Abstract
Nuclear factor, erythroid 2-like 2 (Nrf2) is a master transcription factor for cellular defense against endogenous and exogenous stresses by regulating expression of many antioxidant and detoxification genes. Here, we show that Nrf2 acts as a key pluripotency gene and a regulator of proteasome activity in human embryonic stem cells (hESCs). Nrf2 expression is highly enriched in hESCs and dramatically decreases upon differentiation. Nrf2 inhibition impairs both the self-renewal ability of hESCs and reestablishment of pluripotency during cellular reprogramming. Nrf2 activation can delay differentiation. During early hESC differentiation, Nrf2 closely co-localizes with OCT4 and NANOG. As an underlying mechanism, our data show that Nrf2 regulates proteasome activity in hESCs partially through proteasome maturation protein (POMP), a proteasome chaperone, which in turn controls the proliferation of self-renewing hESCs, three germ layer differentiation and cellular reprogramming. Even modest proteasome inhibition skews the balance of early differentiation toward mesendoderm at the expense of an ectodermal fate by decreasing the protein level of cyclin D1 and delaying the degradation of OCT4 and NANOG proteins. Taken together, our findings suggest a new potential link between environmental stress and stemness with Nrf2 and the proteasome coordinately positioned as key mediators.
Keywords: Human embryonic stem cells, Nrf2, Pluripotency, POMP, Proteasome
Introduction
Nrf2 is in the subfamily of the basic leucine zipper (bZip) transcription factors containing a cap “n” collar (CNC) structure [1]. The CNC-bZip domain allows Nrf2 to heterodimerize with a small Maf protein, which serves as a transcriptional cofactor. Nrf2 has a well-established role in activating cellular defense against endogenous and exogenous stresses [2, 3]. Normally, Nrf2 protein is sequestered in the cytoplasm by its binding partner KEAP1 and subsequently degraded by the ubiquitin-proteasome system. However, oxidative and xenobiotic stresses disrupt the Nrf2-KEAP1 interaction, and induce the nuclear translocation of Nrf2. Upon translocation, Nrf2 can activate its downstream target genes including NAD(P)H quinone oxidoreductase (NQO-1), Heme oxygenase 1 (HO-1) and the catalytic (GCLC) and modifier (GCLM) subunits of glutamate-cysteine ligase to initiate antioxidant responses. Among the 36 proteosome subunits tested in mouse liver, 19 subunits had an increased expression level upon Nrf2 activation. Specifically, Nrf2 regulates PSMB5 expression by directly binding to its promoter [4]. Nrf2 activator also increased the expression of proteasome subunits α4, β1, β2 and β5 in human fibroblasts [5]. Interestingly, antioxidant and ubiquitin/proteasome-related genes are among the most enriched transcripts and proteins identified in transcriptomic and proteomic studies of embryonic stem cells [6, 7]. Considering that Nrf2 is a common upstream regulator of these genes, Nrf2 has potential to serve as a master regulator of both redox homeostasis and proteostasis in embryonic stem cells. Recent papers suggest Nrf2 to have important functions in controlling proliferation and homeostasis of adult stem cells such as hematopoietic and intestinal stem cells [8, 9]. Furthermore, Nrf2 has been implicated in cancer with the role of promoting tumor growth and resistance to anticancer drugs [10, 11]. However, the specific role that Nrf2 plays in the self-renewal and differentiation process in embryonic stem cells remains elusive.
The proteasome is a non-lysosomal threonine protease that plays a key role in the removal of both ubiquitinated and damaged proteins. The core of the proteasome machinery is the 20S particle, which consists of structural α- and catalytic β-subunits. One 20S core particle can bind to one or two 19S regulatory caps to form the 26S complex. Dysfunction of proteasome-dependent proteolysis is heavily implicated in aging and cell senescence. While loss of proteasome activity has been observed in aging human tissues and senescent primary cells [12], activation of proteasome has been shown to delay senescence and aging in fibroblasts [5, 13]. Recently, it has been reported that human embryonic stem cells (hESCs) have elevated proteasome activity levels compared to their differentiated progenies [14]. Futhermore, immunoproteasomal subunits have been linked with pluripotency in hESCs [15]. We posit that the elevated proteasome activity observed in embryonic stem cells is an important mechanism for maintaining proper cell identity during their indefinite proliferation in vitro. In parallel, this proteasome activity is also likely to regulate embryonic stem cell fate decisions as large-scale modifications in protein profiles are required for major identity changes.
Here, we identify Nrf2 as a novel pluripotency gene. Nrf2 inhibition directly disrupts self-renewal of hESCs and cellular reprogramming, while Nrf2 activation delays differentiation of hESCs. Additionally, Nrf2 plays an important role in maintaining and regulating proteasome activity through POMP in hESCs. Finally, we find that the proteasome plays a key role in the proliferation of self-renewing hESCs, three germ layer differentiation and cellular reprogramming, suggesting the Nrf2-mediated high proteasome activity of hESCs to be an important, functional characteristic. Taken together, these data uncover a previously unrecognized link between the Nrf2-proteasome pathway and the fundamental hallmarks of hESCs, pluripotency and self-renewal.
Materials and Methods
Cell culture
H1 (WiCell), H9 (WiCell) and iPS-IMR90 (a gift from James A. Thompson) cells were maintained on Matrigel (BD Biosciences) with mTeSR1 medium (Stem Cell Technologies). H1, H9 and iPS-IMR90 cells were passaged by mechanical dissociation every 5–6 days. For differentiation, H1, H9 and iPS-IMR90 cells were seeded onto laminin (Sigma)-coated plates and induced to differentiate with hESC culture medium (DMEM/F12, 20% Knockout Serum Replacement, MEM Non-essential Amino Acid Solution, 0.1 mM β-mercaptoethanol) without FGF2. HEK293T cells (CRL-11268) and human dermal fibroblasts derived from normal human breast skin of 28 years old Caucasian female were purchased from ATCC and Cell Applications, inc., respectively, and maintained in DMEM supplemented with 10% fetal bovine serum (FBS).
Lentiviral vector cloning
Wild type and mutant ubiquitin genes were amplified by PCR from pRK5-HA-Ubiquitin-WT (Addgene #17608) and pRK5-HA-Ubiquitin-KO (Addgene #17603), respectively. The amplified fragments were cloned into pWPI vector (Addgene #12254) between the SwaI/PacI sites. Human POMP and KEAP1 cDNAs were synthesized (Genscript) and also cloned into pWPI between the SwaI/PacI site. Synthesized scramble and human Nrf2 shRNAs were introduced into the EcoRI/PacI site of pLKO.3G (Addgene #14748). The target sequences of shRNAs were as followed; scramble shRNA : CCTAAGGTTAAGTCGCCCTCG and Nrf2 shRNA: GCTCCTACTGTGATGTGAAAT.
