Abstract
There is accumulating evidence that mesenchymal stem cells (MSC) have their origin as perivascular cells (PVC) in vivo, but precisely identifying them has been a challenge, as they have no single definitive marker and are rare. We have developed a fluorescent transgenic vertebrate model in which PVC can be visualized in vivo based upon sdf1 expression in the zebrafish. Prospective isolation and culture of sdf1DsRed PVC demonstrated properties consistent with MSC including prototypical cell surface marker expression; mesodermal differentiation into adipogenic, osteogenic and chondrogenic lineages; and the ability to support hematopoietic cells. Global proteomic studies performed by 2-dimensional liquid chromatography and tandem mass spectrometry revealed a high degree of similarity to human MSC and discovery of novel markers (CD99, CD151 and MYOF) that were previously unknown to be expressed by hMSC. Dynamic in vivo imaging during fin regeneration showed that PVC may arise from undifferentiated mesenchyme providing evidence of a PVC – MSC relationship. This is the first model, established in zebrafish, in which MSC can be visualized in vivo and will allow us to better understand their function in a native environment.
Introduction
Mesenchymal stem cells (MSC) are a component of the bone marrow niche and are thought to provide support to hematopoietic cells. Biologic characterization chiefly has been performed only after MSC have been culture expanded from the microenvironment in which they reside. MSC do not originate exclusively from the bone marrow, previous evidence shows that MSC also can be derived from multiple tissue sources[1, 2], suggesting that the ancestral cell of the MSC is common to most if not all tissues. Two previous reports suggested that a cell found on the abluminal surface of blood vessel, a perivascular cell (PVC), is a potential source for MSC[3, 4].
Early evidence for a PVC origin for MSC was provided by Crisan who identified PVC expression of CD146 (melanoma associated adhesion molecule), NG2 (glia cell marker), and platelet-derived growth factor receptor expression-β (PDGF-Rβ) expression. Flow cytometry sorted CD146 positive cells and cultured outgrowths on plastic dishes ultimately give rise to MSC, defined by CD73, CD90, and CD105 expression (and lacking mature hematologic or endothelial marker expression) and their ability to become adipogenic, osteogenic, or chondrogenic cells using in vitro differentiation assays[4]. Zannettino et al. showed that the vasculature of adipose tissue harbors CD146 and 3G5 (beta amyloid) co-expressing PVC which met the definition of MSCs based upon phenotype and in vitro differentiation capacity[5].
SDF1 was originally isolated from cultured bone marrow stromal cells that were consistent with MSC[6]. Recently, Ding et al showed that endothelial and perivascular cells express SDF1 in vivo using a transgenic Sdf1 reporter mouse model[7]. Sugiyama reported that Sdf1 expressing cells are located near sinusoidal endothelial cells or in the endosteal marrow region in a different Sdf1 reporter mouse[8]. However, direct visualization of MSC ancestors as PVC and their isolation has been technically difficult[4].
In our previous work, we described the development an sdf1a:DsRed transgenic zebrafish which was the first animal model that allowed direct visualization of cells that produce high levels of stromal cell-derived factor-1 (Sdf1a) marked by fluorescent reporter expression. Given that Sdf1a is used by a number of developing organ systems for cell migration during development[9–12], we were not surprised to find that our sdf1a:DsRed transgenic zebrafish indicated sdf1a was expressed in multiple organs and cell types including gills, brain, and PVC[13].
Building on these prior observations, we have now prospectively isolated sdf1DsRed PVC and have shown they express genes consistent with PVC. We have established a culture system for zebrafish PVC expansion in vitro and have shown that PVC can be tri-differentiated into mesodermal lineages and support the growth of hematopoietic cells in vitro. Tandem mass spectrometry analysis of cultured sdf1DsRed PVC reveled their proteome to be similar to the human MSC proteome including cell surface proteins characteristic of MSCs. Finally, during fin regeneration after amputation, sdf1DsRed cells contributed to the mesenchyme of the newly developing bony rays prior to re-occupying the “perivascular niche”. The sdf1:DsRed transgenic zebrafish will be a powerful tool to investigate the nature of PVCs and the PVC-MSC connection in further studies.
Materials and Methods
Zebrafish Strains and Fish husbandry
Fish were maintained by the University of Minnesota Zebrafish Core Facility according to standardized procedures[14] and with the approval of the International Animal Care and Use Committee, IACUC. Wild-type fish were obtained from Segrest Farms (Gibsonton, Florida) and bred in-house. The original sdf1a:DsRed transgenic was crossed with the fli1:EGFP as previously reported[15].
Microscopy
Live embryos, juvenile adult fish, and fish during fin regeneration experiments were anesthetized in 100 mg/L tricaine-S solution until no longer responsive when disturbed but still respiring. Fish were imaged using a QImaging Retiga 2000R camera and QCapture™ software on an inverted Leica DMI6000B microscope with a variety of objectives. Scale bars were placed via software integration. For z-stack imaging, confocal laser scanning microscopy was performed using a Zeiss LSM 510 inverted microscope system with a 20× air objective and appropriate filter sets. For z-stack imaging, confocal laser scanning microscopy was performed using a Zeiss LSM 510 inverted microscope system with a 20× air objective. Fish were anesthetized as above and placed in two- or four-well coverslip imaging chambers (Nunc). Excitation and emission filters were as follows: GFP, 488 nm excitation, emission BP 500–530 nm filter; RFP, 543 nm excitation, emission BP 565–615 nm filter.
Tail Digestion
Half of the caudal fin was removed using a sterile razor blade. Each fin was immersed for 10 seconds in 100% ethanol and placed separately in a 24 well plate in 100 µL PBS on ice while other fins were collected. The PBS was removed and replaced with 1:10,000 solution of bleach in PBS for 10 minutes at 25°C. Following bleaching, the fins were washed with 100 µL PBS three times for five minutes. Liberase was added to 0.01M HEPES in DMEM to create the digest media according to the manufacture’s instructions, and 1mL of the digest media was added to each fin sample. Each sample was then mechanically shredded into smaller pieces using sterile scissors and incubated at 32°C for 30 minutes. Following incubation, the samples were dissociated manually using a pipette and placed on a shaker at 4°C overnight. The samples were dissociated again the next day with a pipette and passed through a 40-micron filter prior to downstream analyses.
Cell Culture
sdf1DsRed PVC were isolated by flow cytometry gating on a DsRed positive cell population and maintained zebrafish cell culture media (250 mL L-15, 175 mL DMEM, 75 mL Ham’s 12, 75mg sodium bicarbonate, 0.15M HEPES, 50 units penicillin/50 micrograms streptomycin, 2 mM l-glutamine, 50 mL fetal bovine serum (FBS), 5 ng/ml selenium, 5 ng/ml human bFGF). Stromal cells appeared in about 2 – 3 weeks. The resulting cellular outgrowths were maintained and split at 80 – 90% confluence once or twice per week. Incubator conditions were 4% CO2 and 5% O2 and a temperature of 28°C. Human MSC were thawed from frozen stock and cultured as previously described[16].
Differentiations/Staining
See Supplemental Methods.
In Situ Hybridization
Fins were amputated in sterile, RNase free conditions, embedded in OCT, and flash frozen in dry-ice cooled isopentane. Frozen blocks were either stored at −80°C or immediately sectioned. Sections were taken at 8µm, allowed to dry for 10 minutes, and then fixed in 4% paraformaldehyde in PBS for 1 hour at room temperature (RT). sdf1a in situ hybridization and immunological detection was performed as previously described with the following deviations: slides were not covered with a coverslip during the overnight RNA probe hybridization, and slides were incubated at 37°C during the hybridization step[17].
Hematopoietic Co-culture
The sdf1DsRed cells were grown to confluency in 24-well plate. 50,000 freshly isolated whole kidney marrow (WKM) cells from bactin2:GFP transgenic fish were place onto of the monolayer in zebrafish cell culture media for 16 days. Plastic only control wells contained no monolayer. After 16 days, cells were pipet aspirated and enumerated by counting on a flow cytometer using counting beads. For short-term transplant experiments, all the hematopoietic cells content of a single well (24-well) were transplanted into a recipient zebrafish that had received 20 Gy irradiation 48 hours prior. Seven days after transplant, recipient WKM was harvested and donor cell engraftment was determined by flow cytometry for GFP positive cells. In one experiment, 50,000 freshly isolated WKM cells were used in comparison.
Flow Cytometry
Cells were harvested using trypsin and stained with anti-myoferlin 7D2 (Abcam, ab76746), anti-CD151 conjugated to Allophycocyanin (Biolegend, #350405), or anti-CD99 conjugated to phycoerythrin (Biolegend, #318008) according to the manufacture’s recommendations. Secondary antibodies were used when appropriate (Biolegend, #Poly4053). Cells were stained for 40 minutes at 4 C followed by once the addition of 3 milliliters of PBS, centrifugation at 1000 × g for five minutes and suspension in PBS prior to flow cytometry on a BD FACS Canto Flow Cytometer. Data analysis was performed using FlowJo (Tree Star, Inc, Ashland, OR)
RT-PCR
RNA was prepared using Trizol per the manufacture’s protocol. RT-PCR was performed according to standard methods. Primers used in this study are listed in Supplemental Table 1. Often SYBR®Green (Invitrogen) quantitation was used, but occasionally TaqMan® (ABI) primers were employed, in which case the product number is listed. The melting temperature was 60 C.
