Abstract
Adenovirus protein VII is the major protein component of the viral nucleoprotein core. It is highly basic, and an estimated 1070 copies associate with each viral genome, forming a tightly condensed DNA-protein complex. We have investigated DNA condensation, transcriptional repression, and specific protein binding by protein VII. Xenopus oocytes were microinjected with mRNA encoding HA-tagged protein VII and prepared for visualization of lampbrush chromosomes. Immunostaining revealed that protein VII associated in a uniform manner across entire chromosomes. Furthermore, the chromosomes were significantly condensed and transcriptionally silenced, as judged by the dramatic disappearance of transcription loops characteristic of lampbrush chromosomes. During infection, the protein VII-DNA complex may be the initial substrate for transcriptional activation by cellular factors and the viral E1A protein. To investigate this possibility, mRNAs encoding E1A and protein VII were comicroinjected into Xenopus oocytes. Interestingly, whereas E1A did not associate with chromosomes in the absence of protein VII, expression of both proteins together resulted in significant association of E1A with lampbrush chromosomes. Binding studies with proteins produced in bacteria or human cells or by in vitro translation showed that E1A and protein VII can interact in vitro. Structure-function analysis revealed that an N-terminal region of E1A is responsible for binding to protein VII. These studies define the in vivo functions of protein VII in DNA binding, condensation, and transcriptional repression and indicate a role in E1A-mediated transcriptional activation of viral genes.
The adenovirus nucleoprotein core consists of double-stranded genomic DNA, three highly basic viral proteins VII, V, and μ (mu), as well as protein IVa2 and the 55-kDa terminal protein (1, 8, 33, 42, 52-54, 61). Protein VII is the major protein component of the core, with an estimated 1,070 copies present per virion (20). Along with μ, it is bound noncovalently to the DNA in a sequence-independent manner (2, 6, 36, 55). Protein V contacts the DNA as well and also acts as a bridge between protein VII and the outer capsid (19, 55). Protein IVa2 makes sequence-specific contacts with the viral DNA packaging sequence and is thought to play a role in DNA packaging (64). Salt-extracted preparations of the core contain only DNA and protein VII, suggesting that this protein is the most tightly DNA bound of all core proteins (60). Similarly, Sarkosyl preparations of the core contain predominantly DNA and protein VII (8).
Structural features of the DNA-protein complex within the adenovirus capsid and during infection remain largely unknown. DNA within the capsid is in a highly compact configuration, and electron microscopy studies of purified viral core reveal structures reminiscent of beads on a string, or higher-order chromatin compaction, depending on the method of preparation (8, 18, 45, 49, 50, 60, 63). Nuclease digestion of core preparations results in discrete populations of protected DNA fragments, suggesting a chromatin-like configuration but without a characteristic ladder of repeating units (15, 45, 58, 60). Nuclease sensitivity of viral DNA extracted from infected cells has been reported to change to a state similar to cellular chromatin, although an association of viral DNA with cellular histones has not been demonstrated (16, 17, 56, 57). A protein the size of protein VII could be cross-linked to viral DNA throughout the infectious cycle, including early times, suggesting that the protein VII-DNA interaction is stable within the nucleus and that it might be the initial substrate for early viral transcriptional activation (14). Supporting this suggestion, immunohistochemical staining of early-phase infected cells revealed that protein VII enters the nucleus within the same time frame as viral DNA (28). Protein V has also been shown to enter the nucleus during infection (14, 43), whereas the fate of μ is not known.
We are interested in the role of adenovirus chromatin, and protein VII in particular, in the mechanism of transcriptional activation of the adenovirus early genes. The viral transactivators E1A289 and E1A243 induce early-phase transcription and regulate cellular transcription, and their mechanisms of action have been well studied. Several cellular transcription factors can interact with E1A289 and have been implicated in E1A-dependent transcriptional activation, including ATF2, TATA box binding protein (TBP) (see, however, reference 5), TBP-associated factors, and the mammalian Srb-Mediator complex (5, 7, 22). Others, which interact with both E1A289 and E1A243, have also been implicated in viral activation (21, 47, 62). However, the function of E1A in the context of viral chromatin has not been explored. With the exception of the E1A gene itself, the viral genome is transcriptionally silent until expression of E1A triggers activation, suggesting a role for viral chromatin in transcriptional repression (34). In addition, viral core preparations and reconstituted protein VII-DNA complexes are inactive or poorly active when used as templates for in vitro transcription or DNA replication assays (37, 48, 59). One possible explanation of these findings is that viral core proteins, most notably protein VII, act to repress transcription at early times during infection and that part of the function of E1A is to overcome this repression.
Here we report the ability of protein VII to condense DNA and repress transcription in a nuclear environment. Furthermore, we demonstrate that ectopically expressed protein VII can recruit E1A to chromatin, suggesting that such recruitment takes place during infection in the context of viral chromatin leading to early transcriptional activation. These results are supported by the demonstration that protein VII and E1A can interact directly in vitro. Additionally, chromatin immunoprecipitation assays reveal an in vivo association between protein VII and adenovirus DNA during the early phase of infection when E1A protein is known to be active. These studies define the in vivo functions of protein VII in DNA binding, condensation, and transcriptional repression and indicate a role in E1A-mediated transcriptional activation of viral genes.
MATERIALS AND METHODS
Cells and viruses.
HeLa and HEK 293 cells were grown as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin (all from GIBCO). Phenotypically wild-type adenovirus type 5 dl309 was propagated in HeLa cells and dl520 was propagated in HEK 293 cells to produce virus stocks.
Transfection and infection.
HEK 293 cells were transfected by calcium phosphate precipitation as described previously (4) and incubated for 48 hs before collection. HeLa cells were infected by addition of purified dl309 (35) or dl520 (30) virus to 100-mm plates of 70% confluent cells at 20 PFU/cell and incubated for 12 hs before collection. To make lysates, cells were scraped from plates and placed in lysis/binding buffer (50 mM HEPES, 250 mM NaCl, 10 mM MgCl2, 1 mM CaCl2, 0.2 mM ZnCl2, 0.1% Nonidet P-40, 50 μg of DNase I [Sigma] per ml, 30 μg of phenylmethylsulfonyl fluoride [PMSF] per ml, 1 μg of aprotinin per ml, 1 μg of pepstatin A per ml, 10 μg of leupeptin per ml [pH 7.8]) on ice for 90 min. Lysates were then cleared by centrifugation in a Sorvall MC-12V centrifuge at 12,000 rpm and 4°C for 20 mins.
Plasmids.
To produce pGSTVII and pKE1A289, inserts were generated by PCR with adenovirus type 5 DNA or E1A 13S cDNA as the template. Primers contained 5′ BamHI and 3′ EcoRI sites and the PCR products were cloned into pGEX 3X (pGST-VII) or pGEM 3 (pKE1A289). To produce pGSTE1A289, pGSTE1ACR3, pGSTE1A 139-289, pGSTE1A 1-67, pGSTE1A 1-120, and pGSTE1A 1-139, inserts were generated by PCR with pKE1A289 as template. Primers contained 5′ BamHI and 3′ EcoRI sites, and PCR products were cloned into pGEX-3X (pGSTE1ACR3, pGSTE1A 139-289, pGSTE1A 1-67, pGSTE1A 1-120, and pGSTE1A 1-139) or pGEX-4T-2 (pGSTE1A289). To produce pSP6-VII, a PCR fragment encoding protein VII was cloned into pCMV5.1 (46), which contains an N-terminal HA tag. PCR amplification of the resulting HA-VII sequence was performed using primers containing 5′ EcoRI and 3′ BglII sites, and the PCR product was cloned into pSP64t. To produce pCMV-Flag VII, inserts were generated by PCR using adenovirus type 5 DNA as the template. Primers contained a 5′ BamHI site and FLAG tag-encoding sequences and a 3′ EcoRI site, and the inserts were cloned into pCMV 5 (46). pBtuF-His, a gift from Robert Kadner, contains a His-tagged Escherichia coli BtuF gene in pET-17b (10). pGST CAS YXXP, a gift from Amy Bouton, contains the YXXP domain of CAS in pGEX-2T-K (9). pBS-HRB87F contains a FLAG-tagged Drosophila Hrb87F gene in pBluescript II KS(+) (65). The glutathione S-transferase (GST) fusion plasmids E1AΔ2-36, E1AΔ38-67, E1AΔ73-120, and the double point mutant E1Apm928/961 were gifts from Joseph R. Nevins (38). The plasmid producing His-tagged E1A289 was a gift from Nick LaThangue. All primer sequences are available on request.
