Abstract
Spotted fever caused by spotted fever group rickettsiae (SFGR) is found throughout China. During 2007–2008, 28 human SFGR isolates and 34 rat SFGR isolates including 15 isolates from Rattus fulvescens, 5 isolates from R. edwardsi, 7 isolates from Callosciurus erythraeus roberti and 7 isolates from Dremomys rufigenis) were obtained from L929 cell culture. Previous research indicated that the 62 strains of SFGR mentioned above shared not only the same serophenotype but also 100% of identity sequences of 16S rRNA, gltA, ompA, groEL and 17KD, which enabled us to apply multispacer typing (MST) to the 62 SFGR isolates in the study. Six primer pairs, which were used for typing of Rickettsia rickettsii and Rickettsia conorii, were chosen, and the results exhibited greater nucleotide polymorphisms among the 62 isolates tested. A total of 48 distinct genotypes were identified. The dominant genotype, represented by h3 isolates, accounted for 21.7% (13/60) of the isolates tested, and the remaining 47 genotypes were all unique. Phylogenetic analysis showed that all the 48 genotypes could be classified in the same clade, while the genetically related strain, R. heilongjiangensis, was close but not the same as the cluster. We concluded that the genetically diverse of spotted fever group rickettsiae strains are endemic in Chengmai County, Hainan Province, China.
Keywords: Spotted fever group rickettsiae (SFGR), spotted fever, Chinese isolates, MST, Chengmai County of Hainan Province, China
Introduction
Spotted fever caused by spotted fever group rickettsiae (SFGR) is prevalent throughout China [1–5]. Atypical clinical manifestations, combined with the lack of a rapid laboratory diagnostic method for identifying SFGR, may cause misdiagnosis and lead to the development of severe disease and even death. In Chengmai County, Hainan Province, A typical example of a misdiagnosed case was that of the six-year-old farm child who was successively misdiagnosed as acute appendicitis, cat–scratch or lymphogranuloma and eventfully confirmed as SFGR infection [6]. In such an unusual situation, an active field epidemiological study was undertaken in Chengmai County where the index patient lived from Dec, 2007 to Feb, 2008. Seroepidemiological investigations indicated that the positive rate (1:80 cut off) of the IgG antibodies against the SFGR isolated from the index patient was 46.1% (374/812) among local agrarian adults, 37.5% (6/16) among preschool children, 88.9% (16/18) among domestic dogs and 11.4% (5/44) among wild rats [6]. A total of 28 human SFGR isolates (20 from adult febrile patients and 8 from febrile and preschool children with rash) and 34 rat SFGR isolates (15 Rattus fulvescens, 5 R. edwardsi, 7 Callosciurus erythraeus roberti, 7 Dremomys rufigenis) were obtained from L929 cell cultures (Table 1). Further studies indicated that the sequences of the 16S rRNA (830 bp), gltA (850 bp), ompA (634 bp), groEL (588 bp) and 17KD (289 bp) of these isolates were identical (unpublished data). In addition, the sequences of the 16S rRNA, gltA, ompA and groEL of the SFGR isolates were 100% homologous with those of the R. heilongjiangensis and 99% identical to the 17KD (289 bp) gene of R. heilongjiangensis [6]. First isolated from Dermacentor silvarum ticks collected in Suifenhe City, Heilongjiang Province, the isolate is currently named Rickettsia heilongjiangensis sp. nov. [7] and is broadly distributed in areas of the far east including Russia, Japan and China [2, 6–8]. Phenotyping and mouse serotyping further demonstrated that all the SFGR isolates obtained from Hainan Province were different from R. heilongjiangensis (unpublished data). In order to trace the molecular epidemiological characterizes of the SFGR isolates, we subjected a total of 62 Chinese SFGR isolates to MST based on 6 intergenic spacers.
Table 1.
