Abstract
Endogenous small RNA pathways related to RNA interference (RNAi) play a well-documented role in protecting host genomes from the invasion of foreign nucleic acids. In C. elegans, the PIWI type Argonaute, PRG-1, through an association with 21U-RNAs, mediates a genome surveillance process by constantly scanning the genome for potentially deleterious invading elements. Upon recognition of foreign nucleic acids, PRG-1 initiates a cascade of cytoplasmic and nuclear events that results in heritable epigenetic silencing of these transcripts and their coding genomic loci. If the PRG-1/21U-RNA genome surveillance pathway has the capacity to target most of the C. elegans transcriptome, what mechanisms exist to protect endogenous transcripts from being silenced by this pathway? In this commentary, we discuss three recent publications that implicate the CSR-1 small RNA pathway in the heritable activation of germline transcripts, propose a model as to why not all epialleles behave similarly, and touch on the practical implications of these findings.
Keywords: argonautes, chromatin, CSR-1, epigenetics, piRNA, PRG-1, RNA activation, 22G-RNA, transgenerational inheritance, WAGO-9/HRDE-1
Introduction
Epigenetic inheritance refers to the phenomenon in which gene expression programs can be altered and heritably maintained independent of changes to the underlying DNA sequence.1 The inheritance of epigenetic information is achieved through a variety of mechanisms, including non-coding RNAs,2 post-translational histone modification,1 and DNA methylation in some organisms.3 In recent years, non-coding small RNAs have been shown to be key regulators of epigenetic information, playing critical roles in controlling gene expression in response to external cues, by facilitating changes in genome architecture to alter transcriptional programs4 and protecting host genomes from foreign nucleic acids by silencing transposable elements5-8 and viruses.9-11 The effector component of all small RNA pathways is the Argonaute protein (AGO), which is guided to target transcripts through an association with small RNAs to mediate gene regulatory outcomes.12
In C. elegans, as well as other organisms including Drosophila and mice, the piRNA (Piwi interacting RNA) pathway has been shown to play an important role in the silencing of transposable elements, such as the Tc3 family5-7 (for a review, see refs. 13 and 14). Beyond transposable elements, the piRNA pathway has also been shown to be involved in the silencing of potentially deleterious foreign nucleic acids.15-19 The predominant Piwi-type Argonaute protein, PRG-1 (Piwi Related Gene-1), associates with at least 30 000 unique genomically encoded piRNAs5,7,20,21 (also known as 21U-RNAs). These 21U-RNAs lack fully complementary targets; however, when imperfect base pairing is permitted, PRG-1/21U-RNA complexes are capable of targeting nearly the entire C. elegans transcriptome.16,17 When these complexes encounter foreign transcripts, such as those of transgenes encoding the green fluorescent protein, gfp, the PRG-1/21U-RNA complex initiates the production of anti-sense small RNAs, called 22G-RNAs.15-19 These 22G-RNAs are synthesized by RNA-dependent RNA Polymerases (RdRPs) and are loaded into a set of worm-specific AGOs (WAGO-1/9/10).15-19 The WAGOs then evoke a cascade of cytoplasmic and nuclear gene silencing events, including changes in the local chromatin landscape of the transgene, such as the acquisition of the heterochromatic histone modification H3K9me3.15,18,19,22 Collectively, these observations give rise to an important question: if the piRNA pathway has the capacity to target and potently silence endogenous transcripts expressed in the germline, how do some sequences/transcripts evade the silencing effects of this pathway and maintain their appropriate germline expression profile?
