Abstract
Plant oil content and composition improvement is a major goal of plant breeding and biotechnology. The Puroindoline a and b (PINA and PINB) proteins together control whether wheat seeds are soft or hard textured and share a similar structure to that of plant non-specific lipid-transfer proteins. Here we transformed corn (Zea mays L.) with the wheat (Triticum aestivum L.) puroindoline genes (Pina and Pinb) to assess their effects upon seed oil content and quality. Pina and Pinb coding sequences were introduced into corn under the control of a corn Ubiquitin promoter. Three Pina/Pinb expression positive transgenic events were evaluated over two growing seasons. The results showed that Pin expression increased germ size significantly without negatively impacting seed size. Germ yield increased 33.8% while total seed oil content was increased by 25.23%. Seed oil content increases were primarily the result of increased germ size. This work indicates that higher oil content corn hybrids having increased food or feed value could be produced via puroindoline expression.
Keywords: corn, puroindoline, oil content, germ size, transformation
Introduction
Plant oil is an important renewable resource for both edible and industrial uses. An increase in seed oil content is a major goal of plant breeding and biotechnology (reviewed in Jaworski and Cahoon, 2003). Maize or corn (Zea mays L.) oil is the main co-product of corn wet and dry milling, and contains a high concentration of essential, polyunsaturated fatty acids, which make it an excellent source of energy and essential fatty acids for food and feed uses (Weber 1987). High oil corn is particularly attractive as a feed, due to its high metabolizable energy (Alexander 1988a). Increases in corn seed oil content are possible by traditional breeding. There are several examples of high oil corn populations developed through recurrent selection (Laurie et al. 2004; Alexander 1988b). However, commercial high-oil corn hybrids are associated with reduced seed size and agronomic yield (Dudley and Lambert. 2004). Many researchers have attempted to identify quantitative trait loci (QTLs) for oil content or quality to increase or modify plant oil content via manipulation of various oil biosynthetic pathway genes (Poneleit and Alexander 1965; Alrefai et al. 1995; Song et al. 2004; Beló et al. 2008; Zheng et al. 2008). For example, Zheng et al. (2008) identified a major oil QTL and demonstrated that increasing acyl-CoA:diacylglycerol acyltransferase (DGAT) during corn seed development can achieve relative increases in oil of approximately 40%. The synthesis of storage oils requires the coordinated activity of many enzymes in the lipid biosynthetic pathway. Despite a good understanding of the genes involved, there has been little success at transgenically increasing seed oil content (Clemente and Cahoon 2009). An alternative to modifying the lipid biosynthetic pathway is to modify the expression of lipid binding proteins. Genes whose products physically interact with lipids may be able to increase seed lipid content by decreasing the level of lipid breakdown.
Puroindoline a and b (PINA and PINB, collectively termed PINs) together make up the functional components of the wheat grain hardness locus (Ha) and control whether wheat seeds are soft or hard textured (Giroux and Morris 1998, Wanjugi et al. 2007). They share a similar structure to that of non-specific lipid-transfer proteins (ns-LTPs) (Giroux and Morris 1997; Marion et al. 1994; Le Bihan et al. 1996; Douliez et al. 2000). Both PINs contain a backbone of 10 Cys residues and likely form a tertiary structure comprised of four α-helices separated by loops and stabilized by five disulphide bridges (reviewed in Bhave and Morris 2008). The unique tryptophan-rich domain found in PINs has been demonstrated to have lipid-binding properties (Kooijman et al. 1997) and is the likely active site in terms of PIN function (Feiz et al. 2009a). This motif is believed to be a non-stick agent that enables PINs to bind to starch granule surface lipids preventing adhesion between starch granules and the surrounding protein matrix during seed maturation (Marion et al. 1994; Giroux and Morris 1998). Significant quantitative differences exist in seed polar lipid (glyco- and phospho-lipids) content between wheats varying in PIN content (Greenblatt et al. 1995; Feiz et al. 2009b, Finnie et al. 2010).
Plant lipids are generally stored as triacylglycerols (TAGs) in oil bodies (Huang 1992). Only a limited number of proteins are specifically associated with plant seed lipid bodies (reviewed in Purkrtova et al. 2008). Oleosins are the major proteins associated with oil bodies and maintain oil bodies as small single units preventing their coalescence during seed desiccation (Leprince et al. 1997). Siloto et al. (2006) observed that oleosins are important factors in determining seed oil body size in Arabidopsis. Both PINs and oleosins have lipid-binding properties. However, while oleosins are integral to oil bodies, PINs are normally only associated with the surface of starch granules (Capparelli et al. 2005; Feiz et al. 2009b; Wall et al. 2010).
Here we tested whether the ectopic expression of Pins can improve corn seed oil content. Puroindolines are endosperm specific and are found only in the Triticeae (Gautier et al. 2000).Corn does not express PINs or even contain Pin homologues (Gautier et al. 2000). Therefore, both Pina and Pinb coding sequences were introduced into corn under the control of a corn Ubiquitin promoter, and the effect of PINs on seed composition and oil content was studied.
Materials and Methods
Plasmid constructs
The Pina and Pinb expression vectors, pUbiPina and pUbiPinb, were described in Krishnamurthy and Giroux (2001). They carry the Pina-D1a/Pinb-D1a alleles reported by Gautier et al. (1994) that are found in all soft hexaploid wheats (Morris et al. 2001). In short, PCR products containing the coding sequences of Pina or Pinb were digested with BamHI and ligated into BamHI-digested and phosphatase treated pAHC17 plasmid (Christensen and Quail 1996), downstream of the corn Ubiquitin promoter and intron. All resultant plasmids were sequenced to ensure correct orientation and fidelity of ligation junctions. The construct pBAR184 used for selection (Frame et al. 2000) contains the corn Ubiquitin promoter-Bar gene cassette as a selectable marker conferring resistance to the herbicides bialaphos (4-[hydroxy(methyl-)phosphinoyl]-L-homoalanyl-L-alanyl-L-alanine) and glufosinate (DL-homoalanin-4-yl(methyl) phosphinic acid).
Corn transformation
Corn transformation was conducted in the Iowa State University Plant Transformation Facility according to their standard protocols (Frame et al. 2000). Highly embryogenic corn line Hi-II was used in biolistic transformation. Hi-II is a hybrid of the inbred lines A188 and B73. Embryogenic calli generated from immature embryos of the Hi-II genotype were co-bombarded with the plasmids using a 1:2.5:2.5 molar ratio (pBAR184:pUbiPina:pUbiPinb). Bialaphos-resistant calli were screened by PCR for the Pina and Pinb coding sequences using gene specific primers (Table 1). The method for screening of the transgenic positive and negative lines of each event was described in Zhang et al. (2009). Herbicide-resistant Pina/Pinb PCR positive T0 lines were pollinated using inbred line B73. Herbicide-resistance of T0 and all subsequent progeny plants was tested by painting an individual leaf with 0.1% glufosinate ammonium (Zhang et al. 2009). Herbicide-resistant T1 plants were heterozygous for the transgene locus and were self-pollinated to produce T2 progeny. Individual T2 plants from each transformation event were tested for both Pin genes as well as for the presence of Bar. T2 plants giving progeny (T3 plants) in which more than 12 consecutive sprayed plants were herbicide resistant and Pina/Pinb PCR positive were considered homozygous positive for Bar and Pina/Pinb, and T2 plants having progeny where more than 5 consecutive sprayed plants were herbicide susceptible and Pina/Pinb PCR negative were considered homozygous negative for Bar and Pina/Pinb. T0 and T1 plants were planted in a Montana State University greenhouse. T2 and T3 plants were planted both in the greenhouse at Montana State University and in the field at the University of Florida in 2007. T4 plants were planted in the field at the University of Florida in 2008. Each line was planted in two rows and each row had 15 plants with between row spacing of 90 cm and within row spacing of 30 cm. Ears from individual self pollinated plants were harvested, dried for three days at 37 °C in a forced air incubator, and then maintained at room temperature and ambient humidity (30%). The moisture content of all seeds at the time analyses were performed was ~10%. The same seed source was used for all the seed quality analysis including kernel weight, starch and protein content, germ yield and oil content. All values represent three independently-derived homozygous positive or negative T3 lines in 2007 and T4 lines in 2008 for each event. Seeds from three ears within the same line were pooled.