Chromatin immunoprecipitation (ChIP).
HeLa cells (4 × 107) were infected with adenovirus type 5 dl309 at a multiplicity of infection (MOI) of 50 and incubated for 4 hs. Nuclei were purified by washing the cells once with phosphate-buffered saline (PBS) at 4°C and twice with nuclear isolation buffer (0.3 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl [pH 7.5], 2 mM EDTA, 0.5 mM EGTA, 14 mM β-mercaptoethanol) at 4°C, centrifuging after each wash (2,000 rpm for 15 s in a Sorvall MC-12V microcentrifuge at room temperature). Pelleted nuclei were resuspended in cross-linking buffer (5 mM HEPES [pH 8.0], 50 μM EGTA, 10 mM NaCl, 100 μM EDTA, 1% formaldahyde, 1% methanol) for 15 min at room temperature. Then 125 mM glycine was added to stop the reaction, and nuclei were pelleted as above. The nuclei were washed once in PBS, resuspended in radioimmunoprecipitation assay buffer and sonicated twice for 15 s at 4°C. The lysate was then cleared by centrifugation (13,000 rpm in a Sorvall MC-12V microcentrifuge for 10 min at 4°C).
Anti-protein VII and control antiserum were prebound to protein A-Sepharose beads (Sigma), washed twice in PBS, and incubated overnight at 4°C with nuclear lysate. The beads were washed five times with PBS, NaCl was added to a final concentration of 200 mM, and the mixture was incubated for 6 h at 65°C to reverse the cross-linking. Proteinase K (50 μg) and RNase A (5 μg) were added, and the sample was incubated for 2 h at 37°C and subjected to two extractions with phenol-chloroform-isoamyl alcohol. DNA was ethanol precipitated overnight at −20°C, air dried, and resuspended in water. PCR was performed using Deep Vent polymerase (New England Biolabs) under standard conditions for 35 cycles of 1 min at 97°C, 1.5 min at 55°C, and 2 min at 72°C, using adenovirus- or rDNA-specific primers. Primers for adenovirus are (from 5′ to 3′) CTGCTGAAGACCATTCACGTAGCCAGCC and GCACCTTCCACATCTTCATGGTCATGTC, and primers for rDNA are CCTACCTGGTTGATCCTGCC and GGTCAGCGCCCGTCGG.
Western blotting.
Western blot analysis was performed essentially as described previously (32). Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and probed with either rabbit polyclonal anti-protein VII antibody (see below), mouse monoclonal anti-human tubulin antibody (Boeringer Mannheim), mouse anti-human Ku86 antibody (a gift from Gary Kupfer), mouse anti-E1A M73 antibody (31), mouse monoclonal anti-tetra-His antibody (Qiagen), or F4 mouse anti-human epidermal growth factor receptor antibody (EGFR) (Sigma). Anti-rabbit-horseradish peroxidase (Amersham Pharmacia) or anti-mouse-horseradish peroxidase (Amersham Pharmacia) was used as the secondary antibody, and blots were visualized by enhanced chemiluminescence.
In vitro transcription and translation.
mRNA for oocyte microinjections was made with the Riboprobe in vitro transcription system (Promega) using 5 μg of pKE1A289, pSP6-VII, or pBS-HRB87F linearized with EcoRI, SacI, or NotI, respectively, followed by addition of GpppG Cap analog (New England Biolabs). The mRNA was treated with RQ DNase-I (Promega), phenol-chloroform extracted, ethanol precipitated, and resuspended in water. 35S-labeled, in vitro-translated E1A289 was produced using the TNT coupled reticulocyte lysate transcription-translation system (Promega) with 1.5 μg of pKE1A289.
Oocyte injections.
Xenopus oocytes were isolated and injected as previously described (51), except that the injections were made into the cytoplasm of medium-size oocytes (stages 4 and 5). Twenty-three nanoliters per oocyte of the following transcripts was injected: VII mRNA (0.6 or 1.2 ng/nl), E1A289 mRNA (4 ng/nl), VII + E1A289 mRNAs (1.3 ng/nl each, or 0.5 ng of VII per nl + 1.5 ng of E1A289 per nl), and Drosophila HRB87F mRNA (0.6 or 1.2 ng/nl). Injected oocytes were incubated in modified Barth's solution (29) for 6 or 24 h at 19°C before being collected for Western blot analyses or processed for lampbrush chromosome spreads.
Lampbrush spreads.
Lampbrush spreads were performed as described previously (25-27), except that dithiothreitol was omitted from the 5:1 isolation medium and both dithiothreitol and Ca+ were omitted from the dispersal medium. Using jeweler's tweezers, the germinal vesicle was quickly isolated in isolation medium, transferred to dispersal medium, where adhering yolk was removed by pipetting, and then moved to the slide dispersal chamber, where the nuclear envelope was removed.
Slides were stored at 4°C until ready for centrifugation (∼30 min). They were centrifuged in the large buckets of a Sorvall RT60000B refrigerated tabletop centrifuge in adaptors made of small plastic boxes lined with a foam pad, two slides per box. Centrifugation was carried out at 3,000 rpm for 30 min. Chromosome preparations were fixed with 2% paraformaldehyde in PBS for 90 mins.
Immunofluorescence staining and microscopy.
Lampbrush preparations were rinsed in three changes of PBS and blocked with 10% horse serum-PBS in a humid chamber at 4°C. Primary antibody was added and the spreads were incubated for 1 h at room temperature. After the preparations were rinsed in PBS, the secondary antibody was applied, and the mixtures were incubated for an additional 1 h at room temperature followed by a PBS rinse.
The following antibodies were used: 3F10 (rat) against the HA epitope (Roche Applied Science) diluted 1:250, anti E1A monoclonal antibody M73 diluted 1:250 (31), rabbit polyclonal anti-protein VII antibody (see below) diluted 1:100 and M2 (mouse) antibody against the FLAG epitope (Sigma) diluted 1:100. Secondary antibodies were Cy3-goat anti-rat immunoglobulin G IgG (Amersham Life Sciences) diluted 1:1,000, fluorescein-labeled horse anti-mouse IgG (H+L), and fluorescein-labeled goat anti-rabbit IgG (H+L) (Vector Laboratories), both diluted 1:100.
Slides were mounted in 50% glycerol containing 1 mg of phenylenediamine per ml to prevent fading and 1 μg of 4′,6-diamidino-2-phenylindole (DAPI) (27). Lampbrush chromosomes were viewed with a Zeiss Axioplan fluorescence microscope.
Production of GST fusion proteins.
Plasmids were transformed into E. coli BL21 Codon Plus RP (Stratagene). Cultures (3 ml) were grown overnight at 37°C in Luria-Bertani LB broth with ampicillin (100 μg/ml) and chloramphenicol (50 μg/ml). They were expanded in 500 ml to an optical density at 600 nm of 0.2, the temperature was then reduced to 30°C, and isopropyl-β-d-thiogalactopyranoside (IPTG) (Gibco-BRL) was added to a final concentration of 0.25 mM. After 3 h, the cells were pelleted by centrifugation and resuspended in binding buffer (540 mM NaCl, 2.7 mM KCl, 10.15 mM Na2HPO4, 1.75 mM KH2PO4, 10 mM MgCl2, 1% [vol/vol] Triton X-100, 50 μg of DNase I, 30 μg of PMSF per ml, 1 μg of aprotinin per ml 1 μg of pepstatin A per ml, 10 μg of leupeptin per ml [pH 7.4]). Lysis was by done with a French press at 18,000 lb/in2 at 4°C, and the lysate was cleared by centrifugation in a Sorvall GSA rotor (5,000 rpm for 15 min at 4°C). To purify GST fusion proteins, 1 ml of a 1:1 slurry of glutathione-agarose beads (Sigma) in PBS was added to 10 ml of cleared lysate at 1 mg of protein per ml and the mixture was incubated for 4 h at 4°C. The bead-bound proteins were washed three times in 15 ml of binding buffer.
