Abstract
Schizophrenia (SCZ) and bipolar disorder (BPD) are polygenic disorders with many genes contributing to their etiologies. The aim of this investigation was to search for dysregulated molecular and cellular pathways for these disorders as well as psychosis. We conducted a blood-based microarray investigation in two independent samples with SCZ and BPD from San Diego (SCZ = 13, BPD = 9, control = 8) and Taiwan (SCZ = 11, BPD = 14, control = 16). Diagnostic groups were compared to controls, and subjects with a history of psychosis [PSYCH(+): San Diego (n = 6), Taiwan (n = 14)] were compared to subjects without such history [PSYCH(−): San Diego (n = 11), Taiwan (n = 14)]. Analyses of covariance comparing mean expression levels on a gene-by-gene basis were conducted to generate the top 100 significantly dysregulated gene lists for both samples by each diagnostic group. Gene lists were imported into Ingenuity Pathway Analysis (IPA) software. Results showed the ubiquitin proteasome pathway (UPS) was listed in the top ten canonical pathways for BPD and psychosis diagnostic groups across both samples with a considerably low likelihood of a chance occurrence (P = 0.001). No overlap in dysregulated genes populating these pathways was observed between the two independent samples. Findings provide preliminary evidence of UPS dysregulation in BPD and psychosis as well as support further investigation of the UPS and other molecular and cellular pathways for potential biomarkers for SCZ, BPD, and/or psychosis.
Keywords: ubiquitin, gene expression, biomarkers, micro-array, mRNA
INTRODUCTION
Schizophrenia (SCZ) and bipolar disorder (BPD) collectively affect 1.5% of the population and are considered two of the most severe and debilitating psychiatric disorders [Merikangas et al., 2007; Saha et al., 2008]. Unlike many heritable disorders such as cystic fibrosis, with known genetic etiology and blood, tissue or other confirmatory tests; complex disorders such as SCZ and BPD have proven difficult to categorize and currently have no objective diagnostic tools [Bearden et al., 2004].
While gene association and expression studies have implicated many genes in the etiology of SCZ and BPD, most results have not replicated or been supported by meta-analysis. Therefore, one approach for developing objective diagnostic tools for SCZ and BPD may be to identify differentially expressed gene networks or pathways that exhibit dysregulation between these disorders. Potential candidate genes can then be extracted from the top dysregulated networks and further investigated for their etiological significance. Identification of such biomarkers in SCZ and BPD patients as well as at-risk individual family members, have the potential to revolutionize the treatment of these disorders and improve patient prognosis. This would in turn have a profound effect on global public health by standardizing the process of primary and differential diagnosis, which presently involves considerable time, effort, and uncertainty.
Recent biomarker discovery efforts have begun to exploit the newly developing high-density and differential-display approaches from multiple domains such as transcriptomics, proteomics, and metabolomics to assist in elucidating dynamic molecular signatures for both of these disorders. Utilization of these approaches has predominately involved collection of post-mortem brain tissue, cerebrospinal fluid (CSF), and/or blood. Post-mortem brain tissue has been used to screen custom-made candidate gene cDNA arrays [Vawter et al., 2001; Mimmack et al., 2002] as well as carry out integrated transcriptomics, proteomics, and metabolomics approaches in both SCZ and BPD [Tkachev et al., 2003; Prabakaran et al., 2004]. More recently, CSF samples have been used in metabolomic and proteomic studies of prodromal [Huang et al., 2007] as well as first-onset [Huang et al., 2006] psychosis. Utilization of post-mortem brain tissue and CSF are reflective of pathophysiological changes within the brain in SCZ and BPD. However, the use of these tissues makes it difficult to differentiate whether gene expression changes are related to the disorder or treatment, and are far less accessible than blood. Thus, these compartments may not be the most advantageous for biomarker discovery and subsequent clinical application. Over the past 4 years, we and others [Vawter et al., 2004; Middleton et al., 2005; Zvara et al., 2005; Bowden et al., 2006; Sullivan et al., 2006] have documented the potential utility of blood-based transcriptomic profiling of mRNA abundances by microarray as a source of biomarkers for SCZ and BPD [Tsuang et al., 2005]. Although a blood-based approach shares many of the disadvantages of post-mortem and CSF approaches [Chana et al., 2008] and it remains unclear whether molecular signatures in blood accurately reflect those found in the brain, several investigators have reported that the circulating blood may act as a “sentinel tissue” [Liew, 1999], “neural probe” [Gladkevich et al., 2004], or “surrogate” [Sullivan et al., 2006] for underlying pathophysiology in psychiatric disorders. Furthermore, blood allows for the collection of larger sample sizes, better standardization of technical procedures, and the ability to profile human subjects in a relatively non-invasive manner [Tsuang et al., 2005].