Table 1.
The sequence of primers used for PCR screening and qRT-PCR
| No | Gene | Database accession | Primer sequences |
PCR product size* | |
|---|---|---|---|---|---|
| Forward(5’-3’) | Reverse(5’-3’) | ||||
| 1 | Pina | DQ363911 | GGTGTGGCCTCATCTCATCT | TCACCAGTAATAGCCAATAGTG | 501(795-1296) |
| 2 | Pinb | DQ363913 | AATAAAGGGGAGCCTCAACC | TCACCAGTAATAGCCACTAGGGAA | 507(213-720) |
| 3 | Pina | X69913 | TAGCGAAGTTGTTGGCAGTT | TTGAGCATCGATCTAGCAGG | 109(87-196) |
| 4 | Pinb | X69912 | GAAGTTGGCGGAGGAGGT | TTTTGTGGGCCAGGTGAC | 105(104-229) |
| 5 | Actin | J01238.1 | TCCTGACACTGAAGTACCCGATTG | CGTTGTAGAAGGTGTGATGCCAGTT | 83(498-581) |
1-2 PCR primers; 3-5 qRT-PCR primers. The position of the amplified region within the accession sequence is shown in parentheses
Southern blot and Northern blot analysis
Southern-blot analysis was performed using DNA extracted from young leaf tissue of homozygous transgene positive T3 transgenic lines as previously described (Zhang et al. 2009). Genomic DNA was isolated from young leaves according to Riede and Anderson (1996), and Southern blots performed as as previously described ( Zhang et al. 2009). . The P32-labeled probes were made using the coding sequence of wheat Pina or Pinb as templates as previously described as described by Gautier et al. (1994). The Pina and Pinb probes are specific and do not cross-hybridize (Gautier et al. 1994; Giroux and Morris, 1997,1998). Northern-blot analysis was performed using RNA isolated from 21 days after pollination (DAP) developing corn seeds as previously described (Zhang et al. 2009). RNA was extracted from 21 DAP developing corn seeds using a Trizol protocol (Invitrogen, Carlsbad, CA) and analyzed by Northern-blot hybridization as previously described (Hogg et al. 2004; Zhang et al. 2009).
Quantitative real-time PCR (qRT-PCR)
Transgene and control gene expression in different tissues was quantified by qRT-PCR using a Dynamo SYBR green 2-step qRT-PCR kit (Finnzymes, Espoo, Finland) and a MiniOpticon RT-PCR machine (Bio-Rad, Hercules, CA), according to the manufacturer's instructions. Total RNA was prepared from 21 DAP corn germs and endosperms as well as leaves of three week old plants using Trizol (Invitrogen, Carlsbad, CA) as previously described (Zhang et al. 2009). The RNA samples were treated with RQ1 DNase (Promega, Madison, WI) and cDNA was generated using random hexamers and M-MuLV RNase H+ reverse transcriptase (Finnzymes, Espoo, Finland). The primer sequences used to detect Pina, Pinb, and the control gene Actin 1 are listed in Table 1. Thermocycling conditions were as follows: 15 min at 95 °C, then 40 cycles of 10 s at 94 °C, 25 s at 56 °C and 30 s at 72 °C. Threshold cycle (Ct) values were automatically detected and compared between samples. The specificity and length of PCR products were verified by agarose gel electrophoresis and homogeneity was assessed via melting curve analysis using a temperature increment of 0.2 °C and a hold time of 2 s between 72 and 90 °C. Pina and Pinb expression levels were normalized using the Actin 1 expression level for each RNA sample. The comparative C method (2−ΔΔCt) was used for relative quantification. The fold differences presented are comparisons of homozygous positive transgenic lines and the non-transgenic (NT) parental inbred line B73. The data shown is the average of two independent replicates.
PIN polyclonal antibody production and ELISA determination of PINA and PINB content
PINA and PINB specific polyclonal antibodies were prepared by purifying PINA or PINB from wheat seeds by Triton X-114 (TX-114) fractionation followed by SDS PAGE gel purification. The TX-114 fractionation, SDS-PAGE gel, and antibody production protocols have each been described previously (Krishnamurthy and Giroux 2001; Giroux et al. 2003). For enzyme-linked immunosorbent assay (ELISA), total puroindoline was extracted from 100 mg of mature whole seed meal from three transgenic corn lines, B73, and the soft wheat control (Heron) using TX-114 detergent by modifying the previously described method (Giroux et al. 2003) to include an additional round of TX-114 fractionation. Whole meal powder for each assay was created by grinding a random sample of seeds for each genotype using a Perten 3303 Laboratory Mill (Perten Instruments, Stockholm, Sweden).
Extracted proteins were diluted 1/200 in coating buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) and analyzed by PINA and PINB ELISA using a Immulon 4HBX 96-well plate (Thermo Electron Corporation, Byron Medical, Waltham, MA). The ELISA platewells were coated with 100 μL of the diluted protein samples and the plate was incubated overnight at 4 °C. The wells were washed three times at RT with a phosphate buffered saline solution (PBS) containing Tween-20 (137 mM NaCl, 1.5 mM KH2PO4, 8 mM Na2HPO4, 2.7 mM KCl, pH 7.4, 0.1% (v/v) Tween 20). 300 μL PBS containing 5% (w/v) bovine serum albumin (PBS-BSA) was added and plates were incubated for 2 hrs at RT. After three washes with PBS-Tween, 100 μL of a solution containing polyclonal PINA or PINB specific antiserum (diluted 1:250 in PBS-BSA) was added and plates were incubated overnight at 4 °C. After three RT washes with PBS-Tween, the PIN linked antibodies were revealed by adding 100 μL of a solution containing goat anti-rabbit IgG conjugated with horseradish peroxidase (Millipore, Billerica, MA) (dilution 1:100,000 in PBS-BSA). After 2 hrs at RT, wells were washed eight times with PBS-Tween and then 100 μL TMB (3, 3’, 5, 5’-tetramethylbenzidine) (Pierce Chemical Co, Rockford, IL) was added and plates were incubated for 15 min at RT. Reactions were stopped by addition of 100 μL 0.18 M H2SO4 and absorbance was measured at 450 nm using a SpectraMax Plus384 spectrophotometer (Molecular Devices, Sunnyvale, CA). Sample wells were blanked against transgene negative corn protein extracts, and all measurements were performed in duplicate.