Production of His-tagged proteins.
Plasmids were transformed into BL21 Codon Plus RP E. coli (Stratagene). Cultures were grown, induced, and pelleted as above. The cells were resuspended in lysis buffer (50 mM Na2HPO4, 300 mM NaCl, 10 mM imidazole, 30 μg of PMSF per ml, 1 μg of aprotinin per ml, 1 μg of pepstatin A per ml, 10 μg of leupeptin per ml [pH 8.0]) and lysed as described above. The lysate was cleared by centrifugation as described above. His-tagged proteins were purified by adding 1 ml of a 1:1 slurry of Ni-nitrilotriacetate agarose beads (Qiagen) to 10 ml of cleared lysate at 1 mg of protein per ml and incubated for 4 h at 4°C. The beads were washed three times in 15 ml of wash buffer (50 mM Na2HPO4, 300 mM NaCl, 20 mM imidazole [pH 8.0]). Elution was done with four 0.5-ml aliquots of elution buffer (50 mM Na2HPO4, 300 mM NaCl, 250 mM imidazole [pH 8.0]).
GST pulldown assays. (i) Pulldown of in vitro-translated product.
A 2-μl volume of in vitro-translated 35S-labeled E1A289 was added to 50 μl of bead-bound GST fusion proteins (1:1 slurry in PBS) in 300 μl of binding buffer (50 mM KCl, 40 mM HEPES [pH 7.9], 5 mM 2-mercaptoethanol, 0.1% Tween 20, 0.5% nonfat dry milk, 0.2 mM ZnCl2), the mixture was incubated for 1 h at 4°C with rotation, and the beads were washed four times in 1.4 ml of binding buffer. Some reaction mixes also contained 250 mM NaCl. Some reaction mixes were brought to 100 μg of ethidium bromide per ml for 10 min just before or just after the 1-h incubation with GST-VII beads. Bound proteins were separated by SDS-PAGE and analyzed by phosphorimager analysis. In some reactions, 35S-labeled protein was replaced by a 32P-end-labeled 237-bp DNA fragment produced by HindIII digestion of pEBG, the products were washed as described above, and the bound DNA was analyzed by scintillation counting.
Conditions for binding of His-tagged proteins were identical, except that the binding buffer also contained 250 mM NaCl and 3 μg of His-tagged protein was used. Bound proteins were analyzed by SDS-PAGE followed by Western blotting using an anti-tetra-His mouse monoclonal antibody (Qiagen).
(ii) Pulldown of Flag-VII or E1A from cell lysate.
A 5-μg portion of bead-bound GST fusion proteins (1:1 slurry in PBS) was added to 250 μl of cell lysate (1.5 mg of total protein per ml) as prepared above, the mixture was incubated for 1.5 h at 30°C with rotation and the beads were washed four times in 1.4 ml of lysis/binding buffer. Bound proteins were analyzed by SDS-PAGE followed by Western blotting with appropriate antibodies.
Antibody production.
GST-VII protein was produced as described above, washed four times in 15 ml of binding buffer, and eluted from the beads four times in 0.5 ml of elution buffer (50 mM Tris-Cl, 15 mM reduced glutathione [Sigma] [pH 8.0]). Antibody production in New Zealand White rabbits was conducted by Cocalico Biologicals, Inc. Anti-protein VII was then affinity purified against recombinant protein VII cross-linked to beads followed by extensive preclearing against GST protein bound to glutathione-agarose beads.
RESULTS
Protein VII associates with viral DNA during the early phase of infection.
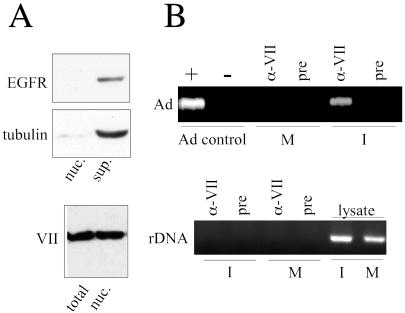
ChIP was used to determine if protein VII associates with adenovirus DNA during the early phase of infection. HeLa cells were infected at a multiplicity of infection (MOI) of 50 and incubated for 4 h. This period represents the early phase of infection, during which E1A protein activates transcription. Isolated nuclei were used for the ChIP analysis to ensure that only infectious genomes would be assayed for the protein VII-DNA association. As controls, Western blot analyses of nuclear preparations were performed with anti-tubulin and anti-EGFR antibodies to demonstrate a lack of contaminating cytoplasmic and plasma membrane components in the isolated nuclei. Moreover, comparison of whole-cell and nuclear lysates demonstrated that virtually all cell-associated protein VII at 4 h postinfection was located in the nucleus (Fig. 1A).
FIG. 1.
ChIP analysis of protein VII during the early phase of infection. Cells were infected with phenotypically wild-type dl309 at an MOI of 50 and incubated for 4 h. Nuclei and postnuclear supernatant were prepared and analyzed as described in Materials and Methods. (A) (Top) Postnuclear supernatant (sup.) and nuclear (nuc.) fractions were analyzed by SDS-PAGE and subjected to Western blotting for tubulin and EGFR to monitor cytoplasmic (tubulin) and plasma membrane (EGFR) contamination of the nuclear preparations. (Bottom) Western blot analysis of total cell lysate (total) and nuclear fraction, using anti-protein VII polyclonal antibody, (B) Nuclei were treated with formaldehyde and sonicated, and the products were subjected to immunoprecipitation with anti-protein VII polyclonal antibody or preimmune serum as described in Materials and Methods. M, lysates from mock-infected cells; I, lysates from dl309-infected cells. α-VII, anti-protein VII antibody; pre, preimmune serum. (Top) PCR analysis for adenovirus-specific DNA. Control PCR amplifications were performed in the presence (+) or absence (−) of purified adenovirus type 5 DNA. (Bottom) PCR analysis for rDNA. Control PCR amplifications were performed with nuclear lysate as the source of DNA.
Polyclonal anti-protein VII antiserum was used in the ChIP assay, and co-precipitated DNA was detected by PCR with primers specific for the E1B region of adenovirus DNA. As shown in Fig. 1B, anti-protein VII antiserum specifically precipitated adenovirus DNA, indicating that protein VII remains bound to the DNA at this stage of infection. Control serum failed to precipitate adenovirus DNA, and no signal was detected from samples derived from mock-infected cells. Precipitates were also assayed using primers specific for rDNA, and no PCR products were observed. This control demonstrates that the anti-protein VII antiserum specifically recognized protein VII-bound DNA and suggests that protein VII is restricted to adenovirus DNA at early times postinfection. Additionally, coding regions of the L2, L3, and E1A genes were assayed by ChIP, as were promoter regions for the E1A, E2A, and E4 genes. All regions gave identical results, as shown here for the E1B region, indicating association of protein VII with viral DNA during the early phase of infection (data not shown).
Protein VII condenses DNA and represses transcription in a nuclear environment.
Adenovirus core particles extracted from virions contain highly condensed DNA. Furthermore, mixing of protein VII and DNA in vitro results in compact DNA-protein structures as viewed by electron microscopy (6; J. Johnson and W. Newcomb, unpublished data). With the exception of the E1A gene, viral DNA that is delivered to the nucleus at the onset of infection is transcriptionally silent until E1A protein is expressed in sufficient amounts (34). Therefore, we reasoned that ectopic expression of protein VII in nuclei might result in condensed, transcriptionally repressed chromatin. To test this hypothesis and to examine the functional properties of protein VII in a nuclear environment, microinjection of Xenopus oocytes was performed.
Stage 4 and 5 Xenopus oocytes contain chromosomes that are highly active transcriptionally. These “lampbrush chromosomes” are easily visible by phase-contrast microscopy and contain numerous loops representing regions of active transcription. Medium-size oocytes containing lampbrush chromosomes were harvested and microinjected with in vitro-transcribed protein VII mRNA or a control mRNA that encoded the Drosophila RNA-binding protein HRB87F. The protein VII mRNA encoded amino acids 25 to 198 of adenovirus type 5 preprotein VII. This portion is identical to the processed form of the protein that is found in mature virions. In addition, the protein contained an N-terminal HA epitope for immunohistochemical detection.
