Abstract
Epithelial-to-mesenchymal transition (EMT) and the reverse process (MET) plays central role in organ developmental biology. It is a fine tuned process that when disturbed leads to pathological conditions especially cancers with aggressive and metastatic behavior. Snail is an oncogene that has been well established to be a promoter of EMT through direct repression of epithelial morphology promoter E-cadherin. It can function in the nucleus, in the cytosol and as discovered recently, extracellularly through secretory vesicular structures. The intracellular transport of snail has for long been shown to be regulated by the nuclear pore complex. One of the Karyopherins, importin alpha, mediates snail import, while importin beta/exportin 1 (Xpo1) or chromosome maintenance region 1 (CRM1) is its major nuclear exporter. A number of additional biological regulators are emerging that directly modulate Snail stability by altering its subcellular localization. These observations indicate that targeting the nuclear transport machinery could be an important and as of yet, unexplored avenue for therapeutic intervention against the EMT processes in cancer. In parallel, a number of novel agents that disrupt nuclear transport have recently been discovered and are being explored for their anti-cancer effects in the early clinical settings. Through this review we provide insights on the mechanisms regulating snail subcellular localization and how this impacts EMT. We discuss strategies on how the nuclear transport function can be harnessed to rein in EMT through modulation of snail signaling.
Keywords: Epithelial-to-mesenchymal transition, EMT, Nuclear Transport, Karyopherin, Importin, Exportin, Xpo1, CRM1, SNAIL, SNAIL2, HMLE-SNAIL
1. Introduction
Epithelial-to-Mesenchymal transition (EMT) is a complex phenomenon in which cancer cells lose their polarity undergo changes from epithelial to mesenchymal morphology thereby achieving plasticity that confers an invasive and metastatic behavior [1]. It is a fine tuned process regulated by a number of proteins that are strategically distributed in the nuclear and cytosolic compartments of cancer cells. A number of different parallel signaling pathways interact in the development of EMT [2]. Over the years, these pathways have been well studied leading to deeper characterization of different EMT promoting proteins and transcription factors (TFs), as well as their localization within and outside the cellular compartments [3]. Major EMT regulating proteins and TFs, such as wnt/β-catenin, notch, TGF-β, Twist and Snail are recognized to undergo nuclear-cytosolic shuttling using specialized transporters: Karyopherins [4]. These observations highlight that the nuclear protein transport process may be playing an integral part in the EMT signaling. The karyopherin importin alpha shuttles in proteins with nuclear localization signal sequence (NLS) [5]. The export of most of the EMT regulating TFs is mediated exclusively by Exportin1/XPO1 [also known as chromosome maintenance region 1 (CRM1)] through nuclear exclusion sequence (NES) recognition [6]. For long it has been fairly well recognized that in addition to transcriptional regulation, the activity of the TFs can be controlled by changing their cellular location which permits rapid response to signals, resulting in a powerful modulation of the biological system [7]. While, disease induced changes in expression of nuclear importer proteins have not been very well characterized, it is fairly unequivocally well recognized that nuclear exporters particularly CRM1, are often aberrantly expressed in cancer [8]. Nevertheless, till date, not many studies have looked into how disturbed nuclear export may interfere with EMT signaling. As discussed below, we propose that the role of aberrant nuclear export is critical to understanding the regulation of EMT through major TFs that they directly target (here a special focus on the Snail protein). As a number of different specific inhibitors of nuclear export are being developed and tested clinically, this may become an attractive therapeutic strategy to interfere in the EMT pathways.
2. The Snail family transcription factors and EMT
The transcription factors (TFs) of the Snail family are best recognized for being the direct repressors of epithelial morphology promoter E-cadherin transcription that drives EMT [9]. Snail family TFs play central role during the embryogenesis processes of both invertebrates and higher order animals where they regulate the cell movements necessary for the formation of the mesoderm [10]. Their involvement in the formation of vertebrate neural crest cells has been appreciated for more than two decades [11]. While Snail induced migratory and invasive behavior in developmental cells is vital for embryonic development, the same becomes problematic when aberrantly activated in later stages especially in pathological states such as cancer [12]. It is well established that enhancement in Snail gene expression in primary tumors promotes cellular motility and the consequent acquisition of metastatic properties [13,14]. On the other hand, in non-transformed cells, the enhancement in snail protein expression induces fibrosis like features [15]. Studies have clearly demonstrated that targeted down-regulation of snail can reverse EMT [16]. Human snail is a 264-amino acid nuclear protein with an amino-terminal basic amino acid-rich domain (SNAG domain) and a carboxyl-terminal DNA-binding domain (zinc finger domain) (Figure 1). The Snail superfamily also include the Scratch proteins [17]. More than 100 targets of Snail have been identified to date from all metazoan groups, with five family members in vertebrates: Snail1, Snail 2, Snail3 and Scratch1 and Scratch2. The domain structures of all Snail and Scratch TFs is conserved, i.e. having a divergent N-terminal half of the protein and a highly conserved C-terminal half as the DNA binding domain (DBD) which contains four to six zinc fingers (ZFs) of the C2H2 type [17] (Figure 1). The expression of Snail family genes is regulated at the transcriptional level by many signaling molecules, including FGF [18], Wnt [19], TGFβ [20] which collectively form the building blocks of the microenvironment that serves as a niche for EMT [21].
Figure 1. Snail structure.
