changing the pattern of input and output promoters between a set of conserved gates.
Results
Construction & characterization of a TetR homolog library
We developed a pipeline to expand the number of available TetR family repressors, to exhaustively measure their activity and orthogonality, and to characterize them in the context of genetic gates (Figure 1b). TetR homologues encompass one of the largest families of transcription factors, with 82,017 members currently annotated in EMBL-EBI35. To build a library of homologues, we started with 73 repressors obtained from a collated list of TetR homologues with known regulatory functions from diverse organisms22 (Figure 1c). Redundant sequences and incomplete entries were excluded from the list. This set contains homologs from 45 distinct prokaryotic species and has an average amino acid identity of 21%. Genes were codon optimized for expression in a set of target organisms and built using DNA synthesis (sequences and sources of each repressor are provided in Supplementary Data Set 1).
For the majority of repressors in the library, the DNA sequences to which they bound were unknown. We adapted an assay based on Cognate Site Identifier (CSI) array analysis in order to determine their operators36. Previously, these arrays had been designed to assay transcription factors that bind to a 4-6 bp operator37. To screen for binding by our repressors, a CSI array was designed to accommodate the inverted-repeat containing operator sequences bound by TetR homologues (described in Online Methods and Supplementary Results, Supplementary Figure 1 and Supplementary Table 1). The array contained a unique putative binding sequence at each of its 2.1 million spots. All possible 28 bp sequences were represented (Figure 2a) with the additional criteria that each sequence must have: 1) a perfect 14 bp inverted repeat, 2) a GC content of ≤51%, and 3) three fixed positions. We recombinantly expressed, purified, and labeled each repressor with a cyanine 5-conjugated antibody and, in the case where binding was observed, applied the repressor to two replicate arrays. Those array features that had high intensity values across two independent arrays (>0.25 COV) were selected and the corresponding sequences analyzed by MEME38 to identify consensus motifs (sequences representing each motif are provided in Supplementary Data Set 2). From this analysis, 10 repressors yielded well-defined motifs (Figure 2b). The operators for McbR, PsrA, QacR, and ScbR had been previously identified, and the array data closely matched sequences from the literature (Supplementary Figure 2). For each of the array-based motifs, the operon encoding the repressor (or the entire genome sequence when available) was analyzed for the presence of sequences similar to the array identified motifs. Sequences sharing significant similarity are illustrated in Supplementary Figure 3. Significant diversity exists among the operator sequences bound by different repressors within the library.
Figure 2. The identification of operator sequences using an in vitro array assay.
(a) The hairpin sequences used to build the array correspond to a 28-mer inverted repeat sequence. N's indicate that all nucleotides are allowed at that position. The arrows above the hairpin mark each 14-mer half site and indicate the axis of symmetry for the palindrome. The variable region is surrounded by GC-clamps on both the 5’ and 3’ ends, and the hairpin contains a GGA loop to induce hairpin formation. The 3’ end is tethered to the array surface via a flexible linker. (b) Operators are shown for those repressors that yielded well-conserved sequence motifs.
Design of synthetic promoters & measurement of crosstalk
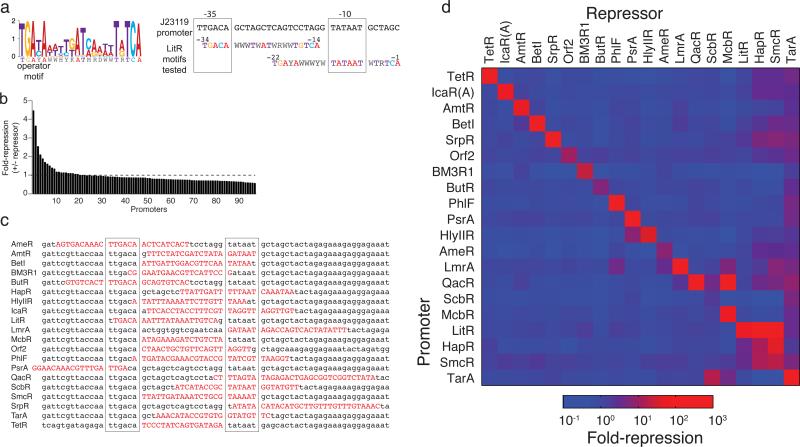
Synthetic promoters were designed to contain operator sequences that were either identified using the array or obtained from the literature (Online Methods). A strong constitutive E. coli promoter (BBa_J23119) was used as a backbone into which an operator was placed39. Promoter libraries were constructed to determine the optimal placement and sequence of the operators. The data from the array were used to determine an “operator motif” the captures the functional diversity of the operator sequence (Figure 3a). Sequences consistent with the motif were constructed using degenerate oligonucleotides and inserted into various positions in the promoter around and between the -35 and -10 sequences. The promoter libraries were then screened in the presence and absence of their cognate repressor by eye or using flow cytometry (Figure 3b and Supplementary Data Set 3). From each library, the promoter that generated the highest dynamic range was identified, sequenced, and then confirmed. At the end of this process, we identified promoters that were responsive to 20 repressors (Figure 3c). This set consists of 10 promoters whose operators were obtained from the CSI array and 10 that were obtained from the literature (Supplementary Table 2).
Figure 3. Design and screening of orthogonal promoters.
(a) Degeneracy in operator sequences (Figure 2b) is converted into a single motif. The LitR motif is shown (W is A/T, H is A/T/C, Y is T/C, K is G/T, M is C/A, R is A/G, and D is A/T/G). The degenerate operator is placed in the BBa_J23119 constitutive promoter spanning either the -35 or -10 element (right panel). (b) The results of screening the LitR promoter library are shown. The fold-repression is calculated as the ratio of fluorescence from the promoter alone and that obtained when the repressor is present and uninduced for a single replicate. (c) The best promoters identified in the screens are shown for each repressor that are part of the final set of 20 repressors. The operator sequence is shown in capital red letters and the Shine-Delgarno as bold letters. Those promoters lacking the Shine-Delgarno sequence contain this sequence adjacent to the 3’ end of the sequence listed; when not shown, the sequence up to the ATG start is identical. (d) The promoters driving YFP expression are carried on a p15a plasmid and the repressors are under HSL-inducible control on a ColE1 plasmid (Supplementary Figure 5 and 4, respectively). The matrix has been sorted by eye, such that the most orthogonal promoters appear at the top and the least at the bottom, and similar patterns of cross-reactivity are clustered together. Repressor expression is induced by 20 μM HSL (except in the case where such concentrations of HSL are toxic, including HapR, Orf2, ScbR, SmcR which were induced with 2 μM, 0.02 μM, 0.2 μM, and 0.2 μM HSL, respectively). The data represent the average of three replicates collected on different days.