Endothelial Co-culture
Co-culture experiments were conducted using human umbilical vein endothelial cells (HUVECs, Lonza C2517A) and previously isolated and culture expanded sdf1DsRed cells from the caudal fin of adult transgenic zebrafish. HUVECs were grown and maintained as instructed by the supplier. Each cell population was grown to approximately eighty percent confluency in their appropriate medium and fluorescently labeled as adherent cells to the flask using CellTracker™ Dyes (Invitrogen) according the manufacture’s instructions. HUVECs were labeled using CellTracker™ Orange (Invitrogen C34551) and cultured sdf1DsRed cells were labeled using CellTracker™ Green (Invitrogen C2925) at a final concentration of 1µM. Following labeling, 15,000 HUVECs and 10,000 culture expanded sdf1DsRed cells were mixed together and grown on 100 µL of Matrigel™ (BD Biosciences) in a 96-well plate with EGM-2 media (Lonza). The cells were imaged 10–16 hours following plating, and the qualitative observations of HUVEC and perivascular cell branching were observed using a fluorescent microscopy as above.
Cross Section Immunohistochemistry
Fish fins were frozen in Tissue-Tek O.C.T compound (Sakura Finetek USA, Torrance, CA) and sectioned at 6 microns on a cryostat. Sections were fixed with a mixture of 3.7% paraformaldehyde, 10% Sucrose for 5 minutes at room temperature. Fins were rehydrated with PBS and blocked with 10% normal donkey serum for 1 hour (Jackson Immunoresearch, West Grove, PA). Primary antibody to DsRed (1:300) (Clontech, Mountain View, CA) was added for 1 hour, washed off with PBS, then mixed with primary antibody to GFP conjugated to FITC (1:200) (Rockland, Gilbertsville, PA) followed by a second secondary antibody which was donkey anti-rabbit-Cy3 (1:500), (Jackson Immunoresearch, West Grove, PA) was applied for 1 hour. Fins were washed with PBS and cover-slipped with hard set DAPI, 4,6-diamidino-2-phenylindole (Vector Labs, Burlingame, CA). Slides were imaged by confocal fluorescence microscopy (Olympus BX61, FV500, Olympus Optical, Tokyo, Japan).
Mass Spectrometry
See Supplemental Methods.
Fin Regeneration
Zebrafish were anesthetized in 100 mg/L tricaine-S solution until it was no longer responsive when disturbed. While anesthetized, the caudal fin was amputated using a sterile razor blade. Photos of the regenerating fin were captured every 24 hours. All photos were taken using an inverted fluorescent microscope as described above. Unless otherwise stated, regeneration images were taken using a 10× objective lens. Fluorescent images were pseudo-colored in Adobe Photoshop CS4 according to the emission of the fluorophore, and multiple color channels within a single field of view were merged together.
Results
Although sdf1DsRed PVC were found in multiple organs as we have previously described[13], we focused on caudal tail isolation, a site of high prominence and visualization (Figure 1, Supplemental Figures 1 and 2). The sdf1DsRed PVC were most evident when we crossed our sdf1a:DsRed transgenic to the fli1:EGFP transgenic zebrafish which robustly illustrates the vasculature composed of EGFP positive endothelial cells[18]. High magnification revealed sdf1DsRed cells notably located on the abluminal surface of various blood vessels including both the central vessels of the bony rays as well as bridging vessels between rays as shown in Figure 1(B–D). This is the first time perivascular cells have been seen using in vivo live fluorescent microscopy in a transgenic animal.
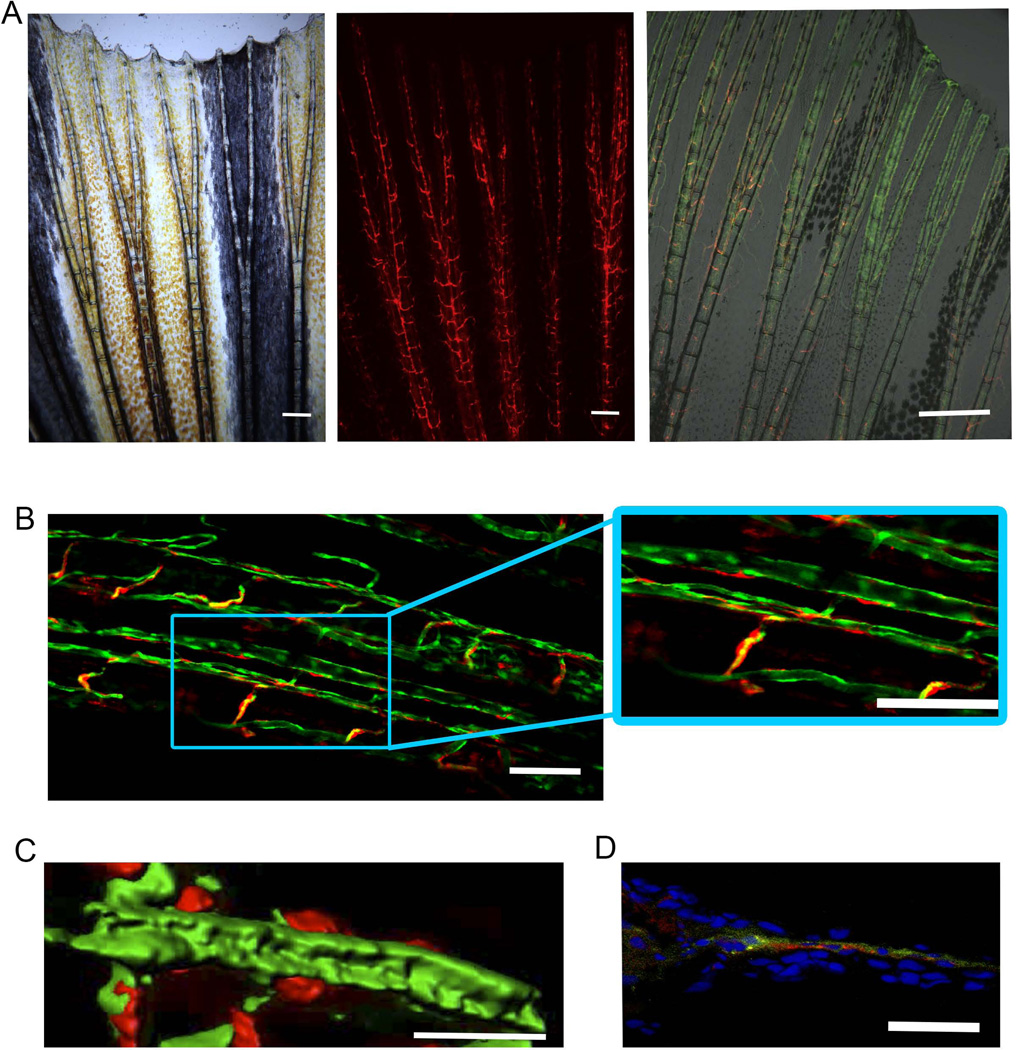
Figure 1.
The sdf1a:DsRed transgene illuminates perivascular cells. (A) Caudal fin of 10-week old sdf1a:DsRed zebrafish at 5×. Scale bar represents 200 microns. Rightmost panel shows live fluorescent microscopy of the tail-fin in a 10-week old fli1:EGFP crossed to sdf1a:DsRed shows a population of sdf1aDsRed cells as perivascular in anatomical location. Scale bar represents 500 microns. (B) Confocal microscopy of a caudal tail fin from a fli1:EGFP × sdf1a:DsRed transgenic zebrafish shows enhanced discrimination of sdf1DsRed cells. Scale bars represent 50 microns. (C) Live confocal microscopy of perivascular cells followed by 3D surface rendering of a z-stack using the Zeiss Zen software. Scale bar represents 20 microns. (D) Immunohistochemistry of fli1:EGFP × sdf1a:DsRed caudal tail fin cross-section. Co-staining was performed with anti-EPG-FITC and anti-DsRed primary antibodies. Anti-DsRed detection was performed using a secondary antibody, donkey anti-rabbit-Cy3. Scale bar represents 50 microns.
The progenitors to the sdf1DsRed PVC start to become visible as early as 3 days post fertilization (dpf) as part of the developing tail fin, but are more clearly evident at 8 dpf (Supplemental Figure 3). The cells appear to originate in the undefined mesoderm of the developing tail fin posterior to the developing spinal cord (which also shows distinct DsRed positivity). As the larval zebrafish continue to undergo tail fin development and the vasculature grows caudally during the juvenile stage of development, distinct PVC can be visualized abluminal to vascular endothelial cells at 21 dpf (Supplemental Figure 3).
MSC from human and murine sources have been proposed to originate from PVC in vivo[3, 4]. Therefore, our primary interest was to determine if sdf1DsRed PVC could be a source for MSC in the zebrafish. Using flow cytometry sorting, we were able to isolate sdf1DsRed cells from the tail fin after enzymatic digestion of the tissue and 8 – 10 % (n = 20 animals) of the cells were DsRed positive (Figure 2). To determine if sdf1DsRed PVC expressed putative markers of PVC other than sdf1, RNA was extracted from flow cytometry-sorted cells obtained from sdf1a:DsRed-fli1:EGFP double transgenic animals followed by quantitative RT-PCR for genes of interest. The previously reported PVC associated genes smooth muscle actin (acta2), chondroitin sulfate proteoglycan (cspg), transgelin (tgln), and cadherin5 (cdh5) were all significantly upregulated in sdf1DsRed positive cells as compared to fli1EGFP endothelial cells, and the fli1 gene showed increased expression in the EGFP+ endothelial cells as expected while cd45 was absent in both cell types (figure 2B). Although there is no single specific marker for either PVC or MSC, expression of these markers have been consistently reported to be expressed by both by PVC and MSC in various studies[19–21].