The PCR primers used in multispacer typing and partially represented sequences deposited in GenBank of isolates tested
| Name of primer | Primer sequences 5'-3' (F/R) | Annealing temperature (°C) | Expected size (bp) | Actual size of the isolates tested | Sequences deposited in GenBank | Reference |
|---|---|---|---|---|---|---|
| dksA-xerC | TCCCATAGGTAATTTAGGTGTTTC/TACTACCGCATATCCAATTA | 54 | 416 | 225 | KJ461702 | [9, 10] |
| KJ461703 | ||||||
| KJ461704 | ||||||
| KJ461705 | ||||||
| KJ461706 | ||||||
| KJ461707 | ||||||
| KJ461708 | ||||||
| KJ461709 | ||||||
| spo0J-abcT1 | AAAGATTTGGAAGAATTAGACTTGAT/TTTGCTTAAACCAACCATTTCA | 50 | 320 | 254 | In process | [9] |
| RR0155-rpmB | TTTCTAGCAGCGGTTGTTTTATCC/TTAGCCCATGTTGACAGGTTTACT | 50 | 290 | 297 | In process | [11] |
| RR0345-tolC | AGAAGCTTCCGGATGTAATA/AGCAAATAAAAACCCTAATAAC | 50 | 238 | 238 | In process | [11] |
| cspA-ksgA | CATCACTGCTTCGCTTATTTT/ATTTCTTTTCTTCCTCTTCATCAA | 50 | 405 | 410 | KF923980 | [11] |
| KF923981 | ||||||
| KF923982 | ||||||
| KF923983 | ||||||
| KF923984 | ||||||
| KF923985 | ||||||
| KF923986 | ||||||
| KF923987 | ||||||
| KF923988 | ||||||
| KF923989 | ||||||
| KF923990 | ||||||
| KF923991 | ||||||
| KF923992 | ||||||
| KF923993 | ||||||
| KF923994 | ||||||
| KF923995 | ||||||
| KF923996 | ||||||
| KF923997 | ||||||
| KF923998 | ||||||
| KF923999 | ||||||
| KF924000 | ||||||
| KF924001 | ||||||
| RR1240-tlc5b | CGGGATAACGCCGAGTAATA/ATGCCGCTCTGAATTTGTTT | 50 | 357 | 366 | In process | [9] |
| RR1372-RR1373 | TCCCGCGCCAGTATCCA/CGGCGGCCAAAATGCTA | 50 | 349 | No product | In process | [11] |
Materials and Methods
Ethics statement
The field epidemiological study conducted in 2007–2008 and the collection protocol were approved by the China CDC Institutional Review Board (No. 201103). Written consent was obtained from participants before the blood sampling. Parents provided written informed consent on behalf of all child participants. In addition to the sampling of blood, the written consent also included the subsequent laboratory assay including serological tests, molecular detection and related etiology studies if a bacteria was isolated. The forests and farmland where the wild rats were captured were belongs to the local government of Xida farm, Chengmai County, and the rat capturing was approved by the Chengmai CDC. All experimental procedures with wild animals were conducted in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals (Publication No 85–23, revised 1985). The Animal Ethics Committee of the Chinese Center for Disease Control and Prevention approved the document on experimental procedures (201104).
Isolates and DNA preparation
All the strains used in the study were isolated from patients living in Chengmai County, Hainan Province and rats collected from Xida farm in the same County between Dec, 2007 and Feb, 2008. Of the 62 strains, 20 (h1–h20) were from febrile adult patients and 8 (h21–h28) from preschool children and another 34 isolates were from wild rats captured in nearby farms and forests. The rats included 15 Rattus fulvescens (m1–m15), 5 R. edwardsi (m16–m20), 7 Callosciurus erythraeus roberti (m21–m27), and 7 Dremomys rufigenis (m28–m34). Previous studies indicated that the sequences of the 16SrRNA, groEL, ompA, gltA and 17 KD were 100% identical and shared the same mouse serotyping.
DNAs for isolates were freshly extracted from L929 cultures using the QIAamp blood and tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The genetically associated rickettsiae R. heilongjiangensis strain 054, preserved in our laboratory, were included in the study as reference strains.