Germ recovery and total oil content
The oil content of whole seeds and germ of transgenic lines was tested by a procedure similar to that of Li et al. (2006). Three T3 lines in 2007 and T4 lines in 2008 of each event were tested. Data were averages of three lines. Germ yield was measured by a small-scale wet milling method adapted from a previously published procedure (Vignaux et al. 2006). Briefly, 10 g of kernels were steeped in 20 mL H2O containing 0.2% (w/v) SO2 and 0.5% (v/v) lactic acid, pH 3.2, 50 °C for 48 hr. Tthe pericarp and germ were then manually removed with forceps. Germs were dried for 2 days at 37 °C using a forced air incubator and weighed. Germ yield was calculated as the ratio of the weight of germ recovered to the total weight of the sample. For oil content tests, about 25 mg of whole seed or germ meal was put in glass tubes (1×10 cm) fitted with Teflon-lined screw caps. The germ was milled in the glass tubes using a Teflon® Pestle. Then 1 mL of 5% (v/v) sulfuric acid in methanol, 25 μL of BHT (0.2% (v/v) butylated hydroxy toluene in methanol), and 300 uL of toluene were added to each tube. One hundred μg glyceryl triheptadecanoate (Sigma-Aldrich, St. Louis, MO) in 20 μg/μL hexane solution was added as an internal TAG standard for each sample to generate 17:0 fatty acid methyl esters (FAME). The mixture was vortexed for 30 s then heated at 90 °C for 90 mins. After cooling to RT, 1.5 mL of 0.9 % NaCl (w/v) aqueous solution and 2 mL of hexane were added and samples were mixed. After being centrifuged at 3,000rpm for 4 min (Medilite, Thermo Electron Corporation, Byron Medical Waltham, MA), the top organic phase containing FAMEs (fatty acid methyl esters) was transferred to a new glass tube. The hexane extraction step was repeated two more times to completely extract FAMEs. Pooled extracts were evaporated under a nitrogen stream and dissolved in 400 μL hexane and analyzed using a GC-2010 dual-FID gas chromatograph (GC) (Shimadzu, Columbia, MD). One uL of each sample (1 μL) was injected into the GC using a Twin-PAL dual rail robot (LEAP Technologies, Carrboro, NC) employing two DB-23 10 m × 0.18 mm i.d. columns (Agilent, Palo Alto, CA) using helium as the carrier gas. The GC was programmed for an initial temperature of 160 °C for 1.5 min, followed by an increase of 20 °C/min to 200 °C and then maintained for a further 1.5 min. Fatty acid composition was analyzed using Shimadzu's GC Solution software, and oil content was calculated based on the internal 17:0 standard.
Polar lipid content
The total lipid was further analyzed for UP6 and UP8 events by quantifying total seed polar lipids. Ten Pin locus homozygous positive or negative T4 seeds of each event were bulked and ground with a mortar and pestle in liquid N2. Total polar lipids were extracted from the whole meal samples as previously described (Welti et al. 2002). Lipid samples were analyzed on an API 4000 electrospray ionization triple quadrupole mass spectrometer (Applied Biosystems, Foster City, CA) at the Kansas Lipidomics Research Center (Kansas State University, Manhattan, KS).
Immunofluorescent localization of PINs
Immunofluorescent localization of PINA and PINB was conducted according to the basic methodologies of Feiz et al. (2009b), with the following modifications. Corn seeds from both parental and transgenic lines were cut with a scalpel longitudinally through the germ. One half of each seed sample was fixed in 2% formaldehyde, 2% glutaraldehyde in 100mM phosphate buffer, pH 7.4 for 48 h. Fixed half seeds were dehydrated with sequential 2 h treatments in 70%, 85%, and 100% (v/v) ethanol/water. Dehydrated seed halves were imbibed in paraffin wax, 4 μm sections were made using a microtome and the sections mounted on microscope slides. Seed sections on slides were de-paraffinized using three washes (3 min each) in 100% xylene, and one final wash in 1:1 xylene:ethanol (3 min). Xylene was removed by successive 3 min washes in 100%, 90%, and 70% ethanol; followed by gently rinsing the slides under tap water for 10 min (Hayat, 2002). Seed sections were blocked in 5% skim milk powder in phosphate-buffered saline/0.1% Tween (PBST) overnight at 4°C. The next morning, seed sections were incubated in a 1/100 dilution of Durotest® mouse anti-puroindoline primary antibody (R-Biopharm AG, Darmstadt, Germany) for 1 h in 2% (w/v) skim milk powder in PBST (Mohammadi et al. 2007). Seed sections, still adsorbed on the surface of the glass slide, were washed for 10 min x 3 in PBST. Slide sections were incubated in a 1/2000 dilution of goat anti-mouse Alexa Fluor® 488-conjugated secondary antibody (Invitrogen, Burlington, Ontario) for 1 h in PBST followed by three 10-minute washes in PBST. Finally, the slide sections were overlaid with Prolong Gold® (Invitrogen) anti-fade reagent and cured at room temperature in the dark for 48 h. Bright field and fluorescent microscopy images were viewed with an Axiophot (Zeiss, Oberkochen, Germany) epifluorescent microscope and captured using a Go-3 CMOS digital camera (QImaging, Surrey, Canada). Overlay images were generated using the ‘color to alpha’ function of the GNU Image Manipulation Program (GIMP), version 2.6.7.
Starch and protein content
Starch content of whole seed meal was measured via a total starch assay method (AACC Method 76-13, 2003) using a Megazyme kit (Megazyme International, Wicklow, Ireland). Protein content (N × 6.25) of whole seed meal was measured using a Leco FP-2000 (Leco Corp, St. Joseph, MI).
Statistical analysis
All mean values represent testing of three independently derived T1 homozygous positive or negative lines for each event. The means of a transgene positive event was compared with its respective transgene negative events or with non-transgenic (NT) parental inbred B73 using a t test. All statistical tests were conducted using GraphPad InStat software (v. 3.06, San Diego, CA)
Results
Development of transgenic corn lines containing Pina and Pinb genes
The transgenic plants were created in Hi-II via biolistic transformation at Iowa State University. Herbicide-resistant T0 lines having both Pina and Pinb were pollinated with pollen from inbred line B73. Resistant T1 plants were selfed to produce segregating T2 progeny. The Bar marker gene segregated in a 3:1 ratio in each of the three independent lines presented here (Table 2). T3 plants were tested using both PCR for the Pin genes as well as herbicide resistance to identify T2 derived lines that were homozygous positive or negative for the transgene locus. There were seven independent transgenic events generated, and three were selected for further analysis on the basis of having good vigor, fertility and being expression positive for both Pina and Pinb (results not shown). The three selected homozygous T3 events each carrying pBAR184, pUbiPina and pUbiPinb were designated UP6, UP8 and UP9. All the seeds analyzed were taken from the central cob region of the ears, and all seeds were equilibrated to the same moisture content.
Table 2.
Pina and Pinb PCR and glufosinate herbicide resistance test results for PIN transgenic corn.
| Eventa | T0 PCR (Pina/Pinb)b | T2 segregation c (resistant/susceptible) | Chi-quare (3:1)d | T3 PCR for Pins (+/−)e | T3 herbicide for Bar (+/−)e |
|---|---|---|---|---|---|
| NT | −/− | 0/5 | 15 | 0/5 | 0/12 |
| UP6 | +/+ | 7/2 | 0.037 | 16/0 | 24/0 |
| UP8 | +/+ | 10/5 | 0.556 | 18/0 | 12/0 |
| UP9 | +/+ | 6/2 | 0 | 16/0 | 10/0 |
All PCR positives T0 plants were crossed with untransformed plants to produce T1 seeds; T1 hemizygous lines were self-pollinated to produce T2 seeds. NT denotes non transgenic (NT) control inbred line B73.
PCR screening was performed using Pina or Pinb specific primer pairs on samples of genomic DNA from each T0 plant
T2 progenies were leaf-painted with 0.1% glufosinate. Seedlings which showed minimal adverse effects were scored as resistant
Chi-square values test the fit of resistant/susceptible of T2 plants to a 3:1 ratio
T3 seedlings were tested with Pina and Pinb PCR screening and herbicide tests to obtain T2:3 seed pools co-segregating homozygous positive or negative for the Pina, Pinb and Bar genes. PCR screening was tested on Bar positive and negative plants
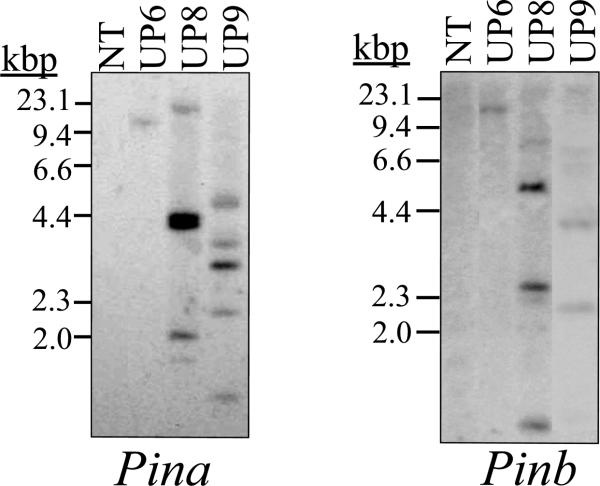
Southern-blot analysis was used to confirm that each line resulted from an independent transformation event. Total genomic DNA was isolated from seedlings and restriction digested with HindIII which cuts once within the pUbiPina and pUbiPinb constructs and not within the Pina and Pinb coding sequences used as hybridization probes. The banding pattern was unique for each transgenic line, indicating that each arose from an independent integration event (Fig. 1).