The diversity in approaches to biomarker discovery in SCZ and BPD may reflect the sparse replication of individual biomarker findings from study to study, albeit other methodological factors (i.e., specimen collection, differential diagnosis, and platform design) should also be considered. Poor reproducibility of individually reported biomarkers from sample to sample should, in turn, guide our consideration of novel approaches to biomarker identification. One such approach is searching for dysregulated molecular and cellular pathways. This type of approach is advantageous in having the potential to reconcile poor biomarker reproducibility between investigations by identifying common biological pathways and pathophysiological outcomes populated by these genes. Thus, we proposed that if such a molecular/cellular pathway discovery process were to be useful it should be replicable in more than one sample. Here we report results of an exploratory pathway analysis using data from two independent (i.e., geographically, ascertainment method, ancestry) blood-based microarray investigations of SCZ, BPD, and psychosis in San Diego and Taiwan in which we found preliminary evidence for disruption of the ubiquitin proteasome system (UPS).
METHODS
San Diego Sample
Subjects were recruited from the University of California, San Diego (UCSD) Psychopharmacology Research Initiatives Center for Excellence (PRICE) participant network as well as from flyers and print advertisements. The Diagnostic Interview for Genetic Studies (DIGS) [Nurnberger et al., 1994] was used to diagnose SCZ or BPD.
Inclusion criteria required participants to: (1) be between the age of 18 and 55 years, inclusive; (2) have at least an eighth-grade education; (3) speak English as their first language; and (4) have no documented evidence of mental retardation. Subjects in the two patient groups (SCZ and BPD) were further required to have met criteria for their primary diagnosis (SCZ or BPD) for at least two years. Exclusion criteria were: (1) substance abuse or dependence in the past year; (2) neurologic problems (e.g., stroke, meningitis); (3) systemic medical illnesses (e.g., heart disease, diabetes); (4) history of head injury with documented loss of consciousness lasting longer than 10 min; (5) pregnancy; or (6) physical disabilities. Subjects in the CNT group were also excluded if they had a personal or family history of a psychotic disorder, BPD, major depressive disorder, or a cluster-A (schizotypal, schizoid, or paranoid) personality disorder.
Whole blood samples (10 ml) were collected in the morning after subjects fasted overnight with EDTA-coated collection tubes and immediately transferred to an RNase-free laboratory, where all subsequent procedures took place. Briefly, each blood sample was passed over a LeukoLOCK™ filter, which was flushed with PBS and then fully saturated with RNAlater® [Gonzales et al., 2005]. Each LeukoLOCK™ filter, containing bound, isolated, stabilized, and purified white blood cells, was sealed and stored in a sterile box at −20°C. Once peripheral blood mononuclear cell (PBMC) samples were acquired from all 34 subjects, the entire batch of samples was processed to isolate mRNA. Eluted mRNA samples were then stored at −20°C until transferred to the GeneChip™ Microarray Core (San Diego, CA) for quality assurance and microarray hybridization. Prior to hybridization, one BPD subject was excluded for low mRNA purity (260:280 nm absorbance <1.7), and one BPD and two CNT subjects were excluded for low RNA integrity (RIN <6.0) [Schroeder et al., 2006]. The remaining 30 (BPD: n = 9; SCZ: n = 13; CNT: n = 8) samples were then transcribed to cDNA and hybridized to GeneChip® Human Exon 1.0 ST Arrays (Affymetrix, Inc., Santa Clara, CA) per the “Whole Transcript (WT) Sense Target Labeling Assay” protocol [Affymetrix, 2006] using 1 μg of total RNA from each sample. All study procedures were approved by the Institutional Review Board at University of California, San Diego.
Taiwan Sample
Peripheral whole blood samples (10 ml) were obtained from healthy control subjects (n = 16) and individuals diagnosed according to DSM-IV criteria with SCZ (n = 11) or BPD (n = 14) as previously described [Tsuang et al., 2005]. All blood samples were collected into sterile violet-capped Vacutainer tubes (Becton, Dickinson and Company, Franklin Lakes, NJ) containing K3 EDTA, temporarily stored at 4°C, and processed within 6 hr of collection. Ascertainment and diagnosis of these subjects according to DSM-IV criteria, collection and preparation of blood samples, separation and lysis of PBMCs, extraction, purification, and hybridization of RNA, quantification of expression levels on cRNA microarrays, and quality-control procedures were all performed by standard methods, which are described in greater detail elsewhere [Tsuang et al., 2005].
Microarray Data Analyses
Three sets of comparisons of diagnostic groups in both samples were performed, as follows: (1) BPD versus CNT; (2) SCZ versus CNT; and (3) subjects with a history of psychosis [PSYCH(+), which included all SCZ subjects and psychotic BPD subjects (n = 6 San Diego, n = 14 Taiwan)] versus subjects with no history of psychosis [PSYCH(−), which included all CNT subjects and the remaining BPD subjects (n = 3 San Diego, n = 0 Taiwan)].
The principal analyses of these data were designed to detect biological pathways that were concordant in two independent samples of BPD and SCZ, as well as to potentially demonstrate the utility of a more robust biomarker discovery method for complex disorders. Raw cell intensity (.CEL) files were generated from gene chip image files using GCOS version 1.0 (Affymetrix, Inc.). Resulting .CEL files were imported into Partek Genomics Suite software (Partek, Inc, St. Louis, MO) using robust multichip average to analyze probe intensities and to determine differential gene expression between diagnostic and control groups. For both samples, analyses compared groups of interest on the mean expression level on a gene-by-gene basis through analyses of covariance (ANCOVAs). ANCOVA models for the San Diego sample included seven factors (diagnostic group, age, sex, ancestry, smoking status, and current use of antipsychotic and/or mood-stabilizers). Whereas, ancestral homogeneity and unavailable smoking status data resulted in inclusion of only five factors (diagnostic group, age, sex, and current use of antipsychotic and/or mood-stabilizers) for the Taiwan sample [for a more detail description of factors, see Tsuang et al., 2005]. Current use of antipsychotic medications was excluded as a covariate for analyses comparing SCZ versus CNT groups in both samples as a result of all SCZ subjects reporting current use, with the exception of one subject in the San Diego sample.