Lentiviral preparation and transduction
Lentiviral vector plasmids were transfected into HEK293T cells along with psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259). After 48h, supernatants were collected, filtered through a 0.45 µm filter and concentrated by ultracentrifugation. Titeration was performed using HEK293T cells transduced with several different dilutions of concentrated viruses.
Competitive cell growth assay
H1 and H9 cells were dissociated using Accutase (Life Technologies) and plated on Matrigel-coated plates with ROCK inhibitor (Y-27632, Millipore). Concentrated viruses were added when cell confluency was around 20–30%. For HEK293T cells and human dermal fibroblasts, concentrated viruses and 8 µg/ml polybrene (Sigma) were added to cells when the confluency was around 20–30%. Two days after transduction, transduced cells were mixed with the same number of untransduced cells. The percentage of GFP+ cells (Transduced cells) was monitored over the course of culturing time by flow cytometry.
RNA isolation and quantitative real-time PCR
Total RNA was extracted with TRIzol Reagent (Life Technologies) and reverse transcribed with SuperScript III First-Strand Synthesis System (Invitrogen). Real-time quantitative PCR was performed according to ABI 7500 system’s protocol with Power SYBR Green PCR Master Mix (Applied Biosystems). GAPDH and β-actin mRNA levels were used as a normalization control. See Supporting Information Table S1 for primers used in this study.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100 and then blocked with 10% FBS. Samples were stained with primary antibodies Oct4 (Santa Cruz), Nanog (R&D), TuJ1 (Covance) and Nrf2 (Abcam) overnight at 4°C and secondary antibodies Alexa Fluor 488-donkey anti-mouse IgG, Alexa Fluor 488-donkey anti-rabbit IgG, Alexa Fluor 555-donkey anti-rabbit IgG, Alexa Fluor 488-donkey anti-goat IgG (Invitrogen).
Western blotting
Cells were lysed in RIPA lysis buffer (Millipore) containing protease inhibitor cocktail (Roche). Protein concentration was determined by Pierce® BCA Protein Assay Kit (Thermo Scientific). Equal amounts of protein were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare Life Sciences). Membranes were blocked for 1h at room temperature with PBST (0.1% Tween 20 in PBS) containing 5% skim milk. Immunoblotting was performed with primary antibodies Nrf2 (Abcam), OCT4 (Santa Cruz), KEAP1 (Millipore), POMP (Abcam), CyclinD1 (Millipore) and β-actin (Sigma) overnight at 4°C. After being washed with PBST, the membrane was incubated with secondary antibodies Alexa Flour 680-goat anti-mouse IgG (Invitrogen) and IRDye® 800CW-goat anti-rabbit IgG (LI-COR) for 1h at room temperature. Protein bands were visualized with LI-COR Odyssey Imaging System (LI-COR).
Flow cytometry analysis
Cells were dissociated into single cells with Accutase (Invitrogen) and stained with primary antibodies for Tra-1–81 (Santa Cruz) and SSEA4 (Santa Cruz) for 1h on ice. Secondary antibody staining was performed for 30 min with R-PE-goat anti-mouse IgG and Alexa Fluor 488-donkey anti-mouse IgG&M. Data were collected with BD Accuri C6 flow cytometry (BD biosciences).
Cellular reprogramming
Human dermal fibroblasts were transduced with pRRL.PPT.SF.hOKSM.idTomato.preFRT vector [16]. On days 1–3 post transduction, cells were fed daily with induction media (DMEM, 10% FBS, 1× Penstrep, 250 µM sodium butyrate, and 50 µg/mL ascorbic acid). On day 4 post transduction, cells were split 1:7 into Matrigel-coated 6-well plates. Cells were fed with induction media until day 6 and switched to ½ induction media + ½ chemically defined Essential 8 media (E8, Life Technologies) with 250 µM sodium butyrate and 50 µg/mL ascorbic acid. On days 7–14, transfected cells were fed daily with E8. For Nrf2 and POMP knock-down, cells were transfected with either 50 nM of control, Nrf2 (HSS107130, HSS181505, Life Technologies) or POMP (HSS147436, HSS147437, Life Technologies) siRNA on day 6 post transduction, using Lipofectamine ® RNAiMAX (Life Technologies).
NeuroD1 differentiation
Human induced neurons (hINs) were generated as described previously [17] but using a tetracycline-inducible lentiviral vector encoding a neural lineage-specific transcription factor, NeuroD1 and an IRES cassette expressing puromycin resistance gene for negative selection of untransduced cells. In brief, on day −3, H9 hESCs were dissociated into single cells by Accutase and plated onto Matrigel coated 6-well plates with 5×105 cells/well in mTESR1. On day −2, cells were transduced with (1) concentrated virus expressing rtTA and (2) concentrated virus expressing a NeuroD1/puromycin resistance gene in fresh mTESR1. Cells were transduced a second time on day −1. On day 0, transduced H9 hESCs were dissociated with Accutase in the presence of ROCK inhibitor and expanded onto Matrigel coated 6-well plates in B27/N2/DMEM/F12 (Invitrogen). Doxycycline (2g/l, Sigma) was added on day 0 to induce TetO gene expression and was continuously supplemented in the medium throughout experiment. On day 2, puromycin (2ug/ul) was added for selection.
Cell cycle analysis
Cells were dissociated in single cells with Accutase (Life Technologies) and fixed with EtOH for 1h on ice, followed by RNase treatment. Propidium iodide (Sigma, 10 µg/ml) was added into samples and data were acquired by BD Accuri C6 flow cytometry (BD biosciences).
siRNA tansfection to hESCs
Negative control, Nrf2 and POMP siRNAs (100 nM, Invitrogen) were transfected to H9 cells using RNAiMAX (Invitrogen) according to the manufacturer’s instructions.
Proteasome activity assay
Chymotrypsin-like proteasome activity was measured with 20S Proteasome Activity Assay Kit (EMD Millipore) according to the manufacturer’s instructions. Briefly, cells were lysed with Lysis buffer (50 mM HEPES, 5 mM EDTA, 150 mM NaCl and 1% Triton X-100). Protein concentration was assessed with Pierce® BCA Protein Assay Kit (Thermo Scientific). The same amounts of protein were incubated with the substrate (LLVY-AMC for chymotrypsin-like activity) for 1h at 37°C and the free AMC fluorescence was quantified using a 380/460 nm filter set in a Gemini EM Fluorescence plate reader (Molecular Devices). To measure trypsin-like and PGPH-like activities, Boc-LRR-AMC (Boston Biochem) and Z-LLE-AMC (Boston Biochem) were used as substrates, respectively.