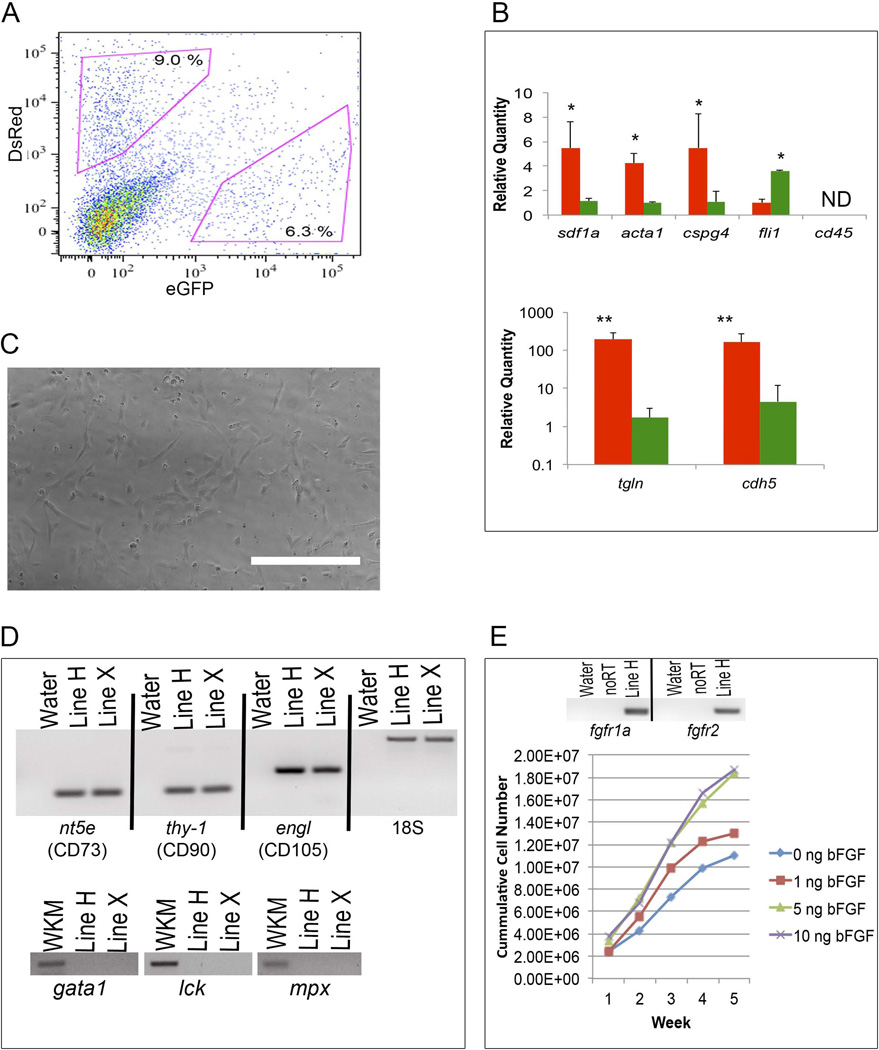
Figure 2.
Perivascular sdf1aDsRed cells express markers of perivascular cells and can be cultured-expanded. (A) The caudal tail fins of adult fli1:EGFP × sdf1a:DsRed animals (n = 8 – 12) were amputated and treated with collagenase to produce a single cell suspension. Shown is a representative scatter plot of green fli1EGFP endothelial cells and sdf1aDsRed perivascular cells. (B) The fli1EGFP endothelial cells and sdf1aDsRed perivascular cells were sorted by flow cytometry and RNA isolated for qRT-PCR. N = 3 experiments, * and ** represent p < 0.05 and p < 0.01 respectively from a Student’s t-test. ND, not detected. (C) Phase-contrast microscopy of culture-expanded perivascular sdf1aDsRed cells 6 weeks after primary isolation. Scale bar represent 100 microns. (D) RNA was extracted from cultured sdf1aDsRed cells and RT-PCR performed for MSC and hematopoietic markers. Shown are the zebrafish gene names as well as the human cluster of differentiation (CD) designations. Whole kidney marrow (WKM) cells were used as a positive control. (E) RT-PCR showing FGF-family gene receptor expression and cumulative sdf1aDsRed cell counts cultured in increasing amounts of bFGF.
A criterion for MSC is plastic adherence after isolation from the tissue source being tested (whether bone marrow, fat, or umbilical cord blood in origin), and subsequent tissue culture outgrowth[22]. We and others have previously developed successful culture systems for zebrafish stromal cells derived from organ outgrowths[23],[24]. We prospectively sorted sdf1DsRed PVC by flow cytometry based on DsRed expression followed by plating onto tissue culture treated plastic dishes. After 24 hours of culture, sdf1DsRed PVC adhered to the bottom of the culture plate acquiring a flattened appearance, and were of irregular sizes and fibroblastoid in shape. After about one week in culture, phase contrast microscopy visualization revealed that cultured cells continued to have a shape similar to that of MSC - being somewhat flattened and fibroblastoid (Figure 2C). Several (n = 5) independent stromal lines were established. Cultured sdf1DsRed PVC were evaluated for prototypical markers of MSC including ecto-5'-nucleotidase (nt5e), thy-1, and endoglin–like (engl) gene expression (the human homologs being CD73, CD90, and CD105) and found to be expressed (Figure 2D). RT-PCR of sorted sdf1DsRed PVC also showed the presence of nt5e, thy-1, and engl mRNA indicating their expression prior to in vitro culture expansion (Supplemental Figure 4). Cultured sdf1DsRed PVC did not express hematopoietic markers including cd45, gata1, lck and mpx.
MSC express receptors for fibroblast growth factor (FGFRs) and are growth responsive to FGF[25]. Our expansion culture medium contained 5 ng/mL human recombinant basic FGF. Therefore, we next determined whether culture expanded sdf1DsRed PVC were growth responsive to FGF. Similar to what has been reported for MSC, Figure 2E shows the culture-expanded sdf1DsRed PVC expressed fgfr1a and fgfr2 and displayed a dose-responsive increase in growth with basic FGF supplementation. Unlike human MSC, which have a limited number of population doublings, the zebrafish culture-expanded sdf1DsRed PVC do not undergo senescence with PVCs being in continuous culture for more than 18 months (> 100 population doublings) with no decrease in growth potential (data not shown).
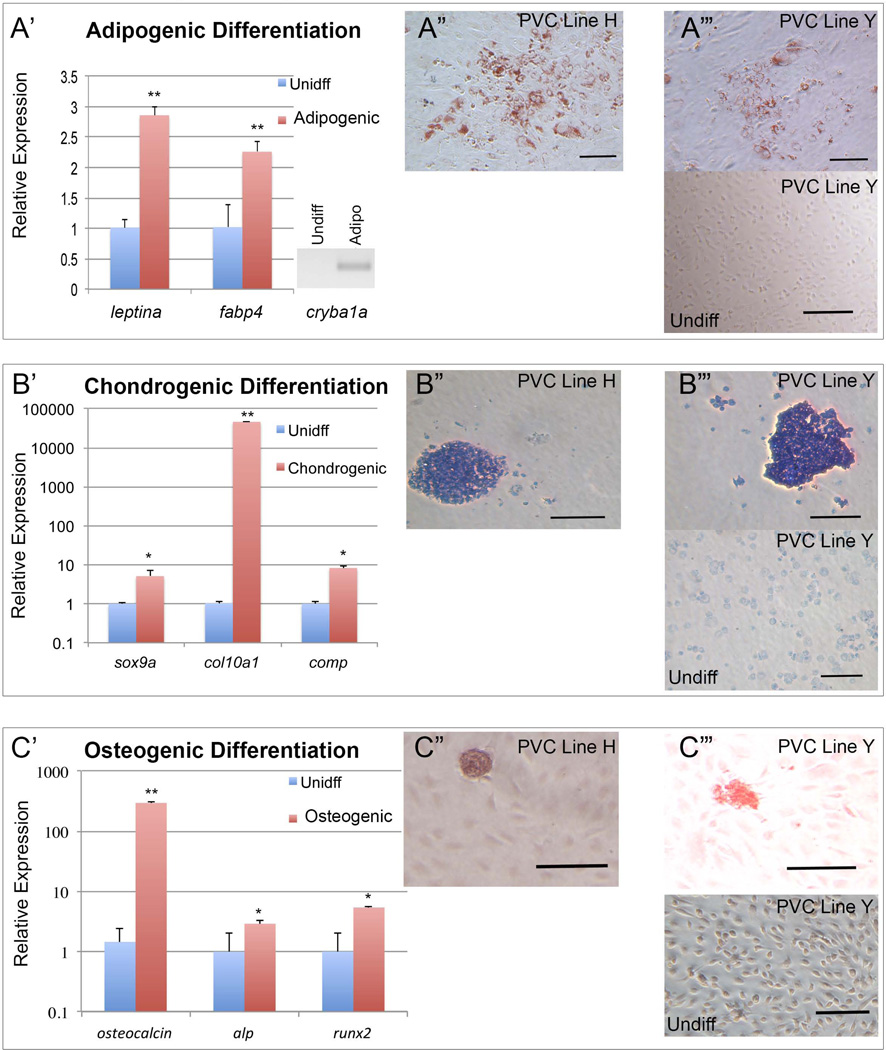
After determining that culture-expanded sdf1DsRed PVC adhere to plastic and express genes similar to MSC, we next performed differentiation of the cells into the classic mesodermal-derived osteogenic, adipogenic, and chondrogenic lineages. As shown in Figure 3A, after 3 to 4 weeks of differentiation in adipogenesis promoting medium, the adipogenic genes of fatty acid binding protein 4 (fabp4) and leptina were highly increased in expression. Additionally, expression of crystalline, beta A1a (cryba1a), was absent in undifferentiated cells, but present after adipogenic differentiation. Furthermore, adipogenic differentiated cells showed classic lipid vacuolization and stained positive with Oil-Red-O. Culture expanded sdf1DsRed PVC placed into chondrogenic conditions for 4 weeks showed increased expression of the chondrocyte-associated genes sox9a, col10a1, and cartilage oligomeric matrix protein (comp) and toluidine blue staining showed glycosaminoglycans were increased (Figure 3B). Differentiation in osteogenic conditions induced the expression of runx2, osteocalcin, and alkaline phosphatase (alp) as determined by qRT-PCR, but Alizarin red staining did not reveal the bright red confluent layer of calcium laden osteocytes which we commonly observe with our human MSC differentiations (Figure 3C)[16]. Instead, we found scattered pink-staining collections of cell clusters akin to bony osteophytes that were absent in undifferentiated cell cultures, which suggests a partial differentiation process.