To measure all possible cross-reactions, we assayed the activity of each repressor against the set of 20 promoters. Repressor expression was controlled by the HSL-inducible PLux promoter in a colE1 plasmid (Supplementary Figure 4). The promoters were fused to yellow fluorescent protein in a p15A plasmid (Supplementary Figure 5). The repressor and promoter plasmids were co-transformed in all combinations. The resulting 400 strains were grown in the presence of inducer, the promoter activity was measured using cytometry, and the fold-repression reported as the ratio between the non-repressor containing control plasmid and the induced repressor. These data were used to construct an orthogonality matrix that shows the specificity of each promoter and repressor (Figure 3d). The repressors are remarkably orthogonal, and a core set of 16 have negligible cross-reactions (TetR, IcaRA, AmtR, BetI, SrpR, Orf2, BM3R1, ButR, PhlF, AmeR, QacR, LmrA, PsrA, HlyIIR, McbR, ScbR, TarA, LitR, HapR, SmcR). Amongst this orthogonal set, the sequence diversity of the DNA-binding region is noteworthy (Supplementary Figure 6)31. Previous work shows that within the recognition region of the DNA binding domain (residues 25-44 in TetR), residues 28 and 37 are particularly important for binding specificity31,33. Out of the set of 16 orthogonal repressors, 11 and 9 different amino acids are represented at these positions, respectively.
Several groups of repressors are not orthogonal and this corresponds to amino acid similarity in their DNA-binding domain (Supplementary Figure 7). The HapR, LitR, and SmcR repressors (all from Vibrio species) interact with each other's promoters and have similar patterns of crosstalk. Similarly, the HlyIIR and PsrA repressors share amino acid identity and bind to similar operators. Surprisingly, the converse is not true, where similarity between promoters is not predictive of crosstalk. This is largely a result of the variability observed in the acceptable spacing distance between the 6-mer repeat of the operator. For example, the LitR and HapR promoters have spacers of 11 bp and 3 bp, but the repressors cross-react equally well with both.
NOT gate construction & response function measurement
The repressors and their synthetic promoters were used to build a library of NOT gates. To measure the response as a function of the activity of an input promoter, the IPTG-inducible PTac promoter was connected to each gate. The output was measured by having the repressible promoter drive the expression of yellow fluorescent protein (YFP). Each NOT gate consists of a 5-UTR, repressor gene, terminator, and synthetic promoter. These parts, along with the PTac inducible system, and YFP were assembled into a p15a plasmid (Supplementary Figure 5).
Ensuring repressor expression levels are within the appropriate range to generate a large output response represents a challenge in converting repressors into gates. Expression levels cannot be changed by varying the dynamic range of the input promoter because, for circuit construction, inputs must be swapped without further modification. Thus, we define the beginning of each gate to be the transcription start site and vary the repressor level by changing the strength of the RBS. Each repressor has unique properties that influence their absolute protein levels (e.g., codon usage, mRNA and protein stability, binding affinity). Hence, the ribosome binding site (RBS) of the repressors had to be individually tuned to maximize the dynamic range. To accomplish this, we used the RBS Calculator (Online Methods) to design a set of sequences that systematically vary the predicted expression level from medium-high to low (because the optimal desired expression level is not known a priori). These were used to design a degenerate oligonucleotide, from which an RBS library was constructed for 19 out of the 20 gates (Supplementary Table 3). These libraries were screened and the RBS that produced the highest dynamic range was selected (Supplementary Table 4).
The response function of a gate captures how the activity of the output promoter changes as a function of the input promoter. This information is critical in determining how gates will operate when connected in a circuit. It is also important that the inputs and outputs are reported in the same units4. As such, we reported these values as relative expression units (REU) (Supplementary Figure 8)39. This is done by normalizing the YFP output values by that measured from a reference standard, and by separately measuring the activity of the PTac input promoter as a function of IPTG (Supplementary Figure 9), using the same reference standard. With these data, it is theoretically possible to know whether the range of the output of one gate is sufficient to serve as an input to the next gate in series.
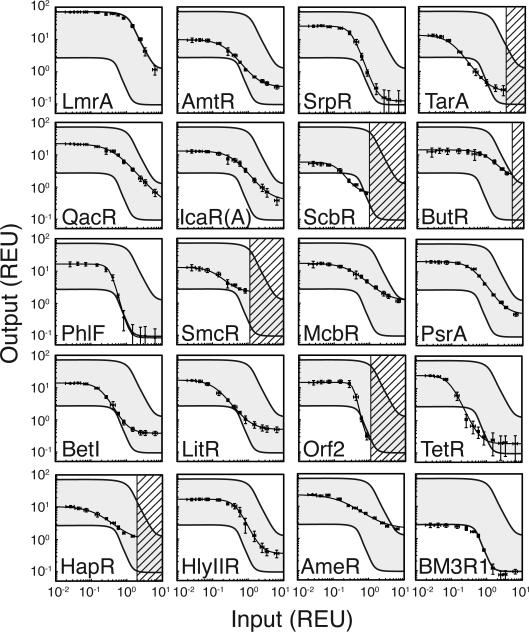
Each gate produces a unique response function (Figure 4). The dynamic ranges of the gates vary from 207-fold (SrpR) to 5-fold (SmcR and ButR) with an average induction of 51.3-fold (Supplementary Table 5). Cytometry distributions are shown for the ON and OFF states, which are narrow and have good separation, even for those that have a smaller dynamic range (Supplementary Figure 10). The response functions can be fit to a Hill function,
| (1) |
where y is the activity of the output promoter, ymin is the minimum output, ymax is the maximum output, n is the Hill coefficient, and K is the threshold level of input where the output is half-maximal. Equation 1 was used to fit the data for each gate and the parameters are shown in Supplementary Table 5.
Figure 4. Response function measurement.
The response functions are measured using the IPTG-inducible PTac promoter as an input and measuring the response of the output promoter. The activity of the input promoter is measured separately using YFP. The activities of the input and output promoters are converted to REU. The response functions of the NOT gates are shown. From left to right, the concentration of IPTG is: 0, 5, 10, 20, 30, 40, 50, 70, 100, 150, 200, 500, and 1000 μM. The error bars show the standard deviation of three experiments performed on different days. As a guide to the eye, the highest (LmrA) and lowest (BM3R1) response functions are shown on each plot with the region between them in grey. The dashed regions indicate the levels of expression beyond which toxicity is observed (Supplementary Figures 11 and 12). The data represent the average of three replicates collected on different days, and error bars correspond to the standard deviation between these measurements.