Alkaline phosphatase assay
Using cells fixed with 4% paraformaldehyde for 15 min, alkaline phosphatase assays were performed with Vector® Red Alkaline Phosphatase Substrate Kit I (Vector laboratories) according to the manufacturer’s instructions.
Statistical analysis
Student’s t-test was used for statistical analysis. P values of less than 0.05 were considered significant.
Chemical list
R,S-Sulforaphane was purchased from LKT laboratories. tert-Butylhydroquinone (t-BHQ) were purchased from Sigma. Lactacystin and epoxomicin were purchased from A.G. Scientific, Inc. Chemicals were reconstituted according to the manufacturer’s instructions.
Results
Nrf2 activity is high in hESCs and helps control self-renewal ability and pluripotency
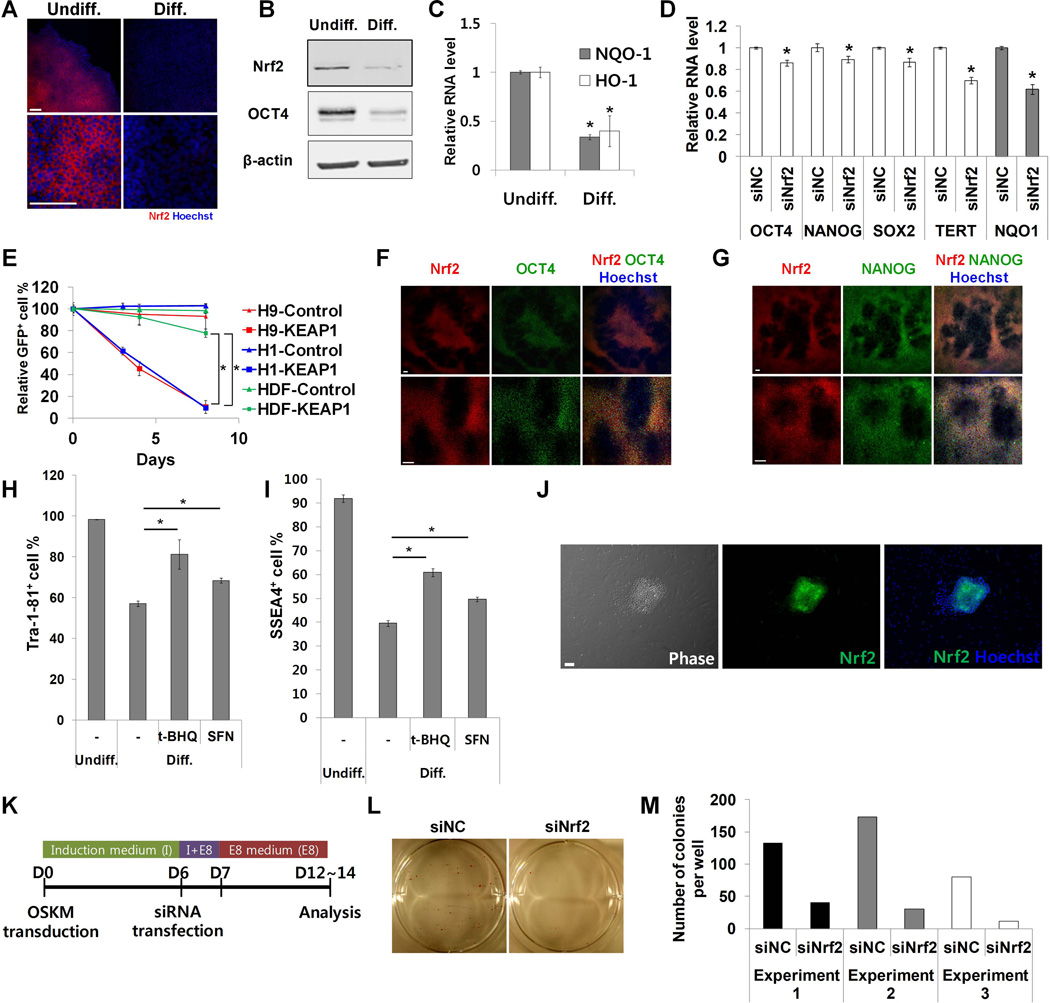
First, we found that hESCs (H1 and H9 cell lines) and iPS-IMR90 [18] induced pluripotent stem cells had high levels of Nrf2 protein that decreased sharply after 8 days of differentiation by removal of FGF2 (Fig. 1 A, 1B; Supporting Information Fig. S1A). Furthermore, a significant amount of Nrf2 protein was detected in the nucleus of hESCs (Supporting Information Fig. S1B). Upon differentiation, total Nrf2 protein level both decreased and was no longer nuclear. Similarly, the expression of well-known Nrf2 target genes, HO-1 and NQO-1, was higher in undifferentiated than in differentiated hESCs and iPS-IMR90 cells (Fig. 1C; Supporting Information Fig. S1C). Nrf2 mRNA level did not dramatically change during differentiation (Supporting Information Fig. S1D), consistent with the regulation of Nrf2 activity being mainly post-transcriptional [19]. Interestingly, the protein and mRNA level of KEAP1 did not significantly change (Supporting Information Fig. S1E, S1F), suggesting the differentiation related regulation of Nrf2 protein and activity level are uncoupled from KEAP1 mRNA expression.
Figure 1. Nrf2 controls self-renewal and pluripotency in hESCs.
(A) Undifferentiated and differentiated H9 cells were immunostained with anti-Nrf2 antibody. (B) Cell lysates from both undifferentiated and differentiated H9 cells were analyzed by western blot. (C) The expression of Nrf2 target genes (HO-1, NQO-1) was analyzed by qPCR in H9 cells during differentiation (n=4). (D) Pluriptency gene expression was analyzed by qPCR in H9 cells transfected with either control or Nrf2 siRNA (n=4). (E) Human ESCs (H1 and H9) and human dermal fibroblasts were transduced with lentiviral vectors expressing KEAP1 together with GFP and then mixed with untransduced cells. The pecentage of GFP+ cells was monitored (n=6 for H9 and HDF, n=3 for H1). Cells transduced with lentiviral vector expressing GFP only were used as control. (F,G) Differentiating H9 cells were co-immunostained with antibodies against Nrf2 and OCT4 (F) or Nrf2 and NANOG (G). (H,I) H9 cells were differentiated with t-BHQ (20 µM) or SFN (3 µM) and immunostained with anti-Tra-1–81 (H) and anti-SSEA4 (I) antibodies, followed by flow cytomety analysis (n=6). (J) Human dermal fibroblasts were reprogrammed with four factors (OCT4, SOX2, KLF4, c-MYC) in feeder-free system. Emerging colonies were immunostained with anti-Nrf2 antibody. (K) Human dermal fibroblasts were transduced with lentiviral vector expressing four factors together with dTomato (Day 0). Cells were transfected with Nrf2 siRNA on day 6 post-transduction and were analyzed on days 12–14. (L,M) Reprogrammed colonies were stained for alkaline phosphatase (AP) activity and AP+ colonies were counted. Error bars represent standard deviation. *: p<0.01 (Student’s t-test), Scale bar, 100 µm, t-BHQ, tert-Butylhydroquinone, SFN, sulforaphane.