Due to the large number of statistical tests to be performed, the probability of committing type-I errors (i.e., finding false-positive results) in this study was greatly inflated. This threat of inflation of the type-I-error rate was addressed statistically, as we controlled family-wise error rates (FWERs) at 10% by estimating the false-discovery rate (FDR) for all comparisons (expressed as q-values, or the proportion of findings at a given significance level that are expected to be false discoveries). In general, this approach followed the guidelines provided by Mirnics et al. [2006], with the exception that we did not filter genes based on any fold-change criterion, since it is not empirically known (nor would it be expected) that any fold-change criterion is either universally applicable to all genes or biologically meaningful.
Verification of Microarray Findings by QRTPCR
Although the primary purpose of this study was to identify dysregulated biological pathways rather than individual transcripts, we did select a subset of dysregulated transcripts from the San Diego sample for verification of the exon array by quantitative reverse-transcriptase polymerase chain reaction (QRTPCR). The subset of transcripts were selected based on discordant differential dysregulation between SCZ and BPD groups, as well as the biological plausibility of the transcript’s involvement in one or more of the disorders evaluated. The levels of expression of the selected transcripts were quantified by QRTPCR in the same PBMC samples used for microarray analyses. Aliquots of each mRNA sample were reversed-transcribed into single-stranded cDNA using SuperScript III First Strand cDNA Synthesis Kit (Invitrogen Corporation, Carlsbad, CA) in 20 μl reactions. Next, 50 ng of cDNA was used as the template in 20 μl TaqMan reactions using ABI universal Master Mix, primers, and probes designed by Applied Biosystems (Foster City, CA). PCR amplification was performed on each sample in triplicate using the DNA Engine Opticon (MJ Research, Inc., Waltham, MA). Gene-specific standard curves for ARF1 (Assay ID Hs00796826_s1), BAT1 (Assay ID Hs00366447_m1), and GDI2 (Assay ID Hs00357525_m1) were generated by purification of amplicon product through sodium citrate precipitation. Purified amplicons were quantified by spectrophotometry and converted to molar concentration using the molar extinction coefficient. Serial tenfold dilutions of known copies of amplicons were prepared and subjected to QRTPCR. Cycle thresholds (CT) were set in the linear range of amplification plot identical to the standard curve and those of the test-subjects. CT values were plotted against the log of molar concentrations and used to determine pmol of mRNA per microgram of total RNA for each specific transcript. Statistical validation of the microarray data was determined by correlating QRTPCR copy number and the raw microarray data for each of the three selected genes.
Pathway Analysis
Following ANCOVAs, the top 100 genes in both samples for each diagnostic group that were nominally dysregulated at P <0.05 were imported into Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems®, Redwood City, CA) to associate dysregulated genes with their representative molecular/cellular pathways. The strategy of selecting the 100 most significant genes was based on previous microarray research [Subramanian et al., 2005] that utilized similar approaches to examine pathways in multiple independent data sets. Use of the top 100 also allowed for standardized analysis and interpretation of pathway data that was manageable across the two independent samples. Canonical pathways that were most significant to the uploaded gene lists were identified by querying the IPA library of canonical pathways. The significance of the association between the datasets and the canonical pathways was measured in two ways: (1) using the Fischer’s exact test we calculated the probability that the association between the gene list and the canonical pathway was explained by chance alone and (2) we calculated a ratio of the number of genes from the gene list that mapped to the pathway divided by the total number of molecules that exist in the canonical pathway. Pathways with a high ratio and a low P-value are indicative of potentially good candidates for further exploration. In addition, pathways occurring across both samples and in both disorders were subjected to a simple probability analysis [Pr = p1p2p3p4/(1 − q1q2q3q4)] to determine the likelihood of the occurrence resulting from chance, in which pn represented the probability that the pathway/function appeared in each of the four independent groups (SCZ and BPD in both samples) given the total possible pathways and qn represented 1 − pn.
RESULTS
Demographics of San Diego and Taiwan Samples
San Diego sample
Table I shows the three groups were evenly matched on age (F(2,26) = 0.240, P = 0.788), sex (χ2(2) = 0.477, P = 0.788), and ancestry (χ2(6) = 7.654, P = 0.264). However, the number of current smokers differed significantly between the groups (P = 0.003), with the CNT group having significantly fewer current smokers than either the SCZ group (χ2(1) = 11.748, P = 0.001) or the BPD group (χ2(1) = 6.296, P = 0.012). The rate of smoking did not differ between SCZ and BPD groups (χ2(1) = 1.119, P = 0.290). In addition, the groups also differed in their rates of use of antipsychotic (P <0.001) and mood-stabilizing medications (P = 0.001). The rate of use of antipsychotic medication was higher in the SCZ group than in either the BPD group (χ2(1) = 4.090, P = 0.043) or the CNT group (χ2(1) = 17.231, P <0.001), and the BPD group also used this class of medications at a higher rate than did the CNT group (χ2(1) = 6.296, P = 0.012). Mood stabilizers were used at a higher rate in the BPD group than in either the SCZ group (χ2(1) = 8.564; P = 0.003) or the CNT group (χ2(1) = 10.578, P = 0.001), but the SCZ and CNT groups did not differ significantly in their rates of use of these medications (χ2(1) = 1.360, P = 0.243).