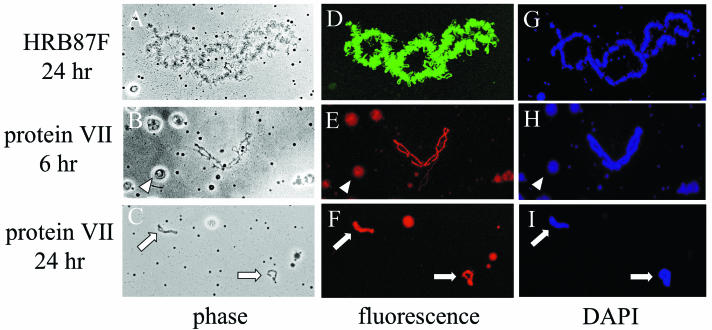
Control microinjections with HRB87F mRNA resulted in normal-appearing lampbrush chromosomes containing numerous transcription loops that were indistinguishable from those of water-injected oocytes (Fig. 2A and data not shown). Staining for HRB87F protein specifically revealed the transcription loops, due to the binding of newly synthesized HRB87F protein to chromosome-associated RNA (Fig. 2D). These data confirmed that the loops visible by phase-contrast microscopy were in fact transcription loops. Protein VII mRNA was microinjected at 0.6 ng/nl (“low”) or 1.2 ng/nl (“high”), and the oocytes were incubated for 6 or 24 h before being analyzed. As seen by phase-contrast microscopy, microinjection of 1.2 ng of protein VII mRNA per nl led to significant chromosomal condensation and a kinked appearance by 6 h postinjection in more than 50% of the oocytes analyzed, and condensation always occurred along with an obvious decrease in transcriptional activity as judged by the disappearance of transcription loops (Fig. 2B, Table 1). The most dramatic condensation and transcriptional repression took place by 24 h postinjection, where effects occurred in 27 of 29 oocytes analyzed (Table 1). We refer to the most severely condensed chromosomes as “commas”; examples are shown in Fig. 2C. All chromosomes are shown at the same degree of magnification.
FIG. 2.
Protein VII binding to lampbrush chromosomes. Microinjection of Xenopus oocytes with in vitro-transcribed mRNAs encoding the indicated proteins was carried out as described in Materials and Methods. Protein VII contained an HA epitope at the N terminus, followed by full-length protein VII. After 6 or 24 h of incubation, oocytes were processed for immunohistochemistry with anti-FLAG (for HRB87F) or anti-HA antibodies as described in Materials and Methods. Samples were also stained with DAPI to identify DNA-containing structures. All chromosomes are shown at the same degree of magnification. Phase, phase-contrast microscopy, fluorescence, fluorescence microscopy of antibody-stained samples; DAPI, fluorescence microscopy of DAPI-stained samples. Arrows indicate comma chromosomes; arrowheads indicate extrachromosomal amplified nucleoli.
TABLE 1.
Protein VII binding, chromosome condensation, and transcriptional repressiona
| Time and mRNA concnb | Total no. of slides | Transcriptional activity (chromosome structure)
|
Level of staining with antibody to protein VII or Hrb87F
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| High (relaxed) | Low (partially condensed) | Inactive (very condensed) | Inactive (commas) | High | Moderate | Low | None | ||
| 6 h | |||||||||
| VII low | 18 | 15 | 2 | 1 | 0 | 1 | 4 | 2 | 11 |
| VII high | 27 | 12 | 4 | 11 | 0 | 7 | 5 | 9 | 6 |
| Hrb87F low | 8 | 8 | 0 | 0 | 0 | 0 | 3 | 4 | 1 |
| Hrb87F high | 7 | 5 | 0 | 2 | 0 | 0 | 2 | 2 | 3 |
| 24 h | |||||||||
| VII low | 11 | 0 | 1 | 9 | 1 | 8 | 0 | 1 | 2 |
| VII high | 18 | 2 | 0 | 13 | 3 | 16 | 0 | 1 | 1 |
| Hrb low | 8 | 8 | 0 | 0 | 0 | 8 | 0 | 0 | 0 |
| Hrb high | 8 | 6 | 2 | 0 | 0 | 6 | 0 | 1 | 1 |
Each slide represents one independently microinjected oocyte. In every case, all chromosomes within an oocyte exhibited identical behavior, and qualitative estimates of binding, condensation, and transcriptional activity were made based on their appearance. Results are compiled from six independent experiments.
Low, 0.6 ng of mRNA per nl; high, 1.2 ng of mRNA per nl.
In the oocytes injected with protein VII mRNA, staining with anti-HA antibody revealed a nearly uniform distribution of protein VII in association with the chromosomes (Fig. 2E and F), and the level of staining generally correlated with the degree of transcriptional repression and condensation (Table 1). By 24 h postinjection, 26 of 29 oocytes injected with protein VII mRNA showed some level of staining. In almost every case for the 6- and 24-h time points, low to moderate staining correlated with some degree of condensation and transcriptional repression and high staining correlated with significant condensation and repression. In very few cases, we observed low to moderate staining without observable condensation or decrease in transcriptional activity. In addition, association of protein VII with nucleoli was observed (Fig. 2E and F), probably due to the binding of protein VII to nucleolar DNA. There was no anti-HA staining of chromosomes from control injections lacking protein VII mRNA (data not shown). For each condition, the proper number of chromosomes per nucleus was confirmed, indicating that the appearance of small chromosomes and commas (as in Fig. 2C) was not the result of fragmentation. DAPI staining showed that the chromosomes associated with protein VII also contained DNA, further confirming their identity (Fig. 2G to I). These data demonstrate the ability of protein VII to bind and condense DNA and repress transcription in a nuclear environment. Since adenovirus core DNA is in a condensed conformation and is transcriptionally silent prior to expression of E1A protein in infected cell nuclei, our results suggest that protein VII plays an important role in these processes and is able to associate stably with DNA in a nuclear environment in addition to its role within the capsid.
Protein VII recruits E1A protein to lampbrush chromosomes.
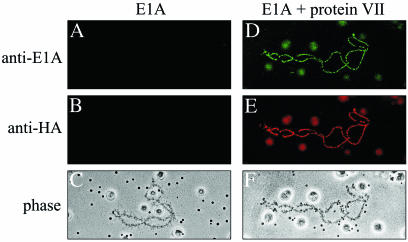
The association of protein VII with adenovirus DNA during the early phase of infection suggested a role for protein VII in E1A-mediated transcriptional activation. To investigate this possibility, E1A mRNA was microinjected into Xenopus oocytes, alone or in the presence of protein VII mRNA (Table 2). Again, protein VII mRNA was injected at two different concentrations, in either the absence or presence of E1A mRNA. The oocytes were incubated for 6 h and then prepared for visualization of lampbrush chromosomes. When E1A mRNA was injected alone, there was no detectable association of E1A protein with lampbrush chromosomes (Fig. 3A; Table 2), despite ample E1A protein expression as determined by Western blot analysis (data not shown). In contrast, when protein VII and E1A mRNAs were coinjected, significant association of E1A with the chromosomes was observed (Fig. 3D; Table 2). This occurred for high (1.3 ng/nl) but not for low (0.5 ng/nl) levels of coinjected protein VII mRNA and correlated well with the efficiency of protein VII association with the chromosomes as determined by antibody staining. When high levels of protein VII mRNA were coinjected, 10 of 13 oocytes exhibited some level of E1A chromosome association, and half of these samples showed moderate to high staining. The presence of E1A did not affect the binding of protein VII to the chromosomes (Fig. 3E; Table 2). These data demonstrate that protein VII and E1A can colocalize on nuclear DNA and that the association of E1A with DNA is dependent on protein VII.
TABLE 2.