To confirm further that high Nrf2 activity is a unique characteristic in hESCs, we compared Nrf2 activity in hESCs with more fully differentiated human induced neurons (hINs). hINs were generated by directly differentiating H9 cells into neurons through over-expressing transcription factor NeuroD1 [17]. hINs showed typical neuronal morphology and high expression of neuron-specific beta III tubulin (TuJ1) (Supporting Information Fig. S1G, S1H). Nrf2 protein and activity levels were dramatically decreased in hINs compared to undifferentiated hESCs (Supporting Information Fig. S1H, S1I).
Intrigued by the enriched Nrf2 protein and activity level in hESCs, we tested whether loss of Nrf2 activity could directly affect the self-renewal ability of hESCs. To down-regulate Nrf2 activity, we first used siRNA against Nrf2. Knockdown of Nrf2 by siRNAs was confirmed by western blot and qPCR (Supporting Information Fig. S1J, S1K). In H9 cells, even the partial loss of Nrf2 activity decreased OCT4 (also known as POU5F1), NANOG, SOX2 and TERT gene expression (Fig. 1D). To more strongly down-regulate Nrf2 activity, we used a lentiviral vector expressing Nrf2 repressor KEAP1 (Kelch-Like ECH-Associated Protein 1), alongside GFP (Supporting Information Fig. S1L). Inhibition of Nrf2 activity due to KEAP1 was confirmed by measuring NQO-1 expression (Supporting Information Fig. S1M). Using this system, we performed a competitive cell growth assay. HESCs (H1 and H9) transduced with control or KEAP1 lentiviral vector were mixed with untransduced cells and the percentage of GFP+ cells (GFP+ %) was monitored over the course of culturing time. KEAP1 overexpression significantly decreased the GFP+ % in hESC cells compared to human dermal fibroblasts (p<0.01) (Fig. 1E). These data suggest that high Nrf2 activity is important for self-renewal in hESCs.
We next examined the role of Nrf2 in differentiation. Since each hESC undergoes differentiation at its own pace, loss of OCT4 and NANOG expression appears highly heterogeneous in differentiating cell populations (Fig. 1 F, 1G). Interestingly, Nrf2 expression closely resembled the single cell pattern of OCT4 and NANOG expression (Fig. 1 F, 1G). Therefore, we hypothesized that Nrf2 down-regulation might be required for differentiation of hESCs. To prevent Nrf2 down-regulation, H9 cells were treated with the Nrf2 activators tert-Butylhydroquinone (t-BHQ) or sulforaphane during differentiation. Treatment of H9 cells with t-BHQ and sulforaphane increased the expression of NQO-1, confirming Nrf2 activation (Supporting Information Fig. S1N). Flow cytometry analysis showed that Nrf2 activation resulted in higher Tra-1–81+ and SSEA4+ cell% (Fig. 1 H, 1I; Supporting Information Fig. S1Q). Furthermore, Nrf2 activation inhibited the decrease of OCT4 and NANOG expression during differentiation (Supporting Information Fig. S1O, S1P). These data suggest that the down-regulation of Nrf2 activity is needed for proper differentiation of hESCs.
To test whether high Nrf2 protein level is reestablished during cellular reprogramming, human dermal fibroblasts (HDF) were reprogrammed in a feeder-free system by introduction of OCT4, SOX2, KLF4 and C-MYC (OSKM). Embryonic stem cell-like colonies started to appear ~12 days post transduction. Colonies had silenced transgene expression and high OCT4, NANOG and alkaline phosphatase levels (Supporting Information Fig. S1S, S1T). Staining with Nrf2 antibody revealed high Nrf2 protein levels in these embryonic stem cell-like colonies of successfully reprogrammed cells (Fig. 1J). Surrounding cells, which were either untransduced fibroblasts or intermediate cells refractory to reprogramming, did not show this reestablished high Nrf2 protein level. Consistent with its uniformity during hESC differentiation (Supporting Information Fig. S1D), Nrf2 mRNA level did not dramatically change during reprogramming (Supporting Information Fig. S1R).
To study the functional role of Nrf2 in reprogramming, HDFs undergoing reprogramming were treated with Nrf2 siRNA 6 days post OSKM transduction and analyzed on day 12–14 (Fig. 1K). Cells treated with negative control siRNA formed many alkaline phosphatase-positive colonies (Fig. 1L, 1M). These colonies showed both high levels of OCT4 and NANOG and silenced transgene expression (Supporting Information Fig. S1U). In contrast, cells treated with Nrf2 siRNA had a much lower efficiency in colony formation (Fig. 1L, 1M), even though a significant number of transduced cells (dTomato+ cells) were still detected (Supporting Information Fig. S1V). These results were confirmed with a second Nrf2 siRNA (Supporting Information Fig. S1W). Together, the data suggest that Nrf2 plays an important role in acquisition of pluripotency during reprogramming.