From the initial characterization of the piRNA genome surveillance pathway, the CSR-1 (Chromosome Segregation and RNAi deficient-1) small RNA pathway was hypothesized to play an important role in protecting germline transcripts from piRNA-mediated silencing.15,17,18 Several key observations implicated CSR-1 as an attractive candidate for antagonizing the piRNA pathway. First, it was previously shown that CSR-1 associated with 22G-RNAs that are antisense to nearly all germline-expressed protein coding genes.23 Surprisingly, despite possessing in vitro endonuclease activity,24 loss of CSR-1 does not lead to an increase in the steady-state levels of its targeted transcripts (as measured by microarray experiments).23 Second, using several 21U-RNA reporter assays, PRG-1 was shown to be required for the initiation, but not maintenance, of piRNA-mediated silencing.17,18 Yet, if the 21U-RNA target sequence was flanked by endogenous CSR-1 target sequences, PRG-1 was then also required for the maintenance of the silent state,16 strongly suggesting that there existed an antagonistic relationship between the two pathways. While rare, small RNA pathways have previously been linked to promoting the expression of targeted loci in mammalian cell culture, through mechanisms that remain largely unclear.25 Together, these observations set the stage for three papers published in late 2013, Seth, et al.,26 Wedeles, et al.,27 and Conine, et al.,28 which demonstrated a role for the AGO, CSR-1, in promoting the expression of germline transcripts.
A C. elegans Argonaute Promotes Germline Transgene Expression
In C. elegans, transgenes have been successfully used as an experimental tool to study the response of an organism to foreign nucleic acids and to identify components implicated in the silencing of foreign DNA, such as those of small RNA-mediated gene silencing pathways.29,30 In one recent description of the piRNA genome surveillance pathway, Shirayama, et al.18 demonstrated that the PRG-1 small RNA pathway initiated the stable epigenetic silencing of single copy transgenes in a process now termed RNAe (RNA epigenetics). RNAe is heritable in a dominant fashion.18 That is, a silent allele, when introduced into a strain possessing an expressed or active allele, resulted in the silencing of the active allele (Fig. 1A). Both Seth, et al.26 and Wedeles, et al.27 exploited aspects of this transgene silencing system to explore the role of CSR-1 in protecting germline transcripts from piRNA-mediated silencing.
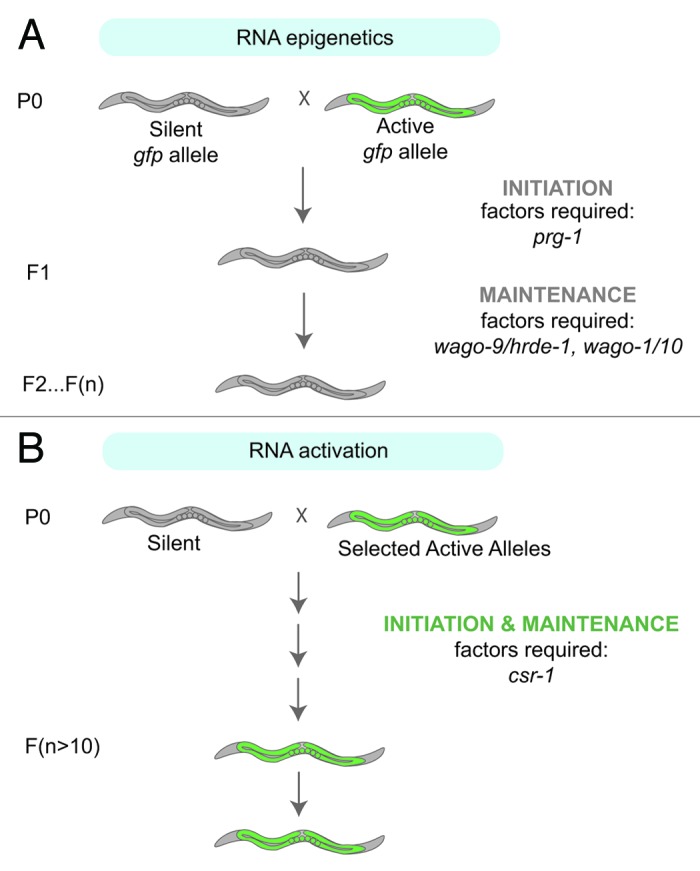
Figure 1. (A) Schematic illustrating RNA epigenetics (RNAe). When a silent allele, such as a gfp transgene, is introduced into a strain possessing an active allele that shares the same DNA sequence, the active allele becomes silenced in a dominant and heritable manner. The initiation of silencing is dependent on the PRG-1/21U-RNA pathway, whereas the maintenance of the silent state requires the WAGO-9/HRDE-1, WAGO-1, WAGO-10 22G-RNA pathway in addition to a number of chromatin factors. (B) Schematic illustrating RNA activation (RNAa). In some rare instances, gfp transgenes have escaped silencing and have been stably licensed as “self.” This trans-activation phenomenon requires the activity of the CSR-1.