Figure 3.
Differentiation of cultured sdf1aDsRed cells into mesodermal lineages. (A’–C’) Cells were cultured in adipogenic, chondrogenic, or osteogenic promoting media for 4 weeks. Relative gene expression was determined by qRT-PCT and ΔΔCT calculation relative to undifferentiated cells as shown. N = 3 experiments, * and ** represent p < 0.05 and p < 0.01 respectively from a Student’s t-test. (A”–C”) Show examples of PVC line H differentiated and stained Oil-Red-O, toluidine blue, or Alizarin red to indicate adipogenic, chondrogenic, and osteogenic cells respectively. (A”’ – C”’) Show examples of PVC line Y differentiated and undifferentiated followed by staining with Oil-Red-O, toluidine blue, or Alizarin red to indicate adipogenic, chondrogenic, and osteogenic cells respectively. Scale bars represents 100 microns in adipogenic and osteogenic images and 500 microns in chondrogenic images.
These data suggest that culture-expanded sdf1DsRed PVC from the zebrafish can give rise to cells with an MSC phenotype in the proper culture conditions. The osteogenic differentiation appears to be incomplete which could be due to a weaker propensity for osteogenic differentiation or possibly due to improper media formulation and lack of zebrafish specific factors driving osteogenic differentiation. Another likely possibility is that even though culture expanded sdf1DsRed PVC appear homogenous during in vitro culture, populations with mixed lineages perhaps with different differentiation potentials are likely present.
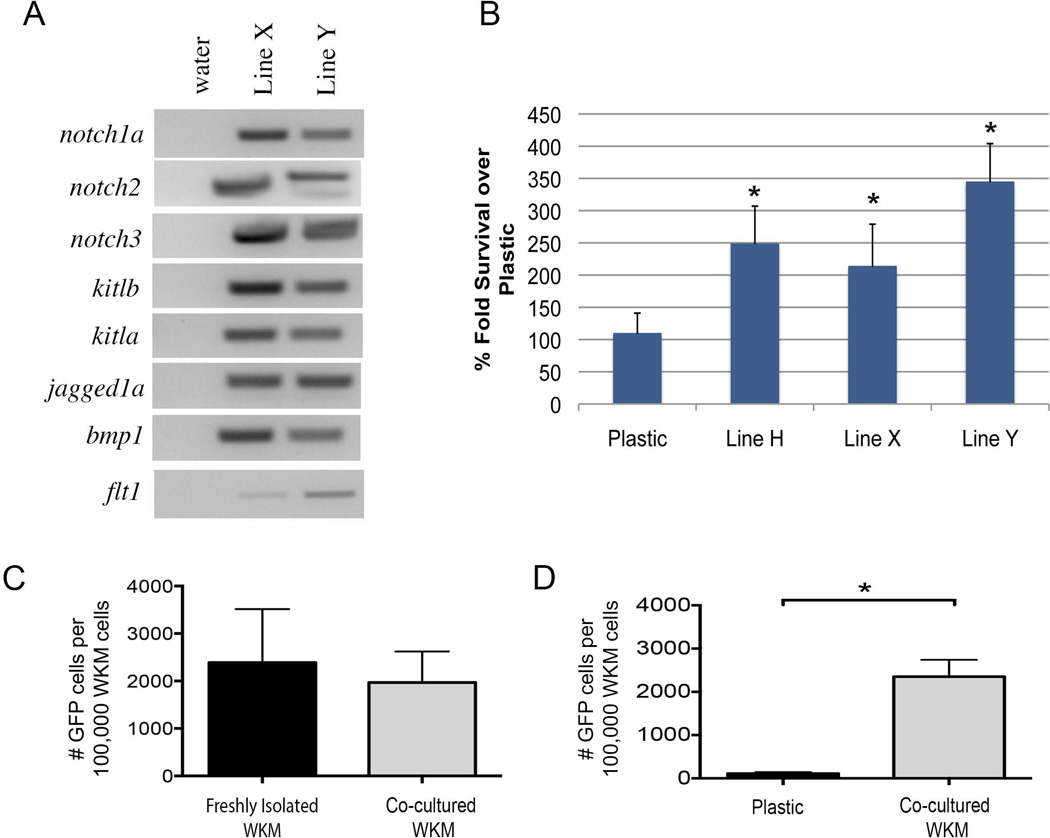
Human MSC have the ability to support the expansion of hematopoietic stem and progenitor cells in vitro[26]. We have previously shown that zebrafish stromal cells have a similar ability[24]. We next explored the capacity of the culture-expanded sdf1DsRed PVC to support hematopoietic cells. Our initial RT-PCR screen for genes expressed by culture-expanded sdf1DsRed PVC showed the expression of several genes important in the growth and maintenance of hematopoietic cells including: several notch family members as well as bmp1, and flt1 (Figure 4A). We next performed co-culture experiments with 50,000 hematopoietic cells from bactin2:GFP transgenic zebrafish seeded onto a monolayer of culture-expanded sdf1DsRed PVC versus hematopoietic cells on plastic alone for 16 days. We found that culture-expanded sdf1DsRed PVC gave a significant survival advantage (2 – 3.5 fold, p < 0.001) to the hematopoietic cells when compared to plastic alone (Figure 4B), though significant hematopoietic cell expansion was not noted. The survival advantage may be through engagement of cell maintenance pathways or because of decreased programmed cells death. The lack of true expansion is most likely due to the absence of supplemental hematopoietic growth factors which are not commonly available.
Figure 4.
Cultured sdf1aDsRed cells support engraftable hematopoietic cells. (A) Genes known to be important for the maintenance of hematopoietic cells were amplified by RT-PCR. Shown are the results from two of three sdf1aDsRed cell lines. (B) 50,000 WKM from bactin2:GFP fish were co-cultured on a monolayer of sdf1aDsRed cells (lines H, X, Y) for 16 days followed by enumeration using a flow cytometry and counting beads (n = 15/group). * indicates p-value < 0.001 from a Student’s t-test. (C) Two days after 20 Gy irradiation, wild-type zebrafish were transplanted with 50,000 bactin2:GFP WKM from freshly isolated donors or previously co-cultured with sdf1aDsRed PVC from Line H. Engraftment analysis was performed 7 days after transplant by flow cytometry for GFP (n = 5 – 7 animals/group). Student’s t-test showed no significant difference between the groups (p = 0.65). (D) WKM from bactin2:GFP transgenics was co-cultured with sdf1aDsRed PVC cells from Line H for 16 days versus cells on plastic alone. The entire contents of a well were used for transplant as above and engraftment determined at seven days post transplant (n = 10 – 12 animals/group). * indicates p-value < 0.001 from a Student’s t-test.
To evaluate short-term engraftment potential, 50,000 hematopoietic cells from bactin2:GFP transgenic zebrafish co-cultured with PVC as before were transplanted into zebrafish that previously received sublethal irradiation (2 days prior), and donor engraftment was assessed 7 days later by flow cytometry (Figure 4C). There was no difference between co-cultured marrow cells and freshly isolated marrow cells in terms of their ability to home and provide short term (p = 0.65). We next compared the short-term engraftment of hematopoietic cells co-cultured on sdf1DsRed PVC versus plastic (as in Figure 4B). After 16 days in culture, the entire content of a well was injected into zebrafish that previously received sublethal irradiation (2 days prior). A significant short-term engraftment advantage (p < 0.001) when measured 7 days after transplant was found for hematopoietic cells in PVC co-culture experiments versus on plastic alone (Figure 4D). These data show that hematopoietic cells when co-cultured with sdf1DsRed PVC cells maintain survival, and conserve their native homing and at least short-term engraftment abilities.