Fig. 1.
Southern-blot analysis for three transgenic lines. Genomic DNA isolated from homozygous T3 seedlings was digested with HinIII. NT denotes non transgenic (NT) control inbred line B73. lines was used as a wildtype control. The 32P-labeled DNA coding sequence of Pina or Pinb was used as a probe for hybridization. The position of DNA molecular size markers is indicated.
Pina and Pinb tissue-specific expression levels
Total RNA was extracted from 21 DAP immature seeds of transgenic lines and control genotypes and analyzed by probing RNA gel blots with Pina and Pinb specific probes (Fig. 2). RNA extracted from developing seeds of the soft wheat control variety Heron was used for quantification of signal intensity. No Pina or Pinb transcripts were detected in non transgenic (NT) control corn inbred B73s. Pina and Pinb RNA expression levels of in transgenic lines were much less than that of the soft wheat control with Pin expression levels in UP8 and UP9 approximately 1-2% that of the soft wheat control variety Heron. UP6 was omitted here, but was analyzed by quantitative real-time PCR (qRT-PCR) along with UP8 and UP9. The size of the Pina and Pinb transcripts in the UP events (~700 bp for both Pina and Pinb) was the same as in the positive control Heron.
Fig. 2.
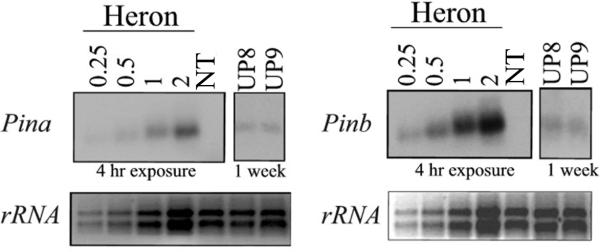
Northern analysis of Pina and Pinb expression in transgenic corn seeds. NT denotes non transgenic (NT) control inbred line B73. Soft wheat cultivar Heron was used as a positive control and RNA of Heron was loaded in a series of increasing amounts to account for varying signal intensities among transformed lines. The control was exposed to 4 hours and the transgenic lines were exposed for one week. A duplicate ethidium bromide stained agarose gel shows discrete bands of rRNA fractionated, indicating a similar loading from lane to lane and a lack of RNA degradation.
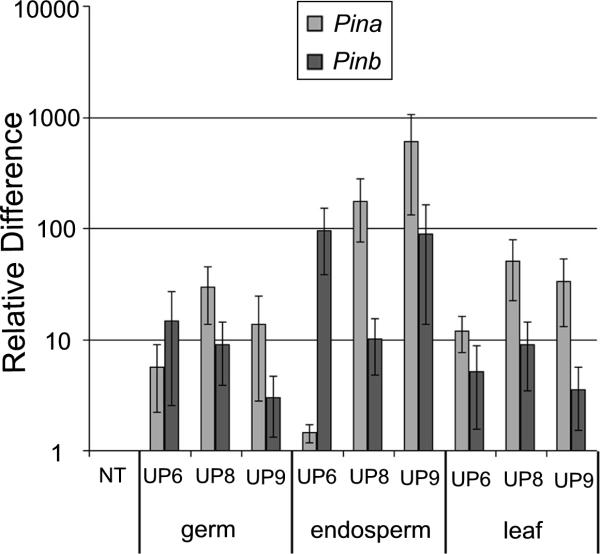
The Pina and Pinb expression in immature embryo, endosperm and leaves of transgenic lines was determined by qRT-PCR (Fig. 3). PCR efficiencies of Actin 1 and Pins genes were approximately equal, and ranged from 85 to 115%. Coefficients of correlation (R2) to evaluate the quality of the standard curve for reference and target genes were between 0.96 and 0.99. No Pin transcripts were detected in non transgenic (NT) control inbred B73. The highest expression levels observed among the PIN transgenic tissues was for Pina in UP8 and UP9 endosperms. The lowest expression level was for Pina in UP6 endosperm. The Pinb expression pattern was similar to Pina for UP8 and UP9 but in UP6 Pinb was expressed at a much high level than Pina. Expression levels in germ, endosperm and leaf were compared. Endosperm had the highest expression levels compared to germ and leaves. The Pina expression levels in germs were the lowest among the different tissues while Pinb expression levels were similar in germ and leaves. Since the comparative C method (2−ΔΔCt) was used for relative quantification, the relative differences are comparisons of homozygous positive transgenic lines and non-transgenic controls inbred B73.. Heron doesn't have the corn Actin 1 gene. However, the expression level of Pina and Pinb in developing seeds of Heron was 50 to 1000 times greater than that in the transgenic corn seeds (data not shown).
Fig. 3.
qRT-PCR analysis of Pina and Pinb expression in transgenic lines. The endogenous corn Actin 1 gene was used as a reference. RNA was prepared from 21 DAP embryos, and endosperms as well as leaves of three week post germination seedlings. NT denotes non transgenic (NT) control inbred line B73. . The relative difference is between the samples and the NT control for either Pina or Pinb. Data are shown are on a logarithmic scale of the relative expression level difference. Error bars denote standard deviations.
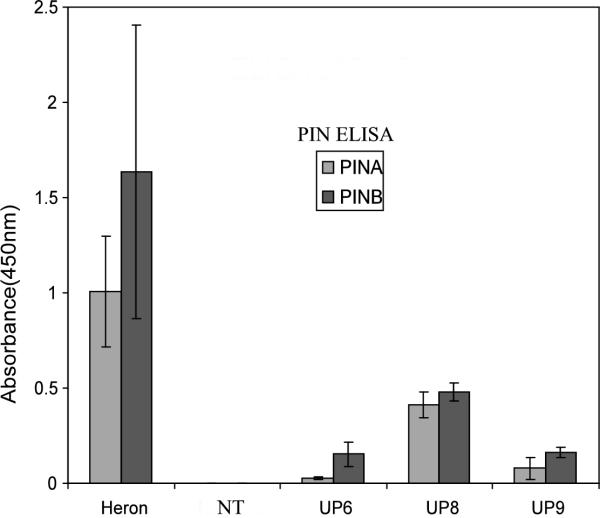
ELISA was performed to quantify PINA and PINB in the transgenic corn seeds. Mature homozygous transgene positive or negative control seeds along with seeds of the soft wheat cultivar Heron were analyzed by ELISA for the presence of PINA and PINB (Fig. 4). The soft wheat Heron, which contains both PINA and PINB, consistently gave strong ELISA reactions for both PINs. All three transgenic events were ELISA positive for both PINA and PINB (Fig. 4). Seeds from UP8 gave the strongest ELISA reactions among the three events for both PINA and PINB while the PINA level in UP6 was lowest among all events. The PINA and PINB levels in the transgenic corn lines were roughly 1/100 to 1/1000 of Heron PINA and PINB levels.
Fig. 4.
ELISA of T3 homozygous transgenic corn seed proteins. Each reading represents an ELISA assay in which a well was loaded with 200 times dilution of 100 mg equivalents of total corn or wheat seed flour meal extracts prepared by TX-114 phase partitioning. Readings are representative of duplicate measurements. Sample wells were blanked against homozygous negative corn protein extracts. Soft wheat cultivar Heron was used as a positive control while the negative control was non transgenic (NT) control inbred line B73.. Error bars denote standard deviations.