PCR primers
Based on previously published data [9, 10], 7 primer pairs used for amplifying the variable intergenic region of spotted fever group rickettsiae were chosen for the study. The sequences and the references of these primers are shown in Table 1. Of these primers, RR0155-rpmB, RR0345-tolC, cspA-ksgA and RR1372-RR1373 were successfully used for typing of Rickettsia rickettsii by Karpathy SE et al. [10], while spo0J-abcT1, RR1240-tlc5b, dksA-xerC), designed by Fournier PE et al., were applied to discriminate the Rickettsia conorii strain [9].
PCR assay
PCR was performed with a SensoQuest LabCycler standard plus (SensoQuest GmbH, Goettingen, Germany) and Taq PCR master mix kits (SBS Genetech Co., Ltd, China) according to the manufacturer’s instructions. A total of 25 μL reaction volume contained 12 μL of sterile water, 2.5 μL 10 × Taq Buffer, 0.5 μL of deoxynucleoside triphosphates (dNTPs, 10 mM), 0.5 μL of Taq DNA polymerase, 5 μL of DNA template, and 1 μL of each paired primer (10 mmol/L) dye. After a 5-min denaturation at 94°C, each reaction underwent 40 cycles of a 40-second denaturation at 94°C, a 30-second annealing incubation, and a 40-second extension at 72°C. This was followed by a final 5-min extension at 72°C. ddH2O was used as a negative control for each PCR preferment, and the DNAs of R. heilongjiangensis strain 054 were used as a positive control. The PCR products were analyzed using 1.0% agarose gel electrophoresis.
Sequencing and analysis
In order to obtain the entire sequences and to avoid the loss of sequence information at the ends of both primers, we cloned the PCR products as follows: the PCR product was purified using a multi-function DNA purification kit (BioTeke Corporation, Beijing, China). Purified PCR products were cloned into a pEASY-T1 cloning vector (Beijing TransGen Biotech Co., Ltd., Beijing, China), and then the recombinant plasmid was transformed into E. coli DH5a competent cells. The positive clones were identified by PCR using the specific primers for each variable intergenic region. Each amplicon was sequenced in both directions with an ABI 3100 genetic analyzer (Applied Biosystems) using BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions.
Each sequence was processed through manual splicing and proofreading using Chromas software (Technelysium, Australia) if any SNPs occurred and then repeatedly underwent PCR amplification and sequencing by another commercial sequencing company. The confirmed sequences were analyzed using the nucleotide blast program (http://blast.ncbi.nlm.nih.gov/). Alignments of sequences were conducted using Megalign software of Lasergen7.1 (DNA star Company, Madison, Wisconsin USA). In order to construct the phylogenetic tree, the sequences of the 6 variable intergenic regions for each isolate and the reference strain of R. heilongjiangensis strain 054 were concatenated in the following order: cspA-ksgA, dksA-xerC, RR0155-rpmB, RR0345-tolC, RR1240-tlc5b and spo0J-abcT1. In addition, the 6 homologous intergenic sequences of the R. montanensis (CP003340), R. philipii (CP003308), R. ricketsii (CP000053) and R. japonic (AP011533), the genomes of which had been sequenced, were downloaded from GenBank for concatenating and added the same phylogenetic analysis. The phylogenetic tree was constructed using the neighbor-joining method. Bootstrap analysis was conducted with 1,000 replicates.
Results
Of the 7 primer pairs tested, 1 primer pair, RR1372-RR1373, failed to yield products. The remaining 6 primer pairs, except RR0345-tolC, produced different sizes of PCR products instead of the expected length [9, 10] (Table 1). Two isolates (h2 and m5) exhibited negative PCR amplification with the spo0J-abcT1 primer pairs (for unknown reasons). In the cspA-ksgA amplification, the PCR products of the m5 isolate were too weak to be sequenced.