Shirayama, et al.18 observed that, unexpectedly, some rare transgenes are able to maintain expression even when exposed to silent alleles. Seth, et al.26 expanded on this intriguing observation by crossing the stably expressed alleles into strains possessing RNAe alleles, and observing that stably expressed alleles have the capacity to license the silenced RNAe transgenes, in a trans-activating process they termed RNAa (RNA activation) (Fig. 1B). Why these transgenes, now referred to as RNAa alleles, are capable of activating RNAe alleles is not entirely clear; however, Seth, et al.26 demonstrated that the trans-activation process is dependent on csr-1 and is mediated by the accumulation of CSR-1-associated small RNAs. Surprisingly, even a 50% reduction of csr-1 limits the ability of trans-activation to occur, supporting a role for CSR-1 in this process.26 Importantly, Seth, et al.26 observed that not all silenced transgenes are activated with the same efficiency. Some silent transgenes required a prolonged exposure to an active transgene before they become fully licensed, while other transgenes failed to activate in trans at all.26 The mechanism behind this slow transfer of activation status is currently unknown, but suggests that there could be a gradual accumulation of small RNAs, small RNA complexes, or chromatin marks that shifts the balance of the epigenetic state from a silent allele to an active one (Fig. 2).
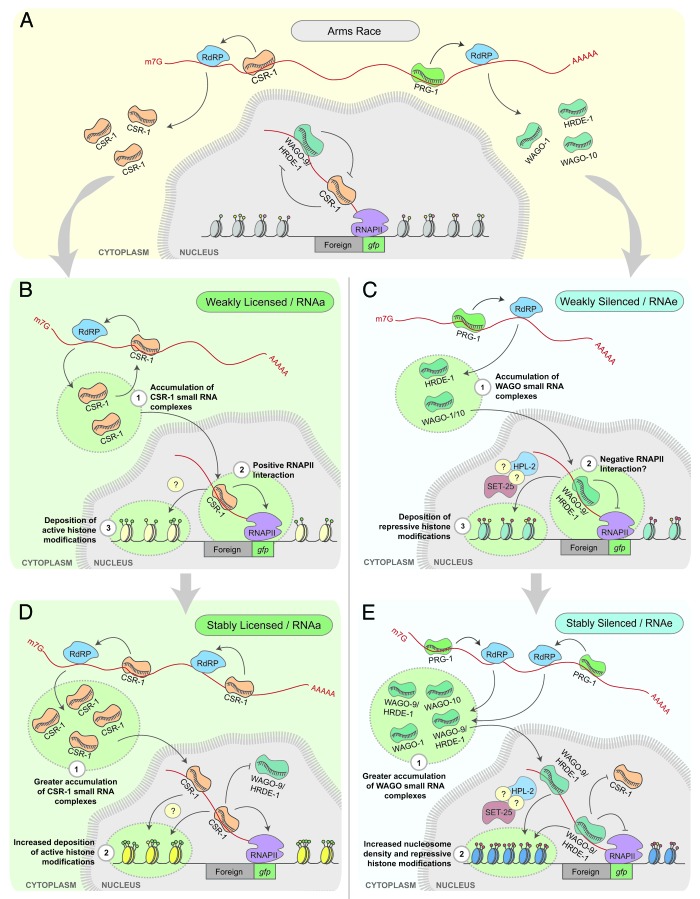
Figure 2. (A) The “arms race” between the PRG-1/WAGO silencing pathway and CSR-1 licensing pathway. (B) Stochastically, when exposed to an RNAa allele, or when CSR-1::λN is tethered to a transcript, CSR-1/small RNA complexes can generate a self-reinforcing loop through the production of additional small RNA complexes. In the nucleus, these complexes can guide the accumulation of active or euchromatin modifications and may positively interact with RNAPII to promote gene expression. (C) The greater accumulation of CSR-1 small RNA complexes and increased deposition of active histone modifications may allow some alleles to stably escape silencing from the PRG-1 pathway. (D) PRG-1 identifies foreign nucleic acids and initiates the production of 22G-RNAs that are loaded into WAGO-9/HRDE-1 complexes. In the nucleus, WAGO-9/HRDE-1 guides the deposition of repressive histone modifications. (E) Some silent alleles are unable to overcome the silencing initiated by PRG-1. We hypothesize this is a result of a greater accumulation of small RNAs in WAGO complexes and a more stable heterochromatin environment at the genomic loci encoding the transcript.