Puroindolines increase germ recovery and oil content without modifying seed size
Data for kernel weight, protein, and starch content are presented in Table 3. Kernel weight, protein, and starch content among the transgene positive lines and their corresponding negative control were similar for UP8 and UP6 (P>0.05). However, UP9 had reduced kernel weight relative to its control lines (0.236 vs 0.275 g, P<0.05) and tended to have higher protein and reduced starch content though the differences observed were not ssignificant (P>0.05).
Table 3.
Means for kernel weight, protein content and starch content of PIN transgenic corn seeds
| Eventa | Sourceb | Kernel weight (g) | Protein content (%)c | Starch content (%)d |
|---|---|---|---|---|
| UP6 | +/+ | 0.248± 0.010(ns) | 10.40±0.20 (ns) | 68.0±5.66 (ns) |
| −/− | 0.258±0.004 | 10.68±0.65 | 68.9±5.16 | |
| UP8 | +/+ | 0.256±0.016 (ns) | 11.93± 1.02(ns) | 69.1± 2.12(ns) |
| −/− | 0.250±0.026 | 11.00±0.52 | 68.1±2.12 | |
| UP9 | +/+ | 0.236±0.046* | 12.09±0.04 (ns) | 66.1±1.41(ns) |
| −/− | 0.275±0.054 | 11.02±1.43 | 68.5±4.03 | |
| NT | NT | 0.265±0.025 | 10.35±0.21 | 68.2±5.23 |
denote significance at P<0.05 in comparisons of homozygous positive (+/+) transgenic lines and the corresponding homozygous negative (−/−) lines. ns, not significant. NT denotesmeans non transgenic (NT) control inbred line B73.
The values presented are the average +/− standard deviation of three lines of each event over two growing seasons
T2 derived T3 seeds with co-segregation homozygous positive(+/+) or homozygous negative(−/−) for the transgene locus and Bar gene
Determined by a Leco FP-2000 (N × 6.25)
Determined by a Megazyme total starch assay kit
Germ recovery (germ weight as a percent of seed weight) along with germ and whole seed oil content for the three events is shown in Table 4. Germ recovery was increased significantly in each of the three transgenic lines relative to their respective controls (P<0.01). The relative increase in germ recovery ranged from 22.62% (UP6, 9.54 vs 7.78 %) to 42.26% (UP9, 12.22 vs 8.59%) with the three events averaging a 33.77% increase. Germ oil content was not significantly different for both UP8 and UP9 (P>0.05) but was for UP6 in which the transgene positive genotypes had higher germ oil content than their negative controls (33.62 vs 30.65 %). Seed oil content was significantly higher for all three events (P<0.001). The oil content of non-transgenic control inbred line B73 was 3.54%, similar to the transgene negative controls (3.79%). The relative increases in seed oil content for UP6. UP8, UP9 were 25.70%, 25.51%, and 24.47%, respectively. Germ fatty acid composition of transgenic and control lines was quantified by gas chromatograph (GC) (Table 5). Stearic, and oleic acids increased while linoleic acid decreased in UP6 (stearic, and oleic acids increased by 41.06% and 31.31%, respectively, while linoleic acid decreased by 11.86%). The same trend was apparent in UP8 in 2007 (data not shown), but did not reach statistical significance over two years while the fatty acid composition of UP9 was relatively unaltered. The lipid profile of the mature seeds of two events (UP6 and UP8) was analyzed and is shown in Table 6. Only total LysoPC was different between the transgenic positive and negative lines (47.27% increase for UP6 and 24.63% increase for UP8).
Table 4.
Means for seed oil traits of PIN transgenic corn.
| Eventa | Source | Germ yield (%)b | Germ oil content (%)c | Germ oil increase(%) | Seed oil content (%)d | Seed oil increase(%) |
|---|---|---|---|---|---|---|
| UP6 | +/+ | 9.54±1.43** | 33.62±0.28* | 9.69 | 4.55±0.35*** | 25.70 |
| −/− | 7.78±0.74 | 30.65±2.51 | 3.62±0.29 | |||
| UP8 | +/+ | 10.86±1.99*** | 34.29± 3.25(ns) | 3.40 | 4.97±0.88*** | 25.51 |
| −/− | 7.96±1.02 | 33.16±4.49 | 3.96±0.52 | |||
| UP9 | +/+ | 12.22±2.04*** | 27.47± 0.76(ns) | 0.81 | 4.73±0.23*** | 24.47 |
| −/− | 8.59± 0.86 | 27.25±3.95 | 3.80±0.09 | |||
| NT | NT | 7.03±0.04 | 30.23±1.45 | 3.54±0.08 |
denote significance at P<0.05, 0.01, and 0.001, respectively, in comparisons of homozygous positive (+/+) transgenic lines and the corresponding homozygous negative (−/−) lines. ns, not significant. NT denotes non transgenic (NT) control inbred line B73.
denote significance at P<0.05, 0.01, and 0.001, respectively, in comparisons of homozygous positive (+/+) transgenic lines and the corresponding homozygous negative (−/−) lines. ns, not significant. NT denotes non transgenic (NT) control inbred line B73.
denote significance at P<0.05, 0.01, and 0.001, respectively, in comparisons of homozygous positive (+/+) transgenic lines and the corresponding homozygous negative (−/−) lines. ns, not significant. NT denotes non transgenic (NT) control inbred line B73.
The values +/− standard deviations presented are the average of three lines of each event over two growing seasons.
Germ yield was determined by a wet milling process and was calculated as the ratio of the weight of germ recovered to the total weight of the sample(Vignaux et al. 2006).
Germ oil content was determined by GC analysis using 17:0 TAG as an internal standard for each sample. It was calculated as the ratio of the total oil content to the weight of germ.
Seed oil content was calculated as the ratio of the oil content to the weight of whole seed meal
Table 5.
Fatty acid composition of PIN transgenic corn germ. The values presented are the average of the percentage of the fatty acid of each event over two years.
| Eventa | Sourceb | Palmitic acidb C16:0(%) | Stearic acidb C18:0(%) | Oleic acidb C18:1(%) | Linoleic acidb C18:2(%) | Linolenic acidb C18:3(%) |
|---|---|---|---|---|---|---|
| UP6 | +/+ | 10.95 ±0.26 | 2.13±0.39* | 27.97±0.23** | 58.04±0.89** | 0.93 ±0.01 |
| −/− | 10.46 ±1.27 | 1.51 ±0.86 | 21.30 ±0.70 | 65.85 ±2.43 | 0.89 ±0.39 | |
| UP8 | +/+ | 11.59 ±1.12 | 1.84 ±0.74* | 21.75 ±4.20 | 64.24 ±6.03 | 0.60 ±0.01 |
| −/− | 11.03 ±1.33 | 1.43 ±0.57 | 21.38 ±5.64 | 65.07 ±7.69 | 1.10 ±0.14 | |
| UP9 | +/+ | 10.36 ±0.69 | 1.45 ±0.69 | 23.54±4.12 | 63.58 ±5.59 | 1.08 ±0.07 |
| −/− | 11.04 ±1.53 | 1.67 ±1.03 | 24.54 ±3.09 | 61.60 ±6.02 | 1.16 ±0.37 | |
| NT | −NT | 10.01 ±0.64 | 1.41 ±0.71 | 23.25 ±3.46 | 63.97 ±5.09 | 1.03 ±0.21 |
denote significance at P<0.05, 0.01, respectively, in comparisons of homozygous positive (+/+) transgenic lines and the corresponding homozygous negative (−/−) lines. NT denotes non transgenic (NT) control inbred line B73.
denote significance at P<0.05, 0.01, respectively, in comparisons of homozygous positive (+/+) transgenic lines and the corresponding homozygous negative (−/−) lines. NT denotes non transgenic (NT) control inbred line B73.