DskA-xerC
A 225 bp amplicon rather than the expected 416 bp length was amplified for the 62 SFGR isolates tested and the reference strain R. heilongjiangensis strain 054 using the dksA-xerC primer pair. Compared to the reference strain R. heilongjiangensis, 8 single nucleotide polymorphisms (SNPs) were identified as shown in Table S1. Eight genotypes (A1-H1) were obtained for the 62 isolates tested based on the genetic diversity (see Table S1). The genotype G1, represented by h3 (isolated from febrile adult patients), accounted for 88.7% (55/62) of the isolates tested while the remaining individual 7 genotypes accounted for 1.6%.
spoOJ-abcT1
Thirteen SNPs were identified in the 254 bp (expected 320 bp) of spo0J-abcT1 amplicons for the 60 SFGR strains (unavailable products of h2 and m5), allowing for the 13 distinct genotypes as shown in Table S2. The dominant genotype (M2), represented by the isolate h3, accounted for 80.0% (48/60) of the 62 isolates tested. The remaining 12 genotypes were all unique, accounting for 1.6%.
RR0155-rpmB
The RR0155-rpmB amplicons had 12 SNPs (Table S3), producing 11 genotypes (A3-K3). The major genotype (genotype K3), represented by h3, accounted for 83.9% (52/62) of the isolates tested while the remaining 10 distant genotypes accounted for 1.6%.
RR0345-tolC
Five rat isolates and 3 human isolates had thymine-to-cytosine transitions respectively (Table S4). Two human isolates (h6 of nucleotide 79 and h8 of nucleotide111) and 2 rat isolates (m7 of nucleotide 53 and m34 of nucleotide 212) had adenine-to-guanine transitions. One rat isolate (m27) had a deletion of thymine at 173, and m22 isolate had an insertion of guanine at nucleotide 5 (Table S4). These genetic differences allowed for the separation of the 62 isolates into 13 different genotypes, and the genotype M4 represented by h3 was dominant, accounting for 80.6% (50/62) of the tested isolates in the study. The remaining 12 distinct genotypes accounted for 1.6%.
cspA-ksgA
Sixty-one isolates aside from m5 isolate (purified PCR products were insufficient for sequencing) generated cspA-ksgA amplicons. A total of 27 SNPs were identified among the 61 isolates, which yielded 22 genotypes (Table S5). Of the 22 genotypes, the genotype V5 represented by the isolate h3 accounted for 60.1% (37/61) of the isolates tested, and the genotype R5 (m26) and genotype T5 (human isolate h4) accounted for 3.2% (2/62). Another19 distinct genotypes accounted for 1.6%.
RR1240-tlc5b
Six human isolates (h1, h5, h6, h4, h15 and h27) and 8 rat isolates (m1, m5, m11, m3, m14, m20, m23, m28) were found to have thymine-to-cytosine transitions (Table S6). Six human isolates (h1, h6, h7, h13, h23 and h25) and 2 rat isolates (m7 of nucleotide 312 and m13 of nucleotide 288) had adenine-to-guanine transitions respectively.
One cytosine-to-thymine transition was identified at nucleotide 248 for h1 human isolate and one guanine-to-adenine transition was found at nucleotide 183 for the m11 rat isolate. Four human isolates, h1, h5, h8 and h23, had a deletion of thymine at nucleotide 300 (Table S6). Two isolates (h6 and m23) had an insertion at nucleotide 301 respectively. All the genetic polymorphisms produced 25 genotypes, and the largest genotype (X6), represented by h3 isolate, accounted for 50.0% of the 62 isolates. The second dominant genotype (W6), represented by h8, accounted for 12.9%, and the remaining individual 23 genotypes accounted for 1.6%.