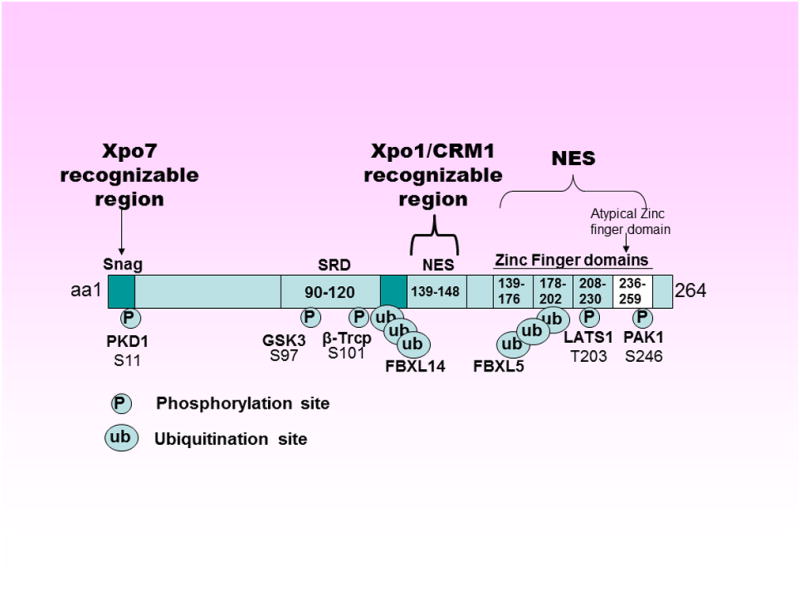
The N-terminal region [amino acid (aa) 1-150] of the Snail protein contains a SNAG (domain (aa 1-9) which includes the consensus sequence PRSFLV found in all Snail family members. This motif is highly conserved among species and also found in several other transcription factors where it is associated with repressive functions. The nuclear exporter Xpo5 recognizes SNAG domain and mediates SNAIL nuclear export. A serine-rich domain (SRD: aa 90-120) and a nuclear export sequence (NES: aa 139-148) are involved in the regulation of Snail protein stability and Xpo1/CRM1 mediated nuclear export, respectively. The C-terminal portion (aa 151-264) contains 3 typical (154-176, 178-202, 208-230) and one atypical (236-259) C2H2-type zinc finger (ZF) domains that serves as a NLS. A number of phosphorylation and ubiquitination sites exist on Snail that collectively regulates its turnover.
3. Cellular localization of snail family proteins
As transcription factors that require sequence specific alignment on DNA for gene regulation, snail proteins must translocate to the cell nucleus in order to be functional. Like all the proteins, snail family members cannot passively diffuse through the nuclear membrane and require a carrier for their nuclear import or export. Their translocation requires energy and in most of the cases, it is mediated importin-β (Impβ/KPNB1) belonging to the karyopherin family proteins [22]. These receptors are 90–130 kDa soluble proteins interacting with the cargo they are going to transport, the transport proteins and the GTPase Ran [23] (Figure 2). The importins mediate transport between the cytoplasm and the nucleus, interacting with their cargoes that carry specific amino acid sequences called nuclear localization signals (NLSs) [24]. These interactions can be direct or they may be mediated by karyopherin family that recognize and bind to the NLS present in many of the proteins imported by Importin β [25]. The directionality of the nuclear transport is imposed by a gradient formed by RanGTP across the nuclear envelope (higher concentration of RanGTP in the nucleus and a lower concentration in the cytoplasm) [26]. Importins exclusively interact with their cargoes in the cytoplasm and relocate them to the nucleus where they interact with RanGTP. The RanGTP binding in the nucleus causes lowering of the affinity of the importins for their substrates leading to their release. The RanGTP-bound importins relocate in the cytosol where, through the involvement of RanGAP and RanBP1, the GTP bound to Ran is hydrolyzed and Ran is released from the importin [27]. The importin can then start a new cycle of nuclear import. Taken together, these findings show that nuclear transporters (both importin and exportins) play central role in the biology of Snail (or other EMT promoting TFs with NES and NLS). Therefore, modulation of nuclear transport proteins should in principle result in alterations in Snail cellular localization and consequently impact snail mediated EMT signaling.
Figure 2. Snail Nuclear Import and Export a Mechanistic Summary.
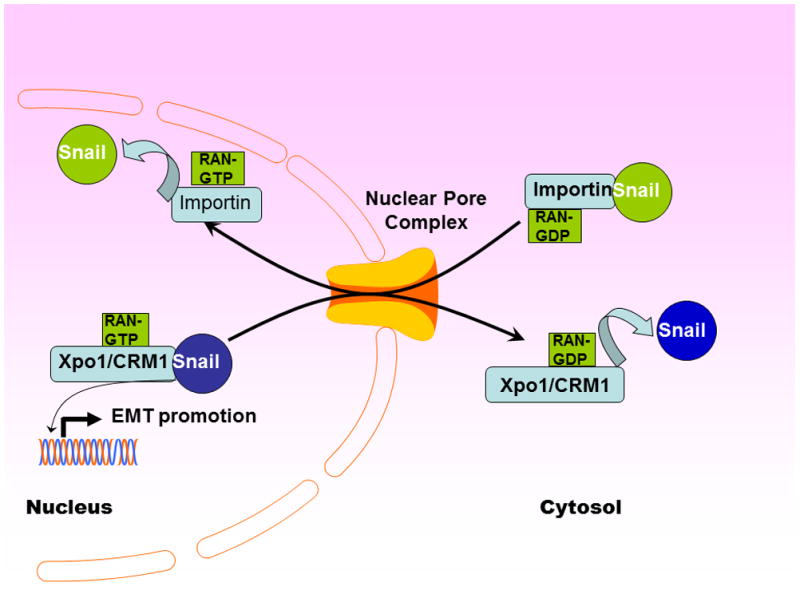
Snail Nuclear import and export are governed by evolutionary conserved nuclear transporters belonging to the Karyopherin family. An energy consuming process, Snail nuclear transport is mediated by nucleotide exchange factors RanGTP.