The values +/− standard deviations presented are the average of the percentage of the total fatty acid of each event over two growing seasons.
Determined by GC analysis
Table 6.
Polar lipid composition of PIN transgenic corn seeds
| Descriptiona | UP6(+/+)b (nmol/mg) | UP6(−/−)b (nmol/mg) | UP8(+/+)b (nmol/mg) | UP8(−/−)b (nmol/mg) |
|---|---|---|---|---|
| Total DGDG | 0.078±0.002 | 0.087±0.018 | 0.087±0.003 | 0.090±0.001 |
| Total MGDG | 0.011±0.002 | 0.014±0.003 | 0.009±0.003 | 0.009±0.000 |
| Total PG | 0.024±0.002 | 0.024±0.004 | 0.034±0.009 | 0.037±0.001 |
| Total lysoPG | 0.000±0.000 | 0.000±0.000 | 0.001±0.001 | 0.000±0.000 |
| Total LysoPE | 0.015±0.000 | 0.012±0.003 | 0.020±0.002 | 0.017±0.001 |
| Total LysoPC | 0.081±0.002* | 0.055±0.015 | 0.110±0.001* | 0.087±0.004 |
| Total PC | 1.692±0.204 | 1.888±0.228 | 1.762±0.159 | 1.921±0.009 |
| Total PE | 0.406±0.036 | 0.448±0.139 | 0.469±0.052 | 0.508±0.005 |
| Total PI | 0.128±0.034 | 0.069±0.028 | 0.159±0.030 | 0.165±0.011 |
| Total PS | 0.013±0.001 | 0.013±0.003 | 0.013±0.001 | 0.013±0.001 |
| Total PA | 0.011±0.002 | 0.017±0.004 | 0.015±0.008 | 0.014±0.000 |
| Total | 2.458±0.277 | 2.628±0.446 | 2.679±0.263 | 2.861±0.007 |
**, ***denote significance at P<0.05, 0.01, and 0.001, respectively, in comparisons of homozygous positive (+/+) transgenic lines and the corresponding homozygous negative (−/−) lines. SD, standard deviation.
The total lipid profile of the matured seeds was analyzed by electrospray ionization triple quadrupole mass spectrometers. Lipid abbreviations: DGDG, digalactosyl- diacylglycerols; MGDG, monogalactosyldiacylglycerol; PG, phosphatidyl glycerol; PC, phosphatidyl choline; PE, phosphatidyl ethanolamine; PI, phosphatidyl inositol; PS, phosphatidyl serine; PA, phosphatidic acid.
The values +/− standard deviations presented are the average of the percentage of the total fatty acid of each event over two growing seasons.
The bright field micrographs of embryos show that the overall cellular shape of oil bodies remained unaltered between the transgenic line and non transgenic control (Fig 5A, 6A). The overall size of the oil bodies between the UP8 and B73 germs was similar. However, the numbers of oil bodies increased in the transgenic lines (Fig. 5E, 6E E), and occupied most of the cell in the transgenic. The corresponding immunofluorescent micrographs show the localization of PINs in UP8 endosperm (Fig 6D, F). PINs localized to the surface of starch granules in endosperm (Fig 6F). However, no PINs were detected in the germ by immunofluorescent analysis even though expression of Pina and Pinb in the germ were confirmed by qRT-PCR (Fig 3).
Fig. 5.
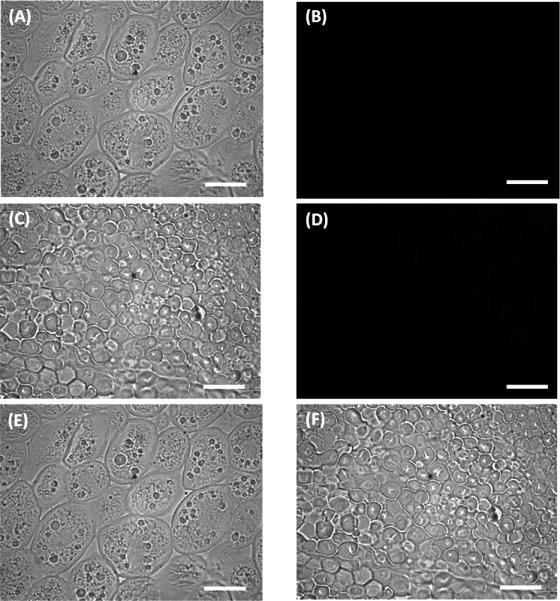
Immunolocalization of puroindoline in untransformed B73 mature corn seed sections incubated with Durotest® mouse anti-puroindoline primary antibody and goat anti-mouse Alexa Fluor® 488 secondary antibody. (a) Bright field micrograph and (b) corresponding immunofluorescent micrograph of germ tissue. (c) Bright field micrograph and (d) corresponding immunofluorescent micrograph of endosperm tissue. (e) Bright field micrograph of untransformed B73 mature corn (a) overlaid with its corresponding immunofluorescent micrograph (b). (f) Bright field micrograph of untransformed B73 mature corn endosperm tissue (c) overlaid with its corresponding immunofluorescent micrograph (d). Immunofluorescent images were exposed for 1s. Scale bar = 25 μm.
Fig. 6.
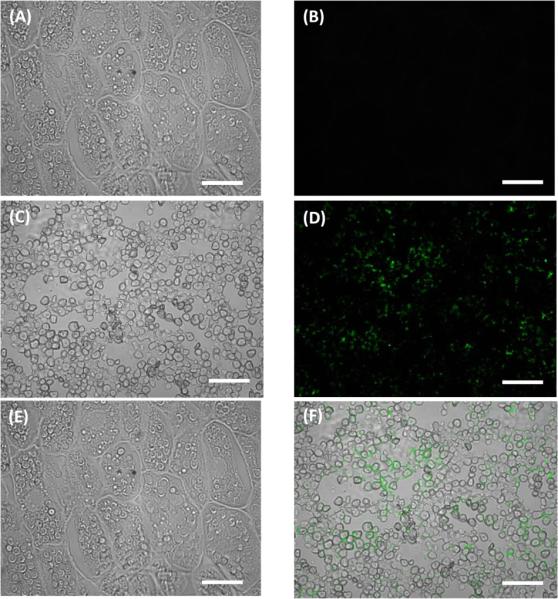
Immunolocalization of puroindoline in UP8 mature corn seed sections incubated with Durotest® mouse anti-puroindoline primary antibody and goat anti-mouse Alexa Fluor® 488 secondary antibody. (a) Bright field micrograph and (b) corresponding immunofluorescent micrograph of germ tissue. (c) Bright field micrograph and (d) corresponding immunofluorescent micrograph of endosperm tissue. (e) Bright field micrograph of UP8 mature corn germ tissue (a) overlaid with its corresponding immunofluorescent micrograph (b). (f) Bright field micrograph of UP8 mature corn endosperm tissue (c) overlaid with its corresponding immunofluorescent micrograph (d). Immunofluorescent images were exposed for 1s. Scale bar = 25 μm.
Discussion
Approximately 84% of corn seed oil is located in the germ (Watson 1984) and therefore corn seed oil content is primarily determined by germ oil content and size. Our experiments indicate that both germ size and oil content were increased via the constitutive expression of puroindolines (Table 4). No increase in germ size or oil content was seen in transgenic corn where puroindolines were expressed under the control of a γ-Zein endosperm-specific promoter (data not shown, Zhang et al. 2009). Therefore, the germ size increase we observed here was likely due to the ectopic expression of Pin genes in tissues other than the endosperm. The germ oil concentration of the transgenic lines was similar to that of the negative controls (P>0.05), except in UP6, which had higher germ oil content than the negative control (33.62% vs 30.65%, P<0.05) (Table 4). These results suggest that total oil content in transgenic corn was increased by increasing germ size, not by increasing germ oil content. In contrast, the key corn oil QTL identified by Zheng et al. (2008) controls germ oil concentration and seed oil content, but not germ size. Recurrent selection for increased oil content is typically associated with reduced seed size and agronomic yield (Laurie et al. 2004; Alexander 1988b) whereas we did not observe consistently reduced seed size in our PIN transgenic corn. The typical fatty acid composition of dent corn oil includes 11.0% palmitic, 2.0% stearic, 24.1% oleic, 61.9% linoleic , 0.7% linolenic and other fatty acids (White et al. 2007). Oil content increases in corn are associated with decreased linoleic acid and increased oleic and saturated acid content (Sniegowski and Baldwin 1954). Increased oleic acid and decreased linoleic acid was also found in the IHO × B73 (Wassom et al. 2008) and the ASKC28IB1 (ASK cycle 28 inbred 1) × PH09B (normal-oil inbred line) populations (Zheng et al. 2008). Increased stearic, oleic, and decreased linoleic acid was indeed observed in UP6. However, that trend was not seen in either UP8 or UP9 (Table 5).