Phylogenetic analysis
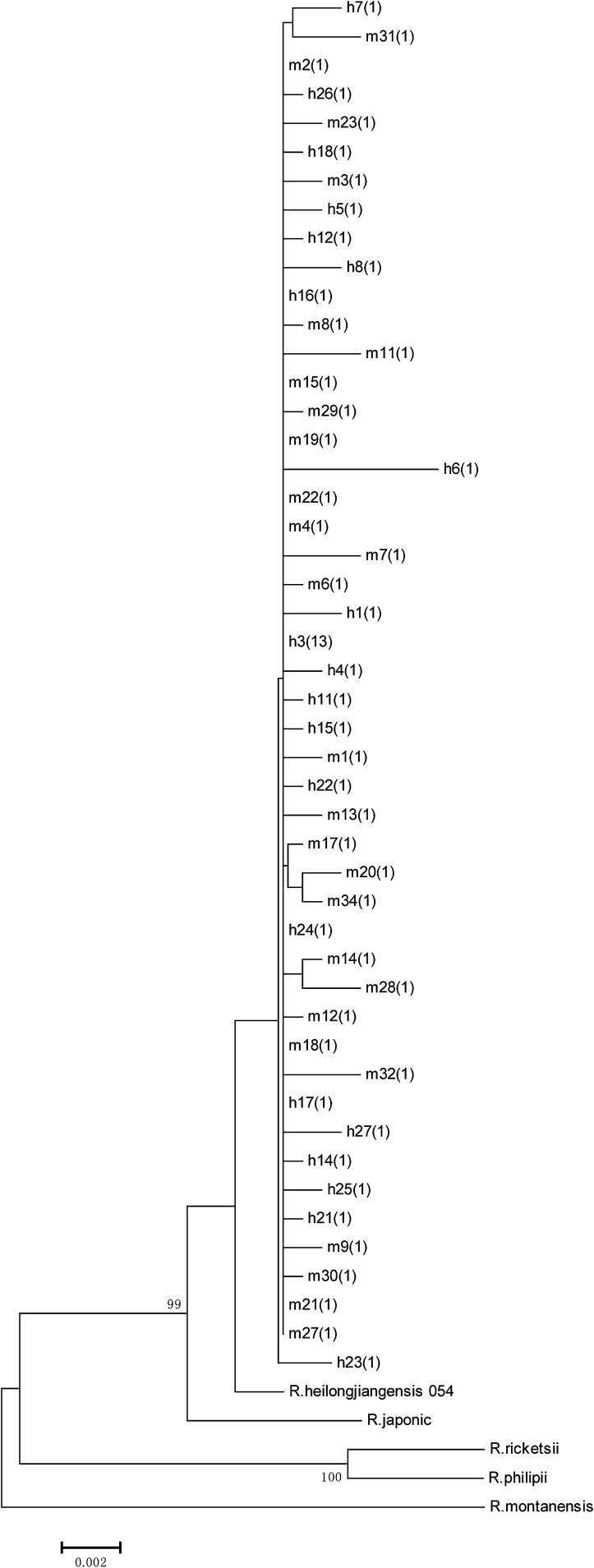
When the sequences of the 6 variable intergenic regions were concatenated and compared, the 62 SFGR could be divided into 48 distinct genotypes. Phylogenetic analysis using the neighbor-joining method showed that all the 48 genotypes could be classified as the same clade (Fig. 1). The genetically related strain, R. heilongjiangensis, was close but not the same as the cluster (Fig. 1). Another 4 reference strains of SFRG, R. montanensis (CP003340), R. philipii (CP003308), R. ricketsii (CP000053) and R. japonic (AP011533), were relatively far from the clade.
Fig. 1.
Phylogenetic relationships of SFGR isolates used in the study. Maximum parsimony phylogenetic relationships of isolates are based on the concatenated sequences of all six intergenic regions. Numbers at the nodes are bootstrap values based on 1,000 bootstrap replicates.
Discussion
The identity and taxonomy of a novel isolate of rickettsia are usually determined by the phenotypic characteristics of serotyping with antisera raised in mice [11] and subsequent molecular characteristics of precise identification and phylogenic classification based on the DNA sequences data of some crucial genes they contain. The sequences of 16S rRNA, 17KD cytoplasmic antigenic protein genes, citrate synthase (gltA), some outer membrane proteins A (OmpA) and B (OmpB) genes, and 120-kDa cytoplasmic antigenic protein genes [12] have proven useful for differentiion of Rickettsia spp. [13–15]. In 2003, Fournier et al. proposed a genetic guidelines for the classification of rickettsial isolates at the genus, group, and species levels by using sequences of the 16S rRNA (rrs) gene and four protein-coding genes, the gltA, ompA, and ompB genes and gene D [7]. According to the taxonomy system, to be classified as a member of the genus Rickettsia, an isolate should exhibit rrs and gltA homology with any of the 20 Rickettsia species studied at a level of ≥ 98.1 and ≥ 86.5%, respectively [7]. However, neither these phenotypic typing methods nor the modern molecular typing methods could differentiate bacteria at the strain level. Recently, the MST method has become indispensable to the differentiation of some strains of rickettsiae and has been applied to identify R. conorii [9], Rickettsia sibirica [4], Rickettsia rickettsii [10] and R. prowazekii [16] as well as some related bacteria such as B. quintana [17] and C. burnetii [18] at the strain level.