Wedeles, et al.27 provided mechanistic evidence for CSR-1 to act in a protective capacity. The authors developed an in vivo RNA tethering assay in which they expressed a gfp transcript followed by five Phage Lambda box b RNA hairpins (gfp::boxb) under the control of a strong germline promoter and 3 UTR. This transgene was integrated as a single copy into a region of the genome not normally targeted by CSR-1 and its 22G-small RNAs. In parallel, another transgene expressing CSR-1 under the control of its own promoter was introduced into the genome. This version of CSR-1 was also engineered to possess a Phage Lambda N anti-termination protein fragment (CSR-1::λN), which recognizes the box b hairpins, and enables CSR-1::λN to specifically bind the gfp::boxb transcript independent of an association with small RNAs.
Consistent with observations made by Shiryama, et al.18 and Seth, et al.,26 when an RNAe allele was crossed into the gfp::boxb strain, the gfp::boxb was silenced. However, if the same cross was performed in the presence of CSR-1::λN tethered to the gfp::boxb transcript, the silencing effect of the previously dominant silent allele was abolished and gfp::boxb remained active and expressed. Thus, CSR-1 was capable of protecting the transgene from silencing by the piRNA pathway. The expression of gfp::boxb was maintained at both the mRNA and pre-mRNA level, suggesting that CSR-1 regulates the expression of the transgene at the transcriptional level. This was further supported by data demonstrating that CSR-1 is enriched at the box b locus27 and its endogenous target gene loci by chromatin immunoprecipitation (ChIP),23 and by the observation that CSR-1 and RNA polymerase II (RNAPII) physically interact.27 It bears noting that, although the predominant activity of CSR-1 in this process may be in the nucleus, this does not preclude CSR-1 from acting in other ways to stabilize the box b transcript.
Using this same tethering assay, Wedeles, et al.27 recruited CSR-1::λN to a previously silenced gfp::boxb locus. Remarkably, after a few generations, the expression the gfp::boxb locus returned to levels comparable to those observed prior to silencing. The observation that the re-activation of the silent allele takes multiple generations supports the hypothesis that gfp small RNAs must accumulate and be loaded into CSR-1 complexes in sufficient quantities in order to re-license the transcript for expression.
Similar to the transactivation phenomenon observed by Seth, et al.,26 when the a fully licensed gfp::boxb transcript tethered by CSR-1::λN was propagated with a previously silenced gfp transgene (RNAe allele, lacking any tethering sites), the fully licensed gfp::boxb transgene was able to activate the silent allele after several generations. This observation supports the hypothesis that gfp small RNAs accumulate and are loaded into CSR-1 complexes for transactivation to occur. Furthermore, Wedeles, et al.27 showed that when CSR-1::λN and a silent allele are introduced to an active gfp::boxb transcript at the same time, the silencing of the active allele occurs to a similar extent as if CSR-1::λN was not present at all. Thus, it appears that even the protective capacity of CSR-1::λN requires the presence of gfp small RNAs preloaded into CSR-1 complexes, or the pre-recruitment of CSR-1::λN to the locus.
