Abstract
IFN-γ plays a central role in the defense against infections and cancer. More recently, however, IFN-γ has also been reported to have immunosuppressive effects in models of autoimmune disease, melanoma and premalignant skin disease. While IL-12 and IL-18 are critical inducers of IFN-γ during infection, the mechanisms that induce IFN-γ in an immunosuppressive context are unknown. Previously, we identified a key role for IFN-γ in mediating the suppression of antigen-specific immune responses in a transgenic mouse model of HPV-associated epidermal hyperplasia, driven by expression of the HPV16 E7 oncoprotein from a keratin 14 promoter (K14E7). We now demonstrate elevated production of IFN-γ, IL-18 and IL-12 by K14E7 transgenic compared to non-transgenic skin. IFN-γ in K14E7 transgenic skin was produced predominantly by CD8+ and CD4+ T cells, which were present in greater number in K14E7 transgenic skin. Production of IFN-γ in K14E7 skin required IL-18 but not IL-12. Our findings show that IL-18 contributes to inducing IFN-γ in an immunosuppressive cutaneous environment caused by viral oncogene-driven hyperplasia.
Keywords: IFN-γ, IL-18, IL-12, T cells, skin, HPV, immunosuppression, tumor immunity
Introduction
IFN-γ is central to the defense against many pathogens as well as to the immune control of cancer (Dunn et al., 2006). During innate immune responses, IFN-γ is predominantly produced by natural killer (NK) cells and natural killer T (NKT) cells, whilst CD4+ Th1 and CD8+ cytotoxic T cells are the main producers of IFN-γ during adaptive responses (Schoenborn and Wilson, 2007). More recently, IFN-γ was reported to suppress effective immune responses in models of experimental autoimmune encephalomyelitis (Willenborg et al., 1999; Minguela et al., 2007; Pastor et al., 2009). Furthermore, in a mouse model of UVB-induced melanoma, IFN-γ-producing macrophages were critical for tumor growth and survival (Zaidi et al., 2011). These findings suggest dichotomous functions of IFN-γ in innate and adaptive immunity.
The inflammatory cytokines IL-18 and IL-12 are potent inducers of IFN-γ during infection, and are crucial for the immune control of intracellular pathogens and viruses (Okamura et al., 1995; Watford et al., 2004; Arend et al., 2008). IL-18 is predominantly expressed by myeloid cells and also by epithelial cells as a precursor protein that requires protease-mediated cleavage, commonly by caspase-1, for biological activity (Arend et al., 2008; Schroder and Tschopp, 2010). IL-12, which consists of the p35 and p40 subunits, is primarily produced by activated myeloid cells. In contrast to an infectious context, the mechanisms that induce IFN-γ in an immune-inhibitory and/or tumor-promoting context are unknown.
Human papillomaviruses (HPV) infect basal epithelial cells of the skin or genital tract, and some HPV types can cause cancer at these sites (zur Hausen, 2002). HPV proteins E6 and E7 inhibit differentiation and apoptosis of epithelial cells, thus driving epithelial hyperplasia and transformation (Moody and Laimins, 2010). Using the K14E7 mouse, a model of HPV16 E7 protein-induced epithelial hyperplasia (Herber et al., 1996), we previously demonstrated that locally produced IFN-γ prevents effective host immune responses against K14E7 hyperplastic skin grafts (Mattarollo et al., 2010). These findings revealed an immunosuppressive role for IFN-γ in K14E7 skin and make this model system suitable to investigate the cytokine milieu that drives expression of IFN-γ in skin expressing the HPV16 E7 oncogene.
Here, we report that the production of IFN-γ, IL-12 and IL-18 was higher in K14E7 transgenic compared to non-transgenic skin. The majority of IFN-γ-producing cells were CD8+ and CD4+ T cells. Blocking IL-12 and IL-18 function showed that IFN-γ production in K14E7 skin required IL-18, but not IL-12. These observations provide insight into the upstream signals that drive IFN-γ production in an immunosuppressive cutaneous environment.