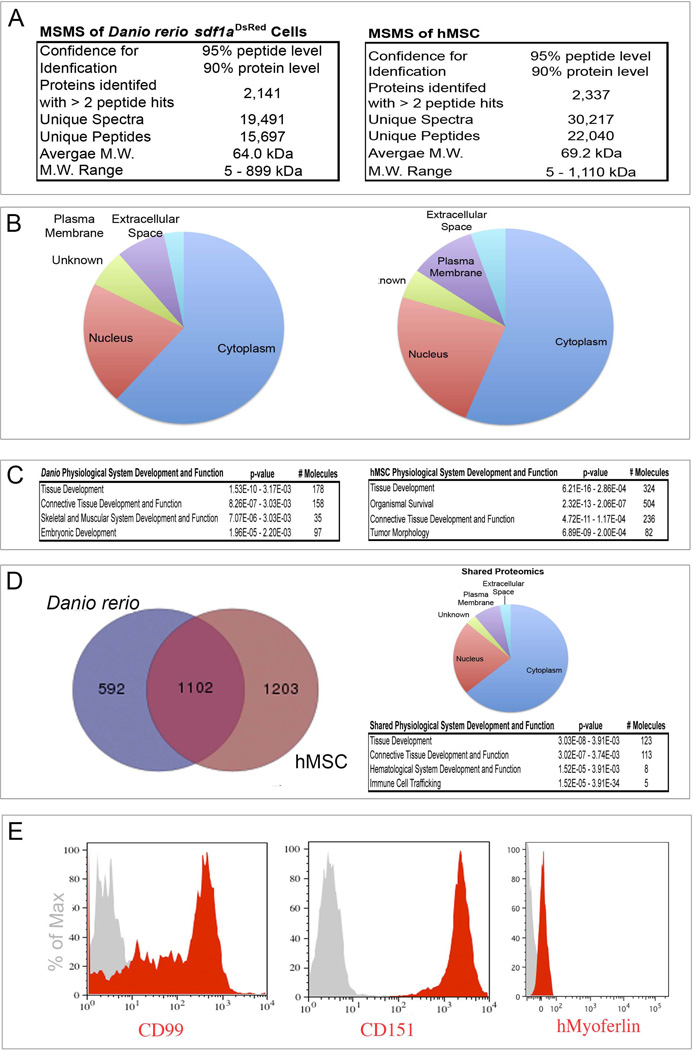
To understand the biology of culture-expanded sdf1DsRed PVC and how they may be related to human MSC, we performed a global proteomic profile analysis using 2-dimensional liquid chromatography and tandem mass spectrometry. We isolated human MSC from three independent donor bone marrow samples for comparative analysis to three independently isolated zebrafish culture-expanded sdf1DsRed PVC (lines H, X, and Y). Whole cell lysates were prepared from confluent T-150 culture flasks, and subject to reversed phase (C18 resin) chromatography at pH 10 (first dimension) with fraction collection followed by reversed phase (C18 resin) chromatography at pH 3 (the second dimension) online with mass spectrometry. MS analysis was performed on an Orbitrap™ mass spectrometer. The resulting protein identifications were derived from those proteins with a 2-peptide minimum identification, a confidence interval (CI) of 95% at the peptide level and a CI of 90% at the protein level (see Supplemental Methods for data analysis). Protein lists were combined and redundant hits eliminated. We collectively identified 2141 proteins in zebrafish culture-expanded sdf1DsRed PVC (from 15697 unique peptides) and 2337 proteins in hMSC (from 22040 unique peptides). The average molecular weight was similar in both proteomes at 64.0 kDa for zebrafish and 69.2 kDa for hMSC with similar ranges (Figure 5). Next, proteins were cross-referenced by Uniprot accession numbers and Ingenuity Pathway Analysis (IPA®) was performed. The percentage of proteins by subcellular location was remarkably similar between the two species with the majority of the proteins identified being of cytoplasmic origin, followed by nuclear proteins, then unknown or plasma membrane proteins, and lastly extracellular proteins. Further analysis of protein relationships based on gene ontology (using GO terms) revealed that many of the zebrafish proteins were involved with tissue development (specifically connective tissue) and skeletal/muscular system development. Similarly, hMSC proteins ranked highest in tissue development (again specifically connective tissue). We next derived gene names from the Uniprot identifications generating subsets of 1694 proteins in the zebrafish, and 2305 proteins in hMSC. We next derived gene names from the Uniprot identifications; final subsets contained 1694 zebrafish proteins and 2305 hMSC proteins. The union of the two data sets showed 1102 common proteins consisting of 65% of the zebrafish culture-expanded sdf1DsRed PVC proteome, and 48% of the hMSC proteome. The subcellular location and gene ontogeny of the common proteins was similar to the original groups. We were curious if uncharacterized hMSC proteins could be discovered using this data, and upon evaluating the union of the two proteomes, we chose three novel cell surface proteins common to both zebrafish and hMSC protein subsets which have not been previously shown to be expressed on hMSC to verify their expression: CD99 (a Ewing’s sarcoma marker), CD151, and myoferlin (a potential modifier of muscular dystrophy). These proteins have been little studied in terms of any relationship to MSC, though we showed confirmation of their expression on hMSC using flow cytometry staining (Figure 5E). These data show that hMSC and zebrafish cultured sdf1DsRed PVC have a high degree of proteomic similarity and new MSC proteins can be discovered by the interrogation of zebrafish sdf1DsRed cells. Furthermore, cultured sdf1DsRed PVC may represent a novel zebrafish source of MSC.
Figure 5.
Proteomics of cultured sdf1aDsRed cells and hMSC. Whole cell lysates were prepared and subjected to 2-dimensional liquid chromatography as described in the supplemental methods. Tandem mass spectrometry was then used to identify peptides on a capillary LC-nanoESI-Orbitrap XL mass spectrometer. Peptide matching to full-length proteins was performed using Sequest (version 27, rev. 12). (A) Absolute numbers of spectra, peptides, and proteins identified. (B) Results of IPA® analysis showing distribution of proteins by cellular compartment. (C) IPA® analysis showing top hits in the “Physiological System and Development” category. (D) Venn diagram showing common proteins identified between cultured sdf1aDsRed cells and hMSC, their cellular compartment, and IPA analysis showing top hits in the “Physiological System and Development” category. (E) Flow cytometry histograms of hMSC for three novel proteins identified in both cultured sdf1aDsRed cells and hMSC. Grey histograms show the isotope control antibody.
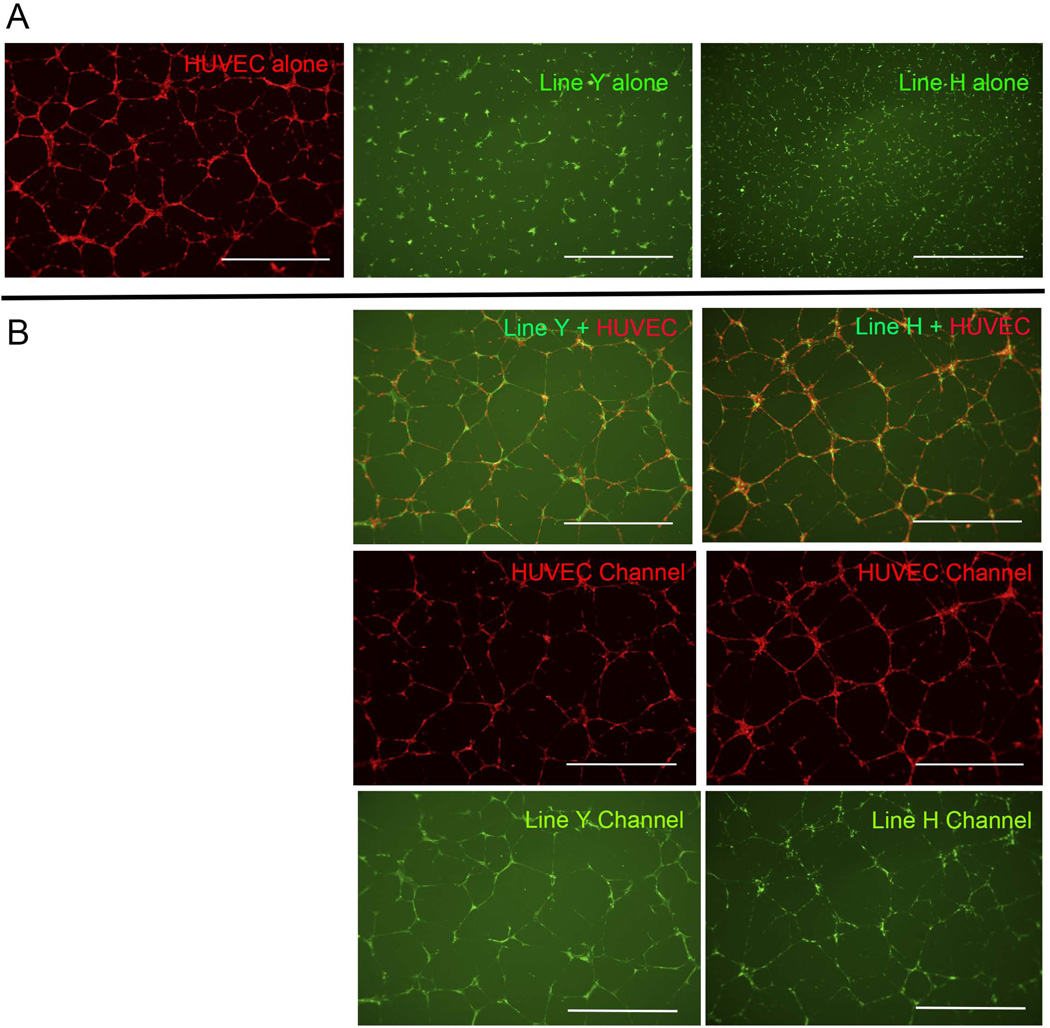
To determine whether or not culture expanded sdf1DsRed PVC could be co-cultured with human endothelial cells to evaluate for morphological changes, we utilized human umbilical vein endothelial cells (HUVECs) in co-culture experiments. Culture-expanded sdf1DsRed PVC showed no cellular organization when cultured on a matrigel substrate alone either in their native media or endothelial growth media (Figure 6A). After the perivascular cells were placed into culture together with HUVECs, the zebrafish culture expanded sdf1DsRed PVC became substantially more networked and co-aligned (becoming peri-endothelial) with the HUVECs (Figure 6B). This experiment shows that endothelial cells can influence the behavior of culture expanded sdf1DsRed PVC.
Figure 6.
Co-culture of sdf1aDsRed cells with HUVECs shows vascular branching. 15,000 HUVECs were labeled with CellTracker™ Orange and 10,000 cultured sdf1aDsRed cells were labeled with CellTracker™ Green. Cells were placed atop 100 µL of matrigel in a 96-well plate in the presence of EGM-2 media for 10 – 16 hours prior to imaging. (A) HUVECs and two sdf1aDsRed cell lines cultured alone. (B) Co-culture of HUVECs with sdf1aDsRed cells and imaged under the appropriate filters. Scale bars represent 1000 microns.