Protein VII recruits E1A to chromosomesa
| mRNA concn at 6 hb | Total no. of slides | Transcriptional activity (chromosome structure)
|
Level of staining for protein VII or E1Ac
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| High (relaxed) | Low (partially condensed) | Inactive (very condensed) | Inactive (commas) | High | Moderate | Low | None | ||
| VII low | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 1 | 7 |
| VII high | 7 | 3 | 1 | 3 | 0 | 5 | 1 | 0 | 1 |
| E1A | 12 | 7 | 3 | 2 | 0 | 0 | 0 | 0 | 12 |
| E1A+VII low | 8 | 6 | 1 | 1 | 0 | 0, 0 | 0, 0 | 0, 3 | 8, 5 |
| E1A+VII high | 13 | 4 | 6 | 2 | 1 | 1, 0 | 4, 5 | 5, 5 | 3, 3 |
Each slide represents one independently microinjected oocyte. In every case, all chromosomes within an oocyte exhibited identical behavior. Results are compiled from two independent experiments.
Low, 0.5 g of mRNA per nl; high, 1.3 ng of mRNA per nl.
For entries with two numbers, the first refers to E1A and the second refers to VII.
FIG. 3.
Protein VII recruits E1A to lampbrush chromosomes. Microinjection of Xenopus oocytes with in vitro-transcribed mRNAs encoding the indicated proteins was carried out as described in Materials and Methods. Protein VII contained an HA epitope at the N terminus, followed by full-length protein VII. After 6 hs of incubation, oocytes were processed for immunohistochemistry with anti-E1A or anti-HA antibodies as described in Materials and Methods. Representative results for 33 oocytes from two independent experiments are shown. In each case, all chromosomes within an individual oocyte showed identical behavior. Both chromosomes shown are at the same degree of magnification. Phase, phase-contrast microscopy.
For injection with E1A mRNA alone, 5 of 12 oocytes exhibited an effect on transcription and chromosome structure in the absence of any association with the chromosomes (Table 2). This effect seemed to occur with greater frequency than the background levels of transcriptional repression (4 of 31 oocytes) observed for injection of control HRB87F mRNA shown in Table 1. This effect is possibly due to E1A binding to cellular factors such as histone acetyltransferases, as has been shown for p300 and P/CAF in mammalian cells (3, 12), and displacing them from transcription complexes, leading to transcriptional repression.
Protein VII and E1A interact in vitro.
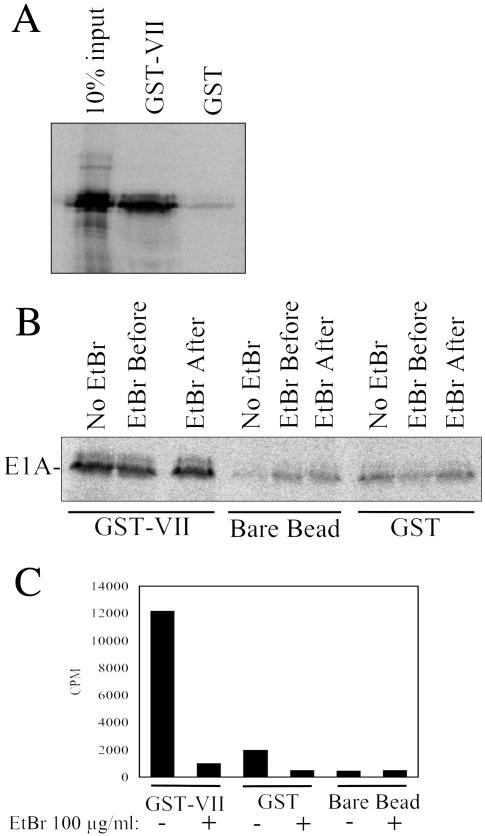
Since protein VII is associated with adenovirus DNA during the early phase of infection, the results shown above suggested that E1A is recruited to the viral DNA by protein VII during infection, possibly by direct protein-protein interactions. To test this hypothesis, GST pulldown experiments were performed. GST-protein VII bound to agarose beads was incubated with in vitro-translated, 35S-labeled E1A289, and after being washed, the bound products were analyzed by SDS-PAGE. Figure 4A shows that E1A289 bound efficiently to GST-protein VII. Little background binding of E1A289 to GST alone was observed.
FIG. 4.
Protein VII binds to E1A in vitro. (A) Equal amounts of GST-protein VII or GST alone were immobilized using glutathione-agarose beads and incubated with 35S-labeled E1A289 as described in Materials and Methods. The reaction products were washed and analyzed by SDS-PAGE. 10% input indicates the amount of control 35S-labeled E1A protein loaded on the gel, whereas 100% was added to the binding-reaction mixtures. (B) The protein VII-E1A interaction is resistant to ethidium bromide (EtBr). GST pulldown assays of 35S-labeled E1A289 were performed as described in Materials and Methods in the presence or absence of 100 μg of ethidium bromide per ml. Ethidium bromide was added to the reactions before (EtBr Before) or after (EtBr After) addition of 35S-labeled E1A; Bare Bead indicates that 35S-labeled E1A289 was added to glutathione-agarose beads without prior GST or GST-protein VII immobilization. (C) GST pulldown assay of a 32P-labeled 237-bp DNA fragment in the presence or absence of 100 μg of ethidium bromide per ml. The assay was performed as described in Materials and Methods.
Since both protein VII and E1A (13) are capable of nonspecific binding to DNA, it was necessary to rule out the notion that the observed interaction between the two proteins was mediated by contaminating DNA in the GST pull-down reactions. To achieve this, binding reactions were performed in the presence of 100 μg of ethidium bromide per ml, which disrupts DNA-protein interactions (39). Addition of ethidium bromide either before or after incubation of GST-protein VII with E1A failed to inhibit binding (Fig. 4B). To demonstrate that this amount of ethidium bromide efficiently blocked DNA-protein interactions, the GST-protein VII agarose beads were incubated with a 32P-labeled, 237-bp DNA fragment in the presence or absence of 100 μg of ethidium bromide per ml. As expected, GST-protein VII was able to bind the 32P-labeled DNA fragment in vitro (Fig. 4C). Importantly, ethidium bromide addition resulted in nearly complete inhibition of binding. These data demonstrate that the observed interaction between protein VII and E1A shown in Fig. 4A was not mediated by a DNA bridge.
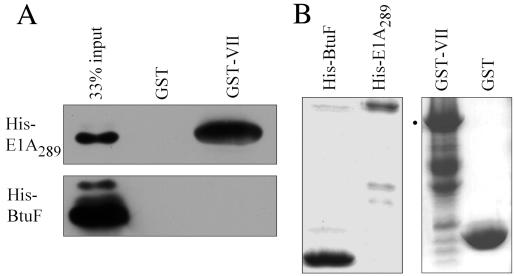
To test if the E1A-protein VII interaction is direct, GST-protein VII and His-tagged E1A289 were first purified from bacteria and shown to be of the appropriate size (Fig. 5B). They were then used in a pulldown assay, and the bound products were analyzed by SDS-PAGE (Fig. 5A). Strong association between GST-protein VII and E1A289 was observed, whereas there was no binding of E1A to GST alone. Also, there was no association between GST-protein VII and a control His-tagged protein, BtuF. These data demonstrate specific, direct in vitro binding of E1A289 to protein VII and nicely parallel the results obtained using the Xenopus system.
FIG. 5.
The E1A-protein VII interaction is direct. GST-protein VII pulldown assays of His-tagged E1A289 or His-tagged BtuF were performed as described in Materials and Methods. (A) The reaction products were washed and analyzed by SDS-PAGE and visualized by Western blot analysis with an anti-His antibody. 33% input indicates that 33% of the protein added to the binding-reaction mixtures. (B) Aliquots of input His-BtuF, His-E1A, GST-protein VII, and GST were analyzed by SDS-PAGE and stained with Coomassie blue. The dot indicates the position of full-length GST-protein VII. Minor breakdown products of GST-protein VII were also observed.