Results
Elevated IFN-γ production in K14E7 compared to wild type skin
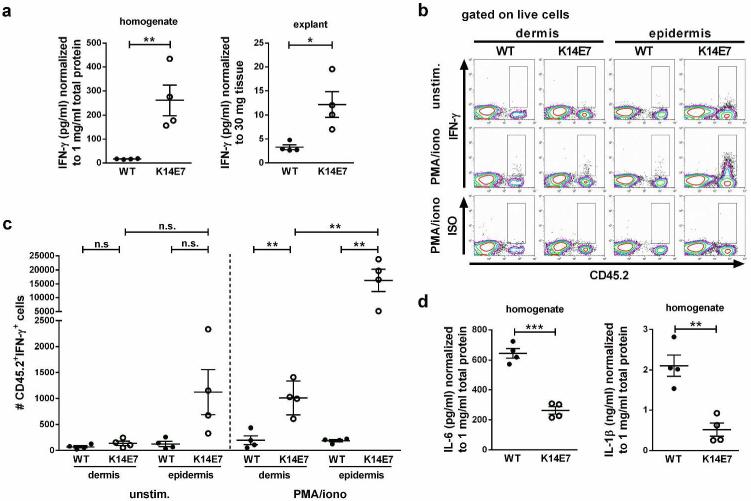
Given the role for IFN-γ in suppressing effective host immune responses against K14E7 skin grafts (Mattarollo et al., 2010), we first determined whether IFN-γ production was different between wild type and K14E7 skin. To this end, we analyzed IFN-γ concentrations in supernatants from whole skin homogenate and from in vitro cultured skin explants. IFN-γ concentrations in supernatants from wild type skin homogenates and explants were very low or below the detection limit, whereas IFN-γ was readily detectable and significantly elevated in supernatants from K14E7 skin (Figure 1 a). Flow cytometric analysis of unstimulated dermal and epidermal cell suspensions further corroborated our findings on the differential expression of IFN-γ in wild type and K14E7 skin. Wild type dermal and epidermal samples contained similar, low numbers of IFN-γ-producing cells (Figure 1b,c). While the number of IFN-γ-producing cells was comparable in the dermis from K14E7 and wild type mice, there were more IFN-γ-producing cells in K14E7 compared to wild type epidermis (9-fold increase compared to wild type epidermis) (Figure 1b,c). Further, stimulation of skin cell suspensions with PMA and ionomycin demonstrated that numbers of cells with the capacity to produce IFN-γ were significantly elevated in the dermis and epidermis of K14E7 mice compared to wild type mice (5- and 84-fold, respectively) (Figure 1b,c). Moreover, the majority of the cells with the capacity to produce IFN-γ in K14E7 skin were found in the epidermis (16-fold higher numbers than in the dermis of K14E7 mice) (Figure 1b,c). Together, these results demonstrate that IFN-γ production is increased in skin of K14E7 compared to wild type animals, and that the majority of IFN-γ-producing cells in K14E7 skin are located in the epidermis. In contrast to IFN-γ, IL-1β and IL-6 concentrations were significantly reduced in K14E7 compared to wild type skin homogenates (Figure 1d), suggesting a specific modulation of the cytokine environment rather than a general elevation of inflammatory cytokine expression in K14E7 skin.
Figure 1. Elevated production of IFN-γ in K14E7 compared to wild type skin.
a IFN-γ concentrations in wild type and K14E7 skin homogenates and supernatants of skin explants (4 mice per group). Data points indicate individual mice analyzed in at least 2 independent experiments. b,c Flow cytometric analysis of cells isolated from dermis and epidermis of wild type and K14E7 mice, unstimulated or stimulated with PMA and ionomycin. b Total live cells gated for CD45.2+IFN-γ+ cells. Plots are representative of 4 independent experiments (2 mice per strain per experiment). c Numbers of CD45.2+IFN-γ+ cells in wild type and K14E7 dermis and epidermis obtained from 4 independent experiments. Bars indicate means±SEM. d IL-1β and IL-6 concentrations in wild type and K14E7 skin homogenates (4 mice per group).
CD8 and CD4 T cells are the main producers of IFN-γ in K14E7 skin
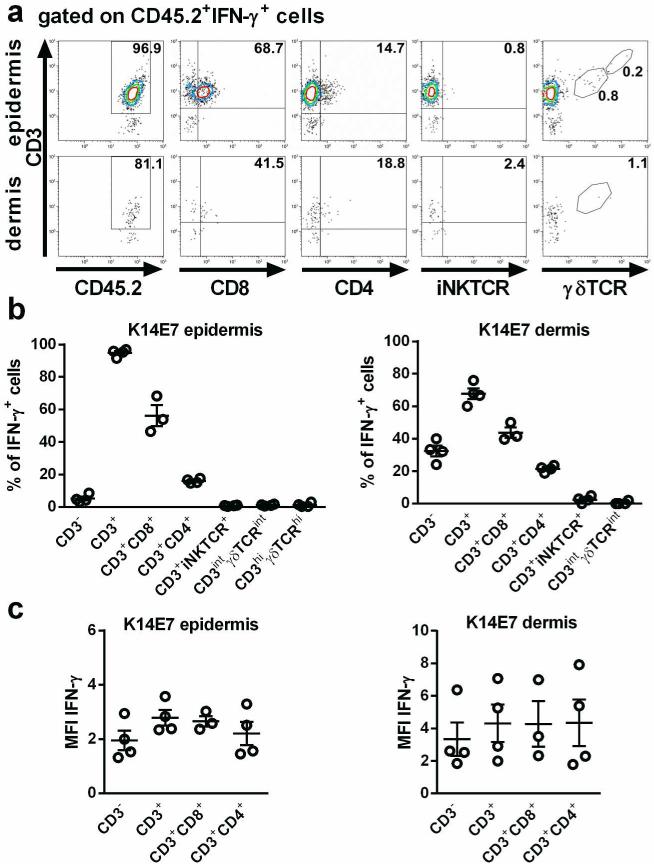
We next determined which cells are the major producers of IFN-γ in K14E7 skin. To identify IFN-γ-producing cell populations in dermal and epidermal cell suspensions stimulated with PMA and ionomycin, hematopoietic (CD45.2+) IFN-γ+ cells were gated for non-T cells (CD3−) and T cell (CD3+) subsets. The vast majority of IFN-γ producing cells in K14E7 epidermis were CD3+ T cells (94.8%±1.2% (mean±SEM)), with CD8+ T cells (56.2%±6.4%) and, to a lesser extent, CD4+ T cells (16.0%±0.7%), as the predominant IFN-γ producing subsets (Figure 2a,b). In contrast, few IFN-γ-producing cells were epidermal γδ T cells (CD3hiγδTCRhi) (1.3%±0.6%), dermal γδ T cells (CD3intγδTCRint) (1.2%±0.3%) and iNKT cells (0.8%±0.1%). Findings in K14E7 dermis were similar, with 67.6%±3.3% of IFN-γ producing cells being CD3+ T cells, 43.7%±3.2% CD8+ T cells, 21.5%±1.0% CD4+ T cells, 2.5%±1.0% iNKT cells, and 0.5%±0.5% dermal γδ T cells (Figure 2a,b). The mean fluorescence intensity of intracellular IFN-γ was similar among the cell subsets analyzed, indicating that they produced comparable amounts of IFN-γ (Figure 2c). In summary, these findings show that the major producers of IFN-γ in K14E7 skin are CD8+ and, to a lesser extent, CD4+ T cells.