TABLE I.
San Diego and Taiwan Samples: Descriptive Statistics by Diagnostic Group
| Variable | San Diego
|
Taiwan
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BPD (n = 9) | SCZ (n = 13) | CNT (n = 8) | Test statistic | P | BPD (n = 14) | SCZ (n = 11) | CNT (n = 16) | Test statistic | P | |
| Age: mean, years (s.d.) | 42 (8) | 44 (9) | 45 (7) | F(2,27) = 0.240 | 0.788 | 44 (13) | 31 (7) | 39 (11) | F(2,38) = 4.635 | 0.016 |
| Sex: female, n (%) | 2 (22) | 4 (31) | 3 (38) | χ2(2) = 0.477 | 0.788 | 11 (79) | 6 (55) | 7 (44) | χ2(2) = 3.829 | 0.147 |
| Ancestry: n (%) | χ2(6) = 7.654 | 0.264 | — | 1.000 | ||||||
| European | 7 (78) | 5 (39) | 5 (63) | — | — | — | ||||
| African | 1 (11) | 6 (46) | 1 (12) | — | — | — | ||||
| Hispanic | 0 (0) | 2 (15) | 1 (12) | — | — | — | ||||
| Asian | 1 (11) | 0 (0) | 1 (12) | — | — | — | ||||
| Han-Chinese | — | — | — | 14 (100) | 11 (100) | 16 (100) | ||||
| Current smoker: n (%) | 5 (56) | 10 (77) | 0 (0) | χ2(2) = 11.880 | 0.003 | — | — | — | — | — |
| Current medication use: n (%) | ||||||||||
| Antipsychotic | 5 (56) | 12 (92) | 0 (0) | χ2(2) = 17.191 | <0.001 | 11 (79) | 11 (100) | 0 (0) | χ2(2) = 31.521 | <0.001 |
| Mood stabilizer | 7 (78) | 2 (15) | 0 (0) | χ2(2) = 14.534 | 0.001 | 12 (86) | 5 (46) | 0 (0) | χ2(2) = 22.700 | <0.001 |
| History of psychosis: n (%) | 6 (67) | 13 (100) | 0 (0) | χ2(2) = 21.388 | <0.001 | 14 (100) | 11 (100) | 0 (0) | χ2(2) = 41.000 | <0.001 |
Taiwan sample
The sex distribution were similar for patients and controls (χ2(2) = 3.829, P = 0.147) but they differed in mean age (F(2,38) = 4.635, P = 0.016) (Table I). Similar to the San Diego sample, the Taiwan sample’s rate of antipsychotic medication (P <0.001) and mood-stabilizing medication (P <0.001) use significantly differed across the groups. Specifically, use of anti-psychotic medication was significantly higher in the BPD group (χ2(1) = 19.82, P <0.001) and SCZ group (χ2(1) = 26.94, P <0.001) compared to the CNT group. However, BPD and SCZ groups did not differ (χ2(1) = 2.650, P = 0.104). The rate of use of mood-stabilizer medication was higher in the BPD group than in either the SCZ group (χ2(1) = 4.58, P = 0.032) or the CNT group (χ2(1) = 22.82, P <0.001), and the SCZ group also used this class of medications at a higher rate than did the CNT group (χ2(1) = 8.89, P = 0.003).
Gene Expression Changes in Schizophrenia, Bipolar Disorder, and Psychosis
Of the 21,866 transcripts from the Exon 1.0 ST Array read into Partek for the San Diego sample, the number and percentage of nominally significant dysregulated (P <0.05) genes for SCZ, BPD, and PSYCH(+) compared to controls were 557 (2.5%), 917 (4.2%), and 2,155 (9.9%) respectively. For the Taiwan sample 54,675 transcripts from the U133A or U133Plus2.0 Array were read into Partek resulting in 9,639 (17.6%), 2,182 (4.0%), and 1,637 (3.0%) significantly dysregulated genes among BPD, SCZ, and PSYCH(+) groups, respectively. However, due to the large number of comparisons made and the consequent severity of the adjustment for multiple testing, the dysregulation of no individual gene remained significant after correction.
QRTPCR of dysregulated gene expression
Four subjects included in the microarray analyses reported above were not included in the QRTPCR analyses due to either inadequate mRNA quantities (n = 2) or quality (i.e., sample degradation over time; n = 2). Based on criteria described in the Methods Section, three genes identified as differentially expressed in the SCZ and BPD groups of the San Diego sample were selected for QRTPCR analysis in an attempt to verify provide an estimate of the validity of the microarray data.