Nrf2 regulates proteasome activity through POMP
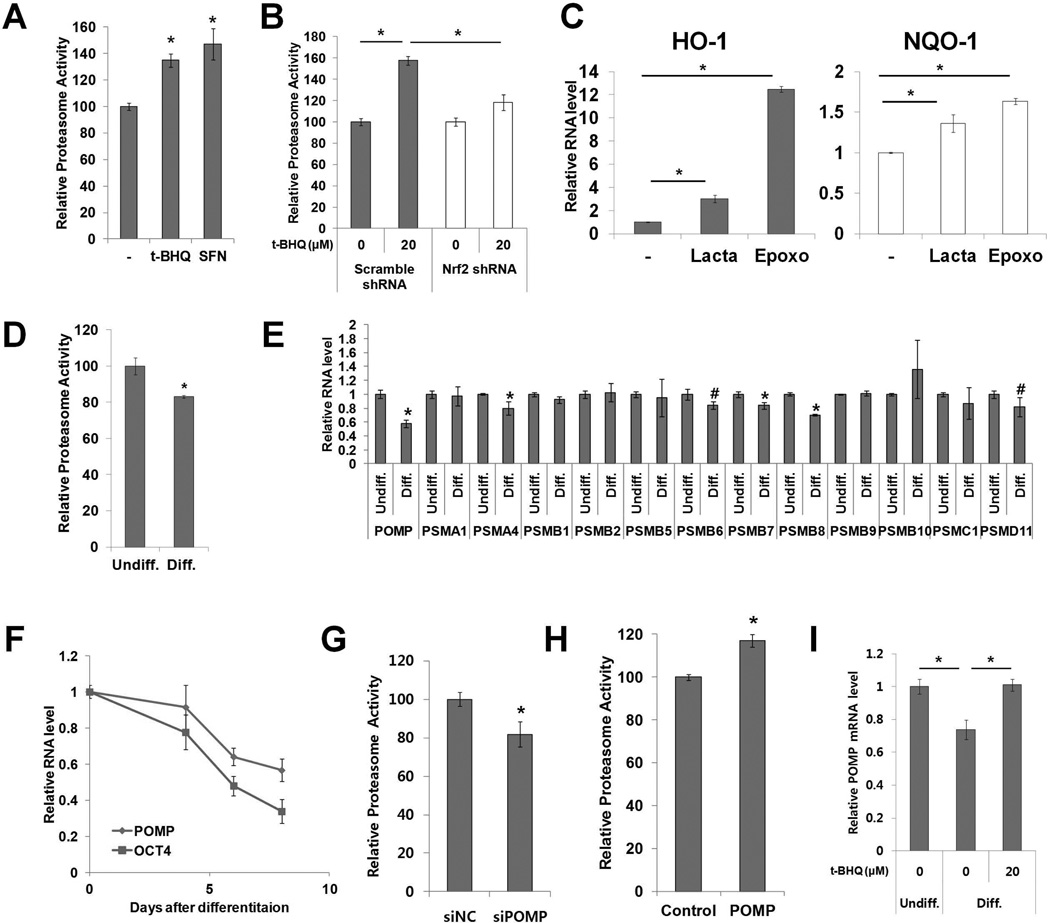
Elevated proteasome activity was recently identified as a characteristic of hESCs [14, 15]. Furthermore, it was reported that the proteasome regulates transcription in tissue-specific gene locus of mouse embryonic stem cells, implicating a functional role for the proteasome in stem cells [20]. Since Nrf2 was found to control the expression of several proteasome subunits in mouse liver, human fibroblasts and lymphoid cells [4, 5, 21], we tested whether Nrf2 regulates proteasome activity in hESCs. We examined the effect of Nrf2 activation rather than its inhibition because many studies have shown that cells are highly capable of compensating for the loss of proteasome activity [22–25]. Nrf2 activation by t-BHQ or sulforaphane increased proteasome activity in both hESCs and iPS-IMR90 cells (Fig. 2A; Supporting Information Fig. S2A, S2B). To test whether the increase of proteasome activity was Nrf2-dependent, H9 cells transduced with lentiviral vector expressing Nrf2 shRNA were treated with t-BHQ. About 50% knock-down of Nrf2 was able to block t-BHQ mediated proteasome activation (Fig. 2B; Supporting Information Fig. S2C). Since Nrf2 itself is subject to proteasome-dependent degradation, we tested whether modulations to proteasome activity in hESCs can directly affect Nrf2 protein and activity levels. We inhibited proteasome activity with proteasome inhibitors (lactacystin and epoxomicin) and observed an increase in Nrf2 protein and target gene expression, suggesting a reciprocal regulation between Nrf2 and the proteasome (Fig. 2C; Supporting Information Fig. S2D).
Figure 2. Nrf2 regulates proteasome activity in hESCs.
(A) H9 cells were treated with Nrf2 inducers (t-BHQ 20 µM or SFN 3 µM) and chymotrypsin-like proteasome activity was measured after 24h (n=5). (B) H9 cells stably expressing Nrf2 shRNA were treated with t-BHQ (20 µM) and chymotrypsin-like proteasome activity was measured after 24h (n=6). (C) H9 cells were treated with proteasome inhibitors (lactacystin 0.5 µM, epoxomicin 30 nM) for 12h. The expression of Nrf2 target genes (HO-1 and NQO-1) was measured by qPCR (n=3). (D) Chymotrypsin-like proteasome activity was measured in undifferentiated and differentiated H9 cells (n=5). (E) The RNA level of various proteasome subunits and related genes was measured by qPCR (n=4). (F) The kinetics of POMP and OCT4 mRNA down-regulation was monitored by qPCR during differentiation (n=3). (G) H9 cells were transfected with POMP siRNA and chymotrypsin-like proteasome activity was measured (n=6). (H) H9 cells were transduced with lentiviral vector expressing POMP cDNA and chymotrypsin-like proteasome activity was measured (n=6). (I) H9 cells were differentiated with or without t-BHQ (20 µM) and the POMP RNA level was measured by qPCR (n=3). Error bars represent standard deviation. *: p<0.01, #: p<0.05 (Student’s t-test), t-BHQ, tert-Butylhydroquinone, SFN, sulforaphane.
Proteasome activity is lower in fully differentiated cells than in hESCs [14, 15]. During the early period of hESC differentiation, we observed a dramatic decrease in Nrf2 activity. To test whether proteasome activity shows a similar decrease during this time frame, we compared the proteasome activity of undifferentiated H9 cells with that of H9 cells differentiated for 8 days by removal of FGF2. Proteasome activity was reduced ~20% in these differentiated H9 cells (Fig. 2D). Searching for a mechanistic link between the decrease in Nrf2 and proteasome activity, we first identified possible proteasome complex subunits responsible for this decrease. After screening expression levels of various proteasome subunits and related chaperones, we observed a significant reduction of gene expression for POMP, PSMA4, PSMB6, PSMB7, PSMB8 and PSMD11, with POMP expression decreasing the most dramatically during early differentiation (Fig. 2E). POMP expression also decreased in differentiated H1 and iPS-IMR90 cells (Supporting Information Fig. S2E). We further analyzed the kinetics of POMP mRNA level down-regulation during H9 cell differentiation and found the pattern to be similar to that of OCT4 (Fig. 2F). To confirm these data in more fully differentiated cells, we compared proteasome activity of H9 cells and hINs. Compared to H9 cells, hINs had significantly decreased proteasome activity and POMP expression (Supporting Information Fig. S2F, S2G).
Considering that POMP is an essential molecular chaperone for proteasome assembly [26], we hypothesized that modulating POMP expression could directly control proteasome activity in hESCs. To examine the effect of POMP loss of function on proteasome activity, H9 cells were transfected with siRNAs targeting POMP. Efficient knock-down of POMP was confirmed by qPCR and western blot (Supporting Information Fig. S2H, S2I). The siRNA-mediated POMP knock-down resulted in a decrease in proteasome activity (Fig. 2G; Supporting Information Fig. S2J). Moreover, POMP overexpression was sufficient to further increase proteasome activity in H9 cells (Fig. 2H). Consistent with previous ChIP-seq study showing that Nrf2 directly binds the promoter region of POMP [21], we found that Nrf2 activation was sufficient to block the down-regulation of POMP expression during early differentiation (Fig. 2I). Taken together, these data suggest that the transcription factor Nrf2 plays a key role in the maintenance and regulation of proteasome activity in hESCs at least partially through POMP.