Figure 2. The majority of IFN-γ producing cells in K14E7 skin are CD8+ and CD4+ T cells.
Flow cytometric analysis of cells isolated from dermis and epidermis of K14E7 mice, stimulated with PMA and ionomycin. a,b Percentage of CD3+, CD3+CD8+ and CD3+CD4+ T cells, iNKT cells and γδ T cells of CD45+IFN-γ+ cells. Plots are representative of 4 independent experiments (2 mice per experiment), whose results are summarized in B. c Mean fluorescence intensity of IFN-γ-producing cell subsets from 4 independent experiments. Bars indicate means±SEM of 4 independent experiments.
Elevated IL-12 production in K14E7 skin is dispensable for IFN-γ production
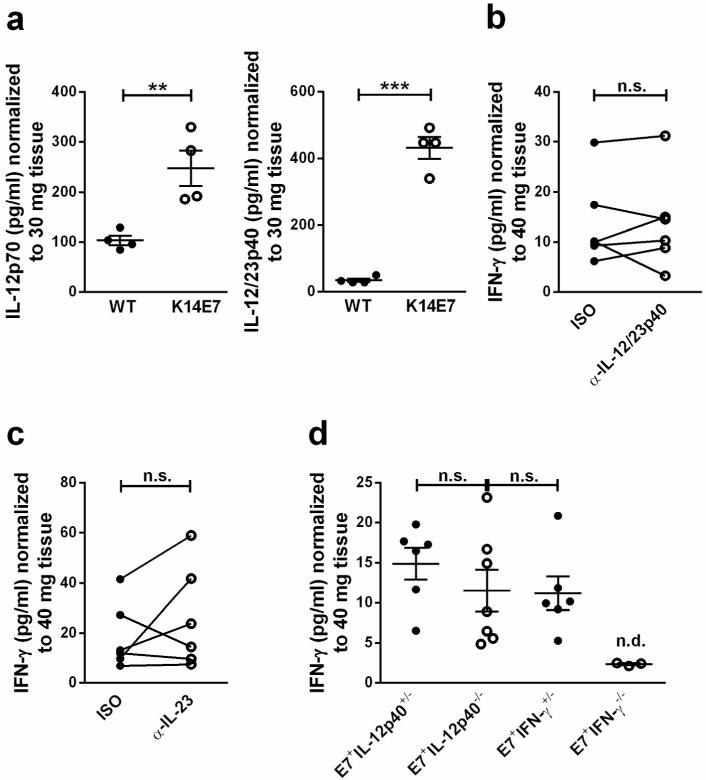
IL-12 and IL-18 play a central role as inducers and enhancers of IFN-γ production (Arend et al., 2008; Watford et al., 2004). It is currently unknown which of these mechanisms drive the induction of IFN-γ in the immunosuppressive environment of K14E7 skin. We first investigated whether IL-12 is differentially expressed in K14E7 compared to wild type skin. IL-12p40 mRNA levels were below the detection limit of the assay in both wild type and K14E7 skin samples (not shown). IL-12p70 and IL-12p40 concentrations were however significantly elevated in supernatants of in vitro cultured K14E7 skin explants (Figure 3a). The observation that IL-12p40 protein but not mRNA was detectable in K14E7 skin is likely due to the fact that RNA was extracted from the tissue at a specific time point, containing the IL-12p40 mRNA present in the tissue at that time. In contrast, in tissue culture, IL-12p40 protein released by skin explants accumulated in the supernatant over a period of 20 h before the protein concentration was determined. This circumstance likely facilitated the detection of IL-12p40 on the protein compared to the mRNA level. To investigate if IL-12 was required for IFN-γ production in K14E7 skin, skin explants were cultured in the presence of a neutralizing anti-IL-12p40 antibody. This did not lead to a reduction of IFN-γ secretion by the K14E7 skin explants (Figure 3b), suggesting that IFN-γ production in K14E7 skin was not driven by IL-12. IL-12p40 is also a component of the IL-23 heteromer but a neutralizing anti-IL-23p19 antibody did also not affect IFN-γ secretion by K14E7 explants (Fig 3c). Consistent with these findings, IFN-γ concentrations in the supernatants of skin explants from K14E7 transgenic IL-12p40−/− and control E7 transgenic IL-12p40+/− mice were similar (Figure 3d), as was ear skin histology (data not shown). Furthermore, K14E7 transgenic IL-12p40 deficient skin grafts were accepted by immunocompetent wild type recipients, indicating that IL-12p40 was not required to locally suppress host immune responses against the E7-expressing graft (Gosmann et al., submitted). Together, these results demonstrate that IL-12 is dispensable for IFN-γ production in K14E7 skin.