Previous studies including phenotyping, mouse serotyping, and MLST molecular typing could not discriminate the 62 SFGR isolates obtained from Chengmai County, Hainan Province. In the present study, we applied the 6 primer pairs (dksA-xerC, spo0J-abcT1, RR1240-tlc5b, RR0155-rpmB, RR0345-tolC, cspA-ksgA) for typing of the 62 Chinese SFGR isolates and successfully differentiated them into 48 distinct genotypes, which demonstrated that striking genetic polymorphisms of SFRG isolates were endemic in the region. Genotypes among the 62 Chinese SFGR isolates were significantly higher than that of R. rickettsii analyzed by Karpathy SE and colleagues using the spo0J-abcT1 (13 genotypes vs 4 genotypes) and RR1240-tlc5b (25 genotypes vs 5 genotypes) primer pairs [10]. Similarly, the 3 amplicons of RR0155-rpmB (11 genotypes), RR0345-tolC (13 genotypes), cspA-ksgA (22 genotypes) also exhibited greater nucleotide polymorphisms among the 62 Chinese SFGR isolates tested.
We were able to identify 27 unique genotypes by analyzing the SNPs identified in the 6 intergenic spacer sequences of the 60 isolates detected in the study. However, no correlation between the phylogenetic groupings and the source of isolates was noticed, and all of the 62 isolates, including 20 isolates recovered from febrile adult patients, 8 from preschool children, 15 from Rattus fulvescens, 5 from R. edwardsi, 7 from Callosciurus erythraeus roberti, and 7 from Dremomys rufigenis, were grouped in the same clade (Fig. 1). Although a great diversity of SFGR isolates was demonstrated and 48 genotypes were identified, the dominant genotype, represented by h3, accounted for up to 21.7% (13/60) of the local isolates tested.
Analyzing the SNPs identified in the study, we found that the thymine-to-cytosine transitions shared the highest percentage (45.6%, 52/114) followed by the guanine-to-adenine transitions (32.5%, 37/114) (Table 2). A total of 15 (13.8%) indel events (insertion/deletion) were observed. In addition, three adenine-to-thymine transversions and 1 thymine-to-adenine transversion were detected (Table 2 and Table S1–S3).
Table 2.