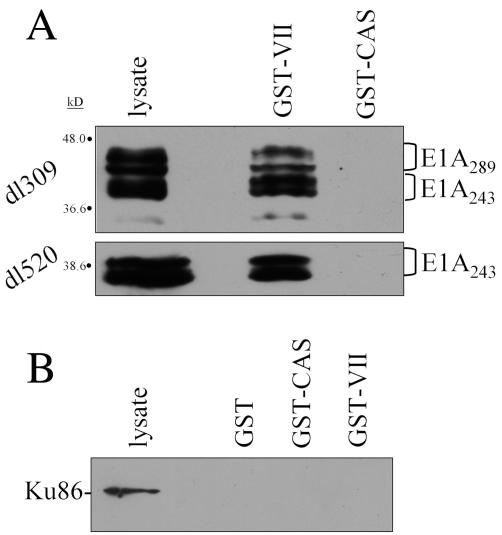
To examine the interaction between protein VII and native E1A proteins, extracts were produced from adenovirus-infected HeLa cells and used in pulldown experiments with GST-protein VII. Cells were infected with either phenotypically wild-type dl309, which produces E1A289 and E1A243 proteins, or dl520, which synthesizes only E1A243. Typically, E1A289 is primarily responsible for activation of viral transcription due to the presence of an internal 46-amino-acid region, CR3, which is not part of E1A243 (22). However, E1A243 is able to activate viral transcription potently in some assays (21, 22, 41, 47). As shown in Fig. 6A, native E1A289 and E1A243 proteins were efficiently retained by GST-protein VII and there was no binding to control protein GST-CAS-YXXP (9). As a further control, pulldown samples were analyzed for the presence of Ku86, an endogenous cellular DNA-binding protein, and no association with GST-protein VII was observed (Fig. 6B). These data demonstrate that protein VII can associate with both major forms of E1A and that the CR3 region of E1A is not required for the association.
FIG. 6.
Interaction of protein VII with native E1A289 and E1A243. HeLa cells were infected for 12 h with the indicated adenoviruses. Extracts were prepared and assayed by GST protein-VII pulldown as described in Materials and Methods. Following SDS-PAGE, Western blot analysis was performed with anti-E1A monoclonal antibody M73 (A) or anti-Ku86 monoclonal antibody (B). GST-CAS, GST-CAS-YXXP (9).
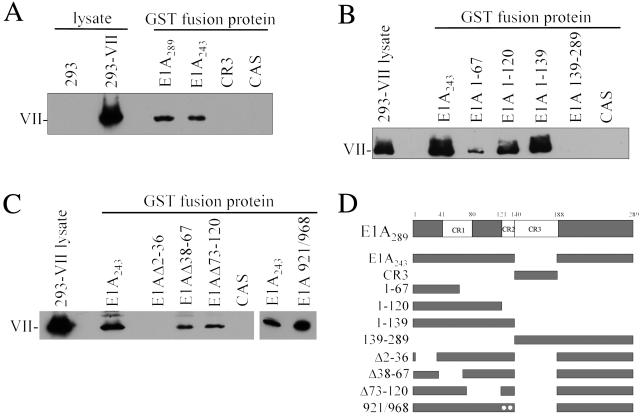
To examine the association of E1A with protein VII produced by human cells, 293 cells were transfected with a FLAG-tagged protein VII gene driven by the cytomegalovirus promoter-enhancer. Transfected cells were harvested, and extracts were prepared for use in pulldown assays with wild-type and mutant forms of GST-E1A. As shown in Figure 7A, protein VII synthesized in 293 cells associated with both E1A289 and E1A243 but not with the CR3 region of E1A (amino acids 140 to 188). The result with CR3 was expected since E1A243, which lacks CR3, binds efficiently to protein VII (compare Fig. 7A with Fig. 6A). Also, these results indicate that although 293 cells produce endogenous E1A protein, this level is not high enough to influence the observed binding of protein VII to GST-E1A proteins in the pulldown assay. To determine the region of E1A that is responsible for binding to protein VII, a series of mutant GST-E1A proteins were produced. The results of these experiments are shown in Fig. 7B and C, and the mutants are presented schematically in Fig. 7D. GST-E1A139-289, which contains CR3 plus the entire C-terminal domain of E1A, failed to bind protein VII, demonstrating that binding sites for protein VII reside solely within the N-terminal 138 amino acids (Fig. 7B). Interestingly, two regions within the N-terminal 138 amino acids proved to be required for efficient binding. First, a limited amount of binding was detected with GST-E1A1-67, suggesting a possible role for this region (Fig. 7B). Consistent with this, deletion of amino acids 2 to 36 in the context of the full-length E1A243 protein completely abolished binding (Fig. 7C). Using equivalent amounts of the different mutant GST-E1A proteins, binding was easily detectable with GST-E1A1-120 and GST-E1A1-139. Since binding of either E1A1-120 or E1A1-139 was much greater than that of E1A1-67, a role for amino acids 68 to 139 is indicated. Mutant GST-E1A921/968 showed binding equivalent to the wild-type E1A protein. This mutant contains two amino acid substitutions within CR2 that abolish binding to the retinoblastoma (Rb) protein (38). Therefore, the Rb-binding domain of E1A is not required for association with protein VII. Deletion of residues 38 to 67 or 73 to 120 reproducibly resulted in somewhat diminished binding, but these proteins still bound efficiently (Fig. 7C). Taken together, these results indicate at least two regions within the N-terminal 139 amino acids that play a role in binding to protein VII. One region lies at the extreme N -terminus, since deletion of residues 2 to 36 abolishes binding and residues 1 to 67 support detectable, albeit weak, binding. The other region lies within amino acids 68 to 139, because addition of these residues to the fragment from residues 1 to 67 greatly enhance binding. It is possible that amino acids throughout the N-terminal 139 amino acids contribute to a structure required for efficient binding to protein VII.
FIG. 7.
E1A N-terminal domain binding to protein VII. (A to C) Human 293 cells were transfected with a cytomagalovirus-driven protein VII gene. Extracts were prepared and assayed by GST pulldown with the indicated GST-E1A mutants as described in Materials and Methods. Equivalent amounts of each GST-E1A protein were used, as judged by SDS-PAGE and staining with Coomassie blue (data not shown). The reaction products were separated by SDS-PAGE and analyzed by Western blotting using an affinity-purified anti-protein VII polyclonal antibody. For loading controls, extracts from untransfected or protein VII-transfected 293 cells were also included. These are indicated as 293 lysate or 293-VII lysate, respectively. For each panel, the amount of these lysates represented 10% of the amount added to the binding-reaction mixtures. (D) Schematic representation of E1A mutants. The GST moiety is not shown. The two dots in the 921/968 mutant represent amino acid substitutions at adenovirus nucleotides 921 and 968, which render E1A protein unable to associate with Rb protein.
DISCUSSION
The structure and fate of adenovirus genomic DNA during infection remains largely unknown. Questions persist as to the function of the viral chromatin and the way in which this complex relates to the cellular transcriptional and DNA replication machinery. Whereas the composition of the virion nucleoprotein core is known, its structure has not been elucidated. More mysterious are the structure and composition of viral chromatin in the infected cell nucleus. The identification of viral and cellular proteins that associate with adenovirus DNA in the nucleus and information regarding their function are far from complete.
The ChIP data presented here indicate that protein VII remains associated with the DNA during the early phase of infection, supporting a role for this protein during the early nuclear steps of virus replication. We also can visualize protein VII at early times in discrete nuclear “dots” whose number corresponds to incoming infectious particles (Y. Xue, J. S. Johnson, and D. A. Engel, unpublished data), further supporting an ongoing role for this protein during infection. It is known that protein VII condenses adenovirus DNA in the intact virion and that the genome is transcriptionally silent during infection prior to E1A protein expression. Our results therefore suggest that protein VII mediates transcriptional repression in the nucleus prior to E1A expression. Furthermore, they suggest that the mechanism of transcriptional activation by E1A may involve reversal of this repression in addition to the well-documented activities of E1A as a transcriptional activator.