3.1 Nuclear import mechanisms of snail family proteins
The regulatory mechanisms that promote Snail nuclear import and enhance its stability have been well investigated. Among the earliest studies, Yamasaki and colleagues utilized a number of different fusion proteins containing a green fluorescent protein (GFP) to generate a series of the Snail fragments to analyze their subcellular localization [28]. In their studies, the fusion of the four zinc fingers to GFP led to the targeting of GFP to the nucleus, indicating that the zinc finger domain is sufficient for nuclear localization. More convincing evidence came from experiments where an in vitro transport system was used and the nuclear import of Snail was reconstituted by importin in the presence of Ran and NTF2. This approach further demonstrated that Snail binds directly to importin in a zinc finger domain-dependent manner. These results indicated that zinc finger domain of Snail functions as a nuclear localization signal and Snail can be transported into the nucleus in an importin-mediated manner. Interestingly, the above studies also highlighted that all four zinc fingers are necessary for efficient nuclear localization, because removing of any one zinc finger alone or in combination resulted in a decreased nuclear accumulation. It is likely that all four fingers are required for the coordination of the structure of the carboxyl terminal domain to interact efficiently with the nuclear import machinery and actually function as the NLS of Snail. It should be noted that there remains a possibility that deletion of any zinc finger domains or disruption of the ternary structure of zinc fingers may cause a loss of DNA binding ability, resulting in a loss of nuclear retention. Therefore, strategies disrupting the ternary complex or blocking access of the importins to the 4 zinc finger domains can certainly be postulated to influence the nuclear retention and the importin binding activity that may provide opportunity for targeted inhibition of nuclear localization of snail.
3.2 Nuclear exclusion mechanisms of snail
Several mechanisms can explain the effect of the exportin recognizable domains in Snail on its own subcellular localization. These domains may act as an anchor linking Snail to specific cytosolic proteins; they may inhibit its nuclear import, or mediate its nuclear export. It is well known that the most common mechanism of nuclear export of proteins in eukaryotic cells is through Xpo1/CRM1-dependent systems. Most exportable proteins have a hydrophobic Leu-rich sequence and snail is no exception. By searching in this direction, the nuclear exclusion sequence (NES) of mouse Snail protein (aa residues 132-143) was discovered more than a decade ago [29]. It was shown that the specific Snail Leu-rich amino acid sequence is similar to those described for in other CRM1-targeted export proteins. These studies identified the sequence LGQLPKQLARLS, between the aa residues 132-143 of murine Snail, that was shown to perfectly match the consensus sequence previously defined for established NESs [LX(1-3)LX(2-3)LXL]. Interestingly, replacement of Leucine with Valine, as commonly observed to occur in human Snail (Leu135 to Val), has been observed in some other NESs, as well as longer spaces among leucines [30]. This putative NES is not present in Slug, a Snail homolog, whose localization is found to be exclusively nuclear. Deletion of part of this sequence (residues 138-151, i.e removal of last two leucines) from a GFP-N-terminal domain fusion protein was sufficient to impair Snail’s exclusion from the nucleus. Further confirmation came from Xpo1/CRM1 inhibition studies using a natural product derived specific inhibitor Leptomycin B (LMB) that block the formation of the CRM1/Ran-GTP/nuclear protein complex necessary for export. In their experiments, treatment of transfected RWP-1 cells with LMB caused rapid translocation of Snail from the cytosol to the nucleus within few minutes. Similar results were obtained when the effect of LMB on the location of other exclusively cytosolic Snail constructs was studied. In the presence of this drug, both the fragment containing the complete regulatory domain (amino acids 1 to 151) and the one containing the NES (82 to 151) were detected mostly in the nucleus.
Aside from CRM1, studies have shown that exportin 5 or Xpo5, which is a major dsRNA exporter (especially pre-miRNA exporter), can also mediate Snail nuclear export [31]. However, nuclear export by Xpo5 does not occur through the typical CRM1 recognizable NES in the zinc finger domains (discussed above). Rather, Xpo5 recognizes a snag domain in Snail protein that serves as a NES mediating the nuclear export. Interestingly, Xpo5 mediated snail export essentially requires binding of eukaryotic elongation factor 1 A (eEF1A). When Xpo5 binds aminoacyl-tRNAs (aa-tRNAs), this complex can recruit and co-transport eEF1A that can interact with the snag domain for E-cadherin interacting (bound snail).
4. Snail nuclear transport modulators and their impact on EMT
There are a number of mechanisms that either enhance or retard both snail nuclear import and export processes (Figure 3). The importin α is recognized to act as a negative regulator of snail import. Importin α is known to act as an adaptor for importin β1 during different protein nuclear import processes [32]. In addition to its adaptor function, several different studies have convincingly demonstrated that importin α in many instances inhibits the nuclear import of different proteins. Thus, the inhibition of Snail nuclear import may be one of the characteristic features of importin α. It is speculated that negative biological regulators or chemical agents that enhance the expression of importin α may suppress snail nuclear localization leading to down-regulation of the protein activity.
Figure 3. Snail Regulatory Mechanisms.