The SNPs of the 62 isolates tested
| SNPsa | ||||
|---|---|---|---|---|
| Tb-Cc Transitions (position of SNP) | Ad-Ge transitions (position of SNP) | A-T/G-A transitions (position of SNP) | Transversion (position of SNP) | Insersion/deletion (position of SNP) |
| m8 (60) | m9 (99) | |||
| m28 (126) | m30 (191) | m30 of T deletion (199) | ||
| h27 (140) | ||||
| m2 (179) | ||||
| m14 (182) | ||||
| m32 (61) | m32 (44) | m18 of C-T (50) | m22 of A deletion (15) | |
| m7 (132) | h13 (90) | m4 of T deletion (247) | ||
| m1 (135) | m31 (101) | |||
| h7 (174) | h4 (218) | |||
| m30 (190) | ||||
| m28 (221) | ||||
| m7 (25) | h12 (50) | h6 of C-T (81) | h22 of A-T (232) | m29 of G deletion (16) |
| h11 (48) | m9 (260) | |||
| m11 (95, 175) | ||||
| h6 (187) | ||||
| m12 (214) | ||||
| h21 (247) | ||||
| m23 (69) | m7 (53) | |||
| m14 (81) | h6 (79) | m22 of G deletion (5) | ||
| h4 (83) | h8 (111) | m27 of T deletion (173) | ||
| m5 (100, 192) | m34 (212) | |||
| h26 (105) | ||||
| m8 (119) | ||||
| h8 (159) | ||||
| m34 (172) | ||||
| m32 (119) | m3 (29) | m6 of G-A (150) | h27 of A-T (298) | |
| m31 (126, 198) | h23 (55) | h24 of G-A (239) | h7 of A-T (98) | m8 of A deletion (405) |
| h6 (203) | h8 (69) | |||
| m12 (260) | m31 (98) | |||
| m29 (261) | h2 (140) | |||
| h1 (268) | m32 (178, 317) | |||
| h23 (273) | h6 (217) | |||
| m20 (367) | m13 (234) | |||
| h18 (236) | ||||
| m20 (244, 340) | ||||
| m26 (340) | ||||
| m34 (340) | ||||
| h20 (355) | ||||
| h4 (361) | ||||
| m18 (361) | ||||
| h1 (255) | ||||
| m5 (31, 211) | h13 (49) | h1of C-T (248) | m17 of T-A) (220) | h6 of T insersion |
| m11 (34) | h25 (57) | 1 (G-A) | m23 of T insersion (301) | |
| m1 (42) | h1 (78) | m11 of C-T (183) | h1 and h5 and h8 and h23 of T deletion (300 ) | |
| h14 (51) | h7 (83, 280) | m2 of G deletion (311) | ||
| m28 (55, 204) | h23 (186) | |||
| h5 (72, 327) | h6 (208, 273) | |||
| h27 (103) | m13 (288) | |||
| h15 (133) | m7 (312) | |||
| h6 (178) | ||||
| m14 (204) | ||||
| m20 (220) | ||||
| m23 (228) | ||||
| m3 (315) | ||||
| h25 (317) | ||||
Note: a: single nucleotide polymorphisms; b: thymine; c: cytosine; d: adenine; e: guanine;
In addition to the discrimination of the 62 SFGR isolates at the strain level, the Chinese SFGR isolate, R. heilongjiangensis, which was closely related to the 62 isolates based on the sequences of 16S rRNA, gltA, ompA, groEL and 17KD genes but shared different serophenotypes, was further differentiated using the phylogenetic analysis based on the 6 concatenated intergenic sequences of each isolate analyzed.
The second largest island in China, Hainan Province is located in a subtropical region with characteristic lush vegetation, high rat density and various vectors. Previous studies demonstrated that the region was a typical epidemic area of multi-rickettsiosis including murine typhus, spotted fever and scrub typhus [19–21]. The high level of SFGR antibody titer among local residents and the high bacteria isolation rate of febrile patients and wild rats in the region, especially rural areas, could lead to outbreaks of tick-born infectious disease in the future. Tracing endemic pathogen strains in outbreak investigations conducted in further public health events will be essential to control the spread of infectious disease, and the MST typing used in this study will undoubtedly prove to be a powerful tool for identifying discrepancies among epidemiological strains.
Acknowledgments
The work was supported by the National Key Science and Technology Projects of China (2008ZX10004-008 and 2012ZX10004215) and the National Basic Research Program of China (973 Program- 2010CB530206).