While both Seth, et al.26 and Wedeles, et al.27 independently provide a role for CSR-1 in the licensing or protection of germline transcripts, the processes that occur downstream of CSR-1 recruitment to a transcript remain elusive. Specifically, the molecular changes that CSR-1 elicits at its target transgene loci or on target transcripts to act antagonistically to the piRNA pathway are largely unclear. However, there are several possibilities (Fig. 2). For instance, CSR-1 has been shown to possess “slicer” endonuclease activity in vitro toward its target transcripts,24 and thus, may prevent PRG-1 recruitment by cleaving PRG-1-targeted transcripts before WAGO-associated 22G-RNAs can be produced.17 Alternatively, CSR-1 could simply compete with PRG-1 for access to target transcripts (independent of cleavage activity) and other available resources, such as RdRPs (Fig. 2A). Another possibility is that CSR-1 is capable of initiating and reinforcing a positive feedback loop. For example, CSR-1 could recruit specific histone-modifying enzymes that establish a chromatin environment that is more accessible to the transcriptional machinery, thus allowing for a more robust transcriptional program and reinforcing an epigenetic memory of germline transcription (Fig. 2B and C). The transgenerational inheritance of this process suggests that epigenetic information, possibly in the form of 22G-RNAs, is passed along in each generation. The transmission of small RNAs would allow for reinforcement in each generation by further recruitment of CSR-1 and downstream pathway factors. This would be consistent with the mechanisms by which a different small RNA silencing pathway in the worm, the nuclear RNAi (NRDE) pathway, establishes and maintains the heritable accumulation of the heterochromatic histone modification H3K9me3 at targeted genomic loci.31 Specifically, after exposure to RNAi, NRDE pathway small RNAs are detected in the progeny before the presence of H3K9me, suggesting that small RNAs are the primary inherited agent.31
Seth, et al.26 observed that not all tested transgenes could be trans-activated to the same extent or with the same efficiency. Why are some single copy transgenes silenced, while other genetically identical transgenes are not? The observation that RNAa alleles are not capable of activating every silent allele tested in trans supports the notion that an accumulation of gfp small RNAs in CSR-1 complexes is not sufficient to overcome silencing and indicates that there are additional components involved in the silencing/activating of genetic loci. In the instance where an RNAe allele fails to be reactivated, perhaps over time, a greater pool of 22G-RNAs have been loaded into WAGO complexes leading to a more potent/stable heterochromatin environment that becomes increasingly more resistant to change inflicted by the CSR-1 pathway (Fig. 2D and E). Highlighting the importance of this chromatin aspect of RNAe, several chromatin-related factors have been shown to be essential for the maintenance of RNAe, including factors such as the putative histone methyltransferases SET-25, SET-32, and the HP1 homolog, HPL-2.15 It remains unclear which chromatin factors act in the CSR-1 pathway to license germline transcripts.
Multiple C. elegans Argonautes Promote the Expression of Spermatogenesis Genes
While the aforementioned studies utilized the powerful context of transgenes to illustrate a role for CSR-1 in opposing the piRNA pathway to promote gene expression, Conine, et al.28 provided evidence that the CSR-1 pathway is involved in transmitting a heritable memory of paternal gene expression established in sperm by the ALG-3/4 small RNA pathway. Previous work had demonstrated that ALG-3/4 associate with 26G-RNAs to regulate the expression of transcripts important for male fertility and spermatogenesis.32 While ALG-3/4 were initially reported to be necessary for downregulating the expression of many of their targets32 using both mRNA-seq and SILAC experiments, Conine, et al.28 identified a subset of positively regulated ALG-3/4 targets. Interestingly, these positively regulated ALG-3/4 targets were also shown to be targets of the CSR-1 22G-small RNA pathway. Using a combination of immunofluorescence and ChIP experiments, ALG-3/4 were shown to be necessary for the recruitment of CSR-1 to its target gene genomic loci and the accumulation of elongating RNAPII and H3K4me2 at these loci. These chromatin-directed processes are required for establishing an epigenetic memory of paternal transcripts. For instance, in one experiment, homozygous alg-3/4 or csr-1 mutant males were crossed with heterozygous hermaphrodites, and the resultant progeny that did not receive paternal csr-1 were assayed for fertility and further propagated. Remarkably, while initial homozygous mutant fathers were fertile, over multiple generations there was a progressive loss of fertility, indicating that CSR-1 and ALG-3/4 are acting in each generation for the male germline to establish and transmit an epigenetic memory of paternal gene expression. Paternal epigenetic inheritance is likely due to the transmission of CSR-1 22G-small RNAs as the ALG-3/4 26G-RNAs were shown to be depleted in mature sperm.32
Practical Considerations for Germline Transgenes