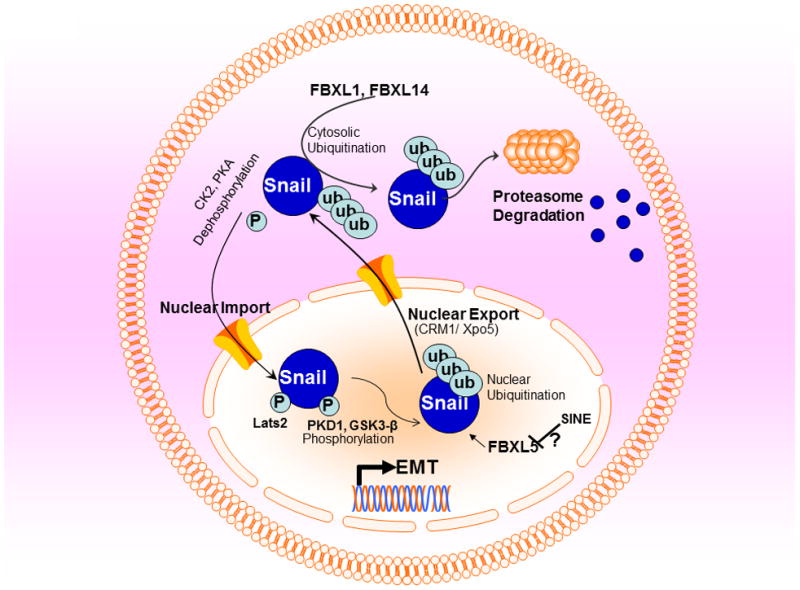
Snail is highly labile protein that is rapidly degraded through proteasomal system. The F-box proteins (components of SCF ubiquitin-ligase complexes namely FBXL1 and FBXL14 enhance the nuclear export and induce ubiquitin-mediated proteolysis of Snail. FBXL5 induces nuclear ubiquitination of snail leading to its nuclear degradation. On the other hand phosphorylation by GSK3-β, PKA1, PDK1 and other can stabilize Snail and induce its nuclear retention.
Phosphorylation has dual effect on snail localization and function [29]. Phosphorylation of a Ser-rich sequence adjacent to the NES of Snail facilitates its export and the expression levels of snail phosphorylation regulators could serve as important regulators of the nuclear localization status of snail. Snail phosphorylation by various kinases such as protein kinase D1 (PKD1) and glycogen synthase kinase-3β (GSK-3β) primed by CK1 can trigger its nuclear export and cytosolic degradation [33]. In this direction, Du and colleagues elucidated the consequence of Snail phosphorylation on EMT signaling [34]. Their studies demonstrated that PKD1 phosphorylates Ser11 (S11) on Snail, triggering its nuclear export via 14-3-3σ binding. On the other hand, Snail S11 mutation causes acquisition of mesenchymal traits and expression of stem cell like markers. Conversely, Snail can be stabilized by small C-terminal domain phosphatase-meditated dephosphorylation or through CK2 and PKA phosphorylation, or O-GlcNAc modification and the nuclear localization of snail can be increased in cancer cells, upon PAK1 phosphorylation (snail phosphorylation mechanisms comprehensively reviewed in [35]). In another important study, using a bioluminescence cell-based assay to evaluate Snail1 protein stability, Zhang and colleagues investigated positive and negative post-translational regulators of total cellular Snail1 protein level [36]. A human kinome RNAi screen identified the protein kinase Lats2 stabilizing cellular Snail1 protein levels post-translationally. In this study Lats2 was shown to modulate snail phosphorylation by interacting with Snail1 in cells and directly phosphorylating Snail1 at T203 in response to multiple signals that activate Lats2, such as TGFβ-induced EMT. Lats2 mediated phosphorylation of Snail1 at T203 occurs in the nucleus and serves to retain Snail1 in the nucleus thereby enhancing protein stability and function. Interestingly a number of studies have shown that de-regulation of Lats2 results cancer invasiveness and poor prognosis [37,38]. Lats2 down-regulation and the consequent enhancement in tumor metastatic potential has also been correlated with disturbed miRNA signaling [39]. In cellular models, it was shown that that Lats2 can affect TGFβ-induced EMT and that this effect depends upon the presence of Snail1. Lats2 has been shown to potentiate Snail1 EMT-promoting function in zebrafish and mouse embryo development models, as well as enhance Snail1’s capacity to regulate tumor cell invasion/migration. While TGFβ mediated phosphorylation and degradation causes Snail turnover. Additionally it has been shown that the activation of Wnt signaling suppresses the activity of TGFβ which results in both Snail and β-catenin stabilization [40]. These findings indicate that there is some cooperation between WNT signaling and other Snail mediated pathways, such as FGF, in the triggering of the EMT. This cooperation has already been highlighted in several developmental systems, such as neural crest mesoderm [41]. For example, when Snail1 activity is maintained, E-cadherin is repressed and is therefore not available to bind β-catenin and form adherens junctions. As a result, β-catenin is available to bind to TCF/LEF and to act as a transcription factor, promoting WNT signaling. Although this situation will only occur concomitantly with an inactive β-catenin degradation system, WNT signaling can increase Snail1 function by preventing its nuclear export and degradation, allowing Snail1 promoted EMT. For example, Yadi et al, showed that the small C-terminal domain phosphatase (SCP) is a specific phosphatase for Snail [42]. SCP interacted and co-localized with Snail in the nucleus and that SCP expression induced its dephosphorylation and stabilization in vitro and in vivo leading to enhancement in EMT and consequent invasive behavior of the tested cells. Thus, it is imperative that the determination of regulation mechanisms of snail import and export will allow better understanding of its context dependent role in epithelial and/or mesenchymal tumors.