Supplementary Material
References
- 1.Fan MY, Yu XJ, Walker DH. Antigenic analysis of Chinese strains of spotted fever group rickettsiae by protein immunoblotting. Am J Trop Med Hyg 1988; 39: 497–501 [DOI] [PubMed] [Google Scholar]
- 2.Parola P, Paddock CD, Raoult D. Tick-borne rickettsioses around the world: emerging diseases challenging old concepts. Clin Microbiol Rev 2005; 18: 719–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu X, Jin Y, Fan M, et al. Genotypic and antigenic identification of two new strains of spotted fever group rickettsiae isolated from China. J Clin Microbiol 1993; 31: 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Jin J, Fu X, et al. Genetic differentiation of Chinese isolates of Rickettsia sibirica by partial ompA gene sequencing and multispacer typing. J Clin Microbiol 2006; 44: 2465–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang JZ, Fan MY, Wu YM, et al. Genetic classification of “Rickettsia heilongjiangii” and “Rickettsia hulinii,” two Chinese spotted fever group rickettsiae. J Clin Microbiol 2000; 38: 3498–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin YM, Zhang LJ, Sun LY, et al. Epidemiological investigation of emerging spotted fever in Chengmai County, Hainan Province. Dis Surveil 2011; 26: 18–22 (in Chinese) [Google Scholar]
- 7.Fournier PE, Dumler JS, Greub G, et al. Gene sequence-based criteria for identification of new rickettsia isolates and description of Rickettsia heilongjiangensis sp. nov. J Clin Microbiol 2003; 41: 5456–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang LJ, Han J, Xu JG, et al. Identification of a new serotype of Rickettsia heilongjiangensis in wild rats from Guangdong Province, China. Clin Microbiol Infect 2009; 15Suppl 2: 338–339 [DOI] [PubMed] [Google Scholar]
- 9.Fournier PE, Zhu Y, Ogata H, et al. Use of highly variable intergenic spacer sequences for multispacer typing of Rickettsia conorii strains. J Clin Microbiol 2004; 42: 5757–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karpathy SE, Dasch GA, Eremeeva ME. Molecular typing of isolates of Rickettsia rickettsii by use of DNA sequencing of variable intergenic regions. J Clin Microbiol 2007; 45: 2545–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philip RN, Casper EA, Burgdorfer W, et al. Serologic typing of rickettsiae of the spotted fever group by microimmunofluorescence. J Immunol 1978; 121: 1961–1968 [PubMed] [Google Scholar]
- 12.Walker DH, Bouyer DH In: Murray PR, ed. Manual of Clinical Microbiology. Washington DC: ASM Press; 2003. pp. 1015–1029
- 13.Fournier PE, Roux V, Raoult D. Phylogenetic analysis of spotted fever group rickettsiae by study of the outer surface protein rOmpA. Int J Syst Bacteriol 1998; 48 Pt 3: 839–849 [DOI] [PubMed] [Google Scholar]
- 14.Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int J Syst Evol Microbiol 2000; 50 Pt 4: 1449–1455 [DOI] [PubMed] [Google Scholar]
- 15.Sekeyova Z, Roux V, Raoult D. Phylogeny of Rickettsia spp. inferred by comparing sequences of ‘gene D’, which encodes an intracytoplasmic protein. Int J Syst Evol Microbiol 2001; 51 Pt 4: 1353–1360 [DOI] [PubMed] [Google Scholar]
- 16.Zhu Y, Fournier PE, Ogata H, et al. Multispacer typing of Rickettsia prowazekii enabling epidemiological studies of epidemic typhus. J Clin Microbiol 2005; 43: 4708–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foucault C, La Scola B, Lindroos H, et al. Multispacer typing technique for sequence-based typing of Bartonella quintana. J Clin Microbiol 2005; 43: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos AS, Tilburg JJ, Botelho A, et al. Genotypic diversity of clinical Coxiella burnetii isolates from Portugal based on MST and MLVA typing. Int J Med Microbiol 2012; 302: 253–256 [DOI] [PubMed] [Google Scholar]
- 19.Lin BH, Sun XJ, Zhan ZN, et al. Investigation of serology and etilogy of spotted fever group rickettsia on Hainan island. Chin J Zoonoses 1999; 5: 90–91 (in Chinese) [Google Scholar]
- 20.Lin BH, Sun XJ, Zhan ZN, et al. Serological and etiological studies on undefined rickettsiosis on Hainan island. J Hainan Med Colleg 1997; 3: 97–102 (in Chinese) [Google Scholar]
- 21.Xu YY Review of the research of rickettsial diseases in Hainan Province. Chin Trop Med 2001; 1: 340–344 (in Chinese) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.