Figure 3. Elevated IL-12 production in K14E7 compared to wild type skin is dispensable for IFN-γ production.
a IL-12p70 and IL-12/23p40 concentrations in supernatants of C57BL/6 and K14E7 skin explants following 20 h of culture (n=4 mice per group). Results are representative of at least 2 independent experiments. b,c IFN-γ concentrations in supernatants of K14E7 skin explants cultured with 10 μg/mL isotype or (b) anti-IL12/23p40 antibody or (c) anti-IL-23p19 antibody (n=6 mice) for 40 h. Results for individual mice obtained in 2 independent experiments are shown. d IFN-γ concentrations in the supernatants of K14E7 transgenic IL-12p40 heterozygous, IL-12p40 deficient, IFN-γ heterozygous (n=6-7 mice each) or IFN-γ deficient (n=3) skin explants (n.d.: below detection limit). Results were obtained in 2 independent experiments. Bars indicate means±SEM.
IL-18 production is elevated in K14E7 compared to wild type skin and acts upstream of IFN-γ production in K14E7 skin
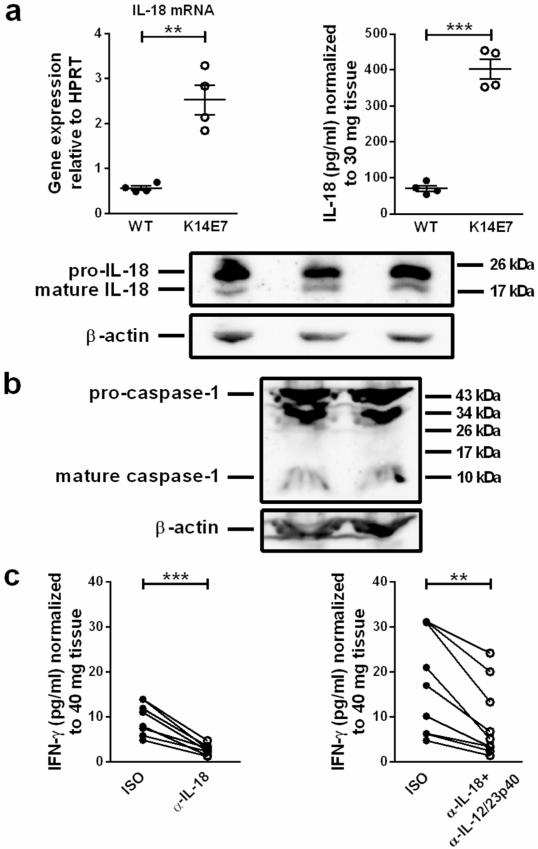
We next investigated whether IL-18 was differentially expressed in K14E7 compared to wild type skin. IL-18 mRNA expression was significantly higher in K14E7 skin (Figure 4a). Moreover, concentrations of secreted IL-18 protein in supernatants of K14E7 skin explants were significantly elevated compared to wild type skin explants, and we confirmed the production of mature IL-18 in K14E7 skin (Figure 4a). In line with this finding, mature caspase-1 was also detectable in K14E7 skin (Figure 4b).
Figure 4. IL-18 production is elevated and induces IFN-γ production in K14E7 skin.
a IL-18 mRNA expression in C57BL/6 and K14E7 skin (4 mice per group). IL-18 concentrations in supernatants of C57BL/6 and K14E7 skin explants (20 h; 4 mice per group). Means±SEM are indicated. a,b Western Blots showing (a) pro-IL-18 (24 kDa), mature IL-18 (18 kDa) (n=3), (b) pro-caspase-1 (45 kDa) and mature caspase-1 (10 kDa) (n=2) in K14E7 skin lysates. β-actin served as loading control. Results are representative of at least 2 independent experiments. c IFN-γ concentrations in supernatants of K14E7 skin explants cultured with 10 μg/mL isotype or anti-IL-18 and/or anti-IL-12/23p40 antibodies for 40 h. Data are paired observations for the same mouse, analyzed in 2 independent experiments (8-9 mice per treatment).
To investigate if IL-18 was required for IFN-γ production in K14E7 skin, skin explants were cultured in the presence of an IL-18-neutralizing antibody. This led to a significant inhibition of IFN-γ production by the explants (Figure 4c). As IL-12 has been reported to enhance IL-18-mediated IFN-γ production (Yoshimoto et al., 1998), we assessed whether simultaneous neutralization of IL-12 and IL-18 inhibited IFN-γ production to a greater extent compared to inhibition of IL-18 alone. Skin explant cultures in the presence of both anti-IL-18 and anti-IL-12 antibodies had significantly reduced IFN-γ concentrations in the culture supernatant, but to no greater extent than IL-18 neutralization alone (Figure 4c; average inhibition 61% (anti-IL-18/IL-12p40) compared to 72% (anti-IL-18)).
We aimed to assess the role for IL-18 in suppressing effective immune responses against the HPV16 E7 antigen by grafting E7 transgenic IL-18 deficient skin onto immunocompetent non-transgenic recipients. However, breeding of K14E7 with IL-18−/− mice failed to produce E7 transgenic IL-18−/− progeny.
In summary, our findings suggest that IL-18, but not IL-12, is required for induction of IFN-γ production in K14E7 skin, and that IL-12 does not enhance IL-18-induced IFN-γ production in K14E7 skin.