High proteasome activity is required for proliferation, proper cell cycle progression, multipotent differentiation and cellular reprogramming
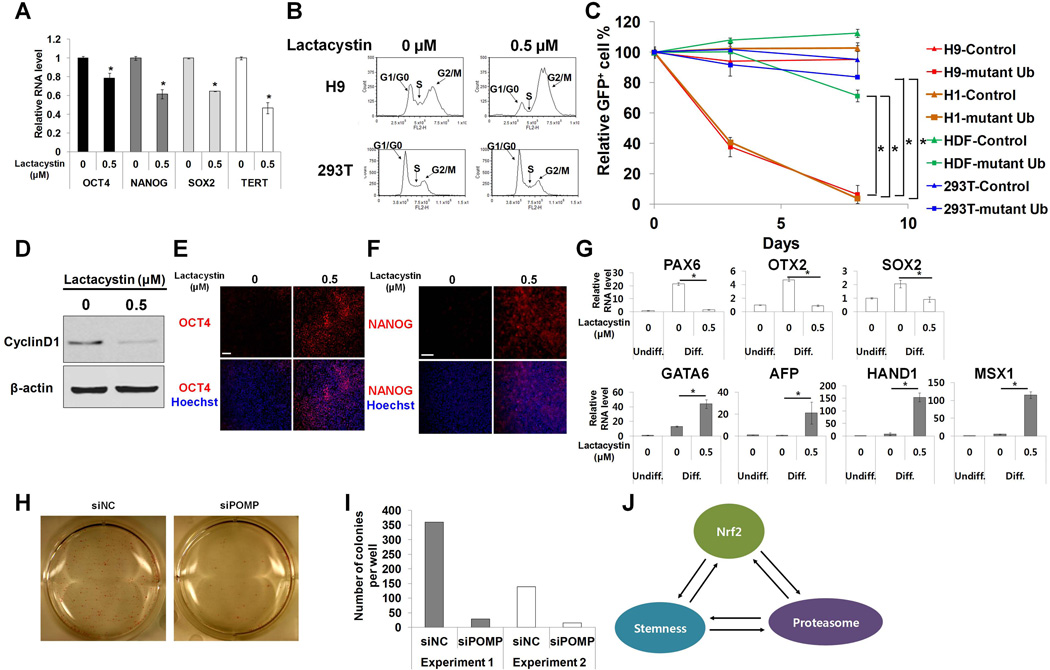
We sought to determine whether high proteasome activity was important for self-renewal and pluripotency in hESCs. To test the effect of proteasome inhibition on the self-renewal ability of hESCs, we utilized specific proteasome inhibitors at non-lethal doses that partially inhibited proteasome activity (Supporting Information Fig. S3A). Lactacystin (0.5 µM) and epoxomicin (30 nM) treatment significantly decreased the pluripotency gene expression in hESCs (Fig. 3A; Supporting Information Fig. S3C). This observation was confirmed in iPS-IMR90 cells treated with lactacystin (Supporting Information Fig. S3D). Self-renewing hESCs have a very fast and unique cell cycle pattern [27]. Since periodic synthesis and degradation of cell cycle regulators are required for cell cycle progression, we hypothesized that high proteasome activity might be necessary for this process. Partial proteasome inhibition by lactacystin (0.5 µM) induced G2/M arrest and depletion of cells in the S phase in both H9 and iPS-IMR90 cells (Fig. 3B; Supporting Information Fig. S3E). However, the same doses of inhibitors had no significant effect on the cell cycle pattern of 293T cells, which have a doubling time similar to that of hESCs [14] (Fig. 3B; Supporting Information Fig. S3B). Epoxomicin, another specific proteasome inhibitor, similarly disrupted the cell cycle of H9 cells (Supporting Information Fig. S3F). To confirm these data, we performed a competitive cell growth assay with cells expressing a mutant ubiquitin that is unable to make ubiquitin chains (Fig. 3C). HESCs (H1 and H9) transduced with lentiviral vector expressing the mutant ubiquitin together with GFP were mixed with untransduced cells. GFP+ % was monitored to observe the effect of impaired ubiquitin-dependent proteasomal degradation on proliferation of self-renewing hESCs. A rapid decrease of GFP+ % was observed in the mutant ubiqutin-transduced hESCs that differed significantly from the effect of mutant ubiquitin in the control, 293T and human dermal fibroblast cells (p<0.01) (Fig. 3C). Taken together, these results suggest that hESCs rely on high proteasome activity to maintain their characteristic cell cycle pattern.
Figure 3. Proteasome activity is important for self-renewal and pluripotency in hESCs.
(A) H9 cells were treated with lactacystin (0.5 µM) for 12h. Pluripotency gene expression was analyzed by qPCR (n=4). (B) H9 cells were treated with lactacystin (0.5 uM) for 12h and the cell cycle pattern was measured by propidium iodide staining. 293T cells were used as control. (C) Cells transduced with lentiviral vector expressing mutant ubiquitin together with GFP were mixed with untransduced cells, and then the percentage of GFP+ cells was monitored by flow cytometry (n=6 for H9, n=3 for H1, 293T and HDF). Cells transduced with GFP only lentiviral vector were used as control. (D) H9 cells were treated with lactacystin (0.5 µM) for 12h. Cell lysates were analyzed by western blot. (E,F) Differentiated H9 cells were immunostained with anti-Oct4 (E) or anti-NANOG (F) antibodies. (G) Germ layer marker expression was analyzed by qPCR (n=4). (H,I) Human dermal fibroblasts were transduced with lentiviral vector expressing four factors (OSKM) together with dTomato (Day 0). Cells were transfected with POMP siRNA on day 6 post-transduction and were analyzed on days 12–14. Reprogrammed colonies were stained for AP activity and AP+ colonies were counted. (J) A schematic diagram depicting interconnectivity between Nrf2, proteasome and stemness. Error bars represent standard deviation; *: p<0.01 (Student’s t-test); Scale bar, 100µm; HDF, human dermal fibroblasts.