The thresholds for the gates are similar with an average of K = 0.4 REU and a range of 0.1 (TarA) to 1.3 REU (ButR). Considering this, all of the NOT gates have sufficiently high ON states (between 3 and 70 REU) to achieve full repression by crossing the threshold required by a downstream circuit. However, the OFF states range between 0.1 and 2.1 REU. Because the OFF states are similar in magnitude to the thresholds, this can be problematic when connecting gates, and can lead to a degradation in the signal as the number of layers in the program increases8.
Gates that exhibit ultrasensitivity generate a large output response with little change in the input signal. This also comes at a cost, where it becomes increasingly difficult to balance the input to span the range required to achieve the maximum response. The cooperativity for the majority of gates is n ≈ 2, which is consistent with that measured for TetR, and a mechanism of dimers binding to a single operator13. Five of the repressors yield gates with n > 3, with the largest being 6.1 for Orf2. This has been observed before with TetR homologues, which can bind with higher cooperativities by assembling as multimers or multiple dimers within a single operator40.
Transcription factors can be toxic and exhibit slow growth when expressed above a critical threshold41. We measured the impact on cell growth by recording the OD600 6 hours post-induction for each NOT gate at various levels of induction (Supplementary Figure 11). The majority of repressors are non-toxic, even when maximally expressed. Six showed toxicity at high input levels: TarA, ScbR, ButR, SmcR, Orf2, and HapR, defined toxicity as >25% reduction of growth (Supplementary Figure 12). In each case, the toxicity occurs after the output promoter has been repressed. The quantification of regions of toxicity enables a designer to build circuits that avoid expression above these levels. Further, it enables a comparison between different biochemistries that can be used for the construction of integrated circuits.
Connection of gates to create integrated circuits
The NOT gates can be converted into multi-input NOR gates by connecting multiple promoters in series to drive repressor expression11. Logic minimization algorithms, such as ESPRESSO42, can convert any arbitrary user-defined circuit into a wiring diagram composed of layered NOR gates43. The wiring diagram can then be replicated by a genetic circuit through assembling a particular pattern of input and output promoters connected to the gates (Figure 5). By changing this assembly pattern, the same set of underlying orthogonal gates can be used to build any arbitrary desired circuit.
Figure 5. Construction and characterization of integrated circuits.
(a) The process of promoter mapping for the assembly of gates into a desired circuit is shown for the NAND circuit. The measured data are grown under conditions of no inducer (−/−), 1 mM IPTG (+/−), 20 μM 3OC6HSL (−/+), and 1 mM IPTG and 20 μM 3OC6HSL (+/+). The bar graph details the measured output levels under all input combinations. Small black bars indicate the predicted output value for the indicated input. The data represent the average of three replicates collected on different days, and error bars correspond to the standard deviation between these measurements. (b) The design, construction, and characterization of the AND circuit is illustrated. Note that when multiple promoters are placed upstream of a repressor, the gate is converted from the NOT to NOR function. The measured data are grown under conditions of no inducer (−/−), 1 mM IPTG (+/−), 100 ng/mL aTc (−/+), and 1 mM and 100 ng/mL aTc (+/+). The bar graph details the measured output levels under all input combinations. Small black bars indicate the predicted output value for the indicated input. The data represent the average of three replicates collected on different days, and error bars correspond to the standard deviation between these measurements.
To demonstrate the assembly of gates, we constructed two simple circuits that perform the AND and NAND logic functions through different permutations of the NOT and NOR gates. The inputs to the circuits consist of different combinations of inducible promoters: PTac (IPTG), PLux (AHL), and PTet (aTc) (Supplementary Figure 13). The NAND gate consists of two NOT gates (based on PhlF and LmrA), which invert the two input signals (Figure 5a). The output of the NOT gates are assembled in series to form an OR gate, which then serves as the output of the circuit. The circuit produces the correct NAND function, with a 6-fold difference between the OFF state (+/+) and the lowest ON state. The OFF state is high, which is consistent with the leakiness of the LmrA promoter.
The AND circuit was constructed by combining three gates (Figure 5b). The PhlF NOT gate (the same as that used for the NAND circuit) serves to invert one of the input promoters. The other input promoter is inverted by the QacR NOT gate. The output promoters of these gates are connected to BetI to form a NOR gate, the output of which drives the expression of YFP. This circuit produces a 4.4-fold response when the ON state (+/+) and the highest OFF state are compared. Flow cytometry histograms for each circuit and the terminal gates are illustrated in Supplementary Figure 14.
To determine whether individually measured response functions of gates (Figure 4) can be used to predict their combined response as a genetic circuit, we developed a simple model of the NAND and AND circuits. This model simply adds the response functions of the inducible inputs and the gates to obtain the response of the circuit as a whole, with no additional fit parameters. The OFF and ON states of inducible promoters that serve as inputs (PTac, PLux, and PTet) were measured independently and converted into REU (Supplementary Figure 15). The (OFF - ON) states of the inducible promoters are: PTac (0.06 - 6.2), PLux (0.7 – 8.2), and PTet (0.07 – 9.8). To determine the predicted function of a circuit, the combinations of signals from the input promoters are tracked through the gates using their response functions (Equation 1). This process is visualized in Supplementary Figure 15.
To model the NAND circuit, the range of PTac is inserted into the PhlF response function, yielding outputs of 16 and 0.1 REU (Supplementary Table 6). Similarly, the range of the PLux input is converted to 61 and 1.4 REU by the LmrA response function. The output of the OR gate is treated as the simple sum of the outputs of the tandem promoters. The predicted values for the four combinations of input states closely matches the experimental data (Figure 5a).
To model the AND circuit, the output of PTac connected to the PhlF gate is the same as reported above (16 and 0.1 REU), and the output of PTet connected to the QacR gate is 20 and 0.4 (Supplementary Table 7). To model the NOR gate, the outputs of these promoters are summed x = x1 + x2 and serve as the input to the BetI response function (Equation 1). As with the NAND circuit, the predicted response closely matches the experimental measurements (Figure 5b). Both circuits have some quantitative differences between the predictions and experimental data. This is likely due to the simplicity of the model, which does not account for changes in genetic context, promoter interference between tandem promoters, plasmid copy number variation6,44, or the growth phase under which the outputs were measured (Supplementary Figure 16).