As mentioned above, Snail is a highly unstable protein that undergoes rapid turnover. In the nucleus, its turnover is decreased while in the cytosol it is rapidly degraded by proteasomes [43]. Post-translational modifications such as phosphorylation, ubiquitination, and lysine oxidation are recognized to collectively influence snail (particularly Snail1) protein stability, sub-cellular localization, and activity [44]. There are two RING finger ubiquitin ligases of the F-box subfamily containing the multimeric complex Skp1-Cullin-Rbx1-F-box (SCF) that have been shown to participate in Snail1 proteasomal degradation. SCFβ-TrCP1/FBXW1 is recognized to polyubiquitinate Snail1 after its phosphorylation by GSK-3β [29]. Snail1 is also targeted by SCFPpa/FBXL14, which is a ubiquitin ligase and acts as a master regulator of the EMT process, as it modulates not only Snail1 but also Snail2, Twist1 and Zeb2 [45]. The two ligases work in very distinct manner. Unlike SCFβ-TrCP1/FBXW1, FBXL14 does not require previous Snail1 phosphorylation by GSK-3β. However, both ligases are present and act exclusively in the cytosol. Additionally, the p53 negative regulator murine double minute 2 (Mdm2), a monomeric ring finger E3, can also degrade the snail family member snail2 [46]. In addition to cytosolic ubiquitination, there are certain mechanisms that negatively regulate nuclear Snail as well. Very recently, using a short hairpin RNA screening, Vinas-Castells and colleagues have identified FBXL5 as a novel nuclear snail ubiquitin ligase [47]. Their investigations pointed towards a pre-dominantly cytosolic localization of FBXL5 protein that could be reversed by LMB, indicating a CRM1 mediated export regulatory mechanism for this Snail regulator. FBXL5 was also shown to be down-regulated by numerous stresses such as γ irradiation and transition metal ion depletion which were correlated with snail stability. Earlier work has identified the FBXL5 binding sequence in Snail1 and Snail2 that were mapped to the carboxy-terminal half of the protein (Snail-CT), mainly to ZnF2 (Figure 1). However, unlike FBXL14 which can regulate other EMT markers such as ZEB1 and Twist1, FBXL5 has more specific effects that are restricted to Snail1 and therefore have confined roles. Collectively, the above described studies clearly show that snail is under a very tight regulation in cancer cells and the over-expression or disturbances in the homeostasis of its direct and indirect regulators can certainly impact its activity and localization, which can then manifest into deregulation of its targeted genes and consequently EMT.
4.1 Modulation of snail induced EMT through exportin inhibition
Given that nuclear transport plays integral role in snail biology and EMT, targeted inhibition of nuclear export and import of the protein is speculated to become an attractive and new form of therapeutic strategy to tame this master regulator. A number of agents have been developed that can target the nuclear exporter CRM1 fairly specifically. The first agent in this series was Leptomycin B (LMB), a natural product derived drug-like compound that was originally developed as an anti-fungal agent [48]. LMB showed potent irreversible inhibitory activity against CRM1. Interestingly, some of the earlier studies utilized LMB to evaluate nuclear ubiquitination mechanisms on snail nuclear retention. These investigations clearly verified that LMB mediated CRM1 inhibition can indeed cause nuclear retention of snail. However, these studies fell short of investigating the impact of such snail retention on the EMT process, especially whether this causes reversal to MET. Additionally, LMB did not make a clinical impact and was discontinued from human application after just a single clinical trial due in part to its associated toxicity [49]. A number of newer agents (analogs of LMB) have been developed that show similar CRM1 inhibitory activity [50]. However, their clinical efficacy is yet to be evaluated. We have developed a potent exportin inhibitor Selinexor that is currently in multiple Phase I clinical trials for both solid tumors and hematological malignancies (Clinical Trial identifiers NCT01986348, NCT02093403, NCT02025985, NCT02088541, NCT02078349, NCT02091245) [51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66]. Additionally, another CRM1 inhibitor, Velidexor, is also being evaluated in canine cancer models [67]. We had earlier shown that these inhibitors can suppress growth of well recognized gemcitabine resistant pancreatic cancer cells with EMT phenotype that over-express CRM1 (unpublished work APA abstract 2013). Recently, using a cellular model system of snail transduced human mammary epithelial (HMLE) cells (HMLE-SNAIL) we also demonstrated in vitro activity of Selinexor, LMB and related compounds in reversing mesenchymal phenotype that was concurrent with growth inhibition and induction of apoptosis [68]. CRM1 inhibitors used at clinically relevant concentrations can reverse EMT phenotype and also restore epithelial markers in these cells. Computational analysis of KPT-185 (an analog of Selinexor) indicates that drug treatment results in global re-organization of cytosolic proteins (majority of which regulate snail and other EMT promoters) into the HMLE-snail cell nucleus. Our laboratory is currently evaluating the molecular mechanism of exportin inhibition and how it reverses EMT. Our initial results indicate that Selinexor treatment results in snail nuclear degradation that is mediated by the nuclear retention of F-Box protein FBXL5. It is recognized that FBXL5 may not be the only repressor that is causing Snail degradation and that CRM1 inhibition by Selinexor may be inducing the relocalization of a number of different important tumor suppressors that direct or indirectly target Snail. Nevertheless, these preliminary studies do indicate that interfering with the nuclear transport can indeed impact EMT signaling and could potentially become a new approach to tame in one of the most difficult to treat, metastatic and EMT harboring tumors.
5. Conclusions and future perspectives
The foundations of metastasis, a major cause of death from cancer, are laid on EMT which has remained a major obstacle to successful anti-cancer therapies. Biologically EMT is a complex process that involves a number of different parallel signaling some of which are known, and others yet to be explored. What is clear is that subcellular localization of various EMT promoting TFs plays central role in this plasticity process and adds another tier of complexity to the already intriguingly multifaceted phenomenon. While proper subcellular localization is critical to any protein function, it is especially important for major TFs that regulate gene expression through sequence specific alignment to DNA for which nuclear retention is vital. Snail is considered as one of the primary drivers of EMT. It is under a very tight regulation by the nuclear compartmentalization mechanisms that regulate its stability and protection from ubiquitination. Aberration in nuclear transport mechanisms, which is commonly found in cancer, shifts this fine-tuned compartmentalization balance leading to mis-localization of the protein which results in many different de-regulatory signaling including EMT. A number of agents have recently been shown to target the nuclear transport machinery with great specificity. Preliminary indications in cellular models do show that such agents (for example Selinexor) can indeed cause reversal of mesenchymal phenotype (to epithelial) and consequent cell death. These findings open some distinct and less explored possibilities for targeting EMT at the nuclear pore. Such approaches may become part of future therapies against aggressive forms of cancers that sustain on EMT, hence may result in better treatment outcomes.