Discussion
IFN-γ is generally regarded as a proinflammatory cytokine. In contrast, we previously reported that IFN-γ promotes local antigen-specific immunosuppression in E7-associated epithelial hyperplasia (Mattarollo et al., 2010), and in a model of UVB-induced melanoma, IFN-γ from macrophages promoted melanocyte immune evasion in skin (Zaidi et al., 2011). The aim of the present study was to identify mechanisms of IFN-γ induction in E7-associated hyperplastic skin, and to establish whether this would assist in understanding the unexpected role of IFN-γ in cutaneous immunosuppression. Here, we report increased IFN-γ, IL-12 and IL-18 production in K14E7 transgenic hyperplastic skin that was not associated with a more general inflammatory response, as IL-1β and IL-6 production were in contrast decreased (Figure 1d), and show that IL-18 but not IL-12 contributed to the production of IFN-γ. In the current study, CD8+ T cells and, to a lesser extent, CD4+ T cells were the predominant producers of IFN-γ in K14E7 transgenic skin and were mostly found in the hyperplastic epidermis. Increased numbers of IFN-γ-producing cells were similarly detected in the hyperplastic epidermis of atopic dermatitis lesions (Hijnen et al., 2013). T cells expressing CCR6, a chemokine receptor involved in cell homing to the epidermis (Mabuchi et al., 2013), are found in K14E7 skin (Choyce et al., 2013), and production of a cognate ligand by hyperplastic skin may therefore similarly contribute to the infiltrate of IFN-γ-producing cells.
IL-18 could induce IFN-γ production in K14E7 transgenic hyperplastic skin by direct action on T cells or indirectly through activation of iNKT cells. iNKT cells, though they constitute only a small percentage of the IFN-γ-producing cells in K14E7 transgenic skin, contribute to E7-specific local immune suppression (Mattarollo et al., 2010), suggesting that they might be a target of IL-18.
E7-reactive CD8+ T cells are eliminated in K14E7 transgenic mice (Frazer et al., 1998) due to E7 expression in the thymus (Herber et al., 1996). Thus, IFN-γ production by CD8+ T cells in K14E7 skin is likely to occur in an antigen-independent manner. Antigen-independent IFN-γ production by memory CD44hi CD8+ T cells in response to IL-12 and IL-18 has been reported in bacterial infection (Berg et al., 2002; Berg et al., 2003; Kambayashi et al., 2003) and in cytokine-stimulated human peripheral blood CD4+ T cells (Munk et al., 2011). However, IFN-γ production in K14E7 skin does not require IL-12, suggesting that different mechanisms may induce IFN-γ in infections and in E7-associated skin hyperplasia.
While IL-18 and IL-12 synergize in inducing IFN-γ production during infection (Yoshimoto et al., 1997; Takeda et al., 1998; Berg et al., 2002; Kambayashi et al., 2003), we unexpectedly found that in E7-induced skin hyperplasia, IL-18 but not IL-12 contributed to IFN-γ production. In a mouse model of hyperplastic epithelium associated with atopic dermatitis, overexpression of IL-18 in serum and IL-12 in skin-draining lymph nodes was also observed, and IL-18 was required for induction of IFN-γ-producing Th1 cells in skin-draining lymph nodes, but the contribution of IL-12 was not assessed (Terada et al., 2006). Molecular mechanisms underlying the synergy of IL-18 and IL-12 in inducing IFN-γ expression include IL-12-mediated up-regulation of IL-18 receptor expression (Yoshimoto et al., 1998), and enhanced binding of IL-18-induced AP-1 to the IFN-γ promoter via a STAT4-mediated mechanism downstream of IL-12 signaling (Nakahira et al., 2002). While the detailed molecular mechanisms of IFN-γ induction in K14E7 transgenic skin remain to be elucidated, IL-12 might not synergize with IL-18 for IFN-γ production in E7-induced epithelial hyperplasia because IL-18 receptor expression is not induced, IL-12 receptor expression is downregulated, or because alternative mechanisms of STAT4 activation, as seen in response to some infections (Buxbaum et al., 2002; Freudenberg et al., 2002), render a requirement for IL-12 redundant. IL-23 can also induce IFN-γ production in T cells (Oppmann et al., 2000) and NKT cells (van de Wetering et al., 2009). However, blocking IL-23p19 in K14E7 transgenic skin explant cultures did not significantly alter IFN-γ production by K14E7 skin explants, ruling out a major contribution of IL-23.
This study demonstrates that IL-18 drives IFN-γ production in hyperplastic skin, contributing to an immunosuppressive environment. In line with this finding, an association between IL-18 promoter polymorphisms and the risk of developing HPV-associated cervical cancer has been reported (Sobti et al., 2008). IL-18 shows antitumor activity in several preclinical animal models, but was not effective in patients with metastatic melanoma (Tarhini et al., 2009; Srivastava et al., 2010). Our findings suggest that for skin-associated malignancy, local blockade of IL-18 rather than systemic administration could facilitate effective anti-tumor immune responses.