Discussion
The ability to manipulate gene regulation is one of the last frontiers in genetic engineering. The implementation of computing in cells has the potential to impact many applications in biotechnology. However, the field has been limited in the size and sophistication of circuits that could be constructed from a small number of characterized transcription factors. Here, we significantly expand the number of repressors that are available for circuit construction. Further, we have rigorously measured the cross reactions to identify a core orthogonal set. Each member of this set is converted into a gate and fully characterized. Finally, we introduced a generalized method by which circuits can be assembled by changing the pattern of input and output promoters to reproduce a wiring diagram composed of NOT and NOR gates. For simplicity, we demonstrate this by building two circuits that perform digital Boolean logic operations. Note that the same approach could be applied to build analog45 and dynamic13 circuits.
The mining effort described here started with 73 homologous repressors and ended with a set of 16 orthogonal gates. By considering all of the possible ways that these gates can be combined, one can imagine a “circuit space” that consists of all the possible wiring diagrams. The size of this space can be estimated by
| (2) |
where n is the size of the orthogonal set and k is the number of repressors in the circuit. This takes into account that: (i) there are up to 2k sensor inputs to the circuit as a whole, (ii) only NOT and 2-input NOR gates are considered, (iii) for a gate, each input can be comprised of one of the circuit inputs, an output from another gate, or be unconnected yielding (3k+1) possibilities, and (iv) functionally redundant or isomorphic circuits are not removed from the set. This estimates that n = 16 orthogonal gates can be used to build N > 1054 possible circuits. This set includes feedback loops and is not limited to digital logic. Each of these circuits can be accessed by permuting the input and output promoters into a particular pattern.
The challenge now becomes achieving a degree of reliability where the gates can be assembled into any of these circuits with a reasonable chance of functioning properly. While we demonstrate this mapping with a few circuits, accessing the potential of the space remains a challenge. The first generation of gates presented here was designed to be simple and consist of a single operator in a constitutive promoter. This simplicity leads to gates that exhibit low cooperativity, a high OFF state, and sensitivity to genetic context44. Further, each gate utilizes the same pair of terminators, which can lead to evolutionary instability for large circuits46,47. Analyzing the gates in different contexts and identifying the failure modes could lead to second generation designs that are engineered to be faster, tunable, and robust by implementing design rules that have emerged from control theory and systems biology44,48-50.
We selected TetR homologues because of their high specificity, stability, and proven capability to operate in synthetic circuits. Other biochemistries could be used to expand the number of orthogonal gates in our library. To be compatible, the only constraint is that the inputs and outputs of each gate must be promoters, thus allowing the gates to be layered. The repressors could be other classes of proteins that bind DNA – such as TALENs or ZFPs – or be the leader RNA that directs Cas9 as part of CRISPRi; a single large circuit could contain mixtures of these biochemistries. Indeed, this may be a mechanism to expand the gate library beyond the informatic limit of any one family. For example, our TetR library already covers 15% of the predicted upper limit on helix-turn-helix repressors34.
The set of orthogonal gates we present is sufficiently large to implement nontrivial circuits of direct relevance to applications in biotechnology, which includes multi-input logic control for environmental or metabolite sensing, timers to control when different genes are expressed, multiple toggle switches for memory, and simple algorithms from control theory. However, now that parts are no longer limiting, it remains a challenge to build large circuits. To this end, computational tools will likely play a more central role in design. Changing the inputs and outputs to gates by rearranging the pattern of input and output promoters is a sufficiently simple operation to be performed by a computer. The co-development of simple schemes for genetic programming, as well as gates designed specifically to be compatible with these schemes, will enable the broader application of genetically encoded algorithms to program cells.
Online methods
Strains and Media
E. coli strain DH10B {F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15ΔlacX74 recA1 endA1 araΔ139 Δ(ara, leu)7697 galU galK λ-rpsL (StrR) nupG} was used for all experiments, except logic gate measurement where DH5α {fhuA2 lac(del)U169 phoA glnV44 Φ80’ lacZ(del)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17} was used, and protein expression/purification where BL21(DE3)pLysS {F− ompT gal dcm lon hsdSB(r −B m −B) λ(DE3) pLysS(cmR)} was used. Cells were grown in Luria Bertani (LB) Miller Broth, M9 minimal media ((6.8 g/L Na2PO4, 3g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, Sigma), 2mM MgSO4, 100uM CaCl2, 0.4% glucose, 0.2% Casamino acids, 340 mg/L thiamine (vitamin B1)), or Super Optimal Broth (SOB). Ampicillin (50 μg/ml), Kanamycin (25 μg/ml), and/or Chloramphenicol (37 μg/ml) were used where appropriate. Isopropyl β-D-1-thiogalactopyranoside (IPTG) or 3OC6-N-(β-Ketocaproyl)-L-homoserine lactone (3OC6HSL) inducers were used as inducers for the various repressor constructs. Each of the newly constructed plasmids was made by the one-step isothermal DNA assembly method or inverse PCR (see below). In all cases, Yellow Fluorescent Protein (YFP)51 was used as the reporter.
Codon optimization and gene synthesis
Repressor coding sequences were optimized for production in E. coli, chloroplasts, and Bacillus subtilis, using multi-parameter gene optimization methods52. Optimized sequences were synthesized by GeneArt, are contained within a pET21a-derived plasmid (where each repressor contains an amino-terminal 6x-histidine tag), and were sequence verified.
Calculation of relative expression units (REU)
REUs were calculated through use of a strain harboring pJ23101-YFP (Supplementary Figure 8), which contains a constitutive promoter (BBa_J23101) followed by a 5’UTR (BBa_B0032) and YFP. A plasmid containing the reference standard was transformed into DH10B cells to result in the in vivo reference strain. The reference strain was grown under conditions identical to an experimental strain (in this work, strains harboring NOT gates or genetic circuits). The mean reference fluorescence of three replicates minus white cell fluorescence was set to 1 REU. The mean fluorescence from experimental strains was divided by the reference standard to obtain their output in REU.
Repressor expression and purification
Plasmids encoding the synthesized repressor were transformed into BL21(DE3)pLysS cells. Single colonies were selected for by growth on LB Miller medium containing ampicillin and chloramphenicol. Cells were inoculated in SOB containing ampicillin and chloramphenicol and grown overnight at 37°C. The following morning, cells were diluted back to an OD600 of 0.1 in 50 mL fresh SOB medium without antibiotics and were induced using 1 mM IPTG once cells reached an OD600 between 0.6 - 0.8. Cells were grown for 6 hours at 37°C at 250 rpm in a shaking incubator, spun down at 4,000 rpm at 4°C, supernatant discarded, and pellets stored at −80°C.