Two lines of evidence support these conclusions. First, microinjection into Xenopus oocytes of in vitro-synthesized mRNA encoding protein VII resulted in efficient association of protein VII with lampbrush chromosomes and dramatic DNA condensation and transcriptional silencing. These results demonstrate dramatically that protein VII is able to bind, condense, and repress genomic DNA in a nuclear environment and suggest that it behaves similarly in the context of adenovirus genomic DNA in the infected cell nucleus. It is worth remembering that Xenopus lampbrush chromatin is constituted with histones. It is not known whether protein VII displaced cellular histones in our experiments or if it entered unoccupied sites on the DNA or both. Due to the high state of transcriptional activity of lampbrush chromosomes, the density of nucleosomes is thought to be relatively low, and it is possible that this configuration allows access to ectopically expressed protein VII (11). It has been suggested from indirect evidence that in adenovirus the DNA converts to a cellular chromatin-like structure during infection, implying that histones displace protein VII from the incoming viral DNA or co-occupy the DNA with protein VII (16, 56, 57). Our ChIP data clearly indicate that complete displacement of protein VII does not occur, because protein VII was associated with viral DNA 4 h postinfection. Considering the ChIP data and the stable association of protein VII with Xenopus chromosomes despite the prior presence of nucleosomal histones, we favor a model in which the protein VII-adenovirus DNA association remains largely intact during infection. Whether cellular histones join the viral chromatin in addition to this is not yet known.
Second, we have observed direct binding of protein VII to E1A in vitro and protein VII-dependent recruitment of E1A to chromatin in vivo. In our pulldown experiments we used native virally expressed E1A and human cell-produced protein VII. These data strongly suggest that a protein VII-E1A interaction is important as an early step in E1A-induced transcriptional activation or relief of transcriptional repression. We also attempted to coimmunoprecipitate E1A and protein VII from infected cells, but these experiments did not give positive results. Several reasons for this are possible, including low levels of the two interacting proteins and low-affinity interactions, which could be important mechanistically. Several mechanistic possibilities exist for the role of protein VII in this relief of transcriptional repression and transcriptional activation. Recruitment of E1A by protein VII might stabilize the interactions between E1A and specific components of the transcriptional machinery, such as ATF-2, that have been implicated in E1A-dependent activation (22). In addition, protein VII binding to E1A may in turn result in recruitment of chromatin-remodeling factors that can alter the structure of the protein VII-DNA complex to allow accessibility to the transcriptional machinery. These could include various types of mammalian complexes including SWI/SNF, histone acetyltransferase complexes, mediator, p400, and TRRAP complexes, each of which has been implicated in aspects of E1A-regulated transcription (3, 7, 12, 24, 40, 44). Our finding that regions of the N-terminal 139 amino acids of E1A are involved in binding to protein VII support this possibility. Our in vitro experiments demonstrated a direct E1A-protein VII interaction. However, in vivo it is possible that this interaction may be mediated in part by cellular E1A-binding proteins or that the binding of these proteins to E1A might affect the interaction with protein VII. Involvement of N-terminal regions of E1A in early-gene transcriptional activation has been reported (21, 47, 62), and the N-terminal 139 amino acids include CR1, CR2, and the “N-terminal” domain, which contain binding sites for transcriptional regulators p300/CBP, P/CAF, GCN5, TRRAP, p400, and Rb (23). It is interesting to speculate that complexes form on adenovirus DNA that include one or more of these proteins along with E1A and protein VII.
Alternatively, binding of E1A to protein VII might result in the release of protein VII from the DNA, leading to chromatin remodeling and relief of transcriptional repression. Our ChIP experiments do not distinguish between full and partial occupancy of protein VII on viral DNA in the nucleus, and so a displacement model is possible. In our microinjection experiments, we observed no evidence for displacement of protein VII from lampbrush chromosomes on expression of E1A. However, given the presumed structural dissimilarities between these and adenovirus chromatin, we cannot rule out E1A-mediated displacement of protein VII in the context of adenovirus chromatin. Previously, UV-cross-linking experiments suggested that protein VII remains associated with adenovirus DNA throughout the infectious cycle (14). Those data and our direct demonstration of a protein VII-DNA interaction in the nucleus of infected cells strongly suggest that complete displacement of protein VII does not take place. This, in combination with the fact that protein VII and E1A can interact, leads us to favor a model in which protein VII plays a continuing role in early-phase transcriptional regulation.
Acknowledgments
This work was supported by Public Health Service grant CA60675 from the National Cancer Institute to D.A.E and by grant MCB-9513589 from the National Science Foundation to A.L.B.
REFERENCES
- 1.Amin, M., A. Mirza, and J. Weber. 1977. Genetic analysis of adenovirus type 2. VII. Cleavage-modified affinity for DNA of internal virion proteins. Virology 80:83-97. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, C. W., M. E. Young, and S. J. Flint. 1989. Characterization of the adenovirus 2 virion protein, mu. Virology 172:506-512. [DOI] [PubMed] [Google Scholar]
- 3.Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374:81-84. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Berk, A. J., T. G. Boyer, A. N. Kapanidis, R. H. Ebright, N. N. Kobayashi, P. J. Horn, S. M. Sullivan, R. Koop, M. A. Surby, and S. J. Triezenberg. 1998. Mechanisms of viral activators. Cold Spring Harbor Symp. Quant. Biol. 63:243-252. [DOI] [PubMed] [Google Scholar]
- 6.Black, B. C., and M. S. Center. 1979. DNA-binding properties of the major core protein of adenovirus 2. Nucleic Acids Res. 6:2339-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, T. G., M. E. Martin, E. Lees, R. P. Ricciardi, and A. J. Berk. 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399:276-279. [DOI] [PubMed] [Google Scholar]
- 8.Brown, D. T., M. Westphal, B. T. Burlingham, U. Winterhoff, and W. Doerfler. 1975. Structure and composition of the adenovirus type 2 core. J. Virol. 16:366-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnham, M. R., M. T. Harte, A. Richardson, J. T. Parsons, and A. H. Bouton. 1996. The identification of p130cas-binding proteins and their role in cellular transformation. Oncogene 12:2467-2472. [PubMed] [Google Scholar]
- 10.Cadieux, N., C. Bradbeer, E. Reeger-Schneider, W. Koster, A. K. Mohanty, M. C. Wiener, and R. J. Kadner. 2002. Identification of the periplasmic cobalamin-binding protein BtuF of Escherichia coli. J. Bacteriol. 184:706-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callan, H. G. 1986. Lampbrush chromosomes. Springer-Verlag KG, Berlin, Germany.