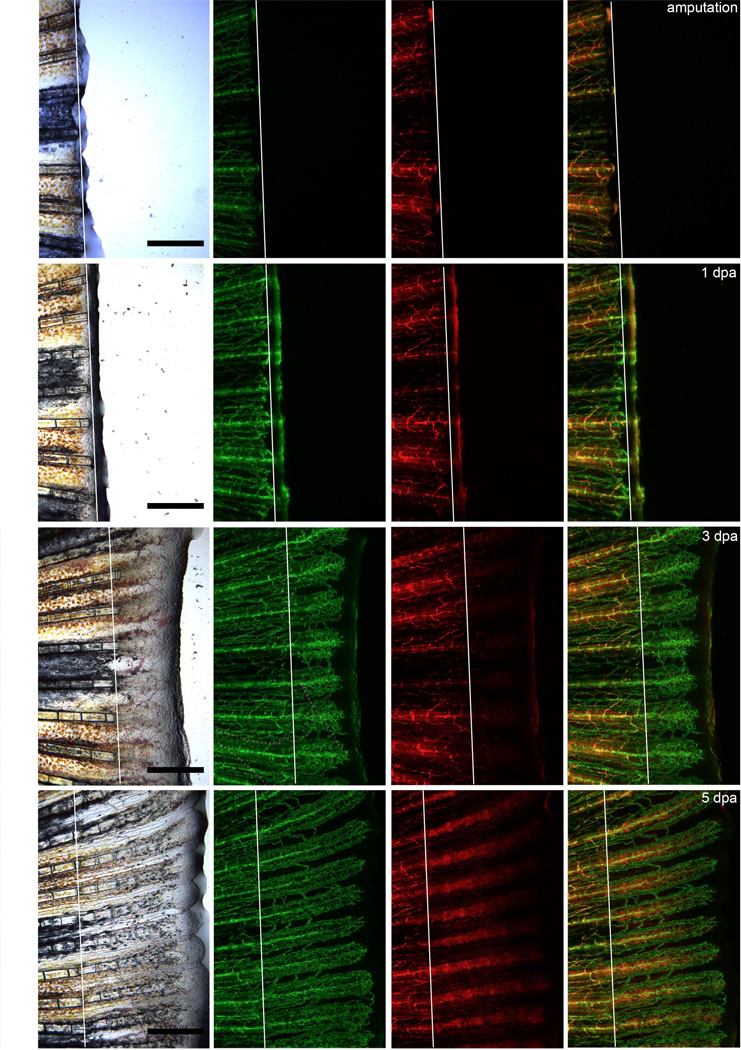
The prior experiments described many of the in vitro properties of culture-expanded sdf1DsRed PVC and their similarity to hMSC. MSC have been used to facilitate the repair of bone and cartilage injuries, but whether this occurs in the de novo setting is unknown[27–29]. We chose to utilize a model of tail fin regeneration to evaluate the behavior of sdf1DsRed PVC in this process. After tail amputation in sdf1a:DsRed × fli1:EGFP transgenic animals, there was sdf1:DsRed expression at the healing edge 1 day post amputation (dpa) as previously described[30]. While rapid vascular regeneration occurred within in 72 hours post amputation, robust DsRed expression with cellular organization became very strong at 4 – 5 dpa (Figure 7) when the sdf1DsRed cells appeared to organize into segments of the regenerating bony ray. Interestingly, it was not until 6–7 dpa when the reappearance of sdf1DsRed cells in a perivascular location was observed around the neovasculature (Supplemental Figure 5). The new PVCs seemed to be derived from the surrounding mesenchyme (versus having migrated from a distal source), which suggests that the newly formed mesenchyme was a source for sdf1DsRed perivascular cells. This model demonstrates the in vivo contribution of sdf1 expressing cells to the regenerating fin structure as well as the derivation of PVC during vasculature regeneration and will be useful in future studies to determine in vivo PVC function, which until now has been difficult in mammalian systems.
Figure 7.
The contribution of sdf1aDsRed cells to fin regeneration. The caudal tail of sdf1aDsRed × fli1:EGFP double transgenic was amputated on day 0 (amputation plane indicated by the white line). The same animal was anaesthetized and imaged using the appropriate filters at the time points given. Photos are representative of the same regeneration experiment performed in ten animals. Scale bars represent 100 microns.
Discussion
The zebrafish is an excellent model to visually observe cellular anatomy given its optical translucence as an embryo as well the recent development of “transparent” adult animals harboring a pigmentation defect[31]. The creation of the sdf1a:DsRed reporter allowed us to visualize DsRed positive, Sdf1a-producing cells. It is well known that SDF1 is expressed by a wide variety of cell types, although its original discovery and purification was from bone marrow stromal cells that were not termed MSC at the time of discovery, the stromal cells most likely fulfilled criteria for MSC[32].
Our sdf1a:DsRed transgenic allows for the observation of distinct PVC in their native environment. We were able to isolate and culture-expand zebrafish sdf1DsRed PVC and showed they fulfilled criteria for classification as MSC (zMSC). One clear difference between zMSC and hMSC is the growth potential of the cells. Human MSC are limited in the number of cell divisions they can undergo with a limit of around 50 population doublings[33–35]. Whereas in zMSC (and other zebrafish stromal cell lines) there is no declination in doubling time nor change in cellular morphology even after 1–2 years in culture reflecting over 100 population doubling times[23, 24]. This phenomenon is most likely related to the constitutive activation of the telomerase gene that we and others have described in zebrafish cells[36–38]. Zebrafish have the ability to regenerate several organ systems repetitively. Even after multiple rounds of tail regeneration, the telomeres seem to maintain their full length and telomerase function is intact[36]. Maintaining “mesenchymal” cells with unlimited cell division potential would certainly contribute to the ability to continually regenerate tissues after damage.
Our data from the proteomic interrogation of both zebrafish and human MSC revealed significant overlap in protein expression. The subcellular distribution was not surprising and corresponded to the native distribution of most proteins (cytoplasmic abundance followed by nuclear and extracellular/membrane localization respectively). The most interesting finding of this analysis was the identification of new cell surface proteins. Expression of CD99 and myoferlin have not been shown to be present on MSC, while CD151 was detected in a prior proteomic interrogation of hMSC microvesicles and may contribute to microvesicle fusion, although other studies showed it was important in angiogenesis[39, 40]. CD99 is a known marker of neuroectodermal cancers including Ewing’s sarcoma but is also expressed in wide variety of other cell types including lymphocytes diminishing its novelty, though its function on MSC is unknown[41, 42]. Myoferlin (MYOF) is an ancient protein involved in internal membrane bound organelle’s ability to fuse with the plasma membrane, and defects in the Ferlin family cause a form of muscular dystrophy due to poor sarcolemma repair[43]. Recent studies have focused on MYOF expression in breast cancer and the regulation of the mesenchymal to epithelial state where expression of MYOF favors a more invasive mesenchymal state[44]. To our knowledge, expression of MYOF has not been shown on MSC. Future exploration of these and other proteins we have determined will be help to broaden our understanding of MSC biology, both in vivo and in vitro.
MSC have had a wide variety of functions ascribed to them including wound healing, vascular healing, and organ damage repair[29, 45, 46]. We visualized the contribution of sdf1DsRed cells to fin regeneration, a model for limb regeneration. It has been specifically shown that assessing gene expression by standard whole-mount in situ hybridization techniques in the regenerating fin is limited due to poor probe penetration through the “mesenchymal” tissue of the fin as it undergoes repair[47]. The advantage to using transgenic reporters like sdf1a:DsRed in our experiments is that visualization of the cellular events is easily accomplished in live animals. One disadvantage to using DsRed is its slow maturation time to form tetramers (24h hours) and its long half-life (4.5 days) which may make it difficult to determine rapid changes in gene expression[48], though the extended DsRed half-life does make it useful to track cellular movement over long periods of time.
The intra-ray tissue of the caudal fin is composed of vessels, nerves, osteoblasts, pigment cells, and “fibroblast-like” cells[49]. The regenerating blastema has been proposed to be derived from intra-ray “mesenchymal” cells or “fibroblast-like” cells[50, 51]. Work by Knopf et al. has demonstrated that the regenerating fin blastema is also partly composed of dedifferentiated osteoblasts. We propose that these fibroblast-like cells are sdf1DsRed mesenchymal cells illustrated in our model of regeneration, though the origin of the regenerating sdf1DsRed mesenchymal cells is still unknown. Possibilities include migration from the injured fin ray stump, dedifferentiation from proximal fin-ray-osteoblasts, or dedifferentiation from proximal fin-ray-epidermal cells. Crossing our transgenic with other reporter lines (osteoblast and epidermal cell reporters) will help answer the question of where the blastemal mesenchymal cells originate.
Our observation of sdf1DsRed cells developing within 1 – 2 dpa in the blastema is consistent with prior reported sdf1 expression in regenerating fins[30, 52]. The blastema sdf1 expression reported by Dufourcq was downregulated at 5 dpa, while our observations suggest that sdf1DsRed cells are quite evident early after amputation, increasing over the 5 dpa course. The prior experiments used in situ hybridization to determine sdf1 expression and given that probe penetration becomes very limited during fin regeneration, we believe sdf1 expression may have been underestimated. Additionally, the long half-life of DsRed allows us to track sdf1 expressing cells over longer periods of time to get a better understanding of the structure changes occurring during regeneration.