Cell pellets were resuspended on ice in 5 mL binding buffer (0.5 M NaCl, 20 mM HEPES pH 8, 5 mM Imidazole, 50 mM Phenylalanine, 50 mM Isoleucine, 10% glycerol, and 0.1 μM DTT) containing protease inhibitors and 0.1% Igepal detergent. Resuspended cells were lysed by sonication at room temperature with a setting of 20% duty cycle and 0.1 second pulses, using two 20 second cycles, followed by a final 10 second cycle, with icing in between. Lysates were clarified by centrifugation at 4°C at 10,000 rpm for 30 minutes. Clarified extracts were then filtered, applied to 0.5 ml Nickel resin (that had been equilibrated with binding buffer for 30 minutes at room temperature using a Nutator), and the resin collected using a gravity flow column. The repressor-bound resin was washed with 5 ml binding buffer, 10 ml wash buffer (0.5 M NaCl, 20 mM HEPES pH 8, 25 mM Imidazole, 50 mM Phenylalanine, 50mM Isoleucine, 10% glycerol, and 0.1 μM DTT), and eluted in 0.5 ml elution buffer (0.5 M NaCl, 20 mM HEPES pH 8, 0.5 M Imidazole, 50 mM Phenylalanine, 50mM Isoleucine, 10% glycerol, and 0.1 μM DTT). To the eluate, 3.5 mL binding buffer was added, applied to a 15 ml microconcentrator and spun down at 4000 x g for 20 minutes at 4°C. The concentrated eluate was stored on ice, the concentration determined using Bradford reagent, distributed into approximately 150 μg aliquots, flash frozen in liquid nitrogen, and stored at −80 °C.
Custom array design and native operator sequence analysis
Analysis of previously characterized operators for the repressors within our library revealed that the repressors bind sequences ranging from 16-55 bp (with an average size of 28 bp) that are predominantly AT-rich, and contain inverted repeats. Based on this information, a custom array that contains AT-rich, 28-mer inverted repeat sequences was developed. To meet the manufacturer-imposed cut-off of 2.1 million probes, motif analysis was carried out (using the native operators) to identify conserved positions that could be fixed within the hairpin. From this analysis, a single motif was identified (Supplementary Figure 1). The alignment of operators representing the consensus sequence is listed in Supplementary Table 1. Those positions that were fixed are as follows (counting back from the axis of symmetry within the hairpin in Figure 2a), 7=A, 13=A, and 14=T. Furthermore, a GC-cutoff of 45% was implemented (and included a portion of those having 51% GC content) to reach 2.1 million distinct probes present on a single array. Arrays were synthesized by Roche Nimblegen and correspond to design file 110308_Hairpin_BS_CGH_HX1.
Repressor binding array experiments and extraction of operator data
Hairpin formation of Nimblegen-synthesized arrays (Figure 2a) was induced by washing arrays in methanol, followed by 30- and 15- minute incubations in 7 M Urea in 1× phosphate buffered saline (PBS) and 1× PBS at 65 °C, respectively, that had been warmed to 65°C, with gentle agitation and protection from light throughout. Arrays were then incubated in non-stringent wash buffer (6.6X SSPE, 0.01% Tween-20) at room temperature for 10 minutes, rinsed in 1× Wash Buffer 3 (Roche Nimblegen), dried using a microarray picofuge, and stored in the presence of desiccant protected from light. Binding reactions (0.25% non-fat dried milk, 0.5 mg/ml BSA, 100 μM DTT, 100 mM NaCl, 50 mM HEPES pH8, 150 μg purified repressor, and 2 μl cy-5 anti-his conjugate in a total volume of 1 mL) were incubated at room temperature for 45 minutes in a light protected microfuge tube. Gaskets were applied to an individual array, the chamber washed with water, protected from light using foil to minimize exposure, and blocked for 75 minutes at room temperature with 2.5% non-fat dried milk. The blocking solution was removed, the chamber washed with buffer containing 100 mM NaCl and 50 mM HEPES pH8, and the binding reaction was hybridized to the array for 1 hour at room temperature using a slow and constant rotating motion. The binding reaction was removed, the chamber washed out with 100 mM NaCl and 50 mM HEPES pH8, and the gasket removed. The array was rinsed with 1× Wash Buffer 3, dried using a microarray picofuge, and imaged using an Agilent DNA Microarray Scanner, Model G2565B. For those purified repressors that exhibited binding, a second array was run to eliminate potential hits that arose due to background or spatial effects. Binding to the array was observed for 34 repressors in the library. For each array, intensity values were assigned to individual sequences using Nimblescan software (Roche Nimblegen). Perl scripts developed by C. Warren (Illumavista Biosciences) were used to extract intensity values across both arrays.
Statistical analysis was carried out for corresponding intensity values; those sequences that met a coefficient of variation cut-off of less than 0.25 for intensities were then averaged. Motif analysis was carried out for those sequences associated with high averaged intensity values using the motif finding algorithm MEME38. Sequences were input into MEME using the background Markov model: A 0.329453127, C 0.170546873, T 0.329453127, G 0.170546873. Further MEME settings included any number of repetitions and searching the given strand only, with minimum and maximum widths between 12 and 28 bases. Sequences representing the operator motifs illustrated in Figure 2b are listed in Supplementary Data Set 2.