- 12.Chakravarti, D., V. Ogryzko, H. Y. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96:393-403. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee, P. K., M. Bruner, S. J. Flint, and M. L. Harter. 1988. DNA-binding properties of an adenovirus 289R E1A protein. EMBO J. 7:835-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee, P. K., M. E. Vayda, and S. J. Flint. 1986. Adenoviral protein VII packages intracellular viral DNA throughout the early phase of infection. EMBO J. 5:1633-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corden, J., H. M. Engelking, and G. D. Pearson. 1976. Chromatin-like organization of the adenovirus chromosome. Proc. Natl. Acad. Sci. USA 73:401-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniell, E., D. E. Groff, and M. J. Fedor. 1981. Adenovirus chromatin structure at different stages of infection. Mol. Cell. Biol. 1:1094-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dery, C. V., M. Toth, M. Brown, J. Horvath, S. Allaire, and J. M. Weber. 1985. The structure of adenovirus chromatin in infected cells. J. Gen. Virol. 66:2671-2684. [DOI] [PubMed] [Google Scholar]
- 18.Ennever, J. F., S. M. Love, and J. A. Harpst. 1985. Ionic effects on the structure of nucleoprotein cores from adenovirus. Biochim. Biophys. Acta 826:67-79. [DOI] [PubMed] [Google Scholar]
- 19.Everitt, E., L. Lutter, and L. Philipson. 1975. Structural proteins of adenoviruses. XII. Location and neighbor relationship among proteins of adenovirion type 2 as revealed by enzymatic iodination, immunoprecipitation and chemical cross-linking. Virology 67:197-208. [DOI] [PubMed] [Google Scholar]
- 20.Everitt, E., B. Sundquist, U. Pettersson, and L. Philipson. 1973. Structural proteins of adenoviruses. X. Isolation and topography of low molecular weight antigens from the virion of adenovirus type 2. Virology 52:130-147. [DOI] [PubMed] [Google Scholar]
- 21.Fax, P., K. S. Lipinski, H. Esche, and D. Brockmann. 2000. cAMP-independent activation of the adenovirus type 12 E2 promoter correlates with the recruitment of CREB-1/ATF-1, E1A(12S), and CBP to the E2-CRE. J. Biol. Chem. 275:8911-8920. [DOI] [PubMed] [Google Scholar]
- 22.Flint, J., and T. Shenk. 1997. Viral transactivating proteins. Annu. Rev. Genet. 31:177-212. [DOI] [PubMed] [Google Scholar]
- 23.Frisch, S. M., and J. S. Mymryk. 2002. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell Biol. 3:441-452. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 25.Gall, J. G. 1998. Spread preparations of Xenopus germinal vesicle contents, p. 3.1-3.3. In D. Spector, D. Goldman, and L. Leinwand (ed.), Cell biology: a laboratory manual, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 26.Gall, J. G., M. Bellini, Z. Wu, and C. Murphy. 1999. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 10:4385-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gall, J. G., C. Murphy, H. G. Callan, and Z. Wu. 1991. Lampbrush chromosomes. Methods Cell Biol. 36:149-166. [PubMed] [Google Scholar]
- 28.Greber, U. F., M. Suomalainen, R. P. Stidwill, K. Boucke, M. W. Ebersold, and A. Helenius. 1997. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 16:5998-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurdon, J. B. 1976. Injected nuclei in frog oocytes: fate, enlargement, and chromatin dispersal. J. Embryol. Exp. Morphol. 36:523-540. [PubMed] [Google Scholar]
- 30.Haley, K. P., J. Overhauser, L. E. Babiss, H. S. Ginsberg, and N. C. Jones. 1984. Transformation properties of type 5 adenovirus mutants that differentially express the E1A gene products. Proc. Natl. Acad. Sci. USA 81:5734-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harlow, E., B. R. Franza, Jr., and C. Schley. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55:533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, p. 471-510. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Hosokawa, K., and M. T. Sung. 1976. Isolation and characterization of an extremely basic protein from adenovirus type 5. J. Virol. 17:924-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, N., and T. Shenk. 1979. An adenovirus type 5 early gene function regulates expression of other early viral genes. Proc. Natl. Acad. Sci. USA 76:3665-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 36.Keller, M., T. Tagawa, M. Preuss, and A. D. Miller. 2002. Biophysical characterization of the DNA binding and condensing properties of adenoviral core peptide mu. Biochemistry 41:652-659. [DOI] [PubMed] [Google Scholar]
- 37.Korn, R., and M. S. Horwitz. 1986. Adenovirus DNA synthesis in vitro is inhibited by the virus-coded major core protein. Virology 150:342-351. [DOI] [PubMed] [Google Scholar]
- 38.Kraus, V. B., J. A. Inostroza, K. Yeung, D. Reinberg, and J. R. Nevins. 1994. Interaction of the Dr1 inhibitory factor with the TATA binding protein is disrupted by adenovirus E1A. Proc. Natl. Acad. Sci. USA 91:6279-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang, S. E., and P. Hearing. 2003. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22:2836-2841. [DOI] [PubMed] [Google Scholar]
- 41.Leff, T., R. Elkaim, C. R. Goding, P. Jalinot, P. Sassone-Corsi, M. Perricaudet, C. Kedinger, and P. Chambon. 1984. Individual products of the adenovirus 12S and 13S EIa mRNAs stimulate viral EIIa and EIII expression at the transcriptional level. Proc. Natl. Acad. Sci. USA 81:4381-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maizel, J. V., Jr., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology 36:126-136. [DOI] [PubMed] [Google Scholar]
- 43.Matthews, D. A., and W. C. Russell. 1998. Adenovirus core protein V is delivered by the invading virus to the nucleus of the infected cell and later in infection is associated with nucleoli. J. Gen. Virol. 79:1671-1675. [DOI] [PubMed] [Google Scholar]
- 44.Miller, M. E., B. R. Cairns, R. S. Levinson, K. R. Yamamoto, D. A. Engel, and M. M. Smith. 1996. Adenovirus E1A specifically blocks SWI/SNF-dependent transcriptional activation. Mol. Cell. Biol. 16:5737-5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirza, M. A., and J. Weber. 1982. Structure of adenovirus chromatin. Biochim. Biophys. Acta 696:76-86. [DOI] [PubMed] [Google Scholar]
- 46.Murphy, D. J., S. Hardy, and D. A. Engel. 1999. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol. 19:2724-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mymryk, J. S., and S. T. Bayley. 1993. Multiple pathways for gene activation in rodent cells by the smaller adenovirus 5 E1A protein and their relevance to growth and transformation. J. Gen. Virol. 74:2131-2141. [DOI] [PubMed] [Google Scholar]
- 48.Nakanishi, Y., K. Maeda, M. Ohtsuki, K. Hosokawa, and S. Natori. 1986. In vitro transcription of a chromatin-like complex of major core protein VII and DNA of adenovirus serotype 2. Biochem. Biophys. Res. Commun. 136:86-93. [DOI] [PubMed] [Google Scholar]
- 49.Nermut, M. V., J. A. Harpst, and W. C. Russell. 1975. Electron microscopy of adenovirus cores. J. Gen. Virol. 28:49-58. [DOI] [PubMed] [Google Scholar]
- 50.Newcomb, W. W., J. W. Boring, and J. C. Brown. 1984. Ion etching of human adenovirus 2: structure of the core. J. Virol. 51:52-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osheim, Y. N., and A. L. Beyer. 1998. EM visualization of transcriptinally active genes after injection into Xenopus oocyte nuclei. Methods Cell Biol. 53:471-496. [DOI] [PubMed] [Google Scholar]
- 52.Prage, L., and U. Pettersson. 1971. Structural proteins of adenoviruses. VII. Purification and properties of an arginine-rich core protein from adenovirus type 2 and type 3. Virology 45:364-373. [DOI] [PubMed] [Google Scholar]
- 53.Rekosh, D. M., W. C. Russell, A. J. Bellet, and A. J. Robinson. 1977. Identification of a protein linked to the ends of adenovirus DNA. Cell 11:283-295. [DOI] [PubMed] [Google Scholar]
- 54.Russell, W. C., W. G. Laver, and P. J. Sanderson. 1968. Internal components of adenovirus. Nature 219:1127-1130. [DOI] [PubMed] [Google Scholar]
- 55.Russell, W. C., and B. Precious. 1982. Nucleic acid-binding properties of adenovirus structural polypeptides. J. Gen. Virol. 63:69-79. [DOI] [PubMed] [Google Scholar]
- 56.Sergeant, A., M. A. Tigges, and H. J. Raskas. 1979. Nucleosome-like structural subunits of intranuclear parental adenovirus type 2 DNA. J. Virol. 29:888-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tate, V. E., and L. Philipson. 1979. Parental adenovirus DNA accumulates in nucleosome-like structures in infected cells. Nucleic Acids Res. 6:2769-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vayda, M. E., and S. J. Flint. 1987. Isolation and characterization of adenovirus core nucleoprotein subunits. J. Virol. 61:3335-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vayda, M. E., K. Leong, and S. J. Flint. 1984. Transcription of adenovirus cores in vitro. Virology 139:152-163. [DOI] [PubMed] [Google Scholar]
- 60.Vayda, M. E., A. E. Rogers, and S. J. Flint. 1983. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 11:441-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber, J. M., G. Khittoo, and A. R. Bhatti. 1983. Adenovirus core proteins. Can. J. Microbiol. 29:235-241. [DOI] [PubMed] [Google Scholar]
- 62.Wong, H. K., and E. B. Ziff. 1994. Complementary functions of E1a conserved region 1 cooperate with conserved region 3 to activate adenovirus serotype 5 early promoters. J. Virol. 68:4910-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong, M. L., and M. T. Hsu. 1989. Linear adenovirus DNA is organized into supercoiled domains in virus particles. Nucleic Acids Res. 17:3535-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, W., J. A. Low, J. B. Christensen, and M. J. Imperiale. 2001. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 75:10446-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zu, K., M. L. Sikes, and A. L. Beyer. 1998. Separable roles in vivo for the two RNA binding domains of Drosophila A1-hnRNP homolog. RNA 4:1585-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]