Acknowledgments
NIH R21 1R21CA16984801 and 1R21CA17597401 to RMM is acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Nakaya Y, Sheng G. EMT in developmental morphogenesis. Cancer Lett. 2013 doi: 10.1016/j.canlet.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 2.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 3.Savagner P. Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays. 2001;23:912–23. doi: 10.1002/bies.1132. [DOI] [PubMed] [Google Scholar]
- 4.Azmi AS. Unveiling the role of nuclear transport in epithelial-to-mesenchymal transition. Curr Cancer Drug Targets. 2013;13:906–14. doi: 10.2174/15680096113136660096. [DOI] [PubMed] [Google Scholar]
- 5.Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–58. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong X, Biswas A, Suel KE, Jackson LK, Martinez R, Gu H, Chook YM. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature. 2009;458:1136–41. doi: 10.1038/nature07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vandromme M, Gauthier-Rouviere C, Lamb N, Fernandez A. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- 8.Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol. 2012;83:1021–32. doi: 10.1016/j.bcp.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–61. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 10.Technau U, Scholz CB. Origin and evolution of endoderm and mesoderm. Int J Dev Biol. 2003;47:531–9. [PubMed] [Google Scholar]
- 11.Baker CV. The evolution and elaboration of vertebrate neural crest cells. Curr Opin Genet Dev. 2008;18:536–43. doi: 10.1016/j.gde.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olmeda D, Jorda M, Peinado H, Fabra A, Cano A. Snail silencing effectively suppresses tumour growth and invasiveness. Oncogene. 2007;26:1862–74. doi: 10.1038/sj.onc.1209997. [DOI] [PubMed] [Google Scholar]
- 14.Olmeda D, Montes A, Moreno-Bueno G, Flores JM, Portillo F, Cano A. Snai1 and Snai2 collaborate on tumor growth and metastasis properties of mouse skin carcinoma cell lines. Oncogene. 2008;27:4690–701. doi: 10.1038/onc.2008.118. [DOI] [PubMed] [Google Scholar]
- 15.Boutet A, De Frutos CA, Maxwell PH, Mayol MJ, Romero J, Nieto MA. Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO J. 2006;25:5603–13. doi: 10.1038/sj.emboj.7601421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azmi AS, Bollig-Fischer A, Bao B, Park BJ, Lee SH, Yong-Song G, Dyson G, Reddy CK, Sarkar FH, Mohammad RM. Systems analysis reveals a transcriptional reversal of the mesenchymal phenotype induced by SNAIL-inhibitor GN-25. BMC Syst Biol. 2013;7:85. doi: 10.1186/1752-0509-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manzanares M, Locascio A, Nieto MA. The increasing complexity of the Snail gene superfamily in metazoan evolution. Trends Genet. 2001;17:178–81. doi: 10.1016/s0168-9525(01)02232-6. [DOI] [PubMed] [Google Scholar]
- 18.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 19.Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ. Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem. 2005;280:11740–8. doi: 10.1074/jbc.M413878200. [DOI] [PubMed] [Google Scholar]
- 20.Cho HJ, Baek KE, Saika S, Jeong MJ, Yoo J. Snail is required for transforming growth factor-beta-induced epithelial-mesenchymal transition by activating PI3 kinase/Akt signal pathway. Biochem Biophys Res Commun. 2007;353:337–43. doi: 10.1016/j.bbrc.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 21.Talbot LJ, Bhattacharya SD, Kuo PC. Epithelial-mesenchymal transition, the tumor microenvironment, and metastatic behavior of epithelial malignancies. Int J Biochem Mol Biol. 2012;3:117–36. [PMC free article] [PubMed] [Google Scholar]
- 22.Pemberton LF, Blobel G, Rosenblum JS. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–9. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 23.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–94. [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–7. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 25.Lee SJ, Matsuura Y, Liu SM, Stewart M. Structural basis for nuclear import complex dissociation by RanGTP. Nature. 2005;435:693–6. doi: 10.1038/nature03578. [DOI] [PubMed] [Google Scholar]
- 26.Rollenhagen C, Muhlhausser P, Kutay U, Pante N. Importin beta-depending nuclear import pathways: role of the adapter proteins in the docking and releasing steps. Mol Biol Cell. 2003;14:2104–15. doi: 10.1091/mbc.E02-06-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–94. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki H, Sekimoto T, Ohkubo T, Douchi T, Nagata Y, Ozawa M, Yoneda Y. Zinc finger domain of Snail functions as a nuclear localization signal for importin beta-mediated nuclear import pathway. Genes Cells. 2005;10:455–64. doi: 10.1111/j.1365-2443.2005.00850.x. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez D, Montserrat-Sentis B, Virgos-Soler A, Guaita S, Grueso J, Porta M, Puig I, Baulida J, Franci C, Garcia de Herreros A. Phosphorylation regulates the subcellular location and activity of the snail transcriptional repressor. Mol Cell Biol. 2003;23:5078–89. doi: 10.1128/MCB.23.14.5078-5089.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lischka P, Rosorius O, Trommer E, Stamminger T. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 2001;20:7271–83. doi: 10.1093/emboj/20.24.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mingot JM, Vega S, Cano A, Portillo F, Nieto MA. eEF1A mediates the nuclear export of SNAG-containing proteins via the Exportin5-aminoacyl-tRNA complex. Cell Rep. 2013;5:727–37. doi: 10.1016/j.celrep.2013.09.030. [DOI] [PubMed] [Google Scholar]