Recently, it was reported that cell cycle plays an important role in determining the differentiation capacity of hESCs, with cyclin D1 identified as a key promoter of ectoderm induction [28]. Because our data showed that proteasome inhibition disrupted the cell cycle pattern of hESCs, we examined the direct effect of proteasome inhibition on cyclin D1 expression. Partial proteasome inhibition via lactacystin dramatically decreased cyclin D1 protein level in hESCs (Fig. 3D), which implicates proteasome activity as a potential regulator of cellular differentiation. Furthermore, only selective pluripotency factors such as OCT4 and NANOG, but not SOX2, are known to be ubiquitinated in embryonic stem cells [29]. Considering that OCT4 and NANOG are strong mesendoderm inducers, selective protein degradation pathways could play a role in directing balanced differentiation. To examine the function of the proteasome in differentiation, H9 cells were induced to differentiate by removal of FGF2, in the presence or absence of lactacystin. High levels of OCT4 and NANOG proteins were observed in lactacystin-treated cells, suggesting proteasome inhibition delayed the degradation of OCT4 and NANOG proteins (Fig. 3E, 3F). Proteasome inhibition via POMP knockdown during differentiation also showed similar accumulation of OCT4 and NANOG (Supporting Information Fig. S3G, S3H). We then analyzed the expression of lineage-specific differentiation markers in H9 and iPS-IMR90 cells. Lactacystin treatment strongly inhibited the induction of ectoderm markers (PAX6, OTX2, SOX2), while promoting the expression of mesendoderm markers (GATA6, AFP, HAND1, MSX1) (Fig. 3G; Supporting Information Fig. S3I). Furthermore, POMP siRNA treatment was also sufficient to skew the fate of differentiating H9 cells toward mesendoderm (Supporting Information Fig. S3J). These data are consistent with the previous report showing FOXO4, a regulator of proteasome activity in hESCs, to be necessary for neural differentiation, suggesting an important role of the proteasome in hESC differentiation [14, 30]. These results indicate that high proteasome activity contributes to balancing germ layer derivation by controlling cell cycle and allowing proper degradation of OCT4 and NANOG proteins.
Intrigued by the important role of proteasome in proliferation and differentiation of human embryonic and induced pluripotent stem cells, we hypothesized that high proteasome activity might be a key criteria for successful reprogramming. Human dermal fibroblasts were treated with POMP siRNA 6 days post OSKM transduction and analyzed on day 12–14. POMP knock-down dramatically decreased the number of colonies formed even though a significant number of positively transduced cells were still present (Fig. 3 H, 3I; Supporting Information Fig. S3K). These results were confirmed with another POMP siRNA (Supporting Information Fig. S1W). Furthermore, most colonies in the POMP siRNA-treated group had cobblestone-like morphology and weak alkaline phosphatase activity (Supporting Information Fig. S3L), suggesting incomplete reprogramming. These data indicate that proteasome plays an important role in acquisition of pluripotency during reprogramming.
Discussion
Although antioxidant and proteasome-related genes are among the most enriched transcripts and proteins in embryonic stem cells [6, 7], how these genes influence pluripotency and self-renewal is not well understood. In this study, we identified a common upstream transcription factor of these genes, Nrf2, as a master regulator upon which control of cellular stress, proteostasis, and stemness in hESCs all converge. First, we found Nrf2 protein and activity levels were enriched in hESCs. Re-establishment of this high Nrf2 protein level occurred during cellular reprogramming and was required for its success. Furthermore, Nrf2 inhibition disrupted self-renewal while Nrf2 activation blocked differentiation of hESCs. Second, we established a reciprocal relationship between Nrf2 and proteasome activity. While Nrf2 helps maintain and regulate proteasome activity partially through POMP, Nrf2 activity itself is regulated by the proteasome. Finally, we showed that Nrf2-mediated high proteasome activity influenced self-renewal, three germ layer differentiation and cellular reprogramming in hESCs via its role in cell cycle control and balancing the degradation of pluripotency proteins.
Recently, it was suggested that pluripotency in hESCs is a state of optimized balance of lineage specifiers [31, 32]. To re-establish pluripotency, mesendodermal and ectodermal transcription factors were able to replace OCT4 and SOX2 as reprogramming factors, respectively [31, 32]. During germ layer differentiation, this delicate balance between pluripotency factors needs to be further fine-tuned. In one direction, OCT4 and NANOG suppress ectoderm fate and promote mesendoderm fate [33, 34]. In contrast, SOX2 represses mesendoderm differentiation and induces neural ectoderm derivation [33, 34]. Recently, Buckley et al., showed that OCT4 and NANOG proteins, but not SOX2, are actively ubiquitinated in mouse embryonic stem cells [29]. By selectively degrading a subset of the pluripotency proteins, the ubiquitin/proteasome system may contribute to fine-tuning of the pluripotency factor balance, helping to maintain pluripotency or direct differentiation. Consistent with this idea, we showed that proteasome inhibition during differentiation disrupted proper three germ layer derivation by skewing the cell fate toward mesendoderm at the expense of ectoderm fate. We also showed that the reestablishment of pluripotency during reprogramming required proteasome activity, which further implicated the importance of the proteasome in pluripotency. Based on these principles, modulating proteasome activity to shift the balance of lineage specifiers may be developed as a method for directing the differentiation of embryonic stem cells and enriching for tissue-specific progenitors.
Modulating the proteasome activity itself is complex because the proteasome consists of at least 32 subunits. In order for cells to adjust proteasome activity efficiently according to their need, the proteasome has key chaperones and subunits sufficient to modulate proteasome activity by themselves. POMP is a molecular chaperone that is essential for 20S proteasome biogenesis [26, 35]. This protein binds 20S pre-proteasome components and is involved in recruiting β subunits during 20S proteasome assembly. Once the proteasome assembly is completed, POMP is degraded to allow for maturation of proteasome. In this study, we found that POMP expression was high in hESCs and dramatically decreased during early differentiation. Furthermore, modulating POMP expression was sufficient to regulate proteasome activity in hESCs, suggesting POMP-mediated proteasome assembly to be one limiting process in adjusting proteasome activity. Our findings suggest an efficient mechanism of proteasome regulation during development.
Consistent with data showing Nrf2 to be an upstream regulator of proteasome subunits in several cell types, our paper found Nrf2 to be a key regulator of proteasome activity in hESCs by controlling POMP expression. Interestingly, it is well known that Nrf2 protein level is controlled by proteasome-dependent degradation. In hESCs, partial inhibition of proteasome activity resulted in accumulation of Nrf2 protein and up-regulation of Nrf2 activity. Because our data showed that high proteasome activity was very important for self-renewal and pluripotency of hESCs, these cells appear to have established a feedback between Nrf2 and the proteasome to safeguard against unintended loss of proteasome activity. The mutual regulation between the proteasome and its upstream regulator Nrf2 may provide a delicate and stable control of proteasome activity in hESCs.