Materials and Methods
Mice
C57BL/6J (wild type) mice and HPV16 E7 transgenic C57BL/6J mice expressing the E7 oncoprotein under control of the K14 promoter (K14E7; Herber et al., 1996), were from the Animal Resources Centre (Perth, Australia). IL-12p40−/− (Magram et al., 1996) and IL-18−/− (Takeda et al., 1998) mice were obtained from Camile Farah (The University of Queensland Centre for Clinical Research, Brisbane, Australia) and Ben Croker (The Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia), respectively. All mice were maintained under specific pathogen-free conditions. To generate K14E7 mice deficient in IL-12p40 or IL-18, hemizygous K14E7 mice were crossed and the E7 expressing progeny backcrossed with homozygous IL-12p40−/− and IL-18−/− mice. Mice were sex- and age-matched for all experiments. Animal procedures were approved by the University of Queensland Animal Ethics Committee (AEC# 290/10/NHMRC/NIH).
Preparation of skin homogenate
Mouse ears were snap-frozen and homogenized on ice in PBS with protease inhibitor (cOmplete ULTRA Mini, Roche, Basel, Switzerland) using a T 10 basic ULTRA-TURRAX® homogenizer (IKA, Staufen im Breisgau, Germany). Homogenates were centrifuged (18000 rcf, 10 min, 4°C) and supernatant collected. Total protein concentration was determined using BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA).
Skin explant culture
Mouse ears were split and placed dermis side down on DMEM with 10% fetal bovine serum (FBS), penicillin/streptomycin/glutamine (1 mM), Sodium Pyruvate (1 mM) (Gibco, Mulgrave, Australia) and HEPES (10 mM) (Invitrogen, Carlsbad, CA) in 24 well plates. To remove cytokines released as a result of tissue preparation, medium was exchanged after 1 h and another 2 h of culture (37°C, 5% CO2). Medium changes reduced the lactate dehydrogenase (LDH) activity (CytoTox 96 NonRadioactive Cytotoxicity Assay; Promega, Fitchburg, WI) in the supernatant to medium control levels 2 h after the last medium exchange (data not shown). Supernatants were collected 20 h after the last medium replacement for cytokine analysis by ELISA, and low LDH activity in the supernatants was confirmed (data not shown).
For cytokine neutralization in skin explant cultures, culture medium was supplemented with monoclonal antibodies against IL-18 (93-10C) (MBL, Woburn, MA) and IL-12/23p40 (C17.8) (eBioscience, San Diego, CA) or matched isotype control antibodies from the same company. Anti-IL-23p19 neutralizing and matched isotype control antibodies were provided by Lilly Research Laboratories (Indianapolis, IN). The dorsal (or ventral) half from one ear was treated with neutralizing antibody and compared with the dorsal (or ventral) half from the other ear of the same mouse, treated with isotype control antibody. Supernatants were replaced after 20 h with antibody-supplemented medium and collected after another 20 h for analysis by ELISA.
Cytokine ELISA
ELISA (mouse IFN-γ, IL-23 (eBioscience); total IL-18 (MBL); IL-12p70, IL-12/23p40, IL-6, total IL-1β (BD Biosciences, San Jose, CA)) was performed according to the manufacturers’ instructions. Samples undetectable within the assay range were plotted as the detection limit. Cytokine concentrations in supernatants of skin explant cultures were normalized to tissue weight and are presented as pg/mg tissue.
Isolation and stimulation of skin cells
Mouse ears were split into halves and incubated dermis-side down in 2 mg/mL dispase in PBS (Roche, Basel, Switzerland) for 1 h at 37°C. To obtain sufficient cell numbers for analysis, ears from two mice per strain were combined for processing. Dermis and epidermis were separated and incubated in PBS containing 0.3 mg/mL collagenase / 5 μg/mL of DNase (Roche) and 0.25% trypsin / 0.1% EDTA / 5 μg/mL DNase, respectively, for 30 min at 37°C. Resulting tissue fragments were vigorously pipetted and the cell suspension was passed through a 40 μm cell strainer (BD Biosciences).
Up to 1 × 106 of total cells per mL were stimulated with 1 μg/mL PMA and 0.5 μg/mL ionomycin (Calbiochem/EMD4Biosciences, San Diego, CA) in complete medium (supplemented as detailed above) for 4 h at 37°C. After 30 min, monensin (BioLegend, San Diego, CA) and GolgiPlug (BD Biosciences) were added to cultures to inhibit cytokine release from the Golgi/endoplasmic reticulum complex for intracellular cytokine staining.
Flow cytometry
Monoclonal antibodies against mouse CD45.2 (104), CD3e (145-2C11), TCRγδ (GL3), IFN-γ (XMG1.2) and corresponding isotype controls were from BD Biosciences. Anti-CD8a (53-6.7) and CD4 (RM4-5) antibodies were from eBioscience. α-GalCer-loaded CD1d tetramer for detection of invariant NKT cells was kindly provided by Dale Godfrey (University of Melbourne, Australia). Cell suspensions were incubated with Fc block (BD Biosciences) for 15 min, followed by Live/Dead Fixable Aqua Dead Cell Stain (Invitrogen) and antibodies against cell surface markers for 20 min. For intracellular cytokine staining, cells were fixed and permeabilized (BD Biosciences Cytofix/Cytoperm kit). Antibodies against cytokines and matched isotype control antibodies were added for 30 min. All steps were performed at 4°C. Flow Count Fluorospheres (Beckman Coulter, Brea, CA) were used for calculation of cell numbers. The samples were acquired on a Gallios (Beckman Coulter) flow cytometer. Data were analyzed using Kaluza software 1.2 (Beckman Coulter). Doublets and dead cells were excluded from analysis.