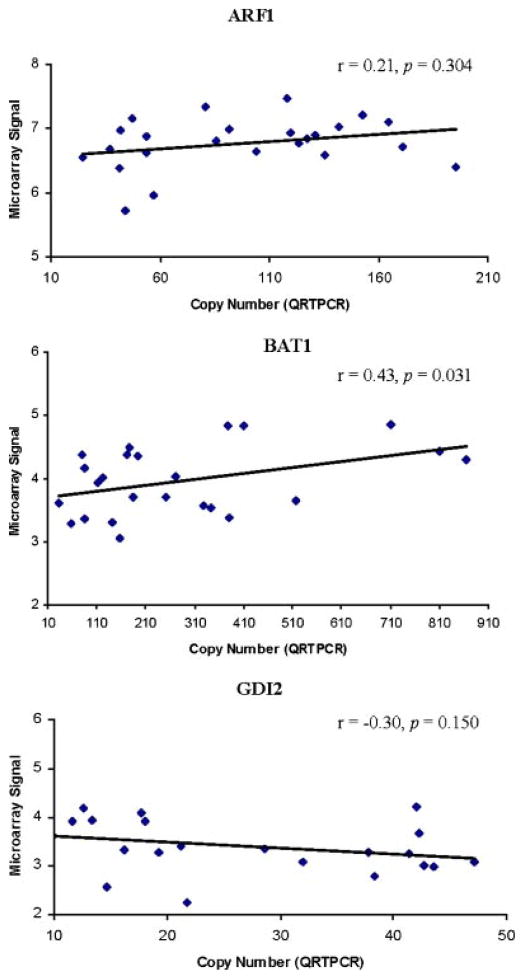
GDI2, BAT1, and ARF1 were selected based on their observed discordant differential expression (i.e., fold-change) between SCZ (GDI2: up-regulated 2.29-fold, P = 0.229; BAT1: down-regulated 2.58-fold, P = 0.001; ARF1: up-regulated 1.62-fold, P = 0.294) and BPD (GDI2: down-regulated 2.23-fold, P = 0.443; BAT1: up-regulated 1.45-fold, P = 0.188; ARF1: down-regulated 1.48-fold, P = 0.710). Pearson’s correlations between each gene’s copy number derived form QRTPCR and raw microarray data showed a significant correlation for BAT1 (r = 0.43, P = 0.031) although not GDI2 (r = −0.30, P = 0.150) or ARF1 (r = 0.21, P = 0.304) (Fig. 1). Thus, only one of the three genes selected for QRTPCR validation was concordant with raw microarray data.
FIG. 1.
Verification of microarray findings by QRTPCR.
Canonical pathways
Table II shows the top ten dysregulated canonical pathways by diagnosis and sample. Of note is that no canonical pathways managed to reach statistical significance in the Taiwan BPD group. However, the protein ubiquitination pathway was listed in the top ten canonical pathways for five of the six groups with two to five genes populating the pathway. The IPA analysis software includes 164 characterized canonical pathways. Therefore, with a total of four independent groups (BPD and SCZ for both samples) the probability of the protein ubiquitination pathway appearing in the top ten for three of the four groups was calculated to be 0.001. Thus, the likelihood these findings represent a chance outcome is considerably low.
TABLE II.
Top Ten Dysregulated Canonical Pathways by Diagnosis and Sample
| Diagnosis | Sample | Canonical pathway | P | Ratio |
|---|---|---|---|---|
| BPD | San Diego | Hypoxia signaling in the cardiovascular system | 0.040 | 2/27 |
| β-Alanine metabolism | 0.050 | 2/99 | ||
| Protein ubiquitination pathway | 0.056 | 3/205 | ||
| Regulation of actin-based motility by Rho | 0.058 | 2/92 | ||
| Integrin signaling | 0.062 | 3/196 | ||
| Actin cytoskeleton signaling | 0.069 | 3/221 | ||
| Parkinson’s signaling | 0.072 | 1/18 | ||
| Estrogen receptor signaling | 0.089 | 2/119 | ||
| Antigen presentation pathway | 0.016 | 1/39 | ||
| Tight junction signaling | 0.158 | 2/163 | ||
| Taiwan | Purine metabolism | 0.061 | 4/418 | |
| T cell receptor signaling | 0.066 | 2/105 | ||
| Inositol metabolism | 0.081 | 1/25 | ||
| Circadian rhythm signaling | 0.119 | 1/32 | ||
| One carbon pool by folate | 0.119 | 1/38 | ||
| B cell receptor signaling | 0.129 | 2/155 | ||
| Ephrin receptor signaling | 0.161 | 2/187 | ||
| ERK/MAPK signaling | 0.174 | 2/185 | ||
| Protein ubiquitination pathway | 0.183 | 2/205 | ||
| Integrin signaling | 0.196 | 2/196 | ||
| SCZ | San Diego | Glutamate metabolism | 0.020 | 2/78 |
| Hypoxia signaling in the cardiovascular system | 0.022 | 2/71 | ||
| D-glutamine and D-glutamate metabolism | 0.037 | 1/27 | ||
| PI3K/AKT signaling | 0.064 | 2/137 | ||
| Cardiac β-adrenergic signaling | 0.067 | 2/136 | ||
| cAMP-mediated signaling | 0.103 | 2/162 | ||
| Purine metabolism | 0.120 | 3/417 | ||
| Protein ubiquitination pathway | 0.131 | 2/205 | ||
| Docosahexaenoic acid (DHA) signaling | 0.131 | 1/45 | ||
| CD27 signaling in lymphocytes | 0.137 | 1/49 | ||
| Taiwan | Chondroitin sulfate biosynthesis | 0.016 | 2/61 | |
| Calcium-induced T lymphocyte apoptosis | 0.017 | 2/62 | ||
| CCR5 signaling in macrophages | 0.023 | 2/87 | ||
| CTLA4 signaling in cytotoxic T lymphocytes | 0.033 | 2/89 | ||
| SAPK/JNK signaling | 0.036 | 2/96 | ||
| T cell receptor signaling | 0.046 | 2/110 | ||
| CD28 signaling in T helper cells | 0.056 | 2/124 | ||
| Cytotoxic T lymphocyte-mediated apoptosis of target cells | 0.071 | 1/27 | ||
| IL-9 signaling | 0.098 | 1/37 | ||