Pluripotency is maintained by a critical balance of pluripotency factors and a unique overall cellular context. As part of this unique cellular context, we found that Nrf2 protein and activity level was highly enriched and was an important component of hESCs. We have shown in this paper that Nrf2 directly regulated proteasome activity in hESCs, a potential mechanism through which Nrf2 acts as a pluripotency gene. However, other cellular processes, such as redox balance, regulated by Nrf2 could also affect pluripotency. Furthermore, Nrf2 directly binds to the genes involved in the WNT, BMP and Notch pathways in mouse embryonic fibroblasts and human lymphoid cells [21, 36]. Given the importance of these pathways in embryonic stem cells, more studies are warranted to further understand the role of Nrf2 in pluripotency.
Our study revealed a previously unrecognized connection between the Nrf2-proteasome pathway and stemness in hESCs (Fig. 3J). In parallel to their conventional roles in cellular defense and housekeeping, Nrf2 and proteasome actively participate in the balance and regulation of self-renewal and pluripotency. Furthermore, the importance of the Nrf2-proteasome pathway has implications on sensescence and degenerative human diseases. Our findings suggest a new potential link between impaired redox homeostasis, proteasome dysfunction and stem cell exhaustion with Nrf2 well-positioned as a central regulator of these hallmarks.
Conclusion
Nrf2, a master regulator of antioxidant and detoxification genes, protects cells against inflammation, toxins and damaged proteins. The central role of Nrf2 as a cellular guardian has attracted pharmaceutical interest in using Nrf2 pathways to treat diseases including inflammation, neurodegeneration and aging. Embryonic stem cells also hold great promise for understanding and treating disease. Given the fundamental abilities of stem cells to remain immortal or differentiate into any cell type, we reasoned that Nrf2 helps protect stem cells in these functions, although the specific roles played by Nrf2 have remained elusive. Here, we discovered the critical importance of the Nrf2-proteasome pathway in both pluripotency and self-renewal of stem cells. Our finding provides mechanistic insights into how impaired redox homeostasis, proteasome dysfunction, and stem cell exhaustion contribute to aging and disease.
Supplementary Material
Acknowledgements
We acknowledge the use of the Laboratory for Stem Cell Biology and Engineering at UC Santa Barbara, which is supported by the California Institute for Regenerative Medicine (CIRM) Grant CL1-00521 and the Dr Miriam and Sheldon G. Adelson Medical Research Foundation and NIH grant R01 MH093661. J.J was supported by a fellowship from the California Institute for Regenerative Medicine (TG2-01151).
Footnotes
Author Contribution Summary
Jiwon Jang: Conception and design, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing
Yidi Wang: Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing
Hyung-Seok Kim: Collection and/or assembly of data
Matthew A. Lalli: Conception and design, Manuscript writing
Kenneth S. Kosik: Conception and design, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Moi P, Chan K, Asunis I, et al. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 3.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwak MK, Wakabayashi N, Greenlaw JL, et al. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapeta S, Chondrogianni N, Gonos ES. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J Biol Chem. 2010;285:8171–8184. doi: 10.1074/jbc.M109.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramalho-Santos M, Yoon S, Matsuzaki Y, et al. "Stemness": transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 7.Baharvand H, Hajheidari M, Ashtiani SK, et al. Proteomic signature of human embryonic stem cells. Proteomics. 2006;6:3544–3549. doi: 10.1002/pmic.200500844. [DOI] [PubMed] [Google Scholar]
- 8.Tsai JJ, Dudakov JA, Takahashi K, et al. Nrf2 regulates haematopoietic stem cell function. Nat Cell Biol. 2013;15:309–316. doi: 10.1038/ncb2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochmuth CE, Biteau B, Bohmann D, et al. Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell. 2011;8:188–199. doi: 10.1016/j.stem.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31:90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Chondrogianni N, Gonos ES. Proteasome dysfunction in mammalian aging: steps and factors involved. Exp Gerontol. 2005;40:931–938. doi: 10.1016/j.exger.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Hwang JS, Chang I, Kim S. Age-associated decrease in proteasome content and activities in human dermal fibroblasts: restoration of normal level of proteasome subunits reduces aging markers in fibroblasts from elderly persons. J Gerontol A Biol Sci Med Sci. 2007;62:490–499. doi: 10.1093/gerona/62.5.490. [DOI] [PubMed] [Google Scholar]
- 14.Vilchez D, Boyer L, Morantte I, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkinson SP, Collin J, Irina N, et al. A putative role for the immunoproteasome in the maintenance of pluripotency in human embryonic stem cells. Stem Cells. 2012;30:1373–1384. doi: 10.1002/stem.1113. [DOI] [PubMed] [Google Scholar]
- 16.Warlich E, Kuehle J, Cantz T, et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19:782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Pak C, Han Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 19.Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szutorisz H, Georgiou A, Tora L, et al. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- 21.Chorley BN, Campbell MR, Wang X, et al. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundgren J, Masson P, Mirzaei Z, et al. Identification and characterization of a Drosophila proteasome regulatory network. Mol Cell Biol. 2005;25:4662–4675. doi: 10.1128/MCB.25.11.4662-4675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meiners S, Heyken D, Weller A, et al. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J Biol Chem. 2003;278:21517–21525. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- 24.Balasubramanian S, Kanade S, Han B, et al. A proteasome inhibitor-stimulated Nrf1 protein-dependent compensatory increase in proteasome subunit gene expression reduces polycomb group protein level. J Biol Chem. 2012;287:36179–36189. doi: 10.1074/jbc.M112.359281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radhakrishnan SK, Lee CS, Young P, et al. Transcription Factor Nrf1 Mediates the Proteasome Recovery Pathway after Proteasome Inhibition in Mammalian Cells. Molecular Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fricke B, Heink S, Steffen J, et al. The proteasome maturation protein POMP facilitates major steps of 20S proteasome formation at the endoplasmic reticulum. EMBO Rep. 2007;8:1170–1175. doi: 10.1038/sj.embor.7401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker KA, Ghule PN, Therrien JA, et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 28.Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckley SM, Aranda-Orgilles B, Strikoudis A, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vilchez D, Boyer L, Lutz M, et al. FOXO4 is necessary for neural differentiation of human embryonic stem cells. Aging Cell. 2013;12:518–522. doi: 10.1111/acel.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shu J, Wu C, Wu Y, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montserrat N, Nivet E, Sancho-Martinez I, et al. Reprogramming of Human Fibroblasts to Pluripotency with Lineage Specifiers. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 33.Thomson M, Liu SJ, Zou LN, et al. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Oron E, Nelson B, et al. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 35.Gallastegui N, Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem Sci. 2010;35:634–642. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Malhotra D, Portales-Casamar E, Singh A, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.