Aside from understanding the contribution of sdf1DsRed mesenchymal cells to the regenerating tail fin bony ray, our model system will useful for interrogating the function of PVC as they are very evident with in vivo imaging. Surprisingly, the PVC in the regenerating fin did not reappear until multiple days after amputation and neovascularization (6 – 7 dpa shown in Supplemental Figure 3). This suggests that PVCs are not required during the immediate development of the newly forming vessels. Our observation that the mesenchymal cells arise before the appearance of PVC suggests that the sdf1DsRed PVC either migrate or differentiate from mesenchymal tissue to adopt an abluminal location on the vessel. Whether the new sdf1DsRed PVC have adopted additional functional properties that are distinct from the sdf1DsRed mesenchymal cells remains unexplored.
We did observe qualitatively that the vasculature prior to PVC presence had a larger diameter, more tortuous appearing, and have increased numbers of branch points as compared to vasculature after PVC appeared. In in vitro endothelial tube assembly models, Stratman et al recently showed that PVC are recruited to endothelial tubes and contribute to basement membrane deposition and endothelial “stabilization” that result in smaller diameter tubes[53, 54]. A second study has shown that PVC can also play a role in “selective pruning” during vasculature development in vitro and in vivo via endosialin (CD248) expression on PVC[55].
Finally, detailed experiments by Bell et al indicate that a critical function of murine brain pericytes is to stabilize the blood-brain-barrier (BBB), and pericyte loss leads to increased BBB permeability with subsequent brain accumulation of protein and other macromolecules[56, 57]. Murine pericytes interact both with astrocytes and endothelial cells via complex signaling networks to maintain vascular integrity with participation of ApoE, Low-density lipoprotein receptor-related protein 1 (Lrp1), and Cyclophilin A (CypA) as important factors. The pericytes identified by Bell et al are analogous in their abluminal anatomical location to the perivascular cells identified in our work, and we observed some sdf1DsRed perivascular cells in the brains of our zebrafish though the former work utilized Pdgfrβ expression to identify pericytes in contrast to our sdf1a promoter transgenic expression. Our own preliminary (and non-quantified) dye-exclusion studies during tail fin regeneration indicted that vascular integrity was restored within a similar to time frame to perivascular re-appearance at about 6 – 7 dpa (data not shown). Whether our sdf1DsRed cells are exactly equivalent to PDGFRβ-expressing pericytes cells is unknown, but the advantages conferred by a zebrafish system will allow further experiments that can broaden the understanding of the complex PVC-endothelium relationship through live imaging studies.
Conclusion
In conclusion, we have developed a model in which PVC can be visualized in an in vivo setting based on sdf1 expression. Prospective isolation and culture of the sdf1DsRed PVC allowed us to demonstrate properties consistent with that of MSC including cell surface marker expression, FGF responsiveness, mesodermal differentiation, and hematopoietic cell support. Proteomic studies revealed a high degree of similarity to hMSC and the description of novel MSC markers (CD99, CD151 and MYOF). Dynamic in vivo imaging of the sdf1DsRed expressing cells show that PVC may arise from undifferentiated mesenchyme, which provides further evidence of the PVC – MSC connection. This model system will allow us to better understand their contribution to vasculature and tissue regeneration processes.
Supplementary Material
Supplemental Figure 1. Fin of 10-week old sdf1a:DsRed zebrafish at 0.73× show DsRed positive network of cells.
Supplemental Figure 2. (A) Hematoxylin and eosin stains of a caudal fin cross-section shows cells in an abluminal position to the central vessel. Scale bars represent 10 microns. (B) In situ hybridization with sdf1a antisense probe shows a perivascular cell around the caudal fin central vessel. Scale bars represent 25 microns.
Supplemental Figure 3. The development of sdf1aDsRed cells in the caudal tail fin. Images represent typical development of sdf1aDsRed × fli1:EGFP double transgenic animals serially imaged at several time days post fertilization (dpf) using appropriate filter sets as described in the methods section. White scale bars indicate 1000 microns; grey scale bar indicates 200 microns.
Supplemental Figure 4. Perivascular sdf1aDsRed cells express markers of perivascular cells and can be cultured-expanded. The caudal tail fins of adult fli1:EGFP × sdf1a:DsRed animals (n = 8 – 12) were amputated and treated with collagenase to produce a single cell suspension. RNA was extracted from sdf1aDsRed cells and RT-PCR performed for the classic MSC markers as indicated. “NO RT” indicates a reaction in which no reverse transcriptase was added and PCR performed for bactin1.
Supplemental Figure 5. The reappearance of sdf1aDsRed perivascular cells at 6 – 7 days post amputation (dpa). Images represent typical tail fin regeneration of sdf1aDsRed × fli1:EGFP double transgenic animals serially imaged at several time days post caudal fine amputation using appropriate filter sets as described in the methods section. Scale bars represent 100 microns.
Supplemental Table 1. Sequences of oligonucleotides used in RT-PCR reactions. For Taqman® reactions, the catalog number of the primer set used is listed.
Acknowledgements
Research reported in this publication was supported by the National Heart, Lung and Blood Institute of the National Institutes of Health under award number K08HL108998, an American Society for Blood and Marrow Transplantation Young Investigator Award, and an American Society for Hematology Scholar Award (TCL). DSL was supported by the New Mexico Spatiotemporal Modeling Center NIH P50GM085273. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would like to acknowledge the use of the Olympus and Zeiss confocal microscopes made available through a National Center for Research Resources Shared Instrumentation Grant (#1 S10 RR16851 and #1 S10 RR16851 respectively).
Footnotes
Author Contribution Summary
TCL financial support, conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
XP collection and/or assembly of data
ACK collection and/or assembly of data
PS collection and/or assembly of data
LH collection and/or assembly of data, data analysis and interpretation
TWM collection and/or assembly of data, data analysis and interpretation
MWS collection and/or assembly of data, data analysis and interpretation
DSL collection and/or assembly of data, data analysis and interpretation
JT manuscript writing, final approval of manuscript
BRB data, data analysis and interpretation, manuscript writing, final approval of manuscript
Conflict of Interest
Each author declares no conflict of interest.
References
- 1.Prunet-Marcassus B, Cousin B, Caton D, et al. From heterogeneity to plasticity in adipose tissues: site-specific differences. Exp Cell Res. 2006;312:727–736. doi: 10.1016/j.yexcr.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 2.da Silva Meirelles L, Caplan AI, Nardi NB. In search of the in vivo identity of mesenchymal stem cells. Stem Cells. 2008;26:2287–2299. doi: 10.1634/stemcells.2007-1122. [DOI] [PubMed] [Google Scholar]
- 3.Corselli M, Chen CW, Crisan M, et al. Perivascular ancestors of adult multipotent stem cells. Arterioscler Thromb Vasc Biol. 2010;30:1104–1109. doi: 10.1161/ATVBAHA.109.191643. [DOI] [PubMed] [Google Scholar]
- 4.Crisan M, Yap S, Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Zannettino AC, Paton S, Arthur A, et al. Multipotential human adipose-derived stromal stem cells exhibit a perivascular phenotype in vitro and in vivo. J Cell Physiol. 2008;214:413–421. doi: 10.1002/jcp.21210. [DOI] [PubMed] [Google Scholar]
- 6.Tashiro K, Tada H, Heilker R, et al. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 7.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugiyama T, Kohara H, Noda M, et al. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Chong SW, Nguyet LM, Jiang YJ, et al. The chemokine Sdf-1 and its receptor Cxcr4 are required for formation of muscle in zebrafish. BMC Dev Biol. 2007;7:54. doi: 10.1186/1471-213X-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalasani SH, Sabelko KA, Sunshine MJ, et al. A chemokine, SDF-1, reduces the effectiveness of multiple axonal repellents and is required for normal axon pathfinding. J Neurosci. 2003;23:1360–1371. doi: 10.1523/JNEUROSCI.23-04-01360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalasani SH, Baribaud F, Coughlan CM, et al. The chemokine stromal cell-derived factor-1 promotes the survival of embryonic retinal ganglion cells. J Neurosci. 2003;23:4601–4612. doi: 10.1523/JNEUROSCI.23-11-04601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doitsidou M, Reichman-Fried M, Stebler J, et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–659. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 13.Lund TC, Glass TJ, Patrinostro X, et al. Stromal cell-derived factor-1 and hematopoietic cell homing in an adult zebrafish model of hematopoietic cell transplantation. Blood. 2011;118:766–774. doi: 10.1182/blood-2011-01-328476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westerfield M. The zebrafish book : a guide for the laboratory use of zebrafish (Brachydanio rerio) Eugene, OR: M. Westerfield; 1993. [Google Scholar]
- 15.Lund TC, Glass TJ, Patrinostro X, et al. Stromal cell-derived factor-1 (SDF-1) and hematopoietic cell homing in an adult zebrafish model of hematopoietic cell transplant. Blood. 2011 doi: 10.1182/blood-2011-01-328476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lund TC, Kobs A, Blazar BR, et al. Mesenchymal stromal cells from donors varying widely in age are of equal cellular fitness after in vitro expansion under hypoxic conditions. Cytotherapy. 2010;12:971–981. doi: 10.3109/14653249.2010.509394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panoskaltsis-Mortari A, Bucy RP. In situ hybridization with digoxigenin-labeled RNA probes: facts and artifacts. BioTechniques. 1995;18:300–307. [PubMed] [Google Scholar]