Our results indicate that cereal seed oil content can be increased by puroindoline expression. However, the mechanism of the increase is not clear. PINs were not detected by immunofluorescent analysis in the germ (Fig. 6E). Post-transcriptional and/or post-translational regulation may have occurred rendering the level of PINs in the germ to be too low to be detected by immunofluorescent analysis or the mechanism may involve expression in tissues outside of the embryo. Fatty acids are generally stored in oil bodies and oleosins are the major proteins associated with oil bodies. The importance of the central hydrophobic domain of oleosins for their correct insertion into lipid bodies was addressed by Van Rooijen and Moloney 1995). The tryptophan-rich domain of PINs has a high affinity for binding to lipids (Kooijman et al. 1997; Wall et al. 2010). Ting et al. (1996) found oleosins are constitutively expressed independent of oil content with high oil corn having larger and more spherical oil bodies relative to low oil content corn. In our transgenic lines, the size of the oil bodies was not significantly different between the seeds expressing PIN and the controls while oil body number was increased (Fig. 5, 6), suggesting that PINs function to increase oil content differently than oleosins.
The increased germ size observed here is likely specific to PIN expression in tissues outside of the endosperm given that Zhang et al. (2009) expressed PINs in the endosperm of transgenic corn using the γ-zein promoter and did not observe any alteration in embryo size or seed oil content. Even though the oil content differences were seen in each of the three transgenic events studied here, it remains possible that explanations other than PINs are responsible for the increased germ size. While seemingly unlikely, it is possible that the insertion site effects are causatory and that the UP events studied have alterations in the expression levels of genes determining germ or oil-content characteristics. Insertion site characterization (Zaidi et al. 2007), along with global microarray profiling could be used to screen for unintended effects. The use of such techniques has aided in the determination of the “substantial equivalence” of a transgenic plant to its non-transgenic counterpart (Abdeen et al. 2010), and may confirm that PIN expression alone results in increased germ size and seed oil content. Compared with previous studies where the goal was to increase or modify oil content via manipulation of the oil biosynthetic pathway (Zheng et al. 2008), our work may yield a new method to increase the seed oil content. The results obtained here should be first validated in other crops. We predict that overexpression of PINs may be useful in increasing oil content of cereal crops via increasing germ size and of oilseed crops by increasing cotyledon size.
Acknowledgments
This work was supported by the Consortium for Plant Biotechnology Research, Dow AgroSciences, USDA-ARS National Research Initiative Competitive Grant Program grants 2004-35301-14538, 2007-35301-18135 and by the Montana Agricultural Experiment Station. Research at the University of Florida was supported by the National Science Foundation, IBN-0444031 and IOS-0815104 and USDA Competitive Grants Program grants 2006-35100-17220 and 2008-35318-18649. The lipid analyses described in this work were performed at the Kansas Lipidomics Research Center. The Kansas Lipidomics Research Center was supported by National Science Foundation (EPS 0236913, MCB 0455318, DBI 0521587), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of National Institute of Health (P20RR16475), and Kansas State University.
References
- Abdeen A, Schnell J, Miki B. Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3. BMC Genomics. 2010;11 doi: 10.1186/1471-2164-11-69. Article Number: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DE. High oil corn: breeding and nutritional properties. 43rd annual corn and sorghum research conference; 1988a.pp. 97–105. [Google Scholar]
- Alexander DE. Breeding special nutritional and industrial types. In: Sprague GF, Dudley JW, editors. corn and corn improvement. American society of agronomy; 1988b. pp. 869–880. [Google Scholar]
- Alrefai R, Berke TG, Rocheford TR. Quantitative trait locus analysis of fatty acid concentrations in maize. Genome. 1995;38:894–901. doi: 10.1139/g95-118. [DOI] [PubMed] [Google Scholar]
- American Association of Cereal Chemists . Approved methods of the American association of cereal Chemists. 10th ed. AACC; 2003. [Google Scholar]
- Beló A, Zheng P, Luck S, Shen B, Meyer DJ, Li B, Tingey S, Rafalski A. Whole genome scan detects an allelic variant of fad2 associated with increased oleic acid levels in maize. Mol Genet Genomics. 2008;279:1–10. doi: 10.1007/s00438-007-0289-y. [DOI] [PubMed] [Google Scholar]
- Bhave M, Morris CF. Molecular genetics of puroindolines and related genes: regulation of expression, membrane binding properties and applications. Plant Mol Biol. 2008;66:221–231. doi: 10.1007/s11103-007-9264-6. [DOI] [PubMed] [Google Scholar]
- Capparelli R, Amoroso MG, Palumbo D, Iannaccone M, Faleri C, Cresti M. Two plant puroindolines colocalize in wheat seed and in vitro synergistically fight against pathogens. Plant Mol Biol. 2005;58:857–867. doi: 10.1007/s11103-005-8270-9. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Trans Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- Clemente TE, Cahoon EB. Soybean Oil: Genetic Approaches for Modification of Functionality and Total Content. Plant Phys. 2009;151(3):1030–1040. doi: 10.1104/pp.109.146282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douliez JP, Michon T, Elmorajani K, Marion D. Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J Cereal Sci. 2000;32:1–20. [Google Scholar]
- Dudley JW, Lambert RJ. 100 Generations of selection for oil and protein in corn. Plant Breed Rev. 2004;24(1):79–110. [Google Scholar]
- Feiz L, Beecher BS, Martin JM, Giroux MJ. In planta mutagenesis determines the functional regions of the wheat puroindoline proteins. Genetics. 2009a;183(3):853–860. doi: 10.1534/genetics.109.106013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiz L, Wanjugi HW, Melnyk CW, Altosaar I, Martin JM, Giroux MJ. Puroindolines co-localize to the starch granule surface and increase seed bound polar lipid content. J Cereal Sci. 2009b;50:91–98. [Google Scholar]
- Finnie SM, Jeannotte R, Morris CF, Giroux MJ, Faubion JM. Variation in polar lipids located on the surface of wheat starch. J Cereal Sci. 2010;51:73–80. [Google Scholar]
- Frame BR, Zhang HY, Cocciolone SM, Sidorenko LV, Dietrich CR, Pegg SE, Zhen SF, Schnable PS, Wang K. Production of transgenic maize from bombarded type II callus: effect of gold particle size and callus morphology on transformation efficiency. In Vitro Cell Dev Biol Plant. 2000;36:21–29. [Google Scholar]
- Gautier MF, Aleman ME, Guirao A, Marioen D, Joudrier P. Triticum aestivum puroindolines, two basic cysteine-rich seed proteins: cDNA analysis and developmental gene expression. Plant Mol Biol. 1994;25:43–57. doi: 10.1007/BF00024197. [DOI] [PubMed] [Google Scholar]