| CXCR4 signaling | 0.099 | 2/168 | ||
| PSYCH | San Diego | Antigen presentation pathway | 0.013 | 2/39 |
| Integrin signaling | 0.014 | 4/196 | ||
| Tight junction signaling | 0.037 | 3/163 | ||
| β-Alanine metabolism | 0.053 | 2/99 | ||
| Protein ubiquitination pathway | 0.060 | 3/205 | ||
| Regulation of actin-based motility by Rho | 0.061 | 2/92 | ||
| Propanoate metabolism | 0.067 | 2/126 | ||
| Actin cytoskeleton signaling | 0.074 | 3/221 | ||
| Parkinson’s signaling | 0.074 | 1/18 | ||
| Fc epsilon RI signaling | 0.085 | 2/102 | ||
| Taiwan | T cell receptor signaling | 0.001 | 4/105 | |
| Protein ubiquitination pathway | 0.001 | 5/205 | ||
| α-Adrenergic signaling | 0.008 | 3/105 | ||
| 14-3-3-mediated signaling | 0.015 | 3/131 | ||
| Erythropoietin signaling | 0.033 | 2/77 | ||
| Estrogen receptor signaling | 0.069 | 2/119 | ||
| Inositol metabolism | 0.078 | 1/25 | ||
| Cardiac β-adrenergic signaling | 0.092 | 2/137 | ||
| One carbon pool by folate | 0.114 | 1/38 | ||
| Synaptic long term depression | 0.118 | 2/162 |
DISCUSSION
This study sought to identify dysregulated molecular and cellular pathways for SCZ, BPD, and psychosis among two independent samples utilizing a blood-based transcriptomic approach. Our principal finding was the consistent dysregulation of the UPS in BPD and psychosis across two samples ascertained in San Diego and Taiwan. Additionally, we identify several other dysregulated molecular and cellular pathways that may aid future biomarker discovery investigations. Although many of these dysregulated pathways are noteworthy, our discussion herein will focus on our principal finding of UPS dysregulation and its potential implications.
The UPS is a highly complex, temporally controlled, and tightly regulated process that plays major roles in a variety of basic cellular processes, specifically degradation of intracellular proteins [Ciechanover et al., 2000]. The UPS has been implicated in a variety of human diseases including several neurodegenerative diseases [Ciechanover and Brundin, 2003] such as Parkinson’s disease [Shimura et al., 2001] and Alzheimer’s disease [Lam et al., 2000]. Furthermore, several SCZ gene expression studies using post-mortem brain tissue have provided evidence of dysregulated expression of several ubiquitin-related genes [Vawter et al., 2001, 2002; Middleton et al., 2002; Konradi et al., 2004; Altar et al., 2005]. Among these studies, three [Vawter et al., 2001; Middleton et al., 2002; Altar et al., 2005] reported ubiquitin carboxyl-terminal esterase L1 (UCHL1) as significantly down-regulated in SCZ. Two studies reported proteasome (prosome, macropain) 26S subunit, ATPase, 6 (PSMC6) as significantly down-regulated in SCZ [Altar et al., 2005] as well as BPD [Konradi et al., 2004]. However, our results did not identify UCHL1 or PSMC6 as significantly dysregulated in either sample, across any of the diagnostic groups. In fact, none of the significantly dysregulated genes identified in the five studies mentioned above were identified in the present study as significantly dysregulated. However, the discordance between previous work and the results presented here is not unexpected given two key methodological differences. First, previous work identifying dysregulated ubiquitin-related genes in SCZ and BPD were conducted on post-mortem tissue from the prefrontal [Vawter et al., 2001, 2002; Middleton et al., 2002] and hippocampal [Konradi et al., 2004; Altar et al., 2005] regions of the human brain. In the current work, we utilized a blood-based approach. Second, previous studies utilized microarray platforms dissimilar to those in the current study. Therefore, it is likely that the discordance observed at the transcript-level across studies results from compartmental and/or array differences.