Quantitative real time PCR (qRT-PCR)
For RNA extraction, mouse ears were homogenized in TRIzol (Invitrogen). Total RNA was purified using the RNeasy Mini Kit (Qiagen, Venlo, Netherlands) and reversely transcribed using Oligo d(T) (Applied Biosystems, Foster City, CA). Primers for SYBR Green qRT-PCR were purchased from Integrated DNA Technologies (IDT, Coralville, IA) and were designed using the PrimerQuest PCR design tool (IDT). Primer sequences: mHPRT FW: 5′ CCCCAAAATGGTTAAGGTTGC 3′; mHPRT RV: 5′ AACAAAGTCTGGCCTGTATCC 3′. Predesigned TaqMan gene expression assays (IDT) were used for mouse IL-12 and IL-18. qRT-PCR was performed using a 7900HT Fast Real-Time PCR System (Applied Biosystems). Data were analyzed using SDS Software 2.4 (Applied Biosystems). mRNA expression relative to the house-keeping gene hypoxanthine-guanine phosphoribosyl-transferase (HPRT) was calculated as 2^(Ct (HPRT) - Ct (gene of interest)).
Western Blot
Mouse ears were snap-frozen and homogenized on ice in RIPA buffer (1% Triton-X, 20 mM Tris pH7.5, 150 mM NaCl, protease inhibitor (Roche), in Milli-Q H2O) using a T 10 basic ULTRA-TURRAX homogenizer. Homogenates were centrifuged (18000 rcf, 10 min), supernatant collected and filtered through a 70 μm cell strainer (BD Biosciences) to remove aggregates (4°C). Total protein concentration was determined as above.
At least 50 μg of denatured skin lysate per sample and Rainbow Molecular Weight Markers (GE Healthcare, Chalfont St Giles, United Kingdom) were separated on 15% (IL-18) or 12% (caspase-1) polyacrylamide gels and transferred onto 0.45 μm polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica, MA). Blots were blocked with 5% skim milk in Tris-buffered saline/0.1% Tween20 (1 h, RT) and incubated with primary antibodies against mouse IL-18 (39-3F; MBL International), caspase-1 p10 (sc-514; Santa Cruz Biotechnology, Dallas, TX), or β-actin (AC-15; Sigma-Aldrich, St. Louis, MO), followed by HRP-conjugated secondary antibody. Blots were developed using SuperSignal West Dura Chemiluminescent Substrate (Thermo Fisher Scientific). Images were acquired with ChemiDoc (Bio-Rad, Hercules, CA).
Statistics
Statistical analysis was performed using Prism (Graphpad Software, La Jolla, CA). Paired Student’s t test was used to compare data from skin explant cultures treated with specific and isotype control antibodies. All other groups were compared by unpaired Student’s t test. Differences were considered significant at p<0.05 and are indicated as *p<0.05, **p<0.01 and ***p<0.001. n.s.: not significant.
Acknowledgements
We thank Lilly Laboratories for providing the IL-23p19 neutralizing antibody, Kon Kyparissoudis and Dale Godfrey (University of Melbourne) for providing the α-GalCer-loaded mouse CD1d tetramer, and the staff of the Biological Research Facility (Brisbane, Australia) for excellent technical assistance and animal care. This work was supported by program grant 080214 from the National Health and Medical Research Council of Australia, NCI grant 5U01CA141583, and funding from the Australian Cancer Research Foundation. Christina Gosmann was recipient of UQIRTA and UQRS postgraduate scholarships from the University of Queensland, and Stephen R. Mattarollo and Antje Blumenthal were in receipt of Balzan Fellowships.
Footnotes
Conflicts of Interest
The authors state no conflict of interest.
References
- Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- Berg RE, Cordes CJ, Forman J. Contribution of CD8+ T cells to innate immunity: IFN-gamma secretion induced by IL-12 and IL-18. Eur J Immunol. 2002;32:2807–2816. doi: 10.1002/1521-4141(2002010)32:10<2807::AID-IMMU2807>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Berg RE, Crossley E, Murray S, et al. Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J Exp Med. 2003;198:1583–1593. doi: 10.1084/jem.20031051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum LU, Uzonna JE, Goldschmidt MH, et al. Control of New World cutaneous leishmaniasis is IL-12 independent but STAT4 dependent. Eur J Immunol. 2002;32:3206–3215. doi: 10.1002/1521-4141(200211)32:11<3206::AID-IMMU3206>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Choyce A, Yong M, Narayan S, et al. Expression of a single, viral oncoprotein in skin epithelium is sufficient to recruit lymphocytes. PLoS One. 2013;8:e57798. doi: 10.1371/journal.pone.0057798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- Frazer IH, Fernando GJ, Fowler N, et al. Split tolerance to a viral antigen expressed in thymic epithelium and keratinocytes. Eur J Immunol. 1998;28:2791–2800. doi: 10.1002/(SICI)1521-4141(199809)28:09<2791::AID-IMMU2791>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Freudenberg MA, Merlin T, Kalis C, et al. Cutting edge: a murine, IL-12-independent pathway of IFN-gamma induction by gram-negative bacteria based on STAT4 activation by Type I IFN and IL-18 signaling. J Immunol. 2002;169:1665–1668. doi: 10.4049/jimmunol.169.4.1665. [DOI] [PubMed] [Google Scholar]