Library screening to identify repressible promoters
A single letter, degenerate code was defined for each position within an array-identified motif based on MEME-identified consensus sequences (Figure 2b) to generate an operator motif (Figure 3a). Degenerate oligonucleotides representing the resulting operator motif were designed to insert the operator motif into a strong, constitutive, synthetic Biobricks BBA_J23119 standard promoter39. Operator motifs were inserted, in various positions, between or around the -35 and -10 elements of the BBA_J23119 promoter using inverse PCR. Specifically, vector sequences were PCR amplified using Phusion DNA polymerase (NEB) along with the degenerate, operator-motif containing oligonucleotides. The resulting product was run on an agarose gel, extracted, and digested with DpnI. The blunted-ended, DpnI-digested product was phosphorylated (T4 Polynucleotide Kinase) and ligated (T4 DNA ligase) in a single reaction at room temperature, transformed into chemically competent DH10B cells, and plated on selective LB medium. Libraries containing individual sequence variants of an operator motif were screened for fluorescence using a blue light transilluminator to ensure that the resulting promoters containing operator motifs retained activity. Those operator motif variants that promoted fluorescence were also screened for repression by co-transformation with the cognate repressor (Figure 3b). Briefly, DH10B cells containing a repressor plasmid expressing the cognate repressor were made competent using the Z-competent cell kit (Zymo Research). Plasmid DNA was prepared, in 96-well format, from individual fluorescent operator motif variants. The resulting plasmid DNA was transformed into Z-competent DH10B cells containing the cognate repressor. Overnights were made from cells containing the fluorescent operator motif reporters only, and cells containing both the reporter co-transformed with the cognate repressor. 1 μl of overnight culture was diluted into 200 μl 1× PBS and flow cytometry was carried out to quantify fluorescence levels in the presence and absence of repressor for the LitR and McbR repressors (and then assessed by eye for all other repressor/reporter screens using a blue light transilluinator). The promoter variant associated with the largest difference in fluorescence in the absence and presence of repressor was selected to be the cognate promoter for a given repressor. Promoters were also constructed using the previously identified operator sequences for the AmtR, BetI, BM3R1, HapR, HlyIIR, IcaR (A), LmrA, PhlF, SmcR, and TetR repressors listed in Supplementary Table 2. Individual operator sequences were inserted into the Biobricks BBA_J23119 standard promoter in various positions surrounding either or both of the -35 and -10 elements. Those promoters that retained constitutive activity were screened for repression by their cognate repressor using the methods outlined above.
Construction and tuning of repressor expression
The reverse engineering feature of the RBS calculator53 (https://salis.psu.edu/software/reverse) was used to identify a weak and a strong RBS sequence for each individual repressor, with the following settings: Free energy model: v1.1 and Organism (16s rRNA): Escherichia coli str. K12 substr. DH10B ACCTCCTTA. Specifically, RBS sequences were reverse engineered using the following 4 RBS sequences to obtain their translation initiation rate for an individual repressor: B0034 GAAAGAGGAGAAATACTAGATG, rbs1 TCACACAGGAAACCGGTTCGATG, rbs2 TCACACAGGAAAGGCCTCGATG, or rbs3 TCACACAGGACGGCCGGATG. Successive, single base substitutions were made until RBSs of the desired strength were obtained. This strategy was used to identify both a weak and strong RBS for a given repressor. The two respective strength RBS sequences were aligned and combined into a single, degenerate RBS (except in the case of TetR where a single RBS was used, Supplementary Table 3). The sequence content based off of the alignment and relative translation initiation strength information for each sequence variant were taken into account when assigning degenerate codes to each position within an RBS. Oligonucleotides were designed to encode the degenerate RBS, which was inserted upstream of the repressor coding sequence, to generate an RBS library. The repressor ORF, reporter fragment, and vector backbone were PCR-amplified using Phusion DNA polymerase (NEB) and fused into a single vector using Gibson assembly to generate a single response function vector (Supplementary Figure 5). The entire 20 μl Gibson reaction was transformed into chemically DH10B cells and plated onto LB selective medium containing ampicillin. Single colonies were inoculated and grown for 6 hours at 37 °C in SOB medium containing ampicillin, in 96-well format, in the presence and absence of 1mM IPTG. Fluorescence levels were quantified using flow cytometry to deduce fold change of the induced and uninduced clones as outlined above. Those clones demonstrating high levels of fluorescence in the absence of inducer, and low levels of fluorescence in the presence of inducer were selected. The RBSs that give rise to the highest fold-change are shown in Supplementary Table 4 and were used for the orthogonality measurements and in the construction of NOT gates.
Measurement of orthogonality matrix
Competent E. coli DH10B cells were made using the Z-competent cell kit (Zymo Research) that contained individual NOT gates (pRF-, Supplementary Figure 5), which serve as the reporter. Cells were transformed with an additional vector containing the repressor (pOrtho-, Supplementary Figure 4), whose expression was controlled by the 3OC6HSL-inducible PLux promoter54, in all possible combinations. Specifically, 10-50 ng of plasmid DNA was incubated with 10-20 μl Z-competent cells on ice for 10 minutes in a 96-well plate. 150 μl SOC broth was added, and cells were outgrown at 37°C for 1 hour with shaking at 1000 rpm in an ELMI shaker (ELMI Ltd) and plated on LB agar. Plated cells were inoculated into LB containing Ampicillin and Kanamycin, and grown overnight at 37°C with shaking at 1000 rpm. The following morning, stationary-phase cultures were diluted 1:200 into LB containing antibiotics, and grown in a 96-well shaking incubator for four hours at 37°C with shaking at 1000 rpm. The cultures were diluted 1:100 into LB containing antibiotics and 20 μM 3OC6HSL, except in the case of HapR, Orf2, ScbR, and SmcR, where 2 μM, 20 nM, 200 nM, and 200 nM 3OC6HSL was used, respectively, due to toxicity. The induced cells were grown at 37°C for 6 hours with shaking at 1000 rpm, and then fluorescence was measured by diluting the induced culture 1:40 in phosphate-buffered saline and carrying out flow cytometry as described below. Induction assays were run in triplicate for each repressor-reporter combination, and a control plasmid for the orthogonality assays (that corresponds to the pOrtho vector lacking a repressor coding sequence) was used as a normalization control to signify the unrepressed state for individual reporters. The data represent the average of three replicates collected on different days.
Measurement of NOT gate response functions
E. coli DH10B cultures containing NOT gate constructs were grown overnight for 16 hours in liquid SOB medium containing ampicillin. The cells were grown in a 96-well shaking incubator at 37°C and 1000 rpm. The next day, stationary-phase cultures were diluted 1:200 into antibiotic-containing minimal M9 media supplemented with glucose, and grown in the 96-well shaking incubator for three hours using the same shaking and temperature settings as the overnight growth. Subsequently, the cultures were diluted 1:700 into antibiotic-containing minimal M9 media supplemented with glucose containing different concentrations of IPTG, and then grown for 6 hours in the shaking incubator to obtain sufficient exponential-phase cell density for cytometric analysis. The IPTG concentrations used were 0 μM, 5 μM, 10 μM, 20 μM, 30 μM, 40 μM, 50 μM, 70 μM, 100 μM, 150 μM, 200 μM, and 1000 μM. At the end of the final growth period, cultures were diluted 1:5 into phosphate-buffered saline. Strains containing the plasmids for the measurement of input promoter activity (Supplementary Figure 9) and the conversion to REU (Supplementary Figure 8) were grown and measured concurrently with these strains. Flow cytometry was performed as described below. The data represent the average of three replicates collected on different days, and error bars correspond to the standard deviation between these measurements. Plasmids encoding all NOT gates (Supplementary Figure 5) and circuits (Figure 5) have been deposited to addgene (http://www.addgene.org/).