The concordance observed at the pathway (i.e., UPS) level across studies indicates potential advantages of searching for dysregulated molecular and cellular pathways rather than individual dysregulated genes. In fact, a broader focus on pathway-level dysfunction is further supported in that genetic functional redundancies within molecular and cellular pathways suggests disruption of any number or combination of genes could result in a common physiological outcome. In the case of the UPS pathway, Ingenuity lists 205 genes that contribute to this pathway. Thus, it is possible given the likely heterogeneity in gene expression between SCZ, BPD, and psychosis groups that genes contributing to the dysfunction of the UPS pathway may vary from one population and/or individual to another. We are not suggesting that transcript-level analysis of complex phenotypes such as those examined here are irrelevant, but that examination of broader molecular and cellular pathways may allow for better targeting of physiological processes that contribute to the onset and course of SCZ, BPD, and psychosis.
Although intriguing, our results should be interpreted in the context of several caveats in addition to those already mentioned. First, sample sizes for both samples were relatively small, which may have prohibited us from detecting effects that would have attained statistical significance in a larger sample. Second, pathway-level analysis was done using the largest knowledge base of biological networks (IPA, Redwood City, CA), a considerable strength. However, IPA is manually curated and relies on previously published findings on mammalian biology from the public literature. Thus, in some cases cellular component annotation can be missing or incomplete due to the lack of information in protein databases to which IPA is linked (e.g., UniProt) and ultimately may underestimate extracellular entities [Pospisil et al., 2006]. In addition, IPA is only one of several algorithms that could be used for pathway analysis of which no single algorithm has been globally accepted or is appropriate in every context [for detailed review, see Bansal et al., 2007]. Thus, replication of the current work using other pathway analysis algorithms may result in different conclusions. Third, we attempted to limit the influence of different subtypes of cells in blood by focusing on leukocytes. However, within this cell category several cell types (i.e., neutrophils, eosinophils, basophils, lymphocytes, monocytes, and macrophages) with varying roles in the blood exist and may influence gene expression profiles [Chana et al., 2008]. Thus, future blood-based studies may find it advantageous to isolate specific lymphocyte subtypes in an effort to achieve greater sensitivity in detecting gene expression changes. Fourth, only one of the three genes selected from the San Diego sample for QRTPCR validation was concordant with raw microarray data. Thus, it is possible that failure to provide complete independent replication was due to a lack of sensitivity of our chosen platform and further advocates for replication of our findings. Finally, in an effort to conserve power, potentially influential covariates (e.g., diet and exercise, dosage and duration of medication usage, length of illness, etc.) were not adjusted for in our analysis. Although we did adjust for current use of antipsychotic and mood-stabilizers, the potential effect of dosage and duration of these medications on gene expression in the current study are unknown. To our knowledge no study to date has estimated the effect of antipsychotic and/or mood-stabilizers on expression at the whole genome level in human leukocytes. However, recent work by Yao et al. [2008] failed to verify several putative biomarkers in the current literature among first-episode SCZ patients, suggesting the effect of these medications on gene expression are of potential concern. Thus, future development of methodologies for assessing medication effects such as treatment of generated lymphoblastiod cells from control patients with antipsychotics followed by assessment of gene expression as well as recruitment of first-episode and/or unmedicated patients may help to elucidate these effects and result in more verifiable biomarkers.
Despite these limitations, our findings support the use of blood-based transcriptomic molecular and cellular pathway exploration and analysis for future biomarker discovery efforts. In addition, our findings provide convergent evidence of UPS pathway dysregulation in BPD and psychosis and support further investigation of the UPS as well as other molecular and cellular pathways for potential biomarkers for SCZ, BPD, and/or psychosis.
Acknowledgments
This work was supported in part by the UCSD Center for AIDS Research Genomics Core, and National Institutes of Health grants P30MH062512 (Igor grant), R01MH079881, R25MH074508, R25MH081482, and R41MH079728 (I.P.E.), R01AG018386, R01AG022381, and R01AG022982 (W.S.K.), R01DA012846, R01DA018662, R01MH065562, R01MH071912, and R21MH075027 (M.T.T.), P50MH081755 (Eric Courchesne/S.J.G.), and a NARSAD Young Investigator Award (S.J.G.).