- Herber R, Liem A, Pitot H, et al. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J Virol. 1996;70:1873–1881. doi: 10.1128/jvi.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijnen D, Knol EF, Gent YY, et al. CD8(+) T cells in the lesional skin of atopic dermatitis and psoriasis patients are an important source of IFN-gamma, IL-13, IL-17, and IL-22. J Invest Dermatol. 2013;133:973–979. doi: 10.1038/jid.2012.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambayashi T, Assarsson E, Lukacher AE, et al. Memory CD8+ T cells provide an early source of IFN-gamma. J Immunol. 2003;170:2399–2408. doi: 10.4049/jimmunol.170.5.2399. [DOI] [PubMed] [Google Scholar]
- Mabuchi T, Singh TP, Takekoshi T, et al. CCR6 is required for epidermal trafficking of gammadelta-T cells in an IL-23-induced model of psoriasiform dermatitis. J Invest Dermatol. 2013;133:164–171. doi: 10.1038/jid.2012.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattarollo SR, Rahimpour A, Choyce A, et al. Invariant NKT cells in hyperplastic skin induce a local immune suppressive environment by IFN-gamma production. J Immunol. 2010;184:1242–1250. doi: 10.4049/jimmunol.0902191. [DOI] [PubMed] [Google Scholar]
- Minguela A, Pastor S, Mi W, et al. Feedback regulation of murine autoimmunity via dominant anti-inflammatory effects of interferon gamma. J Immunol. 2007;178:134–144. doi: 10.4049/jimmunol.178.1.134. [DOI] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Munk RB, Sugiyama K, Ghosh P, et al. Antigen-independent IFN-gamma production by human naive CD4 T cells activated by IL-12 plus IL-18. PLoS One. 2011;6:e18553. doi: 10.1371/journal.pone.0018553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira M, Ahn HJ, Park WR, et al. Synergy of IL-12 and IL-18 for IFN-gamma gene expression: IL-12-induced STAT4 contributes to IFN-gamma promoter activation by up-regulating the binding activity of IL-18-induced activator protein 1. J Immunol. 2002;168:1146–1153. doi: 10.4049/jimmunol.168.3.1146. [DOI] [PubMed] [Google Scholar]
- Okamura H, Tsutsi H, Komatsu T, et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378:88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- Oppmann B, Lesley R, Blom B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- Pastor S, Minguela A, Mi W, et al. Autoantigen immunization at different sites reveals a role for anti-inflammatory effects of IFN-gamma in regulating susceptibility to experimental autoimmune encephalomyelitis. J Immunol. 2009;182:5268–5275. doi: 10.4049/jimmunol.0800681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. 2007;96:41–101. doi: 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Sobti RC, Shekari M, Tamandani DM, et al. Association of interleukin-18 gene promoter polymorphism on the risk of cervix carcinogenesis in north Indian population. Oncol Res. 2008;17:159–166. doi: 10.3727/096504008785114156. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Salim N, Robertson MJ. Interleukin-18: biology and role in the immunotherapy of cancer. Curr Med Chem. 2010;17:3353–3357. doi: 10.2174/092986710793176348. [DOI] [PubMed] [Google Scholar]
- Takeda K, Tsutsui H, Yoshimoto T, et al. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity. 1998;8:383–390. doi: 10.1016/s1074-7613(00)80543-9. [DOI] [PubMed] [Google Scholar]
- Tarhini AA, Millward M, Mainwaring P, et al. A phase 2, randomized study of SB-485232, rhIL-18, in patients with previously untreated metastatic melanoma. Cancer. 2009;115:859–868. doi: 10.1002/cncr.24100. [DOI] [PubMed] [Google Scholar]
- Terada M, Tsutsui H, Imai Y, et al. Contribution of IL-18 to atopic-dermatitis-like skin inflammation induced by Staphylococcus aureus product in mice. Proc Natl Acad Sci U S A. 2006;103:8816–8821. doi: 10.1073/pnas.0602900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering D, de Paus RA, van Dissel JT, et al. IL-23 modulates CD56+/CD3− NK cell and CD56+/CD3+ NK-like T cell function differentially from IL-12. Int Immunol. 2009;21:145–153. doi: 10.1093/intimm/dxn132. [DOI] [PubMed] [Google Scholar]
- Watford WT, Hissong BD, Bream JH, et al. Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4. Immunol Rev. 2004;202:139–156. doi: 10.1111/j.0105-2896.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- Willenborg DO, Fordham SA, Staykova MA, et al. IFN-gamma is critical to the control of murine autoimmune encephalomyelitis and regulates both in the periphery and in the target tissue: a possible role for nitric oxide. J Immunol. 1999;163:5278–5286. [PubMed] [Google Scholar]
- Yoshimoto T, Okamura H, Tagawa YI, et al. Interleukin 18 together with interleukin 12 inhibits IgE production by induction of interferon-gamma production from activated B cells. Proc Natl Acad Sci U S A. 1997;94:3948–3953. doi: 10.1073/pnas.94.8.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto T, Takeda K, Tanaka T, et al. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-gamma production. J Immunol. 1998;161:3400–3407. [PubMed] [Google Scholar]
- Zaidi MR, Davis S, Noonan FP, et al. Interferon-gamma links ultraviolet radiation to melanomagenesis in mice. Nature. 2011;469:548–553. doi: 10.1038/nature09666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]