- Gautier MF, Cosson P, Guirao A, Alary R, Joudrier P. Puroindoline genes are highly conserved in diploid ancestor wheats and related species but absent in tetraploid Triticum species. Plant Sci. 2000;153:81–91. [Google Scholar]
- Giroux MJ, Morris CF. A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch-surface friabilin. Theor Appl Genet. 1997;95:857–864. [Google Scholar]
- Giroux MJ, Morris CF. Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc Natl Acad Sci USA. 1998;95:6262–6266. doi: 10.1073/pnas.95.11.6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux MJ, Sripo T, Gerhardt S, Sherwood JE. Puroindolines: their role in grain hardness and plant defense. In: Stephen EH, editor. In biotechnology and genetic engineering reviews. Vol. 20. Intercept; Hampshire: 2003. pp. 277–290. [DOI] [PubMed] [Google Scholar]
- Greenblatt GA, Bettge AD, Morris CF. Relationship between endosperm texture and the occurrence of friabilin and bound polar lipids on wheat starch. Cereal Chem. 1995;72(2):172–176. [Google Scholar]
- Hayat MA. Microscopy, immunohistochemistry, and antigen retrieval methods: for light and electron microscopy. Kluwer Academic Publishers; New York: 2002. [Google Scholar]
- Huang AHC. Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:177–200. [Google Scholar]
- Jaworski J, Cahoon EB. Industrial oils from transgenic plants. Curr Opin Plant Bio. 2003;6:178–184. doi: 10.1016/s1369-5266(03)00013-x. [DOI] [PubMed] [Google Scholar]
- Kooijman M, Orsel R, Hessing M, Hamer RJ, Bekkers ACAPA. Spectroscopic characterisation of the lipid-binding properties of wheat puroindolines. J Cereal Sci. 1997;26(2):145–159. [Google Scholar]
- Krishnamurthy K, Giroux MJ. Expression of wheat puroindolines genes in transgenic rice enhances grain softness. Nat Biotechnol. 2001;19:162–166. doi: 10.1038/84435. [DOI] [PubMed] [Google Scholar]
- Laurie CC, Chasalow SD, LeDeaux JR, McCarroll R, Bush D, Hauge B, Lai C, Clark D, Rocheford TR, Dudley JW. The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics. 2004;168:2141–2155. doi: 10.1534/genetics.104.029686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan T, Blochet JE, Désormeaux A, Marion D, Pézolet M. Determination of the secondary structure and conformation of puroindolines by infrared and Raman spectroscopy. Biochemistry. 1996;35:12712–12722. doi: 10.1021/bi960869n. [DOI] [PubMed] [Google Scholar]
- Leprince O, van Aelst AC, Pritchard HW, Murphy DJ. Oleosins prevent oil-body coalescence during seed imbibition as suggested by a low-temperature scanning electron microscope study of desiccation-tolerant and -sensitive oilseeds. Planta. 1997;204:109–119. [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J. Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry. 2006;67:904–915. doi: 10.1016/j.phytochem.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Marion D, Gautier MF, Joudrier P, Ptak M, Pezolet M, Forest E, Clark DC, Broekaert W. Structure and function of wheat lipid binding proteins. In: al Cimino S. Martino., editor. Wheat kernel proteins: molecular and functional aspects. Proc Int'l Mtg, Universita Degli Sudi della Tuscia; Viterbo: 1994. pp. 175–180. [Google Scholar]
- Mohammadi M, Zaidi MA, Ochalski PA, Tanchak MA, Altosaar I. Immunodetection and immunolocalization of tryptophanins in oat seeds. Plant Science. 2007;172:579–587. [Google Scholar]
- Morris CF, Lillemo M, Simeone MC, Giroux MJ, Babb SL, Kidwell KK. Prevalence of puroindoline grain hardness genotypes among historically significant North American spring and winter wheats. Crop Sci. 2001;41(1):218–228. [Google Scholar]
- Poneleit CG, Alexander DE. Inheritance of linoleic and oleic acids in maize. Science. 1965;147:1585–1586. doi: 10.1126/science.147.3665.1585. [DOI] [PubMed] [Google Scholar]
- Purkrtova Z, Jolivet P, Miquel M, Chardot T. Structure and function of seed lipid body-associated proteins. C R Biologies. 2008;331:746–754. doi: 10.1016/j.crvi.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Riede CR, Anderson JA. Linkage of RFLP markers to an aluminum tolerance gene in wheat. Crop Sci. 1996;36:905–909. [Google Scholar]
- Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell. 2006;18:1961–1974. doi: 10.1105/tpc.106.041269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski MS, Baldwin AR. Fatty acid compositions of corn oils in relation to oil contents of the kernels. J Am Oil Chem Soc. 1954;31:414–416. [Google Scholar]
- Song XF, Song TM, Dai JR, Rocheford TR. QTL mapping of kernel oil concentration with high-oil maize by SSR markers. Maydica. 2004;49:41–48. [Google Scholar]
- Ting JTL, Lee K, Ratnayake C, Platt KA, Balsamo RA, Huang AHC. Oleosin genes in maize kernels having diverse oil contents are constitutively expressed independent of oil contents. Planta. 1996;199:158–165. doi: 10.1007/BF00196892. [DOI] [PubMed] [Google Scholar]
- Van Rooijen GJ, Moloney MM. Plant seed oil-bodies as carriers for foreign proteins. Biotechnology (NY) 1995;13:72–77. doi: 10.1038/nbt0195-72. [DOI] [PubMed] [Google Scholar]
- Vignaux N, Fox SR, Johnson LA. A 10-g laboratory wet-milling procedure for maize and comparison with larger scale laboratory procedures. Cereal Chem. 2006;83(5):482–490. [Google Scholar]
- Wall ML, Wheeler H, Huebsch MP, Smith JC, Figeys D, Altosaar I. The tryptophan rich domain of puroindoline is directly associated with the starch granule surface as judged by tryptic shaving and mass spectrometry. J Cereal Sci. 2010 in press doi:10.1016/j.jcs.2010.04.002. [Google Scholar]
- Wanjugi HW, Hogg AC, Martin JM, Giroux MJ. The role of puroindoline A and B individually and in combination on grain hardness and starch association. Crop Sci. 2007;47:67–76. [Google Scholar]
- Wassom JJ, Mikkelineni V, Bohn MO, Rocheford TR. QTL for fatty acid composition of maize kernel oil in Illinois high oil × B73 backcross-derived lines. Crop Sci. 2008;48:69–78. [Google Scholar]
- Watson SA. Corn and sorghum starches: Production. In: Whistler RL, BeMiller JN, Paschall EF, editors. Starch: chemistry and technology. Academic Press; Orlando: 1984. pp. 417–418. [Google Scholar]
- Weber EJ. Lipids of the kernel. In: Watson SA, Ramstad PE, editors. Corn: chemistry and technology. American Association of Cereal Chemists; St. Paul, MN: 1987. pp. 311–350. [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses: Role of phospholipase D{alpha} in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- White PJ, Pollak LM, Duvick S. Improving the fatty acid composition of corn oil by using germplasm introgression. Lipid Technol. 2007;19(2):35–38. [Google Scholar]
- Zaidi MA, Cheng XY, Altosaar I. Characterization of left-border flanking sequences of T-DNA integration in transgenic rice (Oryza sativa L.) expressing cry1Ab. Cereal Res Comm. 2007;35(3):1375–1383. [Google Scholar]
- Zhang J, Martin JM, Beecher B, Morris CF, Hannah LC, Giroux MJ. Seed-specific expression of the wheat puroindoline genes improves maize wet milling yields. Plant Biotech J. 2009;7:733–743. doi: 10.1111/j.1467-7652.2009.00438.x. [DOI] [PubMed] [Google Scholar]
- Zheng PZ, Allen WB, Roesler K, Williams ME, Zhang S, Li J, Glassman K, Ranch J, Nubel D, Solawetz W, Bhattramakki D, Llaca V, Deschamps S, Zhong G-Y, Tarczynski MC, Shen B. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat Genet. 2008;40:367–372. doi: 10.1038/ng.85. [DOI] [PubMed] [Google Scholar]