References
- Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, Young TA, Bullard J, Yokoe H, Webster MJ, Knable MB, Brockman JA. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. How to infer gene networks from expression profiles. Mol Syst Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden CE, Reus VI, Freimer NB. Why genetic investigation of psychiatric disorders is so difficult. Curr Opin Genet Dev. 2004;14:280–286. doi: 10.1016/j.gde.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Bowden NA, Weidenhofer J, Scott RJ, Schall U, Todd J, Michie PT, Tooney PA. Preliminary investigation of gene expression profiles in peripheral blood lymphocytes in schizophrenia. Schizophr Res. 2006;82:175–183. doi: 10.1016/j.schres.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Chana G, Glatt SJ, Everall IP, Tsuang MT. Blood and brain gene expression in major psychiatric disorders: A search for biomarkers. In: Turck CW, editor. Biomarkers for psychiatric disorders. New York: Springer; 2008. [Google Scholar]
- Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–446. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: Biological regulation via destruction. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Gladkevich A, Kauffman HF, Korf J. Lymphocytes as a neural probe: Potential for studying psychiatric disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2004;28:559–576. doi: 10.1016/j.pnpbp.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Gonzales J, Kemppainen J, Latham G, Goldrick M. Isolate RNA from white blood cells captured by a novel filter system. Ambion TechNotes. 2005;12:24–25. [Google Scholar]
- Huang JT, Leweke FM, Oxley D, Wang L, Harris N, Koethe D, Gerth CW, Nolden BM, Gross S, Schreiber D, Reed B, Bahn S. Disease biomarkers in cerebrospinal fluid of patients with first-onset psychosis. PLoS Med. 2006;3:e428. doi: 10.1371/journal.pmed.0030428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JT, Leweke FM, Tsang TM, Koethe D, Kranaster L, Gerth CW, Gross S, Schreiber D, Ruhrmann S, Schultze-Lutter F, Klosterkotter J, Holmes E, Bahn S. CSF metabolic and proteomic profiles in patients prodromal for psychosis. PLoS ONE. 2007;2:e756. doi: 10.1371/journal.pone.0000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C, Eaton M, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, Ramage R, Mayer RJ, Layfield R. Inhibition of the ubiquitin-proteasome system in Alzheimer’s disease. Proc Natl Acad Sci USA. 2000;97:9902–9906. doi: 10.1073/pnas.170173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CC. Method for the detection of gene transcripts in blood and uses thereof. 1999. [Google Scholar]
- Merikangas KR, Akiskal HS, Angst J, Greenberg PE, Hirschfeld RM, Petukhova M, Kessler RC. Lifetime and 12-month prevalence of bipolar spectrum disorder in the national comorbidity survey replication. Arch Gen Psychiatry. 2007;64:543–552. doi: 10.1001/archpsyc.64.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Pato CN, Gentile KL, McGann L, Brown AM, Trauzzi M, Diab H, Morley CP, Medeiros H, Macedo A, Azevedo MH, Pato MT. Gene expression analysis of peripheral blood leukocytes from discordant sib-pairs with schizophrenia and bipolar disorder reveals points of convergence between genetic and functional genomic approaches. Am J Med Genet Part B. 2005;136B:12–25. doi: 10.1002/ajmg.b.30171. [DOI] [PubMed] [Google Scholar]
- Mimmack ML, Ryan M, Baba H, Navarro-Ruiz J, Iritani S, Faull RL, McKenna PJ, Jones PB, Arai H, Starkey M, Emson PC, Bahn S. Gene expression analysis in schizophrenia: Reproducible up-regulation of several members of the apolipoprotein L family located in a high-susceptibility locus for schizophrenia on chromosome 22. Proc Natl Acad Sci USA. 2002;99:4680–4685. doi: 10.1073/pnas.032069099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T. Diagnostic interview for genetic studies, rationale, unique features, and training. NIMH genetics initiative. Arch Gen Psychiatry. 1994;51:849–859. doi: 10.1001/archpsyc.1994.03950110009002. discussion 863–864. [DOI] [PubMed] [Google Scholar]
- Pospisil P, Iyer LK, Adelstein SJ, Kassis AI. A combined approach to data mining of textual and structured data to identify cancer-related targets. BMC Bioinformatics. 2006;7:354. doi: 10.1186/1471-2105-7-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, Wayland M, Freeman T, Dudbridge F, Lilley KS, Karp NA, Hester S, Tkachev D, Mimmack ML, Yolken RH, Webster MJ, Torrey EF, Bahn S. Mitochondrial dysfunction in schizophrenia: Evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- Saha S, Chant D, McGrath J. Meta-analyses of the incidence and prevalence of schizophrenia: Conceptual and methodological issues. Int J Methods Psychiatr Res. 2008;17:55–61. doi: 10.1002/mpr.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trocken-bacher A, Schneider R, Mizuno Y, Kosik KS, Selkoe DJ. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: Implications for Parkinson’s disease. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet Part B. 2006;141B:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, Starkey M, Webster MJ, Yolken RH, Bahn S. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Nossova N, Yager T, Tsuang MM, Guo SC, Shyu KG, Glatt SJ, Liew CC. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: A preliminary report. Am J Med Genet Part B. 2005;133B:1–5. doi: 10.1002/ajmg.b.30161. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Barrett T, Cheadle C, Sokolov BP, Wood WH, III, Donovan DM, Webster M, Freed WJ, Becker KG. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull. 2001;55:641–650. doi: 10.1016/s0361-9230(01)00522-6. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, Freed WJ. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: A preliminary study. Schizophr Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Ferran E, Galke B, Cooper K, Bunney WE, Byerley W. Microarray screening of lymphocyte gene expression differences in a multiplex schizophrenia pedigree. Schizophr Res. 2004;67:41–52. doi: 10.1016/s0920-9964(03)00151-8. [DOI] [PubMed] [Google Scholar]
- Yao Y, Schroder J, Karlsson H. Verification of proposed peripheral biomarkers in mononuclear cells of individuals with schizophrenia. J Psychiatr Res. 2008;42:639–643. doi: 10.1016/j.jpsychires.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Zvara A, Szekeres G, Janka Z, Kelemen JZ, Cimmer C, Santha M, Puskas LG. Over-expression of dopamine D2 receptor and inwardly rectifying potassium channel genes in drug-naive schizophrenic peripheral blood lymphocytes as potential diagnostic markers. Dis Markers. 2005;21:61–69. doi: 10.1155/2005/275318. [DOI] [PMC free article] [PubMed] [Google Scholar]