- 18.Lawson ND, Weinstein BM. In Vivo Imaging of Embryonic Vascular Development Using Transgenic Zebrafish. Developmental Biology. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 19.Wein F, Pietsch L, Saffrich R, et al. N-cadherin is expressed on human hematopoietic progenitor cells and mediates interaction with human mesenchymal stromal cells. Stem Cell Res. 2010;4:129–139. doi: 10.1016/j.scr.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Kozanoglu I, Boga C, Ozdogu H, et al. Human bone marrow mesenchymal cells express NG2: possible increase in discriminative ability of flow cytometry during mesenchymal stromal cell identification. Cytotherapy. 2009;11:527–533. doi: 10.1080/14653240902923153. [DOI] [PubMed] [Google Scholar]
- 21.Salem HK, Thiemermann C. Mesenchymal stromal cells: current understanding and clinical status. Stem Cells. 2010;28:585–596. doi: 10.1002/stem.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chamberlain G, Fox J, Ashton B, et al. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 23.Stachura DL, Reyes JR, Bartunek P, et al. Zebrafish kidney stromal cell lines support multilineage hematopoiesis. Blood. 2009;114:279–289. doi: 10.1182/blood-2009-02-203638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund TC, Glass TJ, Somani A, et al. Zebrafish stromal cells have endothelial properties and support hematopoietic cells. Exp Hematol. 2012;40:61–70. e61. doi: 10.1016/j.exphem.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coutu DL, Francois M, Galipeau J. Inhibition of cellular senescence by developmentally regulated FGF receptors in mesenchymal stem cells. Blood. 2011;117:6801–6812. doi: 10.1182/blood-2010-12-321539. [DOI] [PubMed] [Google Scholar]
- 26.Caplan AI, Dennis JE. Mesenchymal stem cells as trophic mediators. J Cell Biochem. 2006 doi: 10.1002/jcb.20886. [DOI] [PubMed] [Google Scholar]
- 27.Erickson IE, Kestle SR, Zellars KH, et al. Improved cartilage repair via in vitro pre-maturation of MSC-seeded hyaluronic acid hydrogels. Biomedical materials. 2012;7:024110. doi: 10.1088/1748-6041/7/2/024110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones V. Meeting educational needs: postgraduate diploma/MSc in wound healing and tissue repair. Journal of wound care. 2001;10:277–279. doi: 10.12968/jowc.2001.10.7.26098. [DOI] [PubMed] [Google Scholar]
- 29.Bruder SP, Fink DJ, Caplan AI. Mesenchymal stem cells in bone development, bone repair, and skeletal regeneration therapy. J Cell Biochem. 1994;56:283–294. doi: 10.1002/jcb.240560809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dufourcq P, Vriz S. The chemokine SDF-1 regulates blastema formation during zebrafish fin regeneration. Development Genes and Evolution. 2006;216:635–639. doi: 10.1007/s00427-006-0066-7. [DOI] [PubMed] [Google Scholar]
- 31.White RM, Sessa A, Burke C, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirozu M, Nakano T, Inazawa J, et al. Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF1) gene. Genomics. 1995;28:495–500. doi: 10.1006/geno.1995.1180. [DOI] [PubMed] [Google Scholar]
- 33.Shay JW, Wright WE. Hayflick, his limit, and cellular ageing. Nature Reviews Molecular Cell Biology. 2000;1:72–76. doi: 10.1038/35036093. [DOI] [PubMed] [Google Scholar]
- 34.Baxter MA, Wynn RF, Jowitt SN, et al. Study of telomere length reveals rapid aging of human marrow stromal cells following in vitro expansion. Stem Cells. 2004;22:675–682. doi: 10.1634/stemcells.22-5-675. [DOI] [PubMed] [Google Scholar]
- 35.Banfi A, Muraglia A, Dozin B, et al. Proliferation kinetics and differentiation potential of ex vivo expanded human bone marrow stromal cells: Implications for their use in cell therapy. Exp Hematol. 2000;28:707–715. doi: 10.1016/s0301-472x(00)00160-0. [DOI] [PubMed] [Google Scholar]
- 36.Lund TC, Glass TJ, Tolar J, et al. Expression of telomerase and telomere length are unaffected by either age or limb regeneration in Danio rerio. PLoS One. 2009;4:e7688. doi: 10.1371/journal.pone.0007688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kishi S, Uchiyama J, Baughman AM, et al. The zebrafish as a vertebrate model of functional aging and very gradual senescence. Exp Gerontol. 2003;38:777–786. doi: 10.1016/s0531-5565(03)00108-6. [DOI] [PubMed] [Google Scholar]
- 38.Elmore LW, Norris MW, Sircar S, et al. Upregulation of telomerase function during tissue regeneration. Experimental biology and medicine. 2008;233:958–967. doi: 10.3181/0712-RM-345. [DOI] [PubMed] [Google Scholar]
- 39.Kim HS, Choi DY, Yun SJ, et al. Proteomic analysis of microvesicles derived from human mesenchymal stem cells. Journal of proteome research. 2012;11:839–849. doi: 10.1021/pr200682z. [DOI] [PubMed] [Google Scholar]
- 40.Bailey RL, Herbert JM, Khan K, et al. The emerging role of tetraspanin microdomains on endothelial cells. Biochem Soc Trans. 2011;39:1667–1673. doi: 10.1042/BST20110745. [DOI] [PubMed] [Google Scholar]
- 41.Dworzak MN, Fritsch G, Buchinger P, et al. Flow cytometric assessment of human MIC2 expression in bone marrow, thymus, and peripheral blood. Blood. 1994;83:415–425. [PubMed] [Google Scholar]
- 42.Rocchi A, Manara MC, Sciandra M, et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J Clin Invest. 2010;120:668–680. doi: 10.1172/JCI36667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Aoki M, Illa I, et al. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat Genet. 1998;20:31–36. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- 44.Li R, Ackerman WEt, Mihai C, et al. Myoferlin depletion in breast cancer cells promotes mesenchymal to epithelial shape change and stalls invasion. PLoS One. 2012;7:e39766. doi: 10.1371/journal.pone.0039766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran N, Li Y, Maskali F, et al. Short-term heart retention and distribution of intramyocardial delivered mesenchymal cells within necrotic or intact myocardium. Cell Transplant. 2006;15:351–358. doi: 10.3727/000000006783981918. [DOI] [PubMed] [Google Scholar]
- 46.Madlambayan GJ, Butler JM, Hosaka K, et al. Bone marrow stem and progenitor cell contribution to neovasculogenesis is dependent on model system with SDF-1 as a permissive trigger. Blood. 2009;114:4310–4319. doi: 10.1182/blood-2009-03-211342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith A, Zhang J, Guay D, et al. Gene expression analysis on sections of zebrafish regenerating fins reveals limitations in the whole-mount in situ hybridization method. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:417–425. doi: 10.1002/dvdy.21417. [DOI] [PubMed] [Google Scholar]
- 48.Mirabella R, Franken C, van der Krogt GN, et al. Use of the fluorescent timer DsRED-E5 as reporter to monitor dynamics of gene activity in plants. Plant physiology. 2004;135:1879–1887. doi: 10.1104/pp.103.038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knopf F, Hammond C, Chekuru A, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Developmental cell. 2011;20:713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Nechiporuk A, Keating MT. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–2617. doi: 10.1242/dev.129.11.2607. [DOI] [PubMed] [Google Scholar]
- 51.Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 2003;226:202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- 52.Laudet V, Bouzaffour M, Dufourcq P, et al. Fgf and Sdf-1 Pathways Interact during Zebrafish Fin Regeneration. PLoS ONE. 2009;4:e5824. doi: 10.1371/journal.pone.0005824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stratman AN, Malotte KM, Mahan RD, et al. Pericyte recruitment during vasculogenic tube assembly stimulates endothelial basement membrane matrix formation. Blood. 2009;114:5091–5101. doi: 10.1182/blood-2009-05-222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stratman AN, Schwindt AE, Malotte KM, et al. Endothelial-derived PDGF-BB and HBEGF coordinately regulate pericyte recruitment during vasculogenic tube assembly and stabilization. Blood. 2010;116:4720–4730. doi: 10.1182/blood-2010-05-286872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simonavicius N, Ashenden M, van Weverwijk A, et al. Pericytes promote selective vessel regression to regulate vascular patterning. Blood. 2012;120:1516–1527. doi: 10.1182/blood-2011-01-332338. [DOI] [PubMed] [Google Scholar]
- 56.Bell RD, Winkler EA, Sagare AP, et al. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron. 2010;68:409–427. doi: 10.1016/j.neuron.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bell RD, Winkler EA, Singh I, et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 2012;485:512–516. doi: 10.1038/nature11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Fin of 10-week old sdf1a:DsRed zebrafish at 0.73× show DsRed positive network of cells.
Supplemental Figure 2. (A) Hematoxylin and eosin stains of a caudal fin cross-section shows cells in an abluminal position to the central vessel. Scale bars represent 10 microns. (B) In situ hybridization with sdf1a antisense probe shows a perivascular cell around the caudal fin central vessel. Scale bars represent 25 microns.
Supplemental Figure 3. The development of sdf1aDsRed cells in the caudal tail fin. Images represent typical development of sdf1aDsRed × fli1:EGFP double transgenic animals serially imaged at several time days post fertilization (dpf) using appropriate filter sets as described in the methods section. White scale bars indicate 1000 microns; grey scale bar indicates 200 microns.
Supplemental Figure 4. Perivascular sdf1aDsRed cells express markers of perivascular cells and can be cultured-expanded. The caudal tail fins of adult fli1:EGFP × sdf1a:DsRed animals (n = 8 – 12) were amputated and treated with collagenase to produce a single cell suspension. RNA was extracted from sdf1aDsRed cells and RT-PCR performed for the classic MSC markers as indicated. “NO RT” indicates a reaction in which no reverse transcriptase was added and PCR performed for bactin1.
Supplemental Figure 5. The reappearance of sdf1aDsRed perivascular cells at 6 – 7 days post amputation (dpa). Images represent typical tail fin regeneration of sdf1aDsRed × fli1:EGFP double transgenic animals serially imaged at several time days post caudal fine amputation using appropriate filter sets as described in the methods section. Scale bars represent 100 microns.
Supplemental Table 1. Sequences of oligonucleotides used in RT-PCR reactions. For Taqman® reactions, the catalog number of the primer set used is listed.