Measurement of genetic circuits
E. coli DH5α cultures containing the plasmids encoding the circuits were grown overnight in liquid SOB medium containing kanamycin and ampicillin (for the 2-plasmid NAND circuit) or kanamycin (for the 1-plasmid AND circuit) in a 96-well incubator at 37°C shaking at 1000 rpm. After 16 hours of growth, cultures were diluted 1:200 into LB medium with antibiotics, and grown in the 96-well shaking incubator for three hours using the same shaking and temperature settings as the overnight growth. Subsequently, the cultures were diluted 1:700 into LB medium with inducers and then grown for 6 hours in the shaking incubator. The inducer concentrations used are: 1 mM IPTG, 20 μM 3OC6HSL, and 100 ng/mL aTc. Cultures were diluted 1:20 into phosphate-buffered saline and fluorescence was measured by flow cytometry as described below.
Cytometry measurement experiments
At the end of growth, cultures were diluted into phosphate-buffered saline with 2 mg/mL kanamycin to arrest cell growth. Cells were analyzed by flow cytometry, using a BD Biosciences LSRII flow cytometer with a blue (488 nm) laser. An injection volume of 10 μL and the flow rate of 0.5 μL/s were used.
Cytometry data analysis
Cells were analyzed using FlowJo (TreeStar Inc., Ashland, OR), and populations were gated on forward scatter area from 100 to 50,000, and on side scatter area from 50 to 50,000. The gated population consisted of thousands of cells. The fluorescence geometric mean of the gated population was calculated, and the mean autofluorescence of a “white cell” control sample was subtracted from the experimental sample's mean. Fold-change is calculated by dividing the mean fluorescence of the ON state by the mean fluorescence of the OFF state (Supplementary Table 5). The data represent the average of three replicates collected on different days, and error bars correspond to the standard deviation between these measurements.
Cellular growth and toxicity data
Repressor toxicity was assessed by comparing the growth of induced, NOT gate-containing cells to the growth of uninduced cells (Supplementary Figure 11). Cells were grown identically to the response function assay. A 100 μL culture aliquot was placed into an optically clear bottom 96 well plate, and the absorbance was measured at 600 nm using a BioTek Synergy H1 Hybrid Microplate Reader. Repressors were considered toxic under conditions where cell growth is less than 75 percent of the uninduced culture growth. The final non-toxic induction point occurs at 200, 150, 100, 70, 70, and 70 μM IPTG for ButR, TarA, HapR, ScbR, SmcR, and Orf2, respectively. If the threshold for toxicity is redefined to a different number, a plot of the maximum induction levels (REU) for a given toxicity threshold is provided (Supplementary Figure 12). The data represent the average of three replicates collected on different days, and error bars correspond to the standard deviation between these measurements.
Supplementary Material
Acknowledgements
We would like to thank Chris Warren of Illumavista Biosciences for aid with array design, array experiments, and data analysis, and for Perl scripts that were used for array data extraction. Research for this publication was conducted with Government support under FA9550-11-C-0028 and awarded by the Department of Defense, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a to A.A.K.N. C.A.V. is supported by Life Technologies, DARPA CLIO (N66001-12-C-4018), Office of Naval Research N00014-13-1-0074, NIH GM095765, and the NSF Synthetic Biology Engineering Research Center (SynBERC, SA5284-11210).
Footnotes
Author Contributions
C.A.V designed and performed experiments, analyzed data, and wrote the manuscript. B.C.S designed and performed experiments, analyzed data, and wrote the manuscript. A.A.K.N designed and performed experiments, analyzed data, and wrote the manuscript. A.T. designed experiments. K.C. aided in project management, managed/performed the codon optimization and DNA synthesis, and edited the manuscript. T.P. aided in project management, managed/performed the codon optimization and DNA synthesis, and edited the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Weiss R, Knight T., Jr. In: DNA Computing Vol. 2054 Lecture Notes in Computer Science. Condon Anne, Rozenberg Grzegorz., editors. Springer; Berlin Heidelberg: 2001. pp. 1–16. Ch. 1. [Google Scholar]
- 2.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nature reviews. Genetics. 2010;11:367–379. doi: 10.1038/nrg2775. doi:10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber W, Fussenegger M. Molecular diversity--the toolbox for synthetic gene switches and networks. Current opinion in chemical biology. 2011;15:414–420. doi: 10.1016/j.cbpa.2011.03.003. doi:10.1016/j.cbpa.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. doi:10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 5.Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nature reviews. Molecular cell biology. 2009;10:410–422. doi: 10.1038/nrm2698. doi:10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 6.Moser F, et al. Genetic Circuit Performance under Conditions Relevant for Industrial Bioreactors. ACS Synthetic Biology. 2012;1:555–564. doi: 10.1021/sb3000832. doi:10.1021/sb3000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson KE, Bashor CJ, Lim WA, Keating AE. SYNZIP Protein Interaction Toolbox: in Vitro and in Vivo Specifications of Heterospecific Coiled-Coil Interaction Domains. ACS Synthetic Biology. 2012;1:118–129. doi: 10.1021/sb200015u. doi:10.1021/sb200015u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. doi:10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabor JJ, Levskaya A, Voigt CA. Multichromatic control of gene expression in Escherichia coli. J Mol Biol. 2011;405:315–324. doi: 10.1016/j.jmb.2010.10.038. doi:10.1016/j.jmb.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu G, et al. Female-specific flightless phenotype for mosquito control. Proc Natl Acad Sci U S A. 2010;107:4550–4554. doi: 10.1073/pnas.1000251107. doi:10.1073/pnas.1000251107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2011;469:212–215. doi: 10.1038/nature09565. doi:nature09565 [pii] 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. doi:10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 13.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. doi:10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 14.Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc Natl Acad Sci U S A. 2003;100:12271–12276. doi: 10.1073/pnas.2135381100. doi:10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. doi:10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 16.Khalil AS, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. doi:10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. doi:10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature biotechnology. 2011;29:149–153. doi: 10.1038/nbt.1775. doi:10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Politz MC, Copeland MF, Pfleger BF. Artificial repressors for controlling gene expression in bacteria. Chemical communications. 2013;49:4325–4327. doi: 10.1039/c2cc37107c. doi:10.1039/c2cc37107c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durai S, Bosley A, Abulencia AB, Chandrasegaran S, Ostermeier M. A bacterial one-hybrid selection system for interrogating zinc finger-DNA interactions. Combinatorial chemistry & high throughput screening. 2006;9:301–311. doi: 10.2174/138620706776843147. [DOI] [PubMed] [Google Scholar]