- 32.Sekimoto T, Miyamoto Y, Arai S, Yoneda Y. Importin alpha protein acts as a negative regulator for Snail protein nuclear import. J Biol Chem. 2011;286:15126–31. doi: 10.1074/jbc.M110.213579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, Cha SY, Ryu JK, Choi YJ, Kim J, Fearon ER, Weiss SJ. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 34.Du C, Zhang C, Hassan S, Biswas MH, Balaji KC. Protein kinase D1 suppresses epithelial-to-mesenchymal transition through phosphorylation of snail. Cancer Res. 2010;70:7810–9. doi: 10.1158/0008-5472.CAN-09-4481. [DOI] [PubMed] [Google Scholar]
- 35.Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–76. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- 36.Zhang K, Rodriguez-Aznar E, Yabuta N, Owen RJ, Mingot JM, Nojima H, Nieto MA, Longmore GD. Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. EMBO J. 2012;31:29–43. doi: 10.1038/emboj.2011.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, Noguchi S. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11:1380–5. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- 38.Huntoon CJ, Nye MD, Geng L, Peterson KL, Flatten KS, Haluska P, Kaufmann SH, Karnitz LM. Heat shock protein 90 inhibition depletes LATS1 and LATS2, two regulators of the mammalian hippo tumor suppressor pathway. Cancer Res. 2010;70:8642–50. doi: 10.1158/0008-5472.CAN-10-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang L, Du WW, Yang W, Rutnam ZJ, Peng C, Li H, O’Malley YQ, Askeland RW, Sugg S, Liu M, Mehta T, Deng Z, Yang BB. MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle. 2012;11:4352–65. doi: 10.4161/cc.22670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JY, Kim YM, Yang CH, Cho SK, Lee JW, Cho M. Functional regulation of Slug/Snail2 is dependent on GSK-3beta-mediated phosphorylation. FEBS J. 2012;279:2929–39. doi: 10.1111/j.1742-4658.2012.08674.x. [DOI] [PubMed] [Google Scholar]
- 41.Bastidas F, De Calisto J, Mayor R. Identification of neural crest competence territory: role of Wnt signaling. Dev Dyn. 2004;229:109–17. doi: 10.1002/dvdy.10486. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y, Evers BM, Zhou BP. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J Biol Chem. 2009;284:640–8. doi: 10.1074/jbc.M806916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–40. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 44.Barbera MJ, Puig I, Dominguez D, Julien-Grille S, Guaita-Esteruelas S, Peiro S, Baulida J, Franci C, Dedhar S, Larue L, Garcia de Herreros A. Regulation of Snail transcription during epithelial to mesenchymal transition of tumor cells. Oncogene. 2004;23:7345–54. doi: 10.1038/sj.onc.1207990. [DOI] [PubMed] [Google Scholar]
- 45.Vinas-Castells R, Beltran M, Valls G, Gomez I, Garcia JM, Montserrat-Sentis B, Baulida J, Bonilla F, de Herreros AG, Diaz VM. The hypoxia-controlled FBXL14 ubiquitin ligase targets SNAIL1 for proteasome degradation. J Biol Chem. 2010;285:3794–805. doi: 10.1074/jbc.M109.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim SO, Kim H, Jung G. p53 inhibits tumor cell invasion via the degradation of snail protein in hepatocellular carcinoma. FEBS Lett. 2010;584:2231–6. doi: 10.1016/j.febslet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Vinas-Castells R, Frias A, Robles-Lanuza E, Zhang K, Longmore GD, Garcia de Herreros A, Diaz VM. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic Acids Res. 2014;42:1079–94. doi: 10.1093/nar/gkt935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida M, Kudo N, Horinouchi S. Leptomycin: a specific inhibitor of protein nuclear export. Tanpakushitsu Kakusan Koso. 1999;44:1379–88. [PubMed] [Google Scholar]
- 49.Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer. 1996;74:648–9. doi: 10.1038/bjc.1996.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakakibara K, Saito N, Sato T, Suzuki A, Hasegawa Y, Friedman JM, Kufe DW, Vonhoff DD, Iwami T, Kawabe T. CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood. 2011;118:3922–31. doi: 10.1182/blood-2011-01-333138. [DOI] [PubMed] [Google Scholar]
- 51.Cheng Y, Holloway MP, Nguyen K, McCauley D, Landesman Y, Kauffman MG, Shacham S, Altura RA. XPO1 (CRM1) inhibition represses STAT3 activation to drive a survivin-dependent oncogenic switch in triple-negative breast cancer. Mol Cancer Ther. 2014;13:675–86. doi: 10.1158/1535-7163.MCT-13-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong Y, El-Gamal D, Dubovsky JA, Beckwith KA, Harrington BK, Williams KE, Goettl VM, Jha S, Mo X, Jones JA, Flynn JM, Maddocks KJ, Andritsos LA, McCauley D, Shacham S, Kauffman M, Byrd JC, Lapalombella R. Selinexor suppresses downstream effectors of B-cell activation, proliferation and migration in chronic lymphocytic leukemia cells. Leukemia. 2014 doi: 10.1038/leu.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner JG, Dawson J, Emmons MF, Cubitt CL, Kauffman M, Shacham S, Hazlehurst LA, Sullivan DM. CRM1 Inhibition Sensitizes Drug Resistant Human Myeloma Cells to Topoisomerase II and Proteasome Inhibitors both In Vitro and Ex Vivo. J Cancer. 