Taken together, these three papers, Seth, et al.,26 Wedeles, et al.,27 and Conine, et al.28 demonstrate an important role for an endogenous small RNA pathway, the CSR-1/22G-RNA pathway, in positively regulating the expression of its target transcripts. In the context of licensing a foreign transcript as self, it remains a mystery as to how some transgenes initially evade piRNA-mediated silencing. Shiryama, et al.18 observed that depending on the size and site of insertion of an epitope tag (such as GFP in the above examples), different transgenes have differences in their ability to become silenced (Table 1). Specifically, they noted that smaller tags (such as Flag), introduced at or near the C terminus of the transgene, were more likely to be expressed and to escape silencing. Based on the observations outlined in the aforementioned papers, it may be useful for researchers wishing to obtain reliable germline expression of transgenes to include regulatory elements (such as the 3′ UTR) from “well-recognized” CSR-1/22G-RNA target genes (such as pie-1). By adding CSR-1 target gene regulatory sequences to a transgene, CSR-1/22G-RNA complexes that recognize the endogenous copy of the gene could be co-opted to the transgene as well. In turn, this could contribute to licensing the transgene, mostly by mistake, whereby small RNAs complementary to the foreign sequences are generated due to the recruitment of CSR-1, and possibly, an RdRP to a flanking “self” sequence. Together, these characteristics may increase the odds of a transgene initially escaping the piRNA-mediated genome surveillance and allowing them to be “accidentally” licensed to enable germline gene expression.
Table 1. Transgene features influence germline expression.
Concluding Remarks
Clearly, the ability to differentiate between self and non-self is a complex process, with multiple layers of regulation and many questions remaining. The ability to force an allele from an epigenetic silent state to an active state reflects an ongoing “arms race” between the PRG-1/WAGO silencing pathway and the CSR-1 licensing pathway (Fig. 2). Perhaps the indecisive nature of these pathways contributes to an evolutionary advantage, providing genomes with the ability to acquire novel and potentially beneficial information that can be drawn upon by subsequent generations.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to members of the Claycomb lab and Mello for helpful discussions on this topic.
References
- 1.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–8. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 2.Stuwe E, Tóth KF, Aravin AA. Small but sturdy: small RNAs in cellular memory and epigenetics. Genes Dev. 2014;28:423–31. doi: 10.1101/gad.236414.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 4.van Wolfswinkel JC, Ketting RF. The role of small non-coding RNAs in genome stability and chromatin organization. J Cell Sci. 2010;123:1825–39. doi: 10.1242/jcs.061713. [DOI] [PubMed] [Google Scholar]
- 5.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–7. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu W, Shirayama M, Conte D, Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–44. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147:1248–56. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanguy M, Miska EA. Antiviral RNA interference in animals: piecing together the evidence. Nat Struct Mol Biol. 2013;20:1239–41. doi: 10.1038/nsmb.2708. [DOI] [PubMed] [Google Scholar]
- 11.Sarkies P, Ashe A, Le Pen J, McKie MA, Miska EA. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res. 2013;23:1258–70. doi: 10.1101/gr.153296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–59. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 13.Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523–34. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 14.Kasper DM, Gardner KE, Reinke V. Homeland security in the C. elegans germ line: Insights into the biogenesis and function of piRNAs. Epigenetics. 2013;9 doi: 10.4161/epi.26647. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–8. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr., Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr., Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31:3422–30. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D, Jr., Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 22.Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–51. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–34. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007;26:5007–19. doi: 10.1038/sj.emboj.7601910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao AL, Slack FJ. RNA-mediated gene activation. Epigenetics. 2013;9 doi: 10.4161/epi.26942. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seth M, Shirayama M, Gu W, Ishidate T, Conte D, Jr., Mello CC. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell. 2013;27:656–63. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev Cell. 2013;27:664–71. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 28.Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D, Jr., Yates JR, 3rd, Mello CC. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell. 2013;155:1532–44. doi: 10.1016/j.cell.2013.11.032. [PMID.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–96. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robert VJ, Sijen T, van Wolfswinkel J, Plasterk RH. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–7. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44:157–64. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:3588–93. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]