- 21.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. doi:10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos JL, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. doi:69/2/326 [pii] 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guss AM, Rother M, Zhang JK, Kulkarni G, Metcalf WW. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea. 2008;2:193–203. doi: 10.1155/2008/534081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dingermann T, et al. RNA polymerase III catalysed transcription can be regulated in Saccharomyces cerevisiae by the bacterial tetracycline repressor-operator system. EMBO J. 1992;11:1487–1492. doi: 10.1002/j.1460-2075.1992.tb05193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lycett GJ, Kafatos FC, Loukeris TG. Conditional expression in the malaria mosquito Anopheles stephensi with Tet-On and Tet-Off systems. Genetics. 2004;167:1781–1790. doi: 10.1534/genetics.104.028175. doi:10.1534/genetics.104.028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatz C, Quail PH. Tn10-encoded tet repressor can regulate an operator-containing plant promoter. Proc Natl Acad Sci U S A. 1988;85:1394–1397. doi: 10.1073/pnas.85.5.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saez E, No D, West A, Evans RM. Inducible gene expression in mammalian cells and transgenic mice. Current opinion in biotechnology. 1997;8:608–616. doi: 10.1016/s0958-1669(97)80037-7. [DOI] [PubMed] [Google Scholar]
- 30.Weber W, et al. A synthetic time-delay circuit in mammalian cells and mice. Proc Natl Acad Sci U S A. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. doi:10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nature structural biology. 2000;7:215–219. doi: 10.1038/73324. doi:10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 32.Helbl V, Tiebel B, Hillen W. Stepwise selection of TetR variants recognizing tet operator 6C with high affinity and specificity. J Mol Biol. 1998;276:319–324. doi: 10.1006/jmbi.1997.1539. doi:10.1006/jmbi.1997.1539. [DOI] [PubMed] [Google Scholar]
- 33.Krueger M, Scholz O, Wisshak S, Hillen W. Engineered Tet repressors with recognition specificity for the tetO-4C5G operator variant. Gene. 2007;404:93–100. doi: 10.1016/j.gene.2007.09.002. doi:10.1016/j.gene.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Itzkovitz S, Tlusty T, Alon U. Coding limits on the number of transcription factors. BMC genomics. 2006;7:239. doi: 10.1186/1471-2164-7-239. doi:10.1186/1471-2164-7-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–312. doi: 10.1093/nar/gkr948. doi:10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren C, et al. Defining the sequence-recognition profile of DNA-binding molecules. Proc Natl Acad Sci U S A. 2006;103:867–872. doi: 10.1073/pnas.0509843102. doi:0509843102 [pii] 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanton BC, et al. Cognate Site Identifier analysis reveals novel binding properties of the Sex Inducer homeodomain proteins of Cryptococcus neoformans. Mol Microbiol. 2009;72:1334–1347. doi: 10.1111/j.1365-2958.2009.06719.x. doi:10.1111/j.1365-2958.2009.06719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–373. doi: 10.1093/nar/gkl198. doi:34/suppl_2/W369 [pii] 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelly JR, et al. Measuring the activity of BioBrick promoters using an in vivo reference standard. J Biol Eng. 2009;3:4. doi: 10.1186/1754-1611-3-4. doi:1754-1611-3-4 [pii] 10.1186/1754-1611-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grkovic S, Brown MH, Schumacher MA, Brennan RG, Skurray RA. The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J Bacteriol. 2001;183:7102–7109. doi: 10.1128/JB.183.24.7102-7109.2001. doi:10.1128/JB.183.24.7102-7109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kittleson JT, Wu GC, Anderson JC. Successes and failures in modular genetic engineering. Current opinion in chemical biology. 2012;16:329–336. doi: 10.1016/j.cbpa.2012.06.009. doi:10.1016/j.cbpa.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Rudell RL. Multiple-valued logic minimization for PLA synthesis. Electronics Research Laboratory, College of Engineering, University of California; 1986. [Google Scholar]
- 43.Brown SV, Z. Fundamentals of Digital Logic with Verilog Design. 2nd edn McGraw-Hill; 2008. [Google Scholar]
- 44.Lou C, Stanton B, Chen YJ, Munsky B, Voigt CA. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nature biotechnology. 2012;30:1137–1142. doi: 10.1038/nbt.2401. doi:10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013;497:619–623. doi: 10.1038/nature12148. doi:10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- 46.Chen YJ, et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 2013;10:659–664. doi: 10.1038/nmeth.2515. doi:10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- 47.Sleight SC, Sauro HM. Visualization of Evolutionary Stability Dynamics and Competitive Fitness of Escherichia coli Engineered with Randomized Multigene Circuits. ACS Synth Biol. 2013;2:519–528. doi: 10.1021/sb400055h. doi:10.1021/sb400055h. [DOI] [PubMed] [Google Scholar]
- 48.Lee TH, Maheshri N. A regulatory role for repeated decoy transcription factor binding sites in target gene expression. Mol Syst Biol. 2012;8:576. doi: 10.1038/msb.2012.7. doi:10.1038/msb.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchler NE, Cross FR. Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol Syst Biol. 2009;5:272. doi: 10.1038/msb.2009.30. doi:10.1038/msb.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bintu L, et al. Transcriptional regulation by the numbers: applications. Current opinion in genetics & development. 2005;15:125–135. doi: 10.1016/j.gde.2005.02.006. doi:10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 52.Fath S, et al. Multiparameter RNA and codon optimization: a standardized tool to assess and enhance autologous mammalian gene expression. PLoS One. 2011;6:e17596. doi: 10.1371/journal.pone.0017596. doi:10.1371/journal.pone.0017596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. doi:10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urbanowski ML, Lostroh CP, Greenberg EP. Reversible acylhomoserine lactone binding to purified Vibrio fischeri LuxR protein. J Bacteriol. 2004;186:631–637. doi: 10.1128/JB.186.3.631-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.