2013;4:614–25. doi: 10.7150/jca.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker CJ, Oaks JJ, Santhanam R, Neviani P, Harb JG, Ferenchak G, Ellis JJ, Landesman Y, Eisfeld AK, Gabrail NY, Smith CL, Caligiuri MA, Hokland P, Roy DC, Reid A, Milojkovic D, Goldman JM, Apperley J, Garzon R, Marcucci G, Shacham S, Kauffman MG, Perrotti D. Preclinical and clinical efficacy of XPO1/CRM1 inhibition by the karyopherin inhibitor KPT-330 in Ph+ leukemias. Blood. 2013;122:3034–44. doi: 10.1182/blood-2013-04-495374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt J, Braggio E, Kortuem KM, Egan JB, Zhu YX, Xin CS, Tiedemann RE, Palmer SE, Garbitt VM, McCauley D, Kauffman M, Shacham S, Chesi M, Bergsagel PL, Stewart AK. Genome-wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT-276. Leukemia. 2013;27:2357–65. doi: 10.1038/leu.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salas Fragomeni RA, Chung HW, Landesman Y, Senapedis W, Saint-Martin JR, Tsao H, Flaherty KT, Shacham S, Kauffman M, Cusack JC. CRM1 and BRAF inhibition synergize and induce tumor regression in BRAF-mutant melanoma. Mol Cancer Ther. 2013;12:1171–9. doi: 10.1158/1535-7163.MCT-12-1171. [DOI] [PubMed] [Google Scholar]
- 57.Tai YT, Landesman Y, Acharya C, Calle Y, Zhong MY, Cea M, Tannenbaum D, Cagnetta A, Reagan M, Munshi AA, Senapedis W, Saint-Martin JR, Kashyap T, Shacham S, Kauffman M, Gu Y, Wu L, Ghobrial I, Zhan F, Kung AL, Schey SA, Richardson P, Munshi NC, Anderson KC. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: molecular mechanisms and therapeutic implications. Leukemia. 2014;28:155–65. doi: 10.1038/leu.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Azmi AS, Al-Katib A, Aboukameel A, McCauley D, Kauffman M, Shacham S, Mohammad RM. Selective inhibitors of nuclear export for the treatment of non-Hodgkin’s lymphomas. Haematologica. 2013;98:1098–106. doi: 10.3324/haematol.2012.074781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Etchin J, Sanda T, Mansour MR, Kentsis A, Montero J, Le BT, Christie AL, McCauley D, Rodig SJ, Kauffman M, Shacham S, Stone R, Letai A, Kung AL, Thomas Look A. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br J Haematol. 2013;161:117–27. doi: 10.1111/bjh.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azmi AS, Aboukameel A, Bao B, Sarkar FH, Philip PA, Kauffman M, Shacham S, Mohammad RM. Selective inhibitors of nuclear export block pancreatic cancer cell proliferation and reduce tumor growth in mice. Gastroenterology. 2013;144:447–56. doi: 10.1053/j.gastro.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inoue H, Kauffman M, Shacham S, Landesman Y, Yang J, Evans CP, Weiss RH. CRM1 blockade by selective inhibitors of nuclear export attenuates kidney cancer growth. J Urol. 2013;189:2317–26. doi: 10.1016/j.juro.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalid O, Toledo Warshaviak D, Shechter S, Sherman W, Shacham S. Consensus Induced Fit Docking (cIFD): methodology validation and application to the discovery of novel Crm1 inhibitors. J Comput Aided Mol Des. 2012;26:1217–28. doi: 10.1007/s10822-012-9611-9. [DOI] [PubMed] [Google Scholar]
- 63.Lapalombella R, Sun Q, Williams K, Tangeman L, Jha S, Zhong Y, Goettl V, Mahoney E, Berglund C, Gupta S, Farmer A, Mani R, Johnson AJ, Lucas D, Mo X, Daelemans D, Sandanayaka V, Shechter S, McCauley D, Shacham S, Kauffman M, Chook YM, Byrd JC. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood. 2012;120:4621–34. doi: 10.1182/blood-2012-05-429506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang K, Wang M, Tamayo AT, Shacham S, Kauffman M, Lee J, Zhang L, Ou Z, Li C, Sun L, Ford RJ, Pham LV. Novel selective inhibitors of nuclear export CRM1 antagonists for therapy in mantle cell lymphoma. Exp Hematol. 2013;41:67–78. doi: 10.1016/j.exphem.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 65.Etchin J, Sun Q, Kentsis A, Farmer A, Zhang ZC, Sanda T, Mansour MR, Barcelo C, McCauley D, Kauffman M, Shacham S, Christie AL, Kung AL, Rodig SJ, Chook YM, Look AT. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia. 2013;27:66–74. doi: 10.1038/leu.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M, Walker A, Klisovic R, Blum W, Caligiuri M, Croce CM, Marcucci G, Garzon R. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood. 2012;120:1765–73. doi: 10.1182/blood-2012-04-423160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.London CA, Bernabe LF, Barnard S, Kisseberth WC, Borgatti A, Henson M, Wilson H, Jensen K, Ito D, Modiano JF, Bear MD, Pennell ML, Saint-Martin JR, McCauley D, Kauffman M, Shacham S. Preclinical evaluation of the novel, orally bioavailable Selective Inhibitor of Nuclear Export (SINE) KPT-335 in spontaneous canine cancer: results of a phase I study. PLoS One. 2014;9:e87585. doi: 10.1371/journal.pone.0087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Azmi AS, Shacham S, Kauffman M, MCcauley D, Aboukameel A, Mohammad RM. Novel activity of selective inhibitors of nuclear export in epithelial-to-mesenchymal transition models [abstract]. Proceedings of the 105th Annual Meeting of the American Association for Cancer Research; 2014 Apr 5–9; San Diego, CA. Philadelphia (PA): AACR; 2014. Abstract nr [LB-185